Significance
Auxin-regulated transcription plays a role in almost every aspect of plant growth and development. Recent structural studies of domains from auxin-activated transcription factors and auxin-degraded repressors have raised fundamental questions about the protein complexes required for auxin response. Here, we leverage the power of a synthetic yeast system to identify and systematically characterize the simplest auxin response unit in the absence of the potentially confounding influence of other family members and interacting pathways.
Keywords: auxin signaling, synthetic circuits, transcription, ARF, PB1
Abstract
Auxin-regulated transcription pivots on the interaction between the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) repressor proteins and the AUXIN RESPONSE FACTOR (ARF) transcription factors. Recent structural analyses of ARFs and Aux/IAAs have raised questions about the functional complexes driving auxin transcriptional responses. To parse the nature and significance of ARF–DNA and ARF–Aux/IAA interactions, we analyzed structure-guided variants of synthetic auxin response circuits in the budding yeast Saccharomyces cerevisiae. Our analysis revealed that promoter architecture could specify ARF activity and that ARF19 required dimerization at two distinct domains for full transcriptional activation. In addition, monomeric Aux/IAAs were able to repress ARF activity in both yeast and plants. This systematic, quantitative structure-function analysis identified a minimal complex—comprising a single Aux/IAA repressing a pair of dimerized ARFs—sufficient for auxin-induced transcription.
Almost every aspect of plant growth and development is regulated by the small hormone auxin. Auxin nuclear response is regulated by the interplay of AUXIN RESPONSE FACTOR (ARF) transcription factors and AUXIN/INDOLE-3-ACETIC ACID repressors (Aux/IAAs, hereafter referred to as IAAs). Extensive genetic and biochemical studies have elucidated the basic framework for auxin-regulated transcription: Auxin promotes ARF transcriptional activity by mediating IAA protein degradation (1–3). Consolidating this generic framework with the diversity of context-specific auxin responses, however, remains a major challenge. Across flowering plants, the large size of the ARF and IAA protein families (4, 5) could provide specificity through combinatorial ARF–IAA complex composition (6, 7). Even so, the coexpression of multiple ARFs and IAAs in a cell and their numerous possible homo- and heterotypic interactions have made it difficult to parse the nature of a minimal auxin transcriptional complex.
ARFs and IAAs interact through a conserved C-terminal domain shared by most members of each family (8). This fold, originally called the III/IV domain, has been reclassified as a Phox and Bem1p (PB1) domain based on recent structural studies of isolated domains from ARF5/MP and ARF7/NPH4 as well as IAA17/AXR3 and PsIAA4 (9–12). Both ARF and IAA PB1 domains fold into two oppositely charged faces (“head” and “tail”) that mediate directional electrostatic interactions. In vitro, isolated PB1 domains aggregate head-to-tail in large homotypic clusters. The propensity of these proteins to aggregate in solution has impeded in vitro characterization of ARFs and IAAs, and crystallization of the PB1 domain required mutations in key interaction residues or optimization of pH conditions (9–12).
These structural insights have challenged the classic model of auxin response where the PB1 domain facilitates a single binary interaction (7). Thus, rather than a single IAA interaction with a single ARF, the PB1 domain could mediate higher order complexes of ARFs and IAAs, further increasing the potential for combinatorial complexity in auxin-regulated transcription. In addition, the PB1 domain allows ARF–ARF and IAA–IAA interactions in yeast two-hybrid (Y2H) and in vitro assays (13–15). Although the functional relevance of these homotypic interactions is not well established, the potential for head-to-tail multimerization through the PB1 domain has led to revised auxin signaling models that posit a role for IAA–IAA PB1 interactions in repression (9) and ARF–ARF PB1 interactions in cooperative DNA binding (10).
In addition to the interactions mediated by the C-terminal PB1 domain, ARFs dimerize through an N-terminal domain flanking the B3 DNA-binding motif (16). Dimerization through this domain is critical for the DNA binding and in vivo function of ARF5. The structure of the ARF5 DNA-binding domain homodimer was solved bound to a short stretch of DNA containing inverted repeats of ARF-binding sites known as Auxin Response Elements (AuxREs). This structure revealed that each isolated DNA-binding domain contacted one of the AuxREs, suggesting that spacing and orientation of AuxREs may contribute to the binding specificity of different ARF dimers. In vitro DNA-binding studies supported this model, as DNA binding by ARF1 and ARF5 was differentially sensitive to the number of nucleotides between AuxREs (16).
These structural studies have raised several fundamental questions about the molecular basis of auxin signaling (7, 8, 17, 18). For example, how does promoter architecture influence ARF-binding specificity? If ARFs are already forming obligate dimers to bind DNA, what is the functional role of ARF–ARF PB1 interactions? Do IAAs compete with other ARFs for binding to ARF PB1 domains? What role, if any, does the potential for PB1 oligomerization play in the function of ARFs or IAAs? In brief, what is the composition of an auxin transcriptional complex?
We assayed structure-guided variants of synthetic auxin response circuits in Saccharomyces cerevisiae (19) to rigorously test the molecular interactions driving auxin-regulated transcription. Yeast auxin response circuits are user-defined circuits comprised of select IAA, ARF, and promoter sequences, combined with defined auxin input levels. These circuits allowed direct, quantitative assessment of the functional importance of homo- and heterotypic interactions between auxin signaling components, while eliminating confounding factors such as feedback or interference by other pathways. Using this system, we demonstrated that ARF activity could be specified by the configuration of binding sites in synthetic promoters. We also found that dimerization in the ARF PB1 domain contributes to the stability of DNA binding. In contrast, we found no evidence to support a significant role for IAA–IAA dimerization in either the rate of their auxin-induced degradation or their effectiveness as repressors.
Results
Promoter Architecture Can Specify ARF Activity.
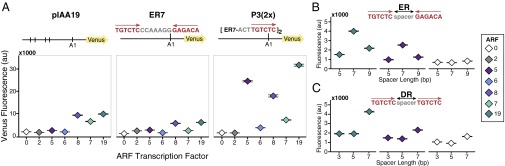
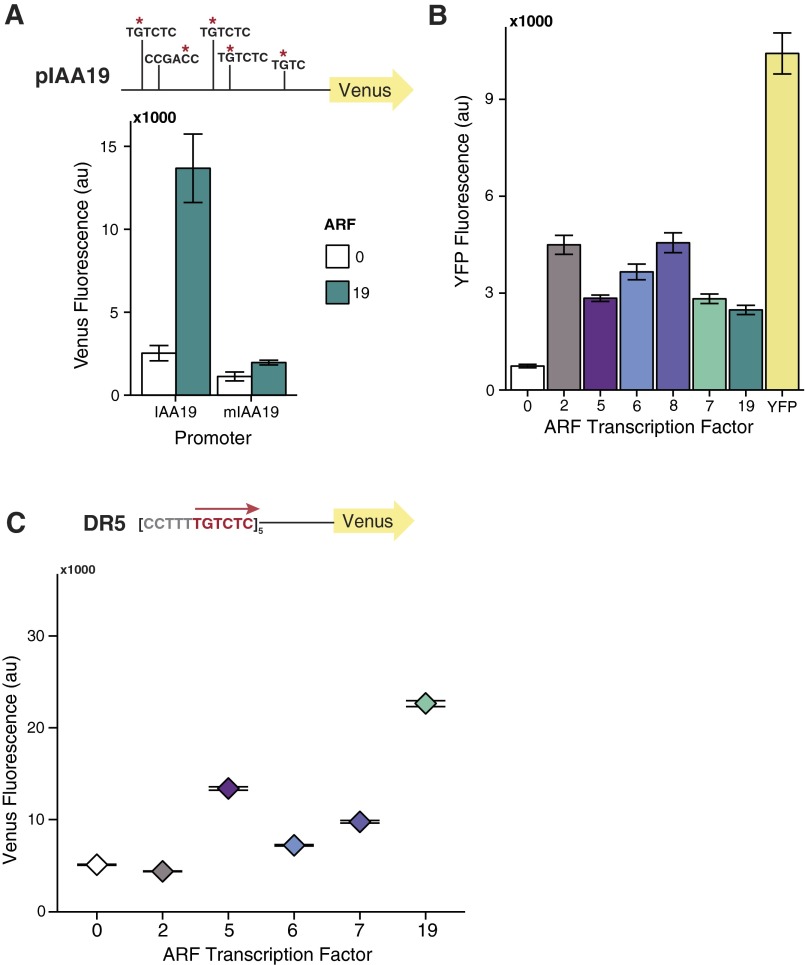
Several lines of inquiry, including recent structural studies, suggest that specificity in auxin responses reflects differences in ARF–DNA interaction. Using flow cytometry, we quantified the activity of a panel of ARFs on the native Arabidopsis IAA19 promoter (pIAA19::Venus) (Fig. 1A). No IAA repressor was present. The ARFs assayed included five activator ARFs (ARF5, ARF6, ARF7, ARF8, and ARF19) and a repressor ARF (ARF2). As observed previously (19), ARF7 and ARF19 were effective activators of the pIAA19::Venus reporter. Although pIAA19 has five AuxREs within 300 nucleotides of the transcriptional start site, ARF8 was the only other ARF tested that effectively activated Venus expression. ARF8 also negatively impacted yeast growth, making further comparisons difficult.
