Significance
The BMI1-containing Polycomb Repressive Complex is an important gene silencer during development, stem cell maintenance, and cancer progression. BMI1 is also involved in DNA damage response, but its downstream mechanism is unclear. Here we identified UBR5 as a downstream factor whose chromatin recruitment is regulated by BMI1. We showed that BMI1 and UBR5 repress the RNA polymerase II (Pol II) elongation and nascent RNA synthesis at UV-induced DNA lesions. UBR5 associates with and regulates the abundance of the histone chaperon complex FACT at damaged chromatin and regulates elongation of Pol II at the lesions. Thus, our studies identify a pathway that regulates the damage-induced transcriptional repression that is governed by BMI1.
Keywords: Polycomb, BMI1, UBR5, FACT, transcription
Abstract
BMI1 is a component of the Polycomb Repressive Complex 1 (PRC1), which plays a key role in maintaining epigenetic silencing during development. BMI1 also participates in gene silencing during DNA damage response, but the precise downstream function of BMI1 in gene silencing is unclear. Here we identified the UBR5 E3 ligase as a downstream factor of BMI1. We found that UBR5 forms damage-inducible nuclear foci in a manner dependent on the PRC1 components BMI1, RNF1 (RING1a), and RNF2 (RING1b). Whereas transcription is repressed at UV-induced lesions on chromatin, depletion of the PRC1 members or UBR5 alone derepressed transcription elongation at these sites, suggesting that UBR5 functions in a linear pathway with PRC1 in inducing gene silencing at lesions. Mass spectrometry (MS) analysis revealed that UBR5 associates with BMI1 as well as FACT components SPT16 and SSRP1. We found that UBR5 localizes to the UV-induced lesions along with SPT16. We show that UBR5 ubiquitinates SPT16, and depletion of UBR5 or BMI1 leads to an enlargement of SPT16 foci size at UV lesions, suggesting that UBR5 and BMI1 repress SPT16 enrichment at the damaged sites. Consistently, depletion of the FACT components effectively reversed the transcriptional derepression incurred in the UBR5 and BMI1 KO cells. Finally, UBR5 and BMI1 KO cells are hypersensitive to UV, which supports the notion that faulty RNA synthesis at damaged sites is harmful to the cell fitness. Altogether, these results suggest that BMI1 and UBR5 repress the polymerase II (Pol II)-mediated transcription at damaged sites, by negatively regulating the FACT-dependent Pol II elongation.
Perturbation of chromatin structures can cause inappropriate gene expression and loss of genome integrity. Polycomb proteins are recognized in all metazoans for their conserved transcriptional repressive function. The canonical Polycomb Repressive Complex 1 (PRC1) contains BMI1, RNF1 (RING1a), RNF2 (RING1b), core components (PC), Polyhomeotic (PH), and CBX proteins (1, 2). BMI1 serves as a key regulatory component of the PRC1 complex, which is required to maintain the transcriptionally repressed state of many genes throughout development via chromatin remodeling and histone modification (1). The only known enzymatic activity of the BMI1-containing PRC1 complex is to monoubiquitinate histone H2A at Lys-119 (K119) residue, which is associated with transcriptional repression (1, 3, 4). However, the E3 ubiquitin ligase activity of RNF2 or the H2AK119-Ub is dispensable for repression of canonical PRC1 target genes during mouse or Drosophila embryonic development, respectively (5, 6), suggesting that the PRC1 complex may also induce gene silencing through other mechanisms (7). A series of studies has suggested that multiple distinct forms of the PRC1 complex with varying components could exist, and each of these may have distinct modes of regulation and functions (reviewed in ref. 2).
In addition to its well-known role as an oncogene, recent evidence suggests that BMI1 participates in the DNA damage response and genome integrity maintenance. BMI1 is known to localize to DNA double-strand break (DSB) sites and facilitates DNA repair (8–10). Additionally, consistent with its role in gene silencing, BMI1 represses local elongation of RNA polymerase II at damaged chromatin (11). How BMI1 or BMI1-induced H2AK119-Ub modulates transcriptional output upon DNA damage remains incompletely understood.
Here we found that the chromatin localization of the HECT E3 ubiquitin ligase UBR5 is largely dependent on the PRC1 components BMI1, RNF1, and RNF2. Similar to BMI1 and PRC1 components, UBR5-depleted cells fail to repress transcription at damaged chromatin. We further show that BMI1- and UBR5-mediated transcription repression involves the FACT histone chaperon complex. Our findings altogether suggest that UBR5 is a downstream effector of the PRC1 components in transcription silencing at damaged chromatin.
Results
UBR5 Chromatin Localization Is Dependent on BMI1, RNF1, and RNF2.
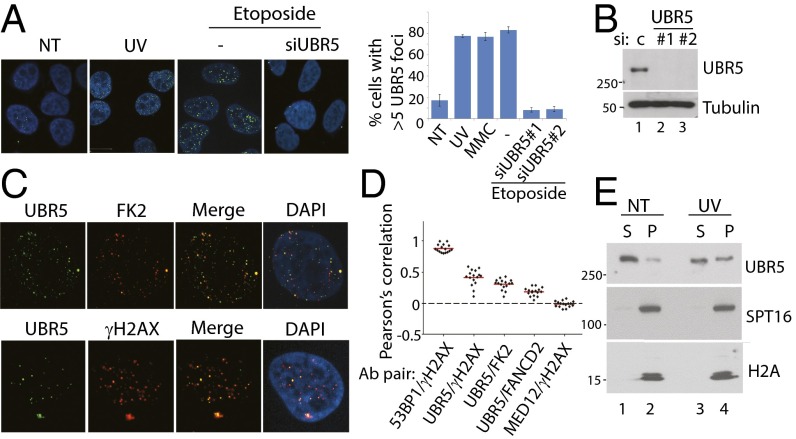
During the course of our studies, we found that endogenous UBR5 proteins form distinct foci in the nucleus, which can be enhanced upon various DNA damaging agent treatments (Fig. 1A). Treatment of two independent siRNAs against UBR5 effectively eliminated the foci, indicating that the foci represent the bona fide UBR5 proteins (knockdown efficiency is shown in Fig. 1B). The foci partially overlap with ubiquitin aggregate (FK2 antibody) and γH2AX, markers of DNA damage (Fig. 1C); however, a significant portion of the foci do not overlap, suggesting that UBR5 in chromatin may have both DNA damage-dependent and -independent functions. The Pearson’s overlap coefficient is quantified using ImageJ software and plotted (Fig. 1D), where the 53BP1–γH2AX overlap is used as a positive control (1 indicates a complete positive relationship), and the MED12–γH2AX overlap as a negative control (−1 indicates a complete negative relationship). Consistent with the immunofluorescence (IF) data, a biochemical fractionation assay shows that a fraction of UBR5 is present in the chromatin-enriched fraction (P), which is increased upon UV damage (Fig. 1E).
