Significance
Most eukaryotic genomes contain abundant transposable elements (TEs), mobile DNA elements that can replicate and move within the genome. Because of the deleterious nature of active TEs, cells have mechanisms to suppress and prevent TE activation, including formation of repressive heterochromatin. In this report, we show that many TEs become activated with age in Drosophila, and this activation is prevented by dietary restriction, an intervention known to extend life span. We also show TE activation is blocked by genetic manipulations that stabilize heterochromatin and increase life span. This study provides evidence that a breakdown in TE silencing and repression may be a contributing factor to aging, and preventing TE activation may be a significant method of ameliorating the diseases of aging.
Keywords: aging, heterochromatin, transposable elements, dietary restriction, silencing
Abstract
Transposable elements (TEs) are mobile genetic elements, highly enriched in heterochromatin, that constitute a large percentage of the DNA content of eukaryotic genomes. Aging in Drosophila melanogaster is characterized by loss of repressive heterochromatin structure and loss of silencing of reporter genes in constitutive heterochromatin regions. Using next-generation sequencing, we found that transcripts of many genes native to heterochromatic regions and TEs increased with age in fly heads and fat bodies. A dietary restriction regimen, known to extend life span, repressed the age-related increased expression of genes located in heterochromatin, as well as TEs. We also observed a corresponding age-associated increase in TE transposition in fly fat body cells that was delayed by dietary restriction. Furthermore, we found that manipulating genes known to affect heterochromatin structure, including overexpression of Sir2, Su(var)3–9, and Dicer-2, as well as decreased expression of Adar, mitigated age-related increases in expression of TEs. Increasing expression of either Su(var)3–9 or Dicer-2 also led to an increase in life span. Mutation of Dicer-2 led to an increase in DNA double-strand breaks. Treatment with the reverse transcriptase inhibitor 3TC resulted in decreased TE transposition as well as increased life span in TE-sensitized Dicer-2 mutants. Together, these data support the retrotransposon theory of aging, which hypothesizes that epigenetically silenced TEs become deleteriously activated as cellular defense and surveillance mechanisms break down with age. Furthermore, interventions that maintain repressive heterochromatin and preserve TE silencing may prove key to preventing damage caused by TE activation and extending healthy life span.
There is mounting evidence that preservation of the epigenetic structure of chromatin is important to maintaining healthy aging, and loss of this chromatin structure may be a hallmark of the aging process (reviewed in ref. 1). In aging yeast, cells lose characteristic silencing in heterochromatic regions, resulting in deleterious age-related consequences such as sterility, and replicative life span is regulated by the histone H4K16 acetylation mark (2, 3). In Caenorhabditis elegans, genetic manipulation of chromatin factors affecting histone methylation and acetylation directly affect life span (4). In Drosophila, the integrity of constitutive repressive heterochromatin declines with age at both the genomic and cellular level (5), with one consequence of these changes being an age-related loss of gene silencing in repressive constitutive heterochromatin regions in multiple tissues of the fly (6). In mice, changes in epigenetic chromatin marks are seen with age, and age-related phenotypes of behavior, memory, and learning are observed on genetic manipulation of certain histone marks (1). A breakdown in heterochromatin organization has also been observed during replicative senescence of human cells in culture, with expression and copy number of transposable elements (TEs) increasing (7, 8).
TEs are mobile genetic elements that make up a large percentage of eukaryotic genomes and that are particularly enriched in regions of heterochromatin (reviewed in ref. 9). Because TEs function by either excising or copying themselves from the genome and integrating into new genomic locations, there is a high potential for deleterious effects on cellular function. Overexpressing TE RNAs is sufficient to cause cytotoxicity, and inhibiting expression of these TEs causes reversal of both senescence and cytotoxic phenotypes (10, 11). TE activation has been observed with age in mice (8, 12), as well as C. elegans (13), yeast (14), Drosophila (15, 16), and senescent human tissue culture cells (7).
To combat the genomic threat of activation of TEs and loss of heterochromatic gene silencing, somatic cells have evolved mechanisms to suppress TE transcription, primarily through establishment and maintenance of repressive heterochromatin at TE sites (17). In Drosophila, the small interfering RNA (siRNA) pathway, through the proteins Dicer-2 and Argonaute 2 (AGO2), recruits the histone H3K9 methyltransferase Su(var)3–9 to catalyze formation of repressive heterochromatin at TE sites (18–20). Mutation of Dicer-2, the double-stranded RNase responsible for generating siRNAs, results in TE reactivation and heterochromatin disorganization (19, 20). The RNA-editing enzyme ADAR (adenosine deaminase acting on RNA) is thought to modify the effectiveness of siRNA-directed heterochromatinization, and therefore the efficacy of TE silencing, by editing the dsRNA substrates used for siRNA biogenesis, thereby preventing proper processing by Dicer-2 (21). Sir2/Sirt1, the founding member of the sirtuin family of proteins, is a protein deacetylase important in maintaining gene silencing in heterochromatin regions of yeast and flies through deacetylation of specific histone lysines required for establishment and spreading of heterochromatin (3, 6, 22). Increasing Sir2 expression has been shown to repress the normal age-related loss of gene silencing in heterochromatin regions in both yeast and flies (3, 6), as well as the loss of gene silencing seen in mammalian embryonic stem cells undergoing genotoxic stress (23). In addition to their roles in heterochromatin formation and maintenance, these pathways have also been linked to aging. Decreasing activity of the siRNA pathway through Ago2 and Dicer-2 mutants leads to de-repression of TE activity and shortened life span (15, 24). Sir2 has a central role in aging, with increased gene dosage shown to extend life span in yeast (2) and in nine of 10 studies in nematodes and flies (25–28). Hypomorphs of Adar, a negative regulator of the siRNA pathway, also exhibit an extension of life span (21). These pathways share an ability to affect heterochromatin formation and life span.