Fig. 1.
Promoter architecture can specify ARF activity. (A) ARFs are differentially active on native and synthetic promoters. Flow cytometry was used to quantify the activity of ARF transcription factors on promoter variants with variable number, spacing, and orientation of binding sites. In each subpanel, yeast strains contain the same Venus reporter construct and differ only by the ARF expressed (2, 5, 6, 8, 7, or 19). “0” indicates absence of an ARF transcription factor. Vertical lines in the first reporter schematic indicate the location of an ARF-binding site (AuxRE) in the native IAA19 promoter. Subsequent panels depict ARF activity when an ER7 or P3(2x) AuxRE variant is inserted at the A1 site in a pIAA19 reporter construct where all of the native AuxREs were mutated (Fig. S1A). All ARFs tested are expressed in yeast at least as well as ARF19 (Fig. S1B). (B) ARF19 and ARF5 exhibit optimal activity when there are seven nucleotides between a pair of AuxRE sites in an ER conformation or (C) DR conformation. When the number of tandem DR5 repeats is increased, all of the activator ARFs exhibit measurable activity (Fig. S1C). The data are shown as median values, with black horizontal bars demarcating 95% confidence intervals around the median.
To distinguish between differences in ARF activation strength and differences in ARF affinity for different AuxRE configurations, we tested the same panel of ARFs against additional synthetic promoters containing previously characterized AuxRE arrays with variable number, spacing, and orientation of sites (20). To limit influence from flanking sequences, promoter variants were generated within a mutated IAA19 promoter where all of the native AuxREs were rendered nonfunctional (Fig. S1A). Into this ARF-insensitive promoter, we introduced a pair of inverted AuxRE repeats spaced by seven nucleotides used in the ARF DNA-binding domain structural studies (16), as well as in early biochemical assays (14). This sequence has historically been called ER7 (20, 21), terminology we are retaining here for clarity. Both ARF19 and ARF8 could activate the ER7-containing promoter to approximately half the extent of the native pIAA19 (Fig. 1A). ARF7, ARF2, and ARF5, which has been shown to bind to this sequence in vitro (16), exhibited only minimal activity.
Fig. S1.
(A) Mutation of AuxRE sites in the IAA19 promoter abolishes ARF19 activity. Red asterisks indicate location of G→A mutations introduced into the 300 bp native IAA19 promoter sequence used for reporter constructs. (B) All ARFs tested are expressed in yeast at least as well as ARF19. Expression of YFP:ARF fusion constructs is shown compared with yeast strains expressing no YFP (0) or YFP alone. The mean from three replicates is shown, with error bars representing SD. (C) All activator ARFs are effective activators of a synthetic promoter containing five direct AuxRE repeats spaced by five base pairs [DR5(5×)]. Fluorescence was measured by flow cytometry. Each yeast strain differs only by the ARF expressed (2, 5, 6, 8, 7, or 19). 0 indicates absence of an ARF transcription factor.
Next, we introduced tandem repeats of the P3 element [P3(2x)], which has been used in both binding assays and transient expression studies (14, 21). Each P3 element is composed of an ER7 sequence separated from an additional AuxRE site by three nucleotides (Fig. 1A). ARF6 could modestly activate the P3(2x) promoter, whereas it was inactive on the ER7 and pIAA19 variants. Introduction of the P3(2x) element also resulted in a dramatic increase of ARF19 activity, more than double the activity observed with the native pIAA19. In contrast, ARF7 showed essentially the same level of activity on the P3(2x) promoter as it had on the native pIAA19. Strikingly, ARF5 strongly activated the P3(2x)-containing promoter, despite exhibiting only minimal activity on the native pIAA19 and ER7 variants.
To determine how the spacing between two binding sites influences ARF-specific activity, we varied the number of nucleotides between a pair of AuxREs in either an inverted repeat (ER) or direct repeat (DR) conformation (Fig. 1 B and C). We quantified activation by ARF19 and ARF5 on ER promoters with five (ER5), seven (ER7), or nine (ER9) base pairs between the two inverted AuxRE sites (Fig. 1B). Consistent with in vitro DNA-binding studies (16), seven-nucleotide spacing between repeats yielded optimal activity for both ARFs. The same trend was observed when comparing ARF activity on DR promoters containing three (DR3), five (DR5), or seven (DR7) base pairs between two direct AuxRE repeats, although the DR7 reporter had the highest background activity (Fig. 1C). Notably, the most widely used auxin transcriptional reporter uses tandem AuxRE repeats with five-nucleotide spacing (DR5) (22–24). When five tandem DR5 repeats replaced the pIAA19 sequence upstream of the mutated A1 site, all of the activator ARFs were active (Fig. S1C). These results suggest that spacing differences alone are unlikely to confer ARF specificity on a target gene. This interpretation is consistent with a recent genome-wide analysis that found a dramatic impact of AuxRE orientation on the binding preferences of ARF2 and ARF5 (25). Seven bases may be the optimal spacing for ARF DNA binding and activity, as this spacing results in minimal DNA torsion or changes in protein conformation (16).
ARFs Activate as Dimers.