Fig. 1.
UBR5 chromatin recruitment is enhanced upon DNA damage. (A) UBR5 forms distinct nuclear foci in HeLa (shown), U2OS, and HCT116 cells in SI Appendix, Fig. S2 following UV (30 J/m2, 1-h recovery), etoposide treatment (2 μM, 12 h), or mitomycin C (MMC) (2 μM, 12 h). n = 80. (B) Confirmation of UBR5 knockdown. (C) The foci partially colocalize with ubiquitin aggregates (FK2) and γH2AX. (D) Pearson’s correlation for overlap from each Ab pair is measured (n = 100). (E) UBR5 becomes enriched in P fraction after UV (S, cyto + nucleoplasm; P, chromatin enriched).
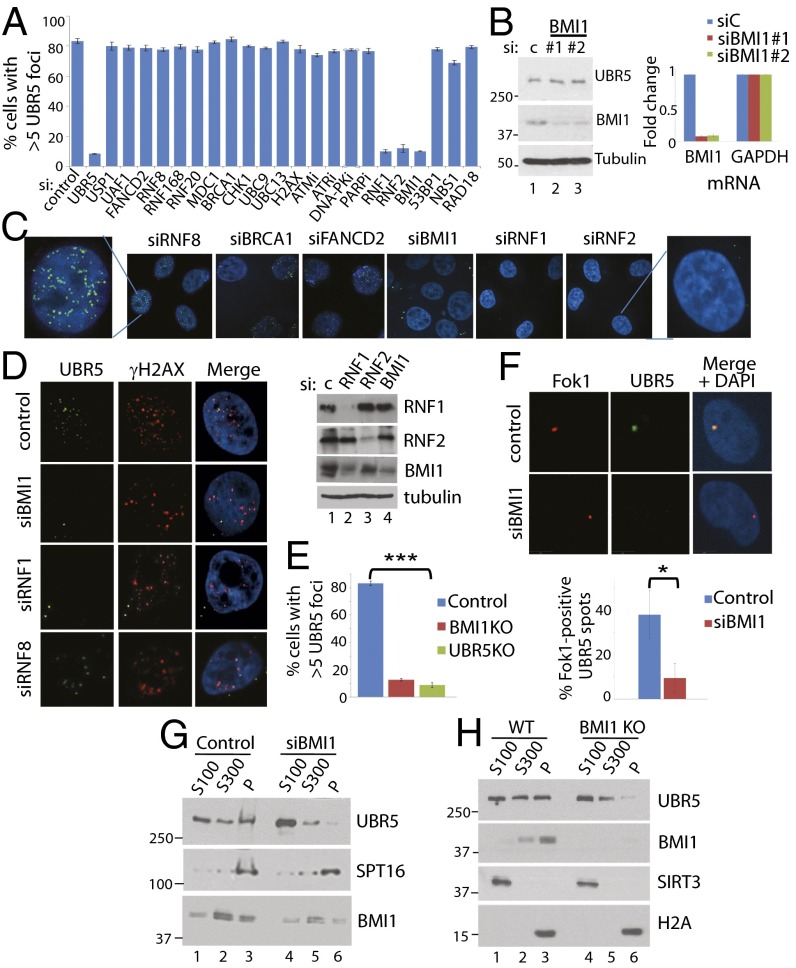
To identify the upstream regulator(s) for the foci formation, we screened siRNAs targeting known regulators of chromatin metabolism and DNA damage response (Fig. 2A). Whereas many canonical DNA damage response factors we tested did not have appreciable effects on the UBR5 foci formation, knockdown of BMI1, RNF1, and RNF2, three integral components of the canonical PRC1 complex, effectively eliminated the foci (two siRNAs were tested for each gene) (Fig. 2A; representative images are shown in Fig. 2 C and D). The knockdown efficiency of selected siRNAs is shown in SI Appendix, Fig. S1. Knockdown of BMI1 did not affect the total UBR5 protein level (Fig. 2B), suggesting that it specifically inhibits the chromatin recruitment. The BMI1 dependence of UBR5 foci was also observed in U2OS and HCT116 cell lines (SI Appendix, Fig. S2). UBR5 foci were also not observed in CRISPR-Cas9 UBR5 KO and BMI1 KO HeLa cells (Fig. 2E). UV-induced foci formation of BMI1 or RNF2 was not affected in UBR5 KO cells (SI Appendix, Fig. S3). UBR5 also localizes at the DSB sites induced by the Fok1 nuclease (Fig. 2F); colocalization of UBR5 and the mCherry–Lac1–Fok1 nuclease is visible upon stabilization of the fusion protein by 4-OHT and Shield-1 (12). The UBR5 foci at Fok1 spots are significantly reduced by BMI1 knockdown. Biochemical fractionation assay further showed that UBR5 in the chromatin fraction (P) is reduced in BMI1 knockdown (Fig. 2G) or KO cells (Fig. 2H). Additional knockdown of a PRC1 member CBX1 did not eliminate the residual UBR5 in the P fraction (SI Appendix, Fig. S4).
Fig. 2.
UBR5 chromatin localization is dependent on the BMI1-containing PRC1 complex. (A) siRNA screen for upstream regulators of UBR5 foci identifies BMI1, RING1a (RNF1), and RING1b (RNF2). HeLa cells were treated with individual siRNAs (20 μM) for 48 h, then 1 μM etoposide was treated for 12 h before fixing. SI Appendix, SI Materials and Methods shows drug treatment. (B) BMI1 knockdown does not reduce total UBR5 protein (Left) or the mRNA (Right) level (qRT-PCR performed in triplicate). (C and D) Some representative images are shown. (E) UBR5 foci were quantified in BMI1 and UBR5 CRISPR-Cas9 KO HeLa cells (n = 100, ***P < 0.0005). (F) Fok1 nucleases at DSBs colocalize with UBR5. Sheild-1 (1 μM) and 4-OHT (1 μM) were treated to PTuner cells for 3 h before fixing (n = 50, *P < 0.01). BMI1 knockdown (G) or KO (H) reduces UBR5 in the chromatin-enriched (P) fractions.