Here we report a characterization of the heterochromatic transcriptome in adult Drosophila tissues and show activation of TEs and increased transposition with age. We provide evidence that the aging process is characterized by a failure to properly control and repress expression of heterochromatin-based genes and TEs in the soma. We show that genetic interventions positively modifying heterochromatin structure or negatively modulating TE activity extend metazoan life span. Finally, we show that attenuating TE activity pharmacologically decreases transposition and extends life span in Dicer-2 mutants. This study offers the first comprehensive characterization of the effect of known life span-extending interventions on heterochromatic regions and explores new interventions for controlling age-associated TE reactivation.
Results
Expression of Genes Located Within Heterochromatin Increases with Age and Is Attenuated Under Dietary Restriction.
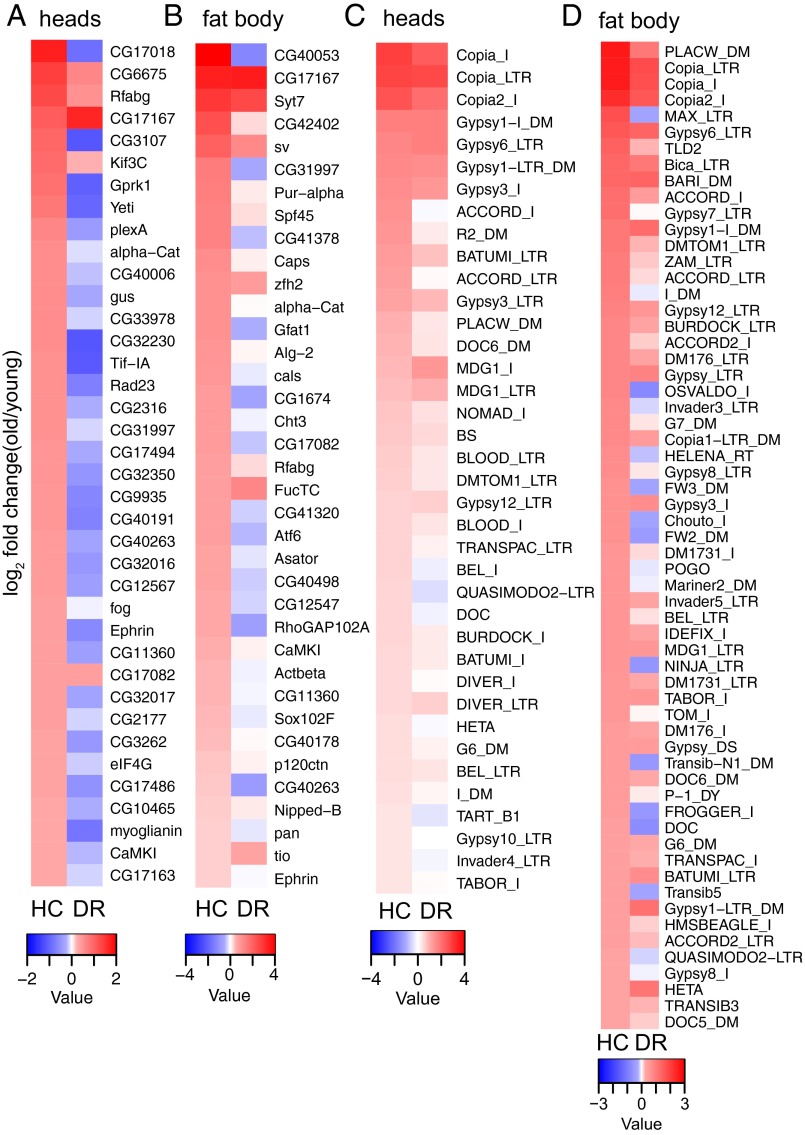
Because heterochromatic regions change in structure with age, specifically in female heads and fat bodies (5), we reasoned that genes located within these regions may also show changes in silencing or gene expression with age. We used RNA-seq to assay the expression of the ∼250 genes natively located within the Drosophila heterochromatin compartments (29) from young (10 d) and old (40 d) female fly heads and fat bodies. The vast majority of cells in the head are neurons and glial cells. The fly fat body is the functional homolog of mammalian adipose tissue and liver, and a central organ in the fly immune system. We found a number of genes located in heterochromatic regions increased expression with age in both wild-type heads and fat bodies (Fig. 1 A and B). Gene ontology analysis reveals no significantly enriched pathways among these genes, suggesting the age-related increase in expression of these genes is likely to be a consequence of loss of heterochromatic silencing, rather than a specific compensatory or correlative response tied to aging.
Fig. 1.