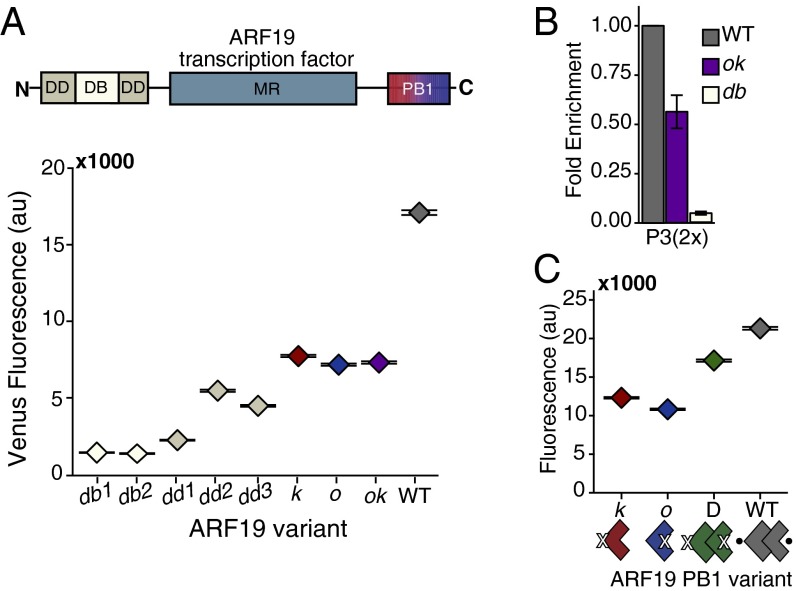
We next assessed the impact of ARF homotypic interactions on transcriptional activity. To maximize the sensitivity of these assays, we focused on the most active ARF-promoter combination: ARF19 and the P3(2x)::Venus reporter. To begin this analysis, we confirmed that ARF19 activity required the homologous residues identified as critical for ARF5 DNA binding (DB) and N-terminal dimerization (DD) in the DNA-binding domain (16). As expected from the ARF5 data, the H138A (db1) and R188A (db2) DNA-binding mutations resulted in the total loss of ARF19 activity (Fig. 2A and Table S1). G247I (dd1), which caused the most severe disruption of dimerization in ARF5, resulted in an almost complete loss of ARF19 activation activity. The A250N (dd2) and A255N (dd3) mutations retained modest levels of ARF19 activity, consistent with the observation that these mutations do not completely disrupt ARF5 dimerization in the DNA-binding domain (16).
Fig. 2.
ARFs activate as dimers. (A) ARF19 requires an intact PB1 domain for full activity. Activation of the P3(2x)::Venus fluorescent reporter by ARF19 variants in yeast was measured using flow cytometry. The data are shown as median values, with black horizontal bars demarcating 95% confidence intervals around the median. DB, DNA binding; DD, dimerization domain; MR, middle region; PB1, C-terminal interaction domain. See Table S1 for specific residues that were mutated in each domain. The diminished activity of the db1, dd1, and ok ARF19 mutant variants is not due to decreased protein levels (Fig. S2A). (B) PB1 mutations decrease DNA binding by ARF19. A ChIP assay was performed on 4×FLAG-ARF19 WT, double PB1 (ok), and DNA-binding mutant (db1) at the P3(2x) locus. Results from three independent replicates are shown normalized to the enrichment of the wild-type ARF19. Error bars represent SE. Relative activity of FLAG-tagged ARFs was similar to untagged strains (Fig. S2B). (C) Activity of ARF19 single PB1 mutants is rescued by coexpressing mutants with complementary interfaces to allow dimerization in the PB1 domain (D = k + o). The data are shown as median values, with black horizontal bars demarcating 95% confidence intervals around the median.
Next, we probed the impact of PB1 interactions on ARF19 activity. To do this, we engineered homologous mutations into ARF19 that were previously shown to compromise one or both PB1 interaction surfaces in ARF7 (9). Specifically, we generated the following ARF19 variants: K962A (k), to disrupt the positively charged face (head); a double mutation D1012A and D1016A in the OPCA motif (o), to disrupt the negatively charged face (tail); and all three mutations in combination, to disrupt interactions at both faces (ok) (Fig. 2A and Table S1). Each PB1 mutant expressed on its own lacks a complementary interface required for PB1 domain interactions. Each variant should therefore provide a similar level of transcriptional activation, representing the efficacy of an ARF19 PB1 monomer. Indeed, all variants exhibited similar diminished activity on the P3(2x)::Venus reporter compared with wild-type ARF19 (Fig. 2A). Early in vitro studies using truncated ARFs indicated that PB1 interactions might be required to stabilize ARF binding to DNA (14). We hypothesized that the observed reduction in transcriptional activation by ARF19 PB1 mutants might reflect reduced binding to the P3(2x) promoter. Chromatin immunoprecipitation assays supported this interpretation (Fig. 2B and Fig. S2B). ARF19 DNA binding was significantly reduced when the PB1 domain was mutated, although this reduced binding was still substantially stronger than what was observed when the DNA-binding domain was mutated.
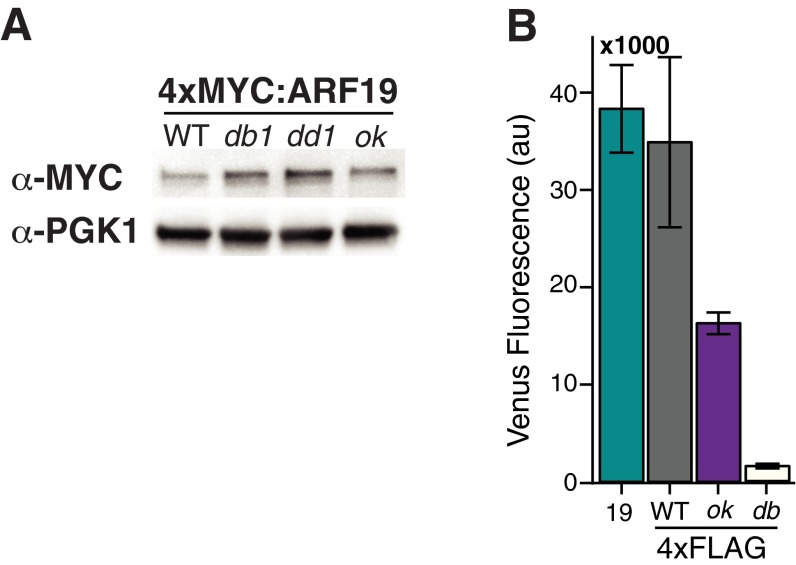
Fig. S2.
(A) Differences in ARF19 mutant variant activity are not due to differences in protein levels. Shown is the Western blot of 4×MYC-tagged ARF19 WT and mutant variants. (B) FLAG-tagged ARF19 variants used in ChIP assays exhibit similar relative activation of the P3(2x)::Venus reporter as untagged constructs in Fig. 2A. The mean from three replicates is shown, with error bars representing SD.
The multimerization potential of the PB1 domain has led to speculation about the composition of the minimal complex required for ARF action. Although ARF19 PB1 monomers showed a significant reduction of activity, we wanted to test the activation potential of ARFs limited to dimers. To do this, we compared expression of the P3(2x)::Venus reporter in yeast expressing wild-type ARF19, either the o or k mutants, or a circuit containing both a k and o variant (Fig. 2C). We expected that wild-type ARF19 had the capacity to form dimers or higher order complexes, whereas o or k mutants could only dimerize within the DNA-binding domain. In contrast, a mixed k and o population could dimerize in the DNA-binding domain and form only a single (dimer) interaction through the PB1 domain. Notably, this reconstituted PB1 dimer was a more effective activator than the monomeric PB1 mutants (Fig. 2C). Thus, dimerization at both N and C termini contributes to optimal DNA binding and activity of ARF19.
IAAs Can Function as Monomers.