UBR5 Is a Regulator of Transcription at Damaged Chromatin.
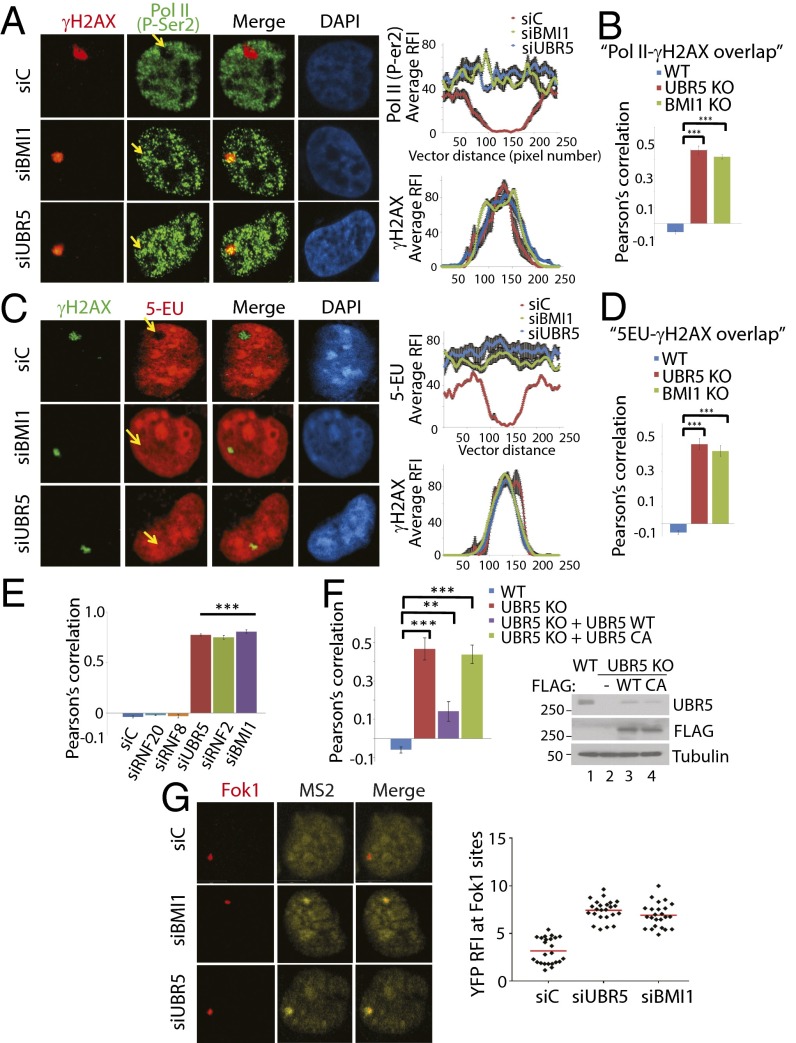
During the DNA damage response, BMI1 suppresses local elongation of RNA polymerase II (Pol II) at damaged chromatin (11). Given the role of BMI1 in transcriptional repression, we sought to ask whether UBR5 acts as a downstream factor of BMI1 to repress transcription at damaged chromatin. We irradiated HeLa cells with UV through a micropore filter, and we observed that the staining of elongating Pol II (anti-CTD P-Ser2 antibody; phosphorylated Ser2 residue of CTD is considered a marker of elongating Pol II) is excluded from γH2AX foci in UV-irradiated spots (Fig. 3A, siC), in line with the notion that transcription is repressed at damaged chromatin. This “exclusion” mechanism is compromised in the BMI1-knockdown cells (siBMI1), consistent with the role of BMI1 in gene silencing at DNA damage sites (11). Interestingly, the same overlap phenotype is observed in the UBR5-knockdown cells (siUBR5). Fig. 3A, Right show measurements of relative fluorescent intensity (RFI) along the UV spots, which highlight that there is no repression of Pol II elongation at the γH2AX spots in the knockdown cells. The same phenotypes were observed in BMI1 and UBR5 KO HeLa cells (Fig. 3B; shown as Pearson’s correlation). A chemical probe, 5-ethynyl uridine (5-EU), is frequently used for detecting global nascent RNA synthesis status using the Click-IT labeling technology; whereas the 5-EU stain is excluded from the γH2AX spots, the 5-EU stain becomes overlapped in both BMI1 and UBR5 knockdown cells (Fig. 3C), or the KO cells (Fig. 3D). These results suggest that RNA synthesis at damaged sites has indeed occurred when UBR5 or BMI1 is depleted. 5-EU was added to dividing cells immediately following UV irradiation, to specifically monitor the nascent RNA synthesis that occurred after the damaging event. Similar phenotypes were observed when the WT, UBR5 KO, and BMI1 KO cells were globally irradiated with UV; the γH2AX and P-Ser2 overlap is shown in SI Appendix, Fig. S5, and the γH2AX and 5-EU overlap is shown in SI Appendix, Fig. S6. Knockdown of RNF2 also led to a near-identical phenotype (Fig. 3E), suggesting that Pol II elongation at damaged sites is regulated by the PRC1 complex activity. Knockdown of RNF20 (which induces H2B ubiquitination) or RNF8 (which induces H1 and H2A ubiquitination) (13) did not show noticeable effects (Fig. 3E). The Pol II overlap with γH2AX in the UBR5 KO cells was largely rescued by introducing UBR5 WT cDNA, but not by UBR5 C2768A catalytic mutant (Fig. 3F; representative images are shown in SI Appendix, Fig. S7), suggesting that the ubiquitinating activity of UBR5 is involved in the transcription repression. The rescue was also seen in the UBR5 knockdown cells (SI Appendix, Fig. S8). Both WT and the C2768A mutant similarly localized to UV-induced lesions (SI Appendix, Fig. S9). UBR5 depletion leads to an elevation of p21 level (14). Transcriptional derepression still occurs in p21−/− HCT116 cells when UBR5 or BMI1 is depleted, indicating that the effects are not caused by elevated p21 proteins (SI Appendix, Fig. S10). ATM and DNA-PK repress transcription at DSB lesions and ATR represses transcription at stalled replication forks (15–18). Inhibition of these kinases led to a varying degree of the P-Ser2 and γH2AX overlap in this assay (SI Appendix, Fig. S11), among which the ATR inhibition was the most effective. This result suggests that ATR may also participate in transcriptional repression in UV-induced lesions. We have further confirmed the transcriptional repressive roles of BMI1 and UBR5 in the reporter cell line described in Tang et al. (12). Consistently, expression of the YFP–MS2 reporter was induced at the Fok1-induced DSB sites in BMI1 and UBR5 knockdown cells (Fig. 3G), but not in the control cells, suggesting that UBR5 represses transcription also at DSB lesions. Altogether, these results suggest that BMI1 and UBR5 induce transcriptional silencing at damaged chromatin.