DR attenuates age-related loss of silencing in heterochromatin genes and TEs. (A and B) Heat map shows log2 fold change (old/young) values for heterochromatin genes in both (A) heads and (B) fat bodies. (Left) Log2 fold change under HC conditions. (Right) Log2 fold change under DR. Heat map shows genes with log2 fold changes greater than 0.263 (1.2× fold change) under HC conditions that are located within annotated heterochromatin regions (29). Ninety-five percent of shown genes in heads and 89% in fat bodies are suppressed by DR. (C and D) Heat map shows log2 fold change (old/young) of TE expression for both HC (Left) and DR (Right) diet in both (C) heads and (D) fat bodies. Heat map shows TEs with log2 fold changes greater than 0.322 (1.25× fold change) under HC conditions. Eighty-seven percent of shown TEs were suppressed by DR in both heads and fat bodies.
Dietary restriction (DR) is an intervention known to extend life span and delay numerous age-related phenotypes. We previously observed that DR can oppose age-related loss of gene silencing of reporter genes in constitutive heterochromatin (6). On this basis, we examined whether the age-dependent increase in expression of native genes in the constitutive heterochromatin regions were also suppressed under a DR regimen. The vast majority (95% in heads, 89% in fat bodies) of heterochromatin genes that increased expression with age on high-calorie food increased less or decreased in expression with age on DR food [high calorie (HC) vs. DR] in both heads (Fig. 1A) and fat bodies (Fig. 1B). Although we observed a similar fraction of euchromatin genes that increased expression with age, this pattern of suppression by DR was not maintained in the most highly up-regulated euchromatin genes (Fig. S1 A and B), suggesting DR has a selective effect on heterochromatin genes. Similar to previous studies using reporter genes in both yeast (2) and flies (6), these data suggest that gene expression in heterochromatic regions becomes dysregulated with age, and that a known life span-extending intervention, DR, can prevent this.
Fig. S1.
Effect of DR on age-related loss of silencing in euchromatin genes. (A and B) Heat map shows log2 fold change (old/young) of top 50 euchromatin genes up-regulated with age in heads (A) and fat bodies (B). (Left) HC diet. (Right) DR. Genes located in euchromatin regions do not show the same consistent DR suppression of age-related increase in expression observed in heterochromatin genes (Fig. 1 A and B).
TE Expression Increases with Age and Is Attenuated Under DR.