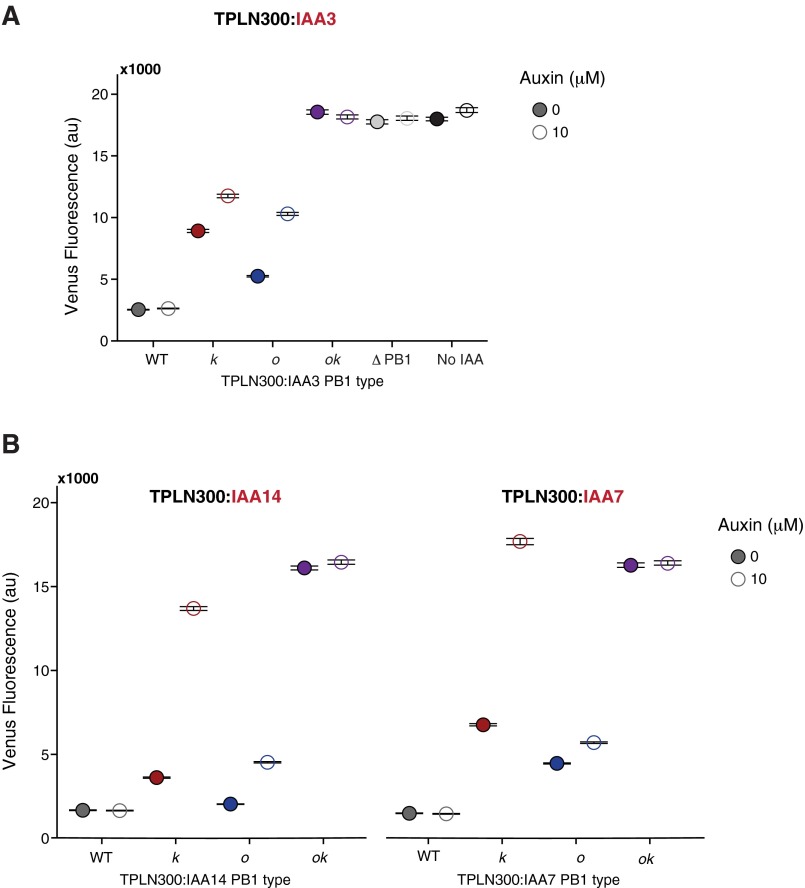
Continuing upstream in auxin response, we sought to examine the functional consequences of limiting IAA PB1 domain interactions. Because the primary functional attribute of the IAA PB1 domain is interaction with an ARF, repression of ARF activity by the IAA is the most straightforward output of this interaction. In a yeast auxin response circuit, where only a single ARF and IAA were expressed, we could directly measure the impact of each PB1 face on IAA repression. An N-terminal fusion of the first 300 amino acids of the TOPLESS (TPL) corepressor to IAA3/SHY2 (TPLN300:IAA3) strongly repressed ARF19 activity in a PB1-dependent manner (Fig. 3A). Mutations on both PB1 interaction surfaces of IAA3 (ok) showed a loss of repression similar to a complete PB1 deletion (ΔPB1). The single interface PB1 IAA mutants (k and o) maintain repression of ARF19, albeit with reduced strength. Similar results were observed for TPLN300:IAA14/SLR and TPLN300:IAA7/AXR2 (Fig. S3).
Fig. 3.
IAAs can function as monomers. (A) Repression of ARF19 activity on the P3(2x)::Venus reporter by IAA3 fused to TPLN300. Single PB1 mutants (k and o) maintain the ability to repress, whereas the double mutant (ok) exhibits a loss of repression equal to the deletion of the PB1 domain (light gray) or absence of IAA3 (white). Similar results were observed for TPLN300:IAA7 and TPLN300:IAA14 (Fig. S3). The data are shown as median values, with black horizontal bars demarcating 95% confidence intervals around the median. (B) TPLN300:IAA3 single PB1 mutants (k and o) can repress ARF19 PB1 monomers and dimers. Mutation of the k PB1 interface reduces the repressive capability of TPLN300:IAA3 on the activity of an ARF19 PB1 dimer. The data are shown as median values, with black horizontal bars demarcating 95% confidence intervals around the median. (C) Single PB1 slr mutants (k and o) maintain the ability to repress ARF activity in plants. Lateral root density (number of lateral roots per millimeter primary root length) was measured in pIAA14::slr PB1 mutant lines. Approximately 20 individuals per line were measured, and the distributions of multiple T2 lines for each genotype are shown. White diamonds mark the mean and the black horizontal line indicates the median root density for each genotype. (D) Relief of TPLN300:IAA3 and TPLN300:IAA14-mediated repression in response to auxin treatment. Auxin responsiveness of TPLN300:IAA single PB1 mutants (k and o) differs between IAAs. Yeast strains were imaged at 100× magnification with a Leica fluorescence microscope 4 h after exposing to either an auxin (10 μM) or mock (95% EtOH) treatment. Flow cytometry was also used to quantify fluorescence (Fig. S3).
Fig. S3.
(A) Repression strength and auxin sensitivity of TPLN300:IAA3 PB1 mutant variants. Each strain was exposed to a 10 μM auxin or mock [95% (vol/vol) EtOH] treatment for 4 h before fluorescence measurements using flow cytometry. (B) Repression strength and auxin sensitivity of TPLN300:IAA14 and TPLN300:IAA7 PB1 mutant variants are similar.
To further refine the functional stoichiometry of ARF–IAA complexes, we assayed IAA repression strength when the PB1-mediated interaction potential of both the IAA and ARF partner were limited. TPLN300:IAA3 single PB1 mutants repressed almost as well as wild-type TPLN300:IAA3 when coexpressed with an ARF19 PB1 mutant with a complementary interaction surface [Fig. 3B; TPLN300:IAA3 o|ARF19 k (red); TPLN300:IAA3 k |ARF19 o (blue)]. TPLN300:IAA3 k mutants were less effective repressors than TPLN300:IAA3 o mutants in circuits where PB1 domains of ARF19 could self-dimerize (green), similar to what was seen in wild-type ARF19 circuits. This difference suggests that although a single face is sufficient for repression, the positive (K) IAA3 PB1 interface may be the preferred surface for heterodimerization with ARF19.
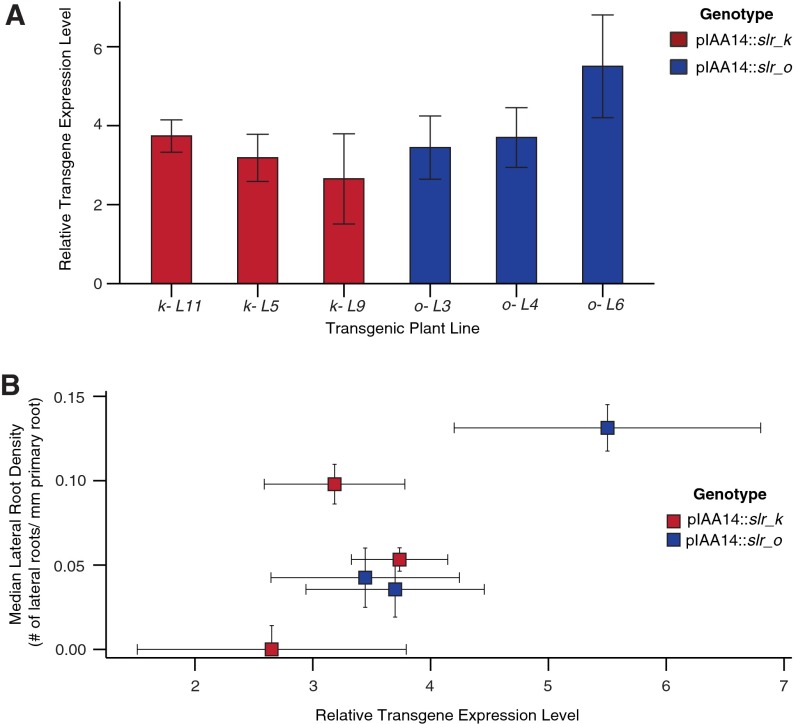
Although a single PB1 interface is sufficient for ARF–IAA interaction in Y2H and in vitro assays (9–12), there is conflicting evidence for whether this limited interaction is sufficient for repression in planta (9, 10). To test if an IAA with a single PB1 interface was also capable of mediating repression in plants, we transformed Arabidopsis seedlings with single and double PB1 mutants of a stabilized form of IAA14/SLR (Fig. 3C). We chose to test slr because of the striking solitary root phenotype for which it was named (26). We and others have previously demonstrated that pIAA14::slr transgenic plants largely recapitulate the phenotype of the slr mutant where the initiation of nearly all lateral roots is inhibited (27, 28). We reasoned that a loss of repression could be quantified by the number of lateral roots formed by slr PB1 mutant transgenic plants. As expected, transgenic lines expressing slr double PB1 (ok) mutants exhibited similar lateral root densities (number of lateral roots per millimeter of primary root) as those expressing a wild-type IAA14, indicating a loss of the dominant negative solitary root phenotype (Fig. 3C). In contrast, multiple independent lines expressing slr single PB1 mutants (k or o) exhibited a significant decrease in lateral root density. Expression levels of the transgene in multiple lines were measured by quantitative PCR (qPCR) and had no correlation with lateral root density (Fig. S4). These results are consistent with our observations in yeast: A single IAA PB1 interface retains the capacity to repress ARF activation.