Fig. 3.
UBR5 depletion phenocopies BMI1 in regulating the transcriptional output at UV-induced lesions. Indicated HeLa cells were locally irradiated with UV (100 J/m2) through 0.4-μM micropore filters, fixed in 1 h, then costained with γH2AX and Pol II P-Ser2 (A) or γH2AX and 5-EU (C). (Right) Measurements of the average RFI along lines across the γH2AX spots. Values were normalized to undamaged regions. Error bars are indicated (n = 10). The γH2AX overlap with Pol II (P-Ser) (B) or 5-EU (D) in WT and KO cells was quantified. Pearson’s correlation was measured using the “co-localization” finder plug-in in ImageJ (n = 100). A score of 1 indicates 100% correlation between red and green pixels; −1, inverse correlation; and 0, no relationship. (E) Indicated HeLa cells were UV irradiated (50 J/m2), and the overlap of P-Ser2 and γH2AX was quantified (n = 150). (F) The P-Ser2 and γH2AX overlap was quantified from the UBR5 KO cells expressing UBR5 WT or C2768A plasmids (n = 50). Western blots are shown on the Right (***P < 0.0005, **P < 0.005). (G) BMI1 and UBR5 knockdowns derepress transcription at DSB sites. Stabilization of the Fok1 nuclease was induced by treatment of 4-OHT and Shield-1 to the PTuner 263 cells, and transcription of the YFP–MS2 reporter was induced by tetracycline 3 h before fixing (n = 35).
UBR5 Associates with BMI1 and the FACT Complex.
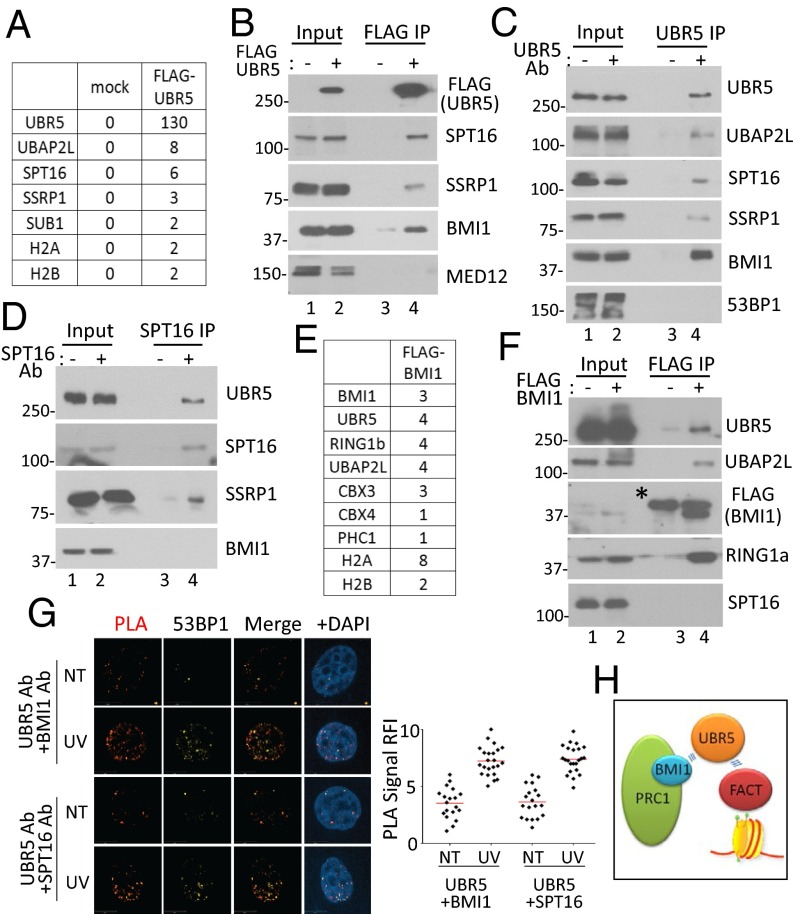
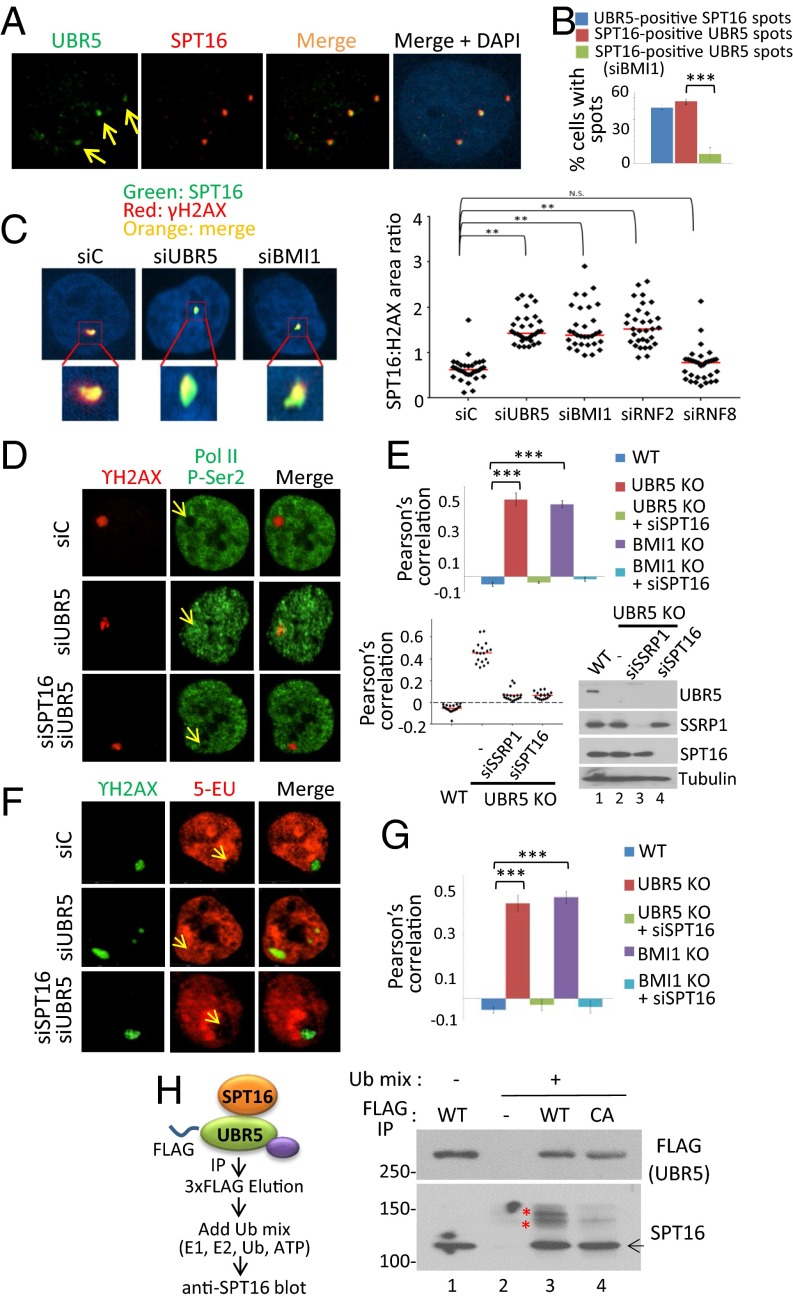
The above results led us to hypothesize that UBR5 may physically interact with certain components of the Pol II complex on the chromatin to regulate the Pol II elongation. With this hypothesis, we investigated the interacting proteins of UBR5 by immunoprecipitating (IP) FLAG–UBR5 from 293T cells followed by mass spectrometry (MS) analysis. The common factors identified from several independent IP-MS experiments included the FACT components SPT16 and SSRP1 and some Pol II-associated factors (Fig. 4A; peptides identified in three IP experiments are shown in SI Appendix, Fig. S12). The interaction between UBR5 and FACT was confirmed by anti-FLAG and anti-UBR5 IPs followed by anti-SPT16 and -SSRP1 Western blots (Fig. 4 B and C); a reciprocal anti-SPT16 IP also showed UBR5 binding (Fig. 4D). Among the proteins identified by MS was UBAP2L, a protein known to interact with BMI1 (19). Because UBAP2L interacts with both BMI1 and UBR5, we blotted for