One of the most notable attributes of heterochromatin is the abundant enrichment of TEs within these regions. The Drosophila genome is rich in retroviral-like long terminal repeat (LTR) and long interspersed nuclear elements (LINEs) (30), and activation of both LINEs and LTRs has been observed in the fly nervous system and fat body during aging (15, 16). Because heterochromatin integrity declines with age in multiple model systems (1), we hypothesized that a loss of heterochromatin in regions highly enriched with TEs would show a correlated increase in TE expression with age. To test this hypothesis, we examined the expression profile of TEs with age and diet in female fly heads and fat bodies, using whole transcriptome RNA-seq. Similar to our observations for native heterochromatin genes, we also found many TEs that increased their expression with age, in both heads and fat bodies (Fig. 1 C and D). We observed a high degree of overlap in the TEs that increased expression most with age between these two tissues, most of which were retrotransposons (Fig. S2A). In both tissue types, as observed with the heterochromatin genes, DR attenuated the age-related increase in expression observed in HC food (Fig. 1 C and D, right “DR” columns). TEs that had the highest increase in expression with age on HC food tended to increase less or even decrease in expression with age under DR conditions (87% lower in DR relative to HC in both heads and fat bodies; Dataset S1). These data suggest that, similar to reporter genes in heterochromatin (2, 6) and native heterochromatin genes (Fig. 1 A and B), TE silencing is lost with age, and DR opposes this.
Fig. S2.
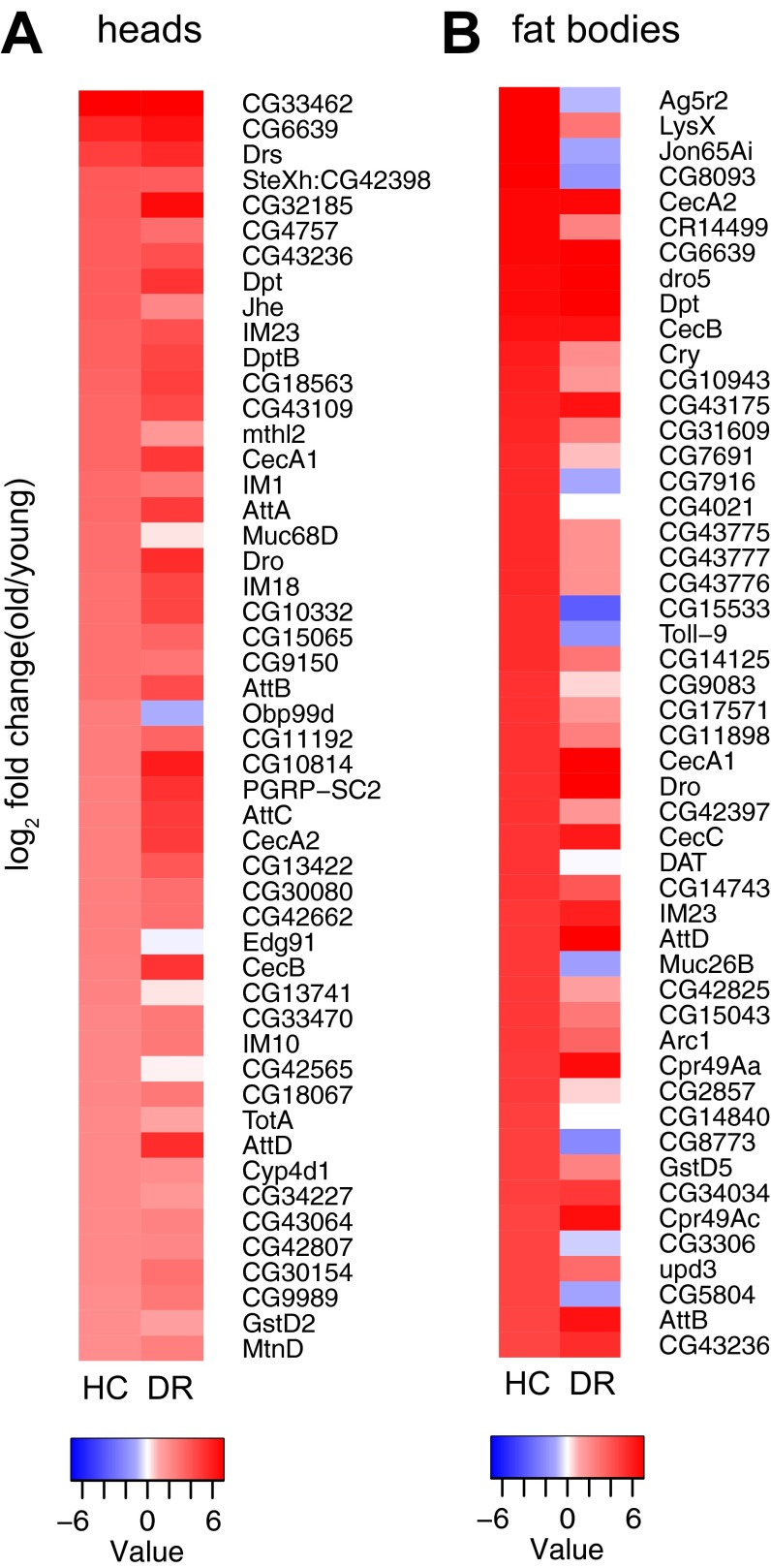
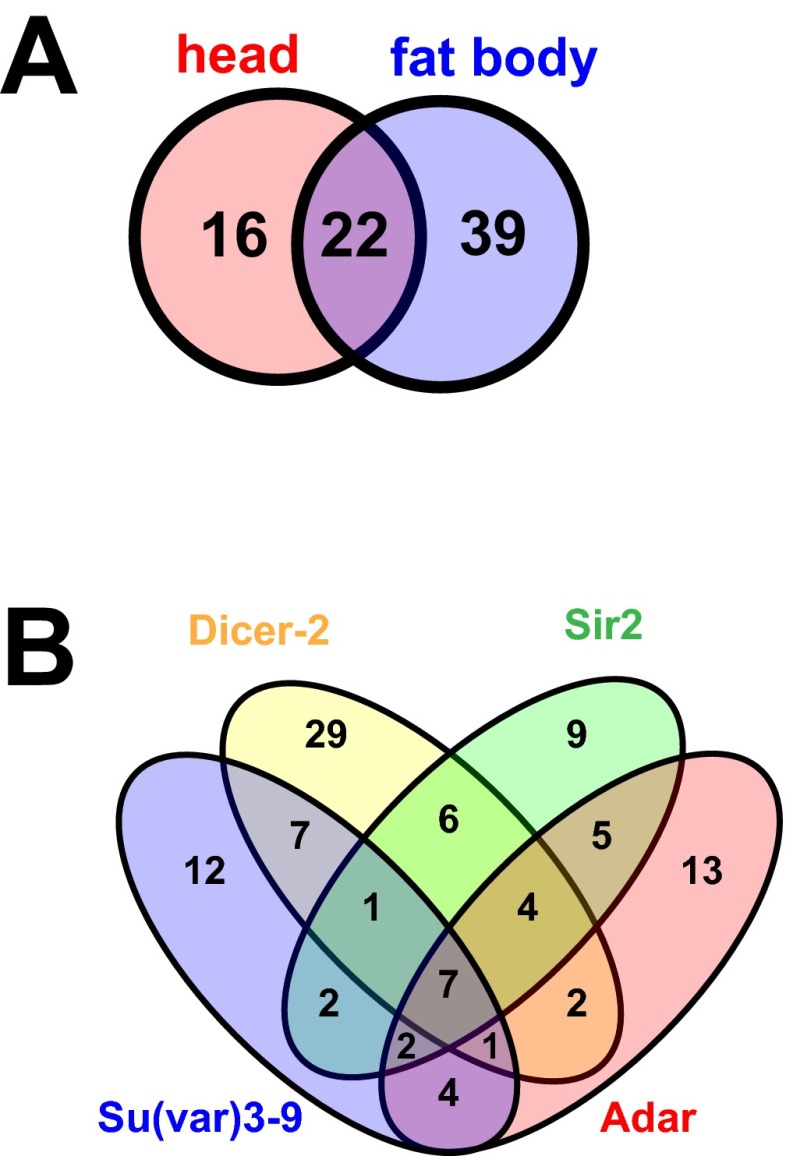
Spectrum of up-regulated TEs shows significant overlap among different tissues and genotypes. (A) Venn diagram indicating overlap of TEs that increase >0.322 log2 fold change in head and fat body libraries (P = 0.0395, Fisher’s exact test). The LTR retrotransposons copia, gypsy6, and accord were among the most highly up-regulated TEs with age in both backgrounds. Copia is the most highly expressed TE in both our head and fat body datasets, and additionally, copia is known to be active in D. melanogaster. (B) Four-way Venn diagram showing overlap of TEs displayed in four genotypes of Fig. 3 A–D. TEs shown are those that increase with age in the control or uninduced condition. Five TEs (seven sequences including Copia_LTR, Copia_I, Gypsy6_LTR, Copia2_I, BATUMI_LTR, BATUMI_I, Gypsy5_I; internal and LTR portions of sequences are annotated separately) appear in all four genetic backgrounds. The fact that there are also differences between the genotypes indicates that effects on TE expression are likely dependent on genetic background and strain-specific genomic location of TEs.