Fig. S4.
(A) Transgene expression levels were determined by qPCR for three independent lines each of pIAA14::slr-o and pIAA14:slr-k. (B) Transgene expression levels in slr single-face PB1 mutant transgenic plants do not correlate to lateral root density. The lateral root densities and corresponding transgene expression levels of three independent lines each of pIAA14::slr-o and pIAA14:slr-k were quantified. Error bars indicate SEM.
ARF–IAA Affinity Tunes Auxin Sensitivity.
In previous work, we demonstrated the importance of auxin-induced IAA degradation rate in tuning cellular sensitivity to auxin (19, 28, 29). In circuits containing only an IAA and AFB receptor, IAA–IAA PB1 interactions had modest or no influence on IAA degradation rates (Fig. S5A). This result is consistent with in vitro assays indicating that IAAs have only weak affinity for homotypic PB1 interactions compared with the strength of heterotypic interactions with ARF PB1 domains (11). At low levels of auxin, we observed a modest slowing of WT IAA degradation rate when ARF19 was coexpressed (Fig. S5A). Thus, the IAA PB1 domain appears to primarily mediate interaction with an ARF.
Fig. S5.
(A) Auxin dose–response of YFP-tagged IAA3 and IAA7 ± ARF19. There is no difference in auxin-induced degradation between WT and ok YFP strains for both IAA3 and IAA7. ARF19 has a modest effect on the auxin-induced degradation of IAA3 and IAA7 when cells were treated with 0.5 μM auxin. Fluorescence of YFP-WT and YFP-ok IAA strains in the presence of the AFB2 auxin receptor was measured 30 min post auxin treatment. Fluorescence levels were normalized to mock treatment levels to compensate for differences in basal expression levels of IAAs. (B) PB1 mutations have little to no effect on the basal expression of IAA3, but the o and ok mutations decrease levels of both IAA7 and IAA14. (C) IAA-ok mutations result in a modest decrease of protein level compared with WT. Shown is the Western blot of YFP-IAA WT and ok variants.
The modest reduction of repression strength in single IAA PB1 mutants was likely a result of a decrease in ARF affinity. We found that we could visualize the relative affinity of the IAA3 PB1 mutant constructs for ARF19 by tracking their cellular localization (Fig. S6). When expressed alone, fluorescence of YFP-IAA3 was diffuse throughout the cell, whereas the coexpression of ARF19 strongly enhanced nuclear localization. In contrast, localization of YFP-IAA3 single and double PB1 mutants remained diffuse even in the presence of ARF19. These results suggest a marked decrease in ARF interaction with just a single IAA PB1 mutation.
Fig. S6.
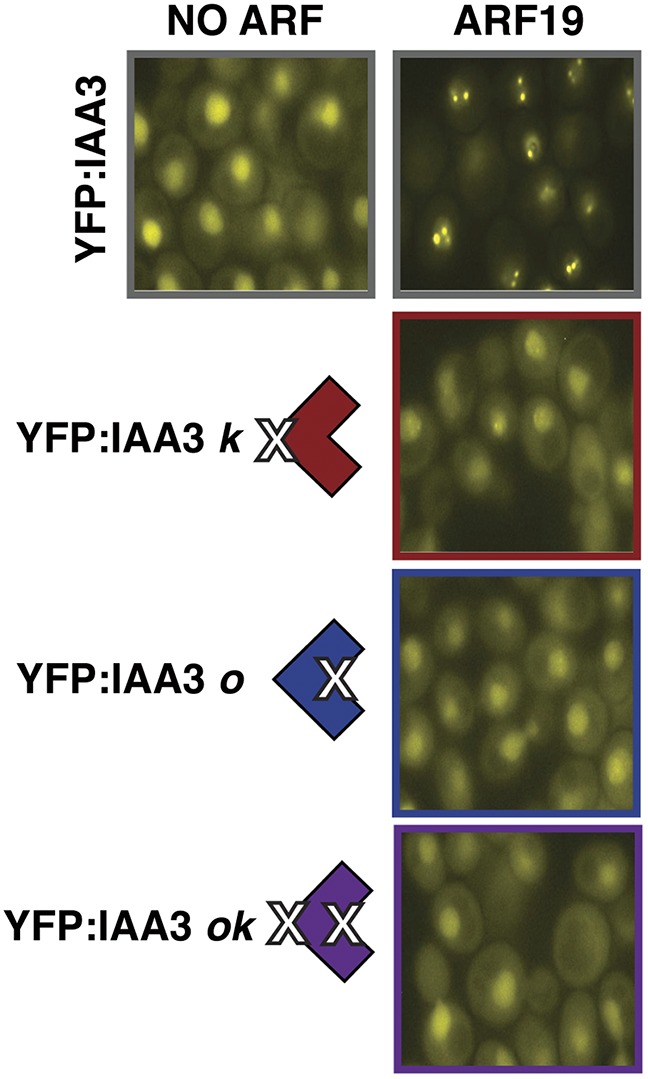
Localization of YFP-tagged IAA3 WT and PB1 mutants in the presence of ARF19. Wild-type IAA3 localization becomes strongly nuclear when ARF19 is coexpressed, whereas localization remains diffuse when a PB1 face is compromised. Yeast were imaged at 100× magnification with a Leica fluorescence microscope.
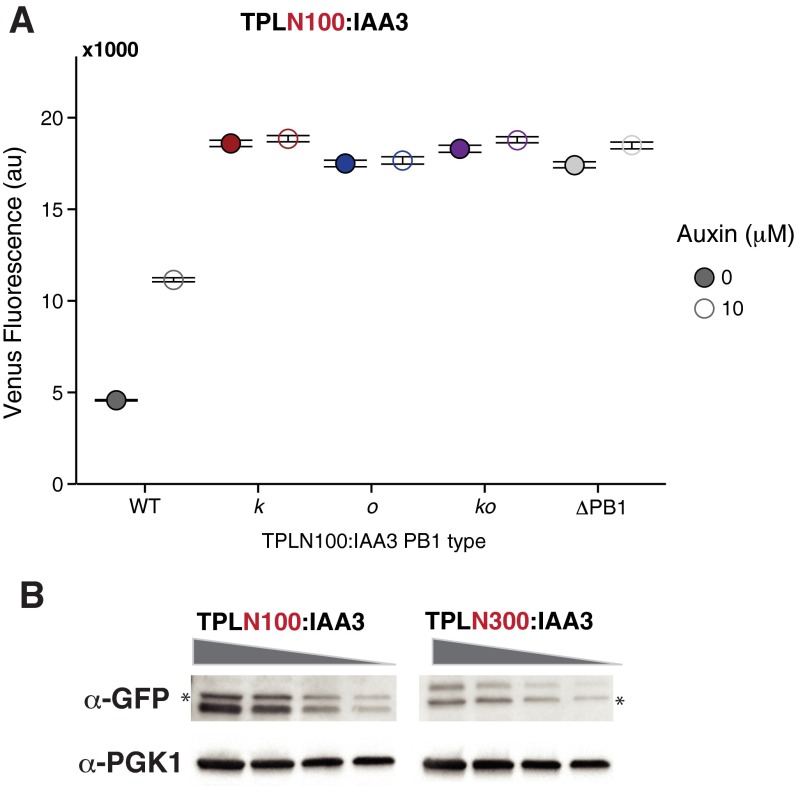
Effective auxin signaling requires a sensitive balance between the strength of IAA interactions with ARFs and with auxin receptors. One measure of this balance is the ease with which repression can be reversed following auxin exposure. We previously demonstrated that only repression by TPLN100:IAA fusions can be relieved by auxin, whereas TPLN300:IAA repression seems to be insensitive to auxin treatment (19). However, even the single interface TPLN100:IAA3 PB1 mutants (k and o) could not repress ARF19 activity (Fig. S7A), impeding our ability to assay auxin response dynamics. Because expression levels of the TPLN300 and TPLN100 fusions are not correlated with their ability to repress (Fig. S7B), the difference in the sensitivity of repression activity to PB1 mutations may reflect different modes of repression enacted by the different TPL truncations. A recent structure of the N-terminal portion of TPL suggests that this result likely reflects the loss of a second TPL dimerization domain in the N100 fragment (30).