BMI1 in the UBR5 IP, and indeed BMI1 was enriched in the eluate of the UBR5 IP (Fig. 4 B and C). This result prompted us to investigate BMI-interacting proteins (pOZ–FLAG–BMI1 expressing HeLa S3 cells) by MS analysis, and interestingly, we found both UBR5 and UBAP2L, along with other PRC1 components, such as RNF2, CBX, and PHC proteins (Fig. 4E). The FLAG–BMI1 IP indeed confirmed the interaction between BMI1 and UBR5 (Fig. 4F). Note that we did not detect interaction between SPT16 and BMI1 in either IP experiment (Fig. 4 D and F). We further validated the interaction analysis using the proximity ligation assay (PLA), which can detect the protein interactions in situ. This assay detected the specific interaction between UBR5 and SPT16, as well as UBR5 and BMI1 (Fig. 4G), supporting the IP results (no signal was detected for either antibody alone) (SI Appendix, Fig. S13). A significant fraction of the PLA spots colocalized with a surrogate damage marker 53BP1 (Fig. 4G and SI Appendix, Fig. S14), suggesting that the UBR5–SPT16 interaction occurs at damaged chromatin. We did not detect an enhanced interaction of UBR5 with BMI1 or SPT16 after UV by IP analysis (SI Appendix, Fig. S15). UV irradiation increased the intensity of the PLA signals (Fig. 4G), suggesting that damage enhanced the interactions. We also observed that SPT16 depletion did not alter UBR5 foci formation (SI Appendix, Fig. S16). Taken altogether, a working model is proposed in Fig. 4H; UBR5 is recruited by PRC1 complex and then “handed over” to the FACT complex, when gene silencing takes place.
Fig. 4.
UBR5 associates with BMI1 and the FACT complex. (A) MS analysis of the FLAG–UBR5 interacting proteins (Materials and Methods). Unique peptides are shown from one IP. Validation of the interactions by anti-FLAG IP (B) and anti-UBR5 IP (C). Either 53BP1 or MED12 is shown as washing controls. (D) Anti-SPT16 IP was blotted with indicated antibodies. (E) pOZ–FLAG–BMI1 stable expressing HeLa S3 cells were used for the anti-FLAG IP (unique peptides from one IP are shown). (F) Validation of the BMI1 IP by Western blotting. (* indicates heavy chain). (G) PLA was performed with or without UV (70 J/m2), and the cells were costained with indicated antibodies. (Right) RFI quantified using ImageJ (n = 25). (H) Model for the interactions.
BMI1 and UBR5 Regulate FACT-Mediated Transcriptional Elongation at UV-Induced Lesions.