TE Transposition Increases with Age and Is Delayed by DR.
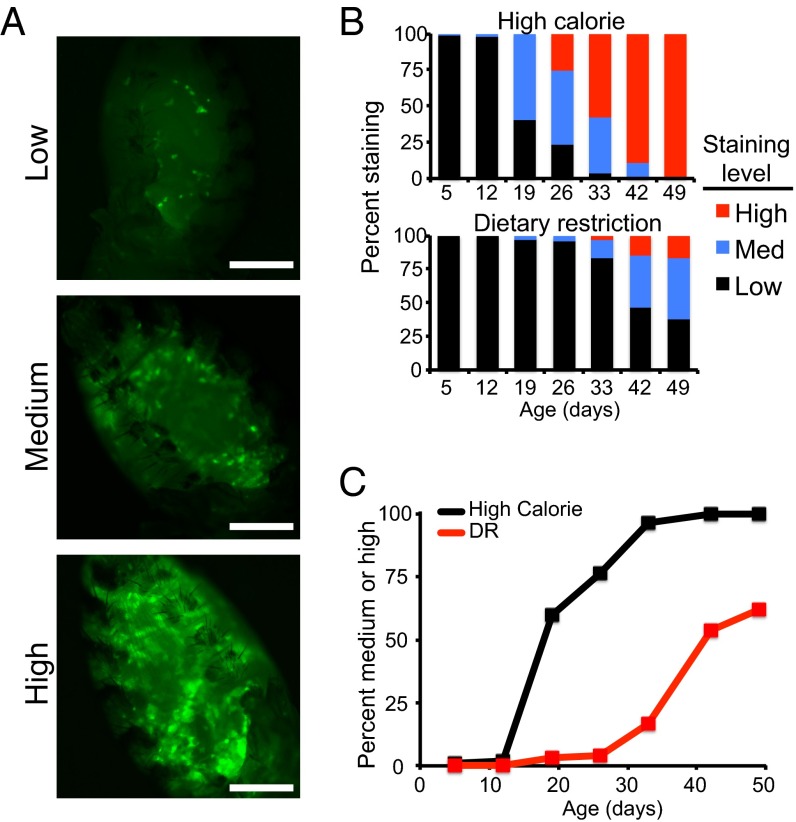
An age-dependent increase in TE expression and transposition in the Drosophila brain associated with decline in neural function has been reported (15). Similar increases in TE expression levels and copy numbers have been observed in aging mouse tissue (8). To assay for age-related transposition in our flies, we used the gypsy-TRAP reporter system (15). This system is designed such that green fluorescent protein (GFP) becomes expressed when an endogenous gypsy retrotransposon jumps into an engineered site containing known hotspot sequences from the ovo locus (31). We monitored flies as they aged and visually sorted them into either low, medium, or high categories based on the number of GFP-positive cells observed in the fat body (Fig. 2A). Longitudinal fly cohorts were assayed for transposition every 7 d on either HC or DR foods (Fig. 2B). We observed a clear age-associated increase in transposition, and DR delayed this increase relative to HC controls (Fig. 2 B and C). Similar results were seen in two additional genetic backgrounds (Fig. S3 A–D). We examined the life spans of these three different genetic background gypsy-TRAP lines and observed that life span correlated with the timing of increased transposition. Lines exhibiting earlier transposition showed shorter life span, and those with later transposition longer life span (Fig. S3E). Similarly, the delay in age-related transposition resulting from DR correlated with the ability of DR to extend life span in each line. To ensure the increased GFP expression we observed with age was caused by transposition rather than age-related disruption in the GAL80/GAL4/GFP reporter system, we examined a gypsy-TRAP line with a mutant ovo locus not susceptible to transposition and observed no significant increase in GFP expression with age (Fig. S3F). Rather than a constant linear increase in GFP expression with time, the gypsy-TRAP assay showed a clear age-associated increase in TE mobilization, consistent with the TE theory of aging, which posits an age-related loss of normal cellular surveillance mechanisms and consequent increase in deleterious TE activity. The relationship between the timing of increased gypsy transposition and life span further supports TE mobilization as an age-associated phenomenon.
Fig. 2.