Fig. S7.
(A) Repression strength and auxin sensitivity of TPLN100:IAA3 PB1 mutant variants. Each strain was exposed to a 10 μM auxin or mock [95% (vol/vol) EtOH] treatment for 4 h before fluorescence measurements were taken using flow cytometry. (B) Protein levels of the TPLN100:IAA3 and TPLN300:IAA3 fusions are not correlated with their ability to repress. Shown is the Western blot of YFP-TPL:IAA3 strains. Gray wedge represents amount of total protein loaded (left to right, 30, 15, 7.5, and 3.75 μg total protein). Asterisks mark a nonspecific band.
As an alternative, we used yeast expressing TPLN300:IAAs to test whether loss of ARF affinity resulting from mutations in a single surface of the IAA PB1 domain could increase auxin sensitivity. Indeed, auxin could relieve repression of TPLN300:IAA k or o mutants (Fig. 3D and Fig. S3A), whereas repression was maintained when TPLN300 was fused to a wild-type IAA. Notably, although the o and k mutations in TPLN300:IAA3 were both moderately auxin-sensitive, there was a marked contrast in the auxin response of the TPLN300:IAA14 single PB1 mutants (Fig. 3D). The TPLN300:IAA14 k variant was quite sensitive to auxin, resulting in an almost total reversal of repression, whereas the strain expressing the TPLN300:IAA14 o variant exhibited only a modest response to auxin. TPLN300:IAA7 PB1variants followed a similar pattern of auxin response as those observed for TPLN300:IAA14 (Fig. S3B). Taken together, our results indicate that although a single IAA PB1 face results in diminished ARF affinity, it is still sufficient for repression.
Discussion
To understand plants, one must understand auxin. The simple structure of the nuclear auxin signaling pathway, and its reliance on highly conserved and modular aspects of eukaryotic biology, has enabled remarkable progress toward this ambitious goal (31). Synthetic auxin response circuits in yeast have made it possible to quantitatively and systematically analyze the requirements for auxin response. In this process, we have validated some of the predicted structure–function connections, called others into question, and defined the functional roles of various molecular interactions driving auxin-regulated transcription (Fig. 4).
Fig. 4.
Key molecular interactions of an auxin transcriptional complex. (A) ARFs (blue) dimerize in both the N-terminal dimerization domain (DD) and the PB1 domain for robust transcriptional activation. Secondary interactions in the PB1 domain stabilize an ARF dimer on the DNA. DB, DNA binding; DD, dimerization domain; MR, middle region; PB1, C-terminal interaction domain. (B) Spacing and orientation of ARF-binding sites may drive conformation of the flexible middle region (MR) upon DNA binding. PB1 interactions likely lock this conformation into place to create binding pockets for specific coactivators. (C) A monomeric IAA (green) can repress ARF activity. The decrease in ARF affinity when a single PB1 face is mutated suggests that an IAA optimally interacts with ARF dimers through both PB1 interfaces. It is possible that the IAA PB1 domain is sandwiched between the PB1 domains of an ARF dimer, creating a scaffold for the recruitment of repression machinery.
Promoter architecture, particularly the number and orientation of AuxREs, strongly influences ARF activation potential. With the exception of ARF8, which led to growth defects in yeast, ARF19 was the only ARF active across all promoter variants. ARF6 and ARF7 were weaker activators in general, whereas ARF5 activity varied considerably with changes in binding-site configuration. The low basal activity of the promoter variants made it difficult to discern any repressive activity of ARF2. Nonetheless, the specific activities we observed for the activator ARFs demonstrate the utility of this system for probing the role of promoter architecture on ARF function. Identification of repressible promoters as well as the coexpression of more than one ARF (i.e., activator and repressor) may lend new insight into repressor ARFs and the functionality of mixed ARF complexes.
Effective activation by ARF19 requires dimerization in both the N-terminal DNA-binding domain and the C-terminal PB1 domain. Our data support a model where an ARF dimer has contacts at both termini (Fig. 4A). The flexibility of ARF binding to direct or inverted repeats suggests that binding-site orientation may drive differential conformation of the flexible middle region (Fig. 4B), potentially leading to the recruitment of specific coactivators (32, 33). Secondary contacts in the PB1 domain could serve to stabilize the conformation of the middle region following DNA binding. Notably, both bHLH and MYB transcription factors have been shown to interact with ARFs through the PB1 domain (34, 35). Coupled with proximal binding sites to the ARFs (36–38), PB1-mediated interactions between an ARF and another transcription factor may provide the same or further enhanced DNA binding as what we observed in ARF–ARF PB1-driven dimers. This would account for ARF binding of single AuxRE sites as well as the integration of regulatory control from other signaling pathways.
The potential for oligomerization via both the ARF and IAA PB1 domains has challenged the classic model of auxin signaling where the PB1 domain mediates a single binary interaction. Our data indicate that the ARFs can activate effectively as dimers but do not rule out the possibility that the PB1 domain could mediate higher order complex formation on more complex promoters. On the other hand, the current study provides strong evidence that IAAs can function as monomers (Fig. 4C). We have demonstrated in both yeast and plants that an IAA with a single PB1 interface can repress ARF activity, despite the associated decrease in affinity for the ARF. Our assays also detected a potential face preference in the PB1 interaction between ARFs and IAAs, a quality that may be dependent on specific ARF–IAA pairs. The differential auxin sensitivity of single PB1 mutants in different IAAs may therefore be indicative of basal differences in affinity for ARF19. Similar reasoning could account for the seemingly contradictory data from previous in planta repression assays (9, 10). Some IAAs may have such low basal affinity for one or more ARFs that any further compromise of this interaction potential through a PB1 mutation renders them poor competitors for wild-type IAAs expressed in the same cells.
Because our yeast assays rely on a TPL fusion for repression, repression strength is primarily a proxy for ARF–IAA interaction strength. Although a TPL:IAA fusion is somewhat artificial in nature, repression in our system is dependent on the PB1 domain to mediate an IAA–ARF interaction. Moreover, the close correspondence of our yeast and plant data indicates that TPL recruitment in planta is not impacted by PB1 mutations in the IAA. Although our studies have shed new light on the nature of IAA–ARF interactions, the mechanistic details of IAA-mediated repression remain elusive. The requirement for a TPL:IAA fusion in yeast, along with the differences in strength and auxin reversibility of TPLN300 and TPLN100 repression, highlight key gaps in our knowledge around TPL recruitment and action. TPL recruitment and its own multimerization could stabilize ARF–IAA interactions, which would explain why IAAs expressed without TPL fusions are poor repressors in yeast. It is also possible that binding of the IAA to the ARF could lead to conformational changes in the flexible middle region of the ARF that blocks recruitment of coactivators (33, 39) or changes the module composition of core transcriptional machinery like the Mediator complex (40). Phosphorylation of the middle region in ARF7 and ARF19 has been shown to inhibit IAA interaction (41), indicating that signal integration from other pathways may take advantage of a link between an IAA interaction and the activity of the ARF middle region.