Based on the above interaction analysis, it is possible that UBR5 may antagonize FACT activity to inhibit the Pol II elongation at damaged sites. Interestingly, a previous report showed that a FACT component SPT16 is localized to the UV-induced lesions and is required for the resumption of Pol II elongation during the recovery from UV damage (20). A possible explanation may be that FACT is transiently inhibited at damaged lesions by a certain activity (possibly UBR5), until the damages are repaired and cleared for Pol II elongation. First, we were able to detect colocalization of UBR5 and SPT16 at locally irradiated UV lesions (Fig. 5A; quantified in Fig. 5B). A significant fraction of the UV-induced UBR5 and SPT16 foci colocalized with γH2AX and 53BP1 (SI Appendix, Fig. S17). Interestingly, we consistently observed that the SPT16 foci sizes at UV lesions become significantly enlarged when UBR5 or BMI1 is knocked down (Fig. 5C; the foci sizes were measured in relation to the γH2AX size, which was not significantly altered). This phenotype was consistently observed in BMI1 and UBR5 KO cells (SI Appendix, Fig. S18). Knockdown of RNF2 led to the similar phenotype, suggesting that the SPT16 chromatin enrichment is regulated by the canonical PRC1 complex. In addition to the foci size, we found that the SPT16 foci kinetics are slightly altered in UBR5 or BMI1 knockdown cells; SPT16 foci at the lesions are visible earlier when BMI1 and UBR5 are depleted (SI Appendix, Fig. S19). These results suggest that BMI1 and UBR5 negatively regulate the enrichment of SPT16 at UV lesions. Next, we asked whether the enrichment of SPT16 is responsible for the elongation of Pol II at the UV lesions. Interestingly, when we knock down SPT16, the derepression of Pol II elongation at UV lesions in the UBR5 and BMI1 knockdown cells is effectively reversed (Fig. 5D; Pearson’s correlation in BMI1 and UBR5 KO cells is shown in Fig. 5E). The rescue of nascent RNA synthesis (5-EU) by SPT16 knockdown was also consistently observed (Fig. 5F, Pearson’s correlation in the KO cells is shown in Fig. 5G). Cells depleted of SPT16 or SSRP1 in WT cells showed no change in the elongation of Pol II at the lesions (SI Appendix, Fig. S20).
Fig. 5.
BMI1 and UBR5 regulate FACT-dependent transcription at UV lesions. (A) A 0.4-μM microfilter was used to locally irradiate UV (70 J/m2) in HeLa cells. Cells were fixed 1 h after UV before staining with the indicated antibodies. (B) Statistics (from A) from three independent experiments (n = 75). (C) Enlargement of SPT16 foci in UBR5 and BMI1 knockdown cells. HeLa cells were irradiated with UV (100 J/m2) through 3-μm micropore filters and fixed 1 h after. The number of pixels in each γH2AX and SPT16 colocalized foci was measured using ImageJ, with each pixel representing an area of 0.2 μm2. The ratio of SPT16 to γH2AX was calculated by dividing the SPT16 area by the γH2AX area. (**P < 0.005, n = 35). Red bars indicate the median value for each set. (D) UV irradiation through 0.4-μm microfilter was applied to HeLa cells and the overlap between γH2AX and P-Ser2 (D) and γH2AX–5-EU (F) was observed. (E) The Pol II–γH2AX overlap in UBR5 KO cells was quantified, shown as Pearson’s correlation (n = 120, ***P < 0.0005). Below, siSPT16 and siSSRP1 were tested in UBR5 KO, with the Western blotting confirming the knockdown. (G) Pearson’s correlation for the 5-EU and γH2AX overlap in UBR5 KO cells (n = 50, ***P < 0.0005). (H) UBR5 ubiquitinates SPT16 in vitro. Scheme of the ubiquitination reaction is shown (Left) (see Materials and Methods). Arrow indicates the unmodified SPT16. * indicates bands that appear in WT, in a Ub mix-dependent manner.
These results suggest that the unscheduled RNA synthesis and Pol II elongation through damaged sites in the UBR5 and BMI1 KO cells is dependent on the FACT activity, and that FACT activity at the lesions is antagonized by UBR5. UBR5 may repress FACT recruitment to UV lesions by direct ubiquitination of SPT16, as immunopurified UBR5 can induce ubiquitination of SPT16 through its catalytic activity-dependent manner (Fig. 5H; the upper band species of SPT16 appears only in the reaction induced by WT UBR5 but not C2768A mutant).
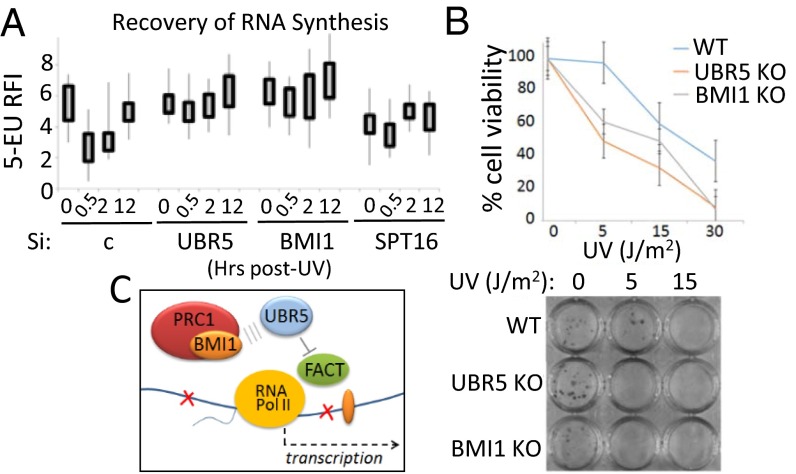
Normally, RNA synthesis is immediately inhibited upon UV irradiation and gradually recovers over time (20). We performed the standard recovery of RNA synthesis (RRS) assay to measure the kinetics of RNA synthesis upon UV treatment in UBR5- and BMI1-depleted cells. Consistent with previous findings, RNA synthesis was suppressed upon UV irradiation (Fig. 6A; 0.5 h post-UV in siC control), which gradually recovered in 12 h. Interestingly, however, the suppression of nascent RNA synthesis was not observed in BMI1 or UBR5 knockdown cells at the same time frame. SPT16 knockdown was used as a control, in which recovery of RNA synthesis is only mildly observed, consistent with a previous study (20). Foci kinetics of UBR5 and SPT16 at the lesions followed a similar trend as 53BP1, although UBR5 disappeared from the lesions a bit earlier than the damage markers 53BP1 and γH2AX (SI Appendix, Figs. S21 and S22, respectively). These results consistently suggest that UBR5 and BMI1 are regulators of UV-induced transcription arrest. Finally, we found that UBR5 KO and BMI1 KO HeLa cells are hypersensitive to UV irradiation compared with the WT HeLa cells (Fig. 6B). This result supports the notion that the failure of RNA synthesis repression may compromise the genome–proteome integrity.
Fig. 6.