DR delays age-related increase in TE transposition. (A) Gypsy-TRAP flies (genotype: w1118; If/+; r4-GAL4, UAS-GFP/tub-ovo-GAL80) were categorized as low (<20 GFP-positive cells), medium (20–100), or high (>100) based on number of GFP-positive cells observed in fat body (representative images shown). The demarcation between low and medium categories is clear; most flies have either fewer than 10 cells staining, or more than 50. (Scale bar, 200 µm.) (B) Percentage of observed flies falling into each category at indicated points for HC (Top) and DR (Bottom) diets (n = ∼120 per cohort). (C) Transposition (defined here as medium- or high-category GFP staining) increases with age and is delayed by DR. Percentage of flies falling into either medium or high categories is displayed with time for both HC and DR diets. Log-rank P < 10−10.
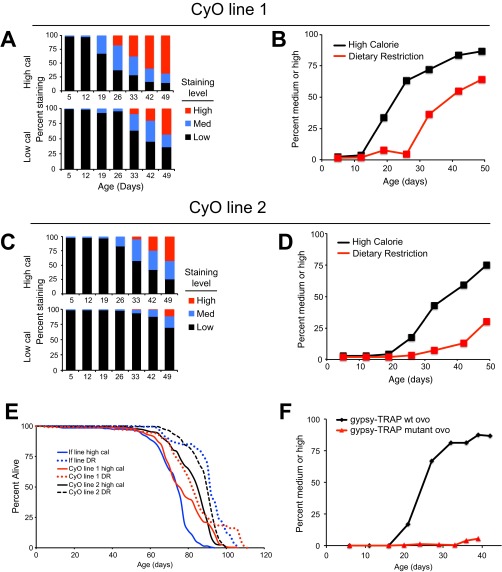
Fig. S3.
DR delays age-related increase in TE transposition in alternate genetic backgrounds. (A and C) Percentage of gypsy-TRAP flies in two independently derived CyO lines (w1118; CyO/+; r4-GAL4, UAS-GFP/tub-ovo-GAL80; see SI Materials and Methods for further details) falling into each GFP staining category at indicated points, for both HC (Top) and DR (Bottom) diets. Data are presented analogously to Fig. 3B (n = 120 HC, 80 DR). (B and D) Percentage of flies falling into medium or high staining categories in these two gypsy-TRAP lines with time, for both HC and DR diets, analogous to Fig. 3C (n = 123 HC, 129 DR). Log-rank P < 10−10 for both lines. (E) Life span of individual gypsy-TRAP lines correlates with onset of increased transposition. Survivorship curves for the three independent gypsy-TRAP genetic backgrounds shown in A–D, as well as Fig 2 B and C. Adult females (n = 250) are shown for each line on HC (solid lines) and DR (dotted) food types. If line: HC mean = 72.3, median = 76; DR mean = 90.9, median = 92. CyO line 1: HC mean = 77.5, median = 76; DR mean = 83.1, median = 82. CyO line 2: HC mean = 82.4, median = 86; DR mean = 88.6, median = 90. All DR life spans are significantly different compared with their respective HC cohort at P < 10−6 (log-rank). All HC life spans are also significantly different from each other (P < 10−6 for If/CyO1 and If/CyO2; P = 0.03 for CyO1/CyO2). (F) Observed increases in GFP expression are not a result of nonspecific age-related changes in GAL80, GAL4, or GFP expression. Percentage of fat body cells staining positive for GFP with age in the normal gypsy-TRAP line (black) as well as a gypsy-TRAP line containing a mutated ovo locus that is not sensitive to gypsy transposon integration (red). Note the mutant gypsy-TRAP line shows no increase in GFP expression with age, indicating GFP expression is reporting increased transposition, rather than general age-related changes.
Age-Related Increases in TE Expression Are Attenuated by Genetic Interventions That Affect Heterochromatin Structure.
Our data demonstrate a correlation between changes in heterochromatin structure with age and expression and transposition of TEs during normal aging. This led us to examine whether genetically manipulating heterochromatin structure and maintenance, either directly or indirectly, could also affect the age-related changes in expression of TEs. To test our hypothesis, we genetically increased the activity of two important systems that maintain repressive heterochromatin in somatic cells: the siRNA pathway and Sir2. Using transgenic fly lines, we increased gene dosage of Sir2, Dicer-2, and Su(var)3–9 and also used an Adar hypomorph, interventions we expect to stabilize heterochromatin. Each of these genes offers a unique and complementary approach to simultaneously manipulating heterochromatic regions and enhancing TE silencing.