The nuclear auxin response pathway is a robust and tunable circuit that underlies almost every aspect of plant growth and development. It is becoming ever more apparent, however, that auxin does not work alone. Our work ex situ has demonstrated the baseline for auxin response at the molecular level, which can now be used as a breadboard for testing additional layers of regulation and integration with other signaling pathways. In addition, a wealth of transcriptome data (42, 43) can now be used to generate hypotheses about the likelihood of binding by a specific ARF through integration of cell type-specific expression maps of auxin signaling components (44) and binding-site preferences from protein binding arrays (16). Yeast auxin response circuits provide a resource to rapidly test these hypotheses and inform experimental designs in planta. By integrating studies in plants, in vitro, in silico, and ex situ we can uncover how evolution has tinkered with the auxin nuclear pathway to accommodate innovations in plant life.
Materials and Methods
See SI Materials and Methods for detailed descriptions of plasmid and yeast strain construction, flow cytometry assays, fluorescent microscopy, Western and ChIP assays, qPCR, and the generation and phenotyping of transgenic Arabidopsis. See Tables S2 and S3 for yeast strains and oligos used in this study.
SI Materials and Methods
Strain Construction.
ARFs were driven by the yeast ADH1 promoter and integrated at the HIS3 locus into a W303-1A MATa strain containing a pIAA19::Venus reporter variant integrated at URA3. Haploid strains were assayed to measure activity of different promoter–ARF combinations. To maintain consistency across circuits of increasing complexity, ARF19 variant strains containing the P3(2x)::Venus reporter were mated to a MATα W814-29B strain containing the AFB2 auxin receptor integrated at LEU2 (YNL 175) for measuring activity of ARF19 point mutants. In experiments testing the activity of ARF19 PB1 dimers, each strain tested contained two genomically integrated copies of ARF19 to ensure a similar metabolic load. A second ARF19 variant was introduced at the HIS3 locus of YNL175 before mating to W303-1A MATa strains containing an ARF19 variant and the P3(2x)::Venus reporter. IAAs were expressed from the yeast GPD promoter and N-terminally tagged with either a TPLN300 or TPLN100 fragment for repression assays or YFP for degradation assays as described previously (19). IAAs were integrated at the TRP1 locus into a MATα W814-29B strain containing the AFB2 auxin receptor integrated at LEU2. For repression assays, IAA strains were then mated to W303-1A MATa strains containing an ARF19 variant and the P3(2x)::Venus reporter. See Table S2 for a complete list of strains used in this study.
Plasmid Construction.
ARF and Aux/IAA point mutants were generated in pDONR vectors through Gibson assembly (45). Integrating yeast expression vectors were then generated through Gateway cloning as previously described (19, 46). For biochemical assays, epitope tags (4×-FLAG or 4×-MYC) were introduced as N-terminal fusions upstream of Gateway recombination sites in the HIS3-targeting single integrating vector (19) before an LR reaction to introduce the ARF19 variant. The 300-bp IAA19 promoter sequence was ordered as a gBlock gene fragment from Integrated DNA Technologies, Inc. (IDT) with a G→A mutation introduced at the second position of each AuxRE site (Fig. S1A). Gibson assembly was used to swap the mutated pIAA19 gene fragment for the native IAA19 promoter sequence upstream of the Venus coding sequence in a URA3 single integrating Gateway vector. AuxRE arrays were ordered as oligos or gBlock gene fragments with Gibson overhangs to introduce the sequences into the mutated pIAA19::Venus reporter construct. See Table S3 for oligo and gene fragment sequences.
Yeast Methods.
Yeast transformations were performed using a standard lithium acetate protocol (47) into MATa W303-1A or MATα W814-29B (gifts from the Gottschling laboratory). Yeast Peptone Dextrose (YPD) and Synthetic Complete (SC) mediums were made according to standard protocols and supplemented with 80 mg·mL−1 adenine. MATa and MATα strains were mated by coinoculating strains at low density into YPD medium and growing overnight at 30 °C with shaking at 220 rpm in a MaxQ 4000 Benchtop Oribital Shaker (Thermo Scientific). Diploids were selected by streaking out 5 μL of culture on SC plates lacking the appropriate amino acids and choosing a single colony to isostreak on YPD plates before glycerol stocking.
Flow Cytometry.
Fluorescence measurements were taken with a custom BD Accuri SORP flow cytometer with a CSampler 96-well plate adapter using an excitation wavelength of 514 nm and an emission detection filter at 545/35 nm. A total of 20,000 events above a 40,000 FSC-H threshold (to exclude debris) were measured for each sample. Data were exported as FCS 3.0 files and processed in R using the flowCore, plyr, and ggplot2 software packages and custom scripts. Scripts and full datasets are available upon request.
ARF Activity and IAA Repression Assays.
A freshly grown colony was inoculated in 500 μL of SC media and grown at 30 °C with shaking at 375 rpm in 2,000 μL Eppendorf Deepwell Plates 96. After 16 h of growth, cultures were either diluted 1:100 (haploid strains) or 1:200 (diploid strains) into fresh SC media. Fluorescence measurements were taken after 6 h of additional growth. For auxin response assays, diluted cultures were grown for 2 h before addition of 10 μM auxin (IAA) or mock treatment [95% (vol/vol) ethanol]. Fluorescence measurements were taken after 4 h of additional growth. The data for at least three independent replicates were pooled and plotted for each strain.
YFP-IAA Expression and Dose–Response Assays.
YFP-IAA strains were grown as described previously (46). Basal YFP-IAA expression was measured in haploid strains lacking the presence of an auxin receptor. For dose–response assays, diploid strains coexpressing the AFB2 auxin receptor and a YFP-IAA variant ± ARF19 or ARF3 were diluted 1:200 into four separate wells containing 500 μL of fresh SC media. After 2 h of additional growth, an auxin dose (0.05, 0.5, or 5 μM IAA) or mock treatment [95% (vol/vol) ethanol] was added to one of the four identical wells for each strain. To compensate for differences between basal YFP-IAA expression levels, dose–response fluorescence measurements were normalized by subtracting background autofluorescence and dividing by fluorescence levels of mock-treated strains.
Western Blotting.
Yeast were grown overnight in 25–50 mL liquid YPD at 30 °C with shaking at 225 rpm. The next morning cultures were diluted to an OD of 0.2 into fresh YPD media and grown until an OD of 1.0–1.5. Cells were then pelleted at 4 °C, rinsed once with TBS, and flash-frozen in liquid nitrogen. Cell pellets were lysed by glass bead disruption in lysis buffer containing protease inhibitors (48). Whole-cell lysate protein concentrations were determined by Bradford assay, and equivalent amounts of protein were loaded onto 4–20% TGX gel (Bio-Rad). Anti-GFP (Sigma) was used to probe for YFP-tagged Aux/IAAs, anti-Myc (Cell Signaling Technology) was used to probe for Myc-ARF variants, and anti-PGK1 (Abcam) was used as a loading control.
Chromatin Immunoprecipitation.