BMI1 and UBR5 are regulators of transcription arrest during UV response. (A) Recovery of RNA synthesis assay shows that repression of nascent RNA synthesis does not occur in BMI1 and UBR5 knockdown HeLa cells upon UV (30 J/m2) irradiation. The incorporation of 5-EU was quantified using ImageJ (RFI, n = 40, experiments done in duplicates). (B) Clonogenic cell survival assay shows UV sensitivity of UBR5 KO and BMI1 KO cells (Materials and Methods). (Bottom) Representative image. (C) Model for the proposed mechanism.
Discussion
Here we described that transcriptional repression at damaged chromatin is regulated through a pathway governed by the BMI1-containing PRC1 complex and UBR5. Previous reports showed that reorganization of nucleosome at damaged sites is crucial for stalled Pol II to resume the elongation during recovery from DNA damage (20, 21). Based on the identification of FACT as an interacting partner of UBR5 (Fig. 4), we focused on studying the role of FACT in transcriptional repression induced by BMI1 and UBR5. Our data suggest that BMI1 and UBR5 antagonize Pol II elongation, at least partly, through repression of SPT16 enrichment at damaged chromatin. Based on the data presented, we propose our working model in which BMI1 and UBR5 function as checkpoint factors to transiently suppress FACT activity and to trigger transcriptional arrest at UV lesions (Fig. 6C). As the temporary arrest of Pol II elongation at the lesions is necessary to facilitate DNA repair (22), the hypersensitivity of the UBR5 and BMI1 KO cells to UV irradiation can be possibly interpreted as failure of transcription arrest at the lesions. In an attempt to address the possibility that enhanced SPT16 recruitment at the lesions is a primary cause of the UV sensitivity, we tested if SPT16 knockdown rescues the UV sensitivity of UBR5 or BMI1 KO cells; however, we did not observe the rescue effects (SI Appendix, Fig. S23). This result is not unexpected, because the FACT complex is also involved in DNA replication (23), thus inhibiting SPT16 would likely result in pleiotropic effects. The UBR5 KO cells display accumulation of γH2AX foci that resolves slower than control cells (SI Appendix, Fig. S24) and an increase in the G2/M population (SI Appendix, Fig. S25). The latter is consistent with a previous report (24), and this could be caused at least partly by transcriptional deregulation.
How might UBR5 negatively regulate the FACT recruitment at damaged chromatin? We show that the ubiquitinating activity of UBR5 is necessary for repressing the Pol II elongation (Fig. 3F), and that UBR5 ubiquitinates SPT16 in a manner dependent on its catalytic activity (Fig. 5H). Whereas it is possible that UBR5 ubiquitinates another unknown factor(s) to achieve transcriptional repression, we propose that SPT16 ubiquitination by UBR5 may be primarily responsible for inhibiting FACT enrichment and Pol II elongation at the lesions. A recent proteomic study identified human SPT16 as a polyubiquitinated protein in response to UV damage (25), and whether this ubiquitination event reflects the process we described remains to be determined. Further studies are needed to define how UBR5-induced ubiquitination of SPT16 influences the FACT enrichment at UV lesions. The catalytic activity of UBR5 did not impact the levels of ubiquitinated H2A and γH2AX (SI Appendix, Fig. S26).
There are emerging insights on how gene repression is regulated in the presence of DNA damage. ATM is a key regulator for DSB-induced gene silencing in a mechanism involving H2A ubiquitination (15). ATM also represses Pol I-meditated transcription in the nucleolus (16, 26, 27), by a mechanism involving the interaction between NBS1 and Treacle protein (28, 29). DNA-PK also induces gene silencing near DSB lesions (17), and a bromodomain protein ZMYND8 and the NuRD complex induce gene silencing at DSB sites (30). At stalled replication forks, ATR induces gene repression through degradation of the ASF1a chaperon (18). Our study particularly pertains to the condition of UV damage-induced transcription repression, but also relates to DSB condition as shown in Fig. 3G. As ATM or ATR inhibition also derepressed the UV-induced transcription repression to some degrees (SI Appendix, Fig. S11), there may be functional cross-talks between these kinases and UBR5 in the gene silencing pathways. A study showed that ATM-mediated phosphorylation of ENL protein recruits BMI1 to DSB lesions and thus represses transcription, placing BMI1 downstream of ATM (31). It is possible that the BMI1–UBR5–SPT16 link we describe is under the control of ATM, during the DSB-induced transcriptional repression. Further studies are needed to determine the possible cross-talk among these pathways.
BMI1 is recruited to damaged sites, and promotes recruitment of DSB factors (8–10). BMI1 depletion induces chromosomal aberrations, reduced DNA repair, and sensitivity to IR (8–11, 32). Consistent with the Chagraoui et al. study (11), our data show that BMI1 represses transcription at damaged sites. As BMI1/RNF2 is known to induce H2AK119-Ub, we considered a possibility that the H2AK119-Ub recruits UBR5 to chromatin; however, we could not convincingly detect the physical association of UBR5 with H2AK119-Ub in IP assays. Our assays did reproducibly detect the association between UBR5 and BMI1 (by MS, co-IP, and PLA). Thus, a model is presented in which BMI1 recruits UBR5 to the damaged chromatin, to induce transcriptional repression (Fig. 6C).
Several UBR5 substrates reported include β-catenin, PEPCK1, RORγt, CDK9, spindle assembly checkpoint factors, BuGZ and Bub3, and DNA damage checkpoint protein ATMIN (33–39). UBR5 was also shown to negatively regulate the amplification of RNF168-mediated ubiquitin signaling at DSB sites (40). Some studies also suggested that UBR5 has a role in negatively regulating transcription; UBR5 regulates miRNA-mediated gene silencing (41) and suppresses the transcript level of ACVRL (42). These reports altogether indicate that UBR5 is a multifunctional protein. Our work suggests that a fraction of UBR5 function is dedicated for transcriptional regulation, whose activity is under control of the BMI1–PRC1 complex.
In conclusion, our work presents a link of BMI1, UBR5, and FACT in transcription repression at damaged chromatin.
Materials and Methods
Cell Line, Plasmids, and Chemicals.
HeLa, 293T, and U2OS cells were grown in DMEM supplemented with 10% bovine serum. The CRISPR-Cas9 KO of UBR5 and BMI1 genes used the guide-RNA synthesized from Santa Cruz Biotechnology.