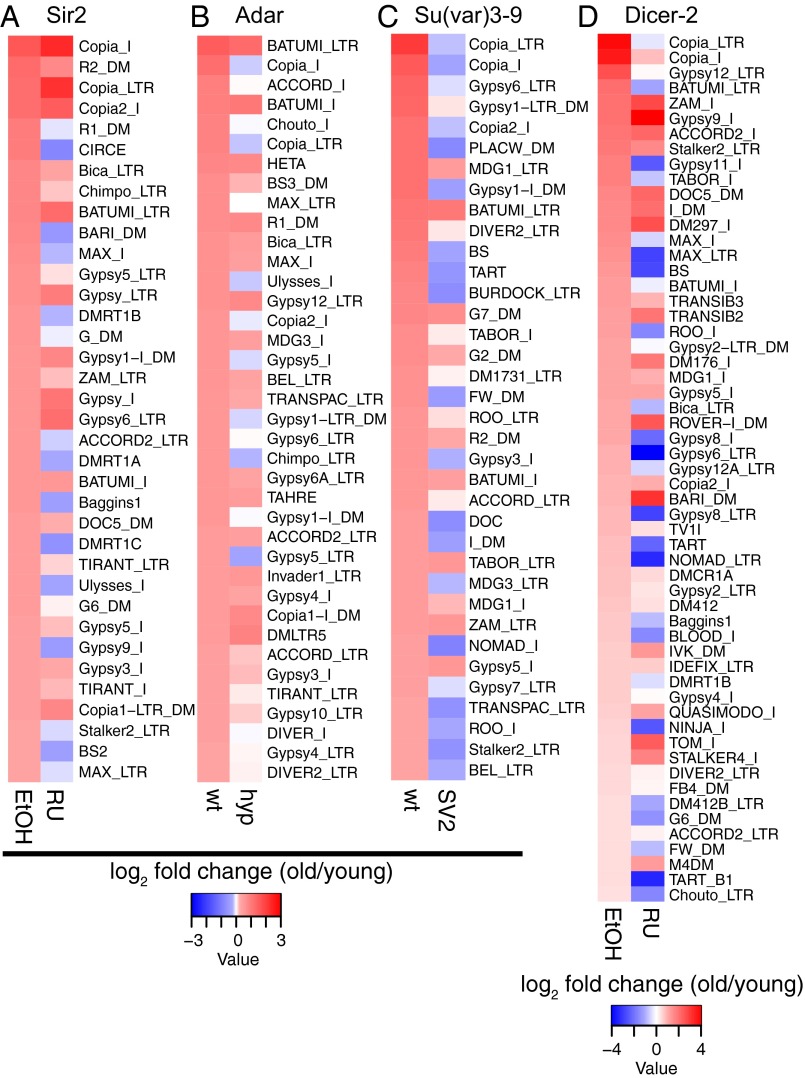
We assayed age-dependent loss of silencing of TEs in each of these various transgenic [Sir2, Su(var)3–9, and Dicer-2] or hypomorphic (Adar) fly lines compared with their respective controls. The TEs that showed the greatest increase in expression with age in the control lines were overwhelmingly attenuated or decreased with age by these genetic interventions [Fig. 3 A–D; Sir2, 72%; Adar, 84%, Su(var)3–9, 92%; Dicer-2, 68%]. Interestingly, we again observed significant overlap in the TEs that increased with age among the different genetic backgrounds (Fig. S2B). Together, these data suggest that increased expression of Sir2, Dicer-2, or Su(var)3–9, as well as decreased gene dosage of Adar, all prevent the age-related loss of silencing of TEs observed in wild-type and control lines, suggesting a common link between heterochromatin factors and proper maintenance of TE silencing.
Fig. 3.
Manipulation of genes in heterochromatin pathways affects TE expression. Heat maps of log2 fold change (old/young) of TEs for the following genotypes and their respective controls: (A) elav-GeneSwitch > UAS-EP2300 (Sir2 overexpression); (B) Adar hypomorph vs. wild-type; (C) Su(var)3–9 overexpression [SV2 and SV3 lines overexpress Su(var)3–9 from a basally expressed UAS promoter 160–180% above control; Fig. S4B]; and (D) tubulin-GeneSwitch > UAS-Dicer-2 (Dicer-2 overexpression). (Left) Control condition. (Right) Transgenic, hypomorph, or RU486-induced condition. The percentage of shown TEs that are suppressed by the indicated genotype are (A) 72%, (B) 84%, (C) 92%, and (D) 68%. All TEs with log2 fold change (old/young) greater than 0.322 (1.25× fold change) in the control condition are shown.