Yeast were inoculated in liquid YPD and grown at 30 °C with shaking at 225 rpm. After 18 h of growth, cultures were diluted 1:50 in fresh YPD to a final volume of 200 mL. Four hours postdilution, cells were treated with 20 mL of fix solution [11% (vol/vol) formaldehyde, 0.1 M NaCl, 1 mM EDTA, 50 mM Hepes–KOH] for 20 min at room temperature with shaking. Cultures were further treated with 36 mL of 2.5 M glycine for 5 min to quench the cross-linking. Cells were then pelleted at 4 °C, washed twice with ice-cold TBS, and flash-frozen in liquid nitrogen. Cells were lysed in breaking buffer [100 mM Tris, pH 8.0, 20% (vol/vol) glycerol, and 1 mM PMSF] before sonication. Samples were processed in a Bioruptor Plus sonication device at 50% for 25 cycles. Following centrifugation (to pellet cellular debris), the supernatant was used for the IP reaction. Anti-FLAG M2 (Sigma) coupled to Protein G Dynabeads (Life Technologies) was used to probe for FLAG-tagged ARF19 variants. Following elution from the beads, samples were incubated overnight at 65 °C to reverse cross-links. DNA was purified using a Qiaquick PCR purification kit (Qiagen). qPCR for three independent replicates was performed using iQ SYBR Green Supermix (Biorad) in a C100 thermocycler fitted with a CFX96 Real-Time Detection System (Biorad).
Generation and Analysis of Transgenic Plants.
A 2-kb fragment of the IAA14 promoter (26) was cloned into pGREEN Gateway-compatible vectors. PB1 mutations were introduced through Gibson assembly in pDONR vectors containing the coding sequence for slr. Constructs were transformed into Col-0 wild-type plants using the floral dip method (49). T2 seeds of transgenic plants were sown on 0.5× Linsmaier and Skoog (LS) medium, 0.8% phytoagar plates supplemented with Kanamycin. Plates were stratified at 4 °C for 2 d and grown in continuous light conditions for 7 dpg. DNA was extracted from transgenic seedlings and sequenced to confirm genotype. Following selection, transformants were transplanted to plain 0.5× LS, 0.8% phytoagar plates and grown vertically in continuous light. Plants were imaged at 14 dpg using an EPSON V500 scanner, and root density was measured in ImageJ. Primary roots were traced using a Wacom Intuos Pro Medium tablet. Immediately after imaging plants were flash-frozen at –80 °C. RNA was extracted from plants using a Spectrum Plant Total RNA kit (Sigma), and 1μg of cDNA was prepared using an iScript cDNA Synthesis kit (Bio-Rad). Samples were analyzed using SYBR Green Supermix (Bio-Rad) reactions run in a CFX96 Real-Time System (Bio-Rad). Relative expression levels were calculated using the formula (Etarget)−CPtarget/(Eref)−CPref (50) and normalized to reference gene At1g13320.
Microscopy.
Yeast strains were grown overnight at 30 °C with shaking at 220 rpm in SC medium. Overnight cultures were diluted 1:200 and grown for an additional 4.5 h. Yeast samples were assembled on glass coverslips with 1% agarose pads placed over the cultures. Strains were immediately visualized using a Leica DMI 3000B microscope fitted with a Leica 100× N-PLAN 1.25 oil objective and illuminated with a Lumencor SOLA light source. Images were captured using Leica LAS AF version 2.6.0 software and a Leica DFC 345FX camera.
Supplementary Material
Acknowledgments
We thank Takato Imaizumi and Eric Klavins for careful reading of our manuscript; Richard Gardner and Toshio Tsukiyama for guidance on ChIP assays; members of the J.L.N. and Imaizumi laboratories for helpful discussions; and Jake Wegesin for the graphics shown in Fig. 4. This work was supported by the Paul G. Allen Family Foundation, the National Science Foundation (MCB-1411949), and the National Institutes of Health (R01-GM107084). E.P.-J. was supported by an NSF Graduate Research Fellowship and the Seattle Chapter of the Achievement Rewards for College Scientists Foundation. B.L.M. received fellowship support from the National Cancer Institute of the National Institutes of Health (F32CA180514).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604379113/-/DCSupplemental.
References
- 1.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 2.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 3.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 4.Paponov IA, et al. The evolution of nuclear auxin signalling. BMC Evol Biol. 2009;9:126. doi: 10.1186/1471-2148-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004;135(3):1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lokerse AS, Weijers D. Auxin enters the matrix—Assembly of response machineries for specific outputs. Curr Opin Plant Biol. 2009;12(5):520–526. doi: 10.1016/j.pbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Wang R, Estelle M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 2012;190:82–88. doi: 10.1016/j.plantsci.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Korasick DA, et al. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA. 2014;111(14):5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanao MH, et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014;5:3617. doi: 10.1038/ncomms4617. [DOI] [PubMed] [Google Scholar]
- 11.Han M, et al. Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci USA. 2014;111(52):18613–18618. doi: 10.1073/pnas.1419525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinesh DC, et al. Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response. Proc Natl Acad Sci USA. 2015;112(19):6230–6235. doi: 10.1073/pnas.1424077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94(22):11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19(3):309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 15.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boer DR, et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156(3):577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Korasick DA, Jez JM, Strader LC. Refining the nuclear auxin response pathway through structural biology. Curr Opin Plant Biol. 2015;27:22–28. doi: 10.1016/j.pbi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinesh DC, Villalobos LI, Abel S. Structural biology of nuclear auxin action. Trends Plant Sci. 2016;21(4):302–316. doi: 10.1016/j.tplants.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc Natl Acad Sci USA. 2014;111(26):9407–9412. doi: 10.1073/pnas.1324147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagen G, Guilfoyle T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol Biol. 2002;49(3-4):373–385. [PubMed] [Google Scholar]
- 21.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276(5320):1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 22.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA. 1999;96(10):5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165(5):1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29(2):153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 27.Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 2005;44(3):382–395. doi: 10.1111/j.1365-313X.2005.02537.x. [DOI] [PubMed] [Google Scholar]
- 28.Guseman JM, et al. Auxin-induced degradation dynamics set the pace for lateral root development. Development. 2015;142(5):905–909. doi: 10.1242/dev.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moss BL, et al. Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol. 2015;169(1):803–813. doi: 10.1104/pp.15.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke J, et al. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci Adv. 2015;1(6):e1500107. doi: 10.1126/sciadv.1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierre-Jerome E, Moss BL, Nemhauser JL. Tuning the auxin transcriptional response. J Exp Bot. 2013;64(9):2557–2563. doi: 10.1093/jxb/ert100. [DOI] [PubMed] [Google Scholar]
- 32.Melcher K. The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional proteins. J Mol Biol. 2000;301(5):1097–1112. doi: 10.1006/jmbi.2000.4034. [DOI] [PubMed] [Google Scholar]
- 33.Wu MF, et al. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin R, et al. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19(8):2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varaud E, et al. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell. 2011;23(3):973–983. doi: 10.1105/tpc.110.081653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berendzen KW, et al. Bioinformatic cis-element analyses performed in Arabidopsis and rice disclose bZIP- and MYB-related binding sites as potential AuxRE-coupling elements in auxin-mediated transcription. BMC Plant Biol. 2012;12:125. doi: 10.1186/1471-2229-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7(10):1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walcher CL, Nemhauser JL. Bipartite promoter element required for auxin response. Plant Physiol. 2012;158(1):273–282. doi: 10.1104/pp.111.187559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F, et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature. 2015;525(7568):269–273. doi: 10.1038/nature14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito J, et al. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc Natl Acad Sci USA. 2016;113(23):6562–6567. doi: 10.1073/pnas.1600739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho H, et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol. 2014;16(1):66–76. doi: 10.1038/ncb2893. [DOI] [PubMed] [Google Scholar]
- 42.Bargmann BO, et al. A map of cell type-specific auxin responses. Mol Syst Biol. 2013;9:688. doi: 10.1038/msb.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis DR, et al. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell. 2013;25(9):3329–3346. doi: 10.1105/tpc.113.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rademacher EH, et al. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011;68(4):597–606. doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- 45.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 46.Havens KA, et al. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160(1):135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 48.Gerace E, Moazed D. Coimmunoprecipitation of proteins from yeast. Methods Enzymol. 2014;541:13–26. doi: 10.1016/B978-0-12-420119-4.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 50.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.