IP and MS Analysis.
Plasmids were transfected to 293T cells at ∼70 confluency, and the harvested cells were lysed with a lysis buffer (25 mM Tris pH 7.4, 0.5% Nonidet P-40, 100 mM NaCl, 0.1 mM EDTA) and the anti-FLAG M2 agarose was added to the cleared lysates for overnight incubation. For the MS sample, the bound proteins were eluted with 4% (wt/vol) SDS and processed using the filter-aided sample preparation method (SI Appendix provides details).
IF and Image Quantification.
Cells (siRNA treated or KO cells) were seeded in 12-well plates onto coverslips, followed by UV irradiation either globally or through micropore filters. Coverslips were washed and fixed for 10 min with 4% PFA. Images were collected by a Zeiss Axiovert microscope equipped with a Perkin-Elmer ERS spinning disk confocal imager using Volocity software. SI Appendix provides the antibody staining in each assay and image quantification methods.
Supplementary Material
Acknowledgments
We thank Dr. Roger Greenberg for the PTuner263 cell line, Dr. Bert Vogelstein for the HCT116 p21−/− cell line, Dr. Charles Watts for sharing the pCMV–Tag2B–UBR5 plasmid through Addgene, Robert Hill for technical support in using confocal microscopy, members of the S.M.S. laboratory, and the University of South Florida’s Center for Drug Discovery and Innovation proteomics facility for MS analysis. This work was supported in part by NIH Grant R15HL126113A1 and a Moffitt–American Cancer Society institutional grant (to Y.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.M. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610735113/-/DCSupplemental.
References
- 1.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6(11):846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 2.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20(10):1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 4.Endoh M, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8(7):e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Illingworth RS, et al. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 2015;29(18):1897–1902. doi: 10.1101/gad.268151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pengelly AR, Kalb R, Finkl K, Müller J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015;29(14):1487–1492. doi: 10.1101/gad.265439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306(5701):1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 8.Ginjala V, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31(10):1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191(1):45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nacerddine K, et al. Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. J Clin Invest. 2012;122(5):1920–1932. doi: 10.1172/JCI57477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chagraoui J, Hébert J, Girard S, Sauvageau G. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc Natl Acad Sci USA. 2011;108(13):5284–5289. doi: 10.1073/pnas.1014263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang J, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20(3):317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorslund T, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527(7578):389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 14.Smits VA. EDD induces cell cycle arrest by increasing p53 levels. Cell Cycle. 2012;11(4):715–720. doi: 10.4161/cc.11.4.19154. [DOI] [PubMed] [Google Scholar]
- 15.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141(6):970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding SM, Boiarsky JA, Greenberg RA. ATM dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. Cell Reports. 2015;13(2):251–259. doi: 10.1016/j.celrep.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19(3):276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 18.Im JS, et al. ATR checkpoint kinase and CRL1βTRCP collaborate to degrade ASF1a and thus repress genes overlapping with clusters of stalled replication forks. Genes Dev. 2014;28(8):875–887. doi: 10.1101/gad.239194.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bordeleau ME, et al. UBAP2L is a novel BMI1-interacting protein essential for hematopoietic stem cell activity. Blood. 2014;124(15):2362–2369. doi: 10.1182/blood-2014-01-548651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinant C, et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell. 2013;51(4):469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155(1):94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci. 2010;35(6):333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Abe T, et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem. 2011;286(35):30504–30512. doi: 10.1074/jbc.M111.264721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz MA, et al. The E3 ubiquitin ligase EDD regulates S-phase and G(2)/M DNA damage checkpoints. Cell Cycle. 2007;6(24):3070–3077. doi: 10.4161/cc.6.24.5021. [DOI] [PubMed] [Google Scholar]
- 25.Elia AE, et al. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol Cell. 2015;59(5):867–881. doi: 10.1016/j.molcel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruhlak M, et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447(7145):730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 27.van Sluis M, McStay B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015;29(11):1151–1163. doi: 10.1101/gad.260703.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen DH, et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat Cell Biol. 2014;16(8):792–803. doi: 10.1038/ncb3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciccia A, et al. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc Natl Acad Sci USA. 2014;111(52):18631–18636. doi: 10.1073/pnas.1422488112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong F, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29(2):197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ui A, Nagaura Y, Yasui A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol Cell. 2015;58(3):468–482. doi: 10.1016/j.molcel.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci. 2010;30(30):10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang T, Cronshaw J, Kanu N, Snijders AP, Behrens A. UBR5-mediated ubiquitination of ATMIN is required for ionizing radiation-induced ATM signaling and function. Proc Natl Acad Sci USA. 2014;111(33):12091–12096. doi: 10.1073/pnas.1400230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cojocaru M, et al. Transcription factor IIS cooperates with the E3 ligase UBR5 to ubiquitinate the CDK9 subunit of the positive transcription elongation factor B. J Biol Chem. 2011;286(7):5012–5022. doi: 10.1074/jbc.M110.176628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutz S, et al. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature. 2015;518(7539):417–421. doi: 10.1038/nature13979. [DOI] [PubMed] [Google Scholar]
- 36.Hay-Koren A, Caspi M, Zilberberg A, Rosin-Arbesfeld R. The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol Biol Cell. 2011;22(3):399–411. doi: 10.1091/mbc.E10-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, He X, Feng D, Zhu X, Zheng Y. RanGTP aids anaphase entry through Ubr5-mediated protein turnover. J Cell Biol. 2015;211(1):7–18. doi: 10.1083/jcb.201503122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scialpi F, Mellis D, Ditzel M. EDD, a ubiquitin-protein ligase of the N-end rule pathway, associates with spindle assembly checkpoint components and regulates the mitotic response to nocodazole. J Biol Chem. 2015;290(20):12585–12594. doi: 10.1074/jbc.M114.625673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang W, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gudjonsson T, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150(4):697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 41.Su H, et al. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Mol Cell. 2011;43(1):97–109. doi: 10.1016/j.molcel.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen HW, et al. A functional genomic approach reveals the transcriptional role of EDD in the expression and function of angiogenesis regulator ACVRL1. Biochim Biophys Acta. 2013;1829(12):1309–1319. doi: 10.1016/j.bbagrm.2013.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.