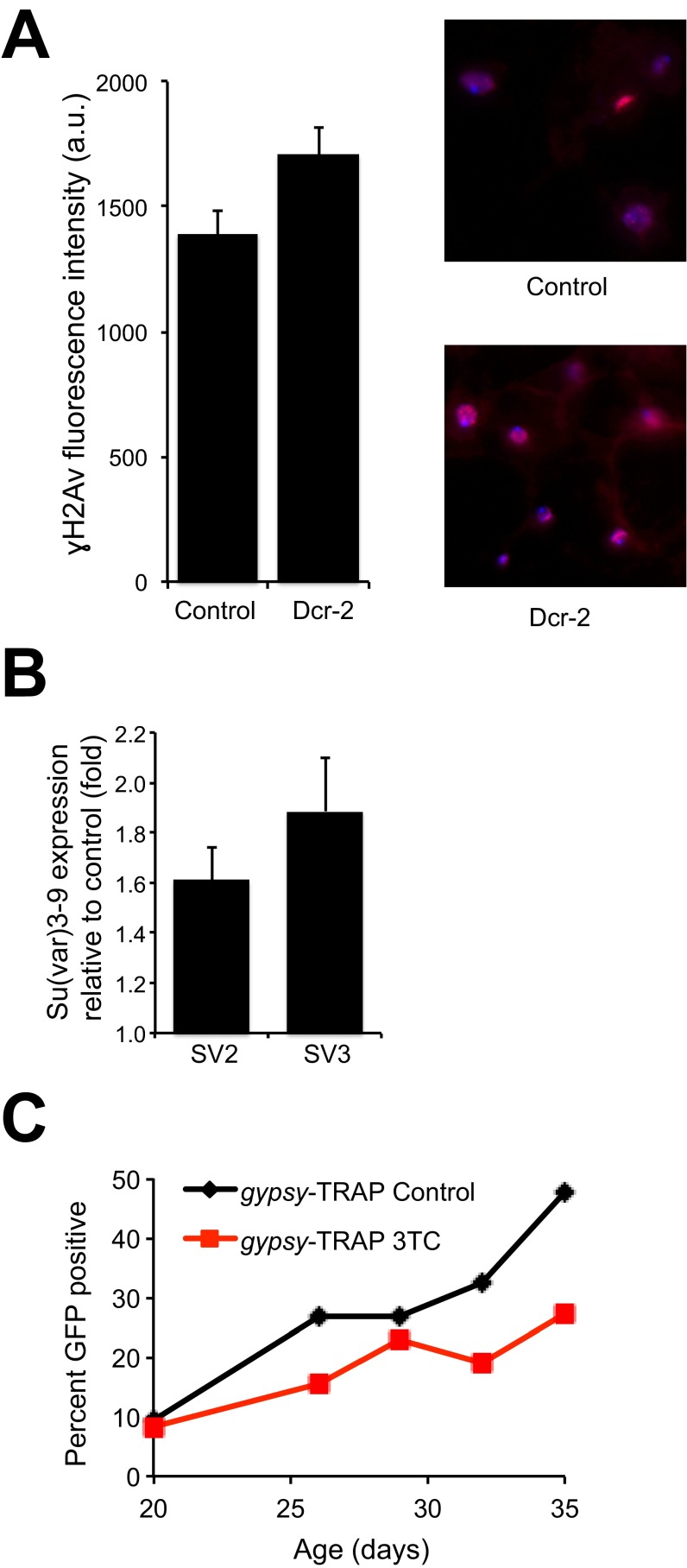
TE activation is known to disrupt gene expression and destabilize the genome. Increased DNA damage has been observed in aging fat body nuclei, as assayed by γH2Av immunostaining (16). Our finding of a repression of TE expression with increased Dicer-2 expression led us to investigate whether genomic integrity was similarly compromised in animals with disrupted RNAi/heterochromatin pathways and increased TE activity. Similar to depletion of AGO2 in fat body cells (16), we observed an increase in double-strand breaks in Dicer-2 mutants compared with controls in fat body nuclei (Fig. S4A), suggesting deleterious genomic consequences when proper TE silencing is disrupted.
Fig. S4.
(A) Dicer-2 mutants have increased double-strand breaks in nuclei compared with controls. Relative nuclear γH2Av fluorescence intensity in fat body cells from w1118 controls or Dicer-2L811fsX mutants. Dicer-2 mean fluorescence is 23% higher than control (n = 50; t test P = 0.027). Representative images from each genotype are shown. (B) Expression level of Su(var)3–9 relative to control for SV2 and SV3 lines, assayed by quantitative PCR, using GAPDH1 as a reference. Error bars represent SD. (C) 3TC treatment delays age-related increase in transposition. Time course of GFP expression in gypsy-TRAP flies either untreated (control) or treated with 10 µM 3TC, starting at day 20 of adulthood. Percentage of GFP-positive (>100 cells) fat bodies for each condition is shown.
Genetic Interventions That Affect Heterochromatin Structure Also Affect Life Span.
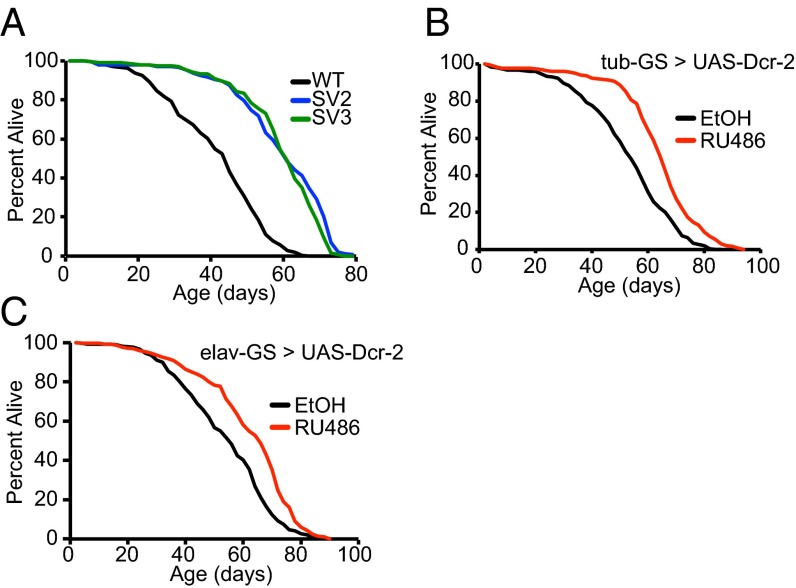
In addition to their effects of delaying age-related heterochromatin gene and TE silencing loss, both increased Sir2 expression and decreased Adar expression have been shown to extend life span in flies (21, 25), whereas Dicer-2 and Ago2 mutants are short-lived (15, 24). Given our findings presented earlier, we examined whether increased expression of Su(var)3–9 or Dicer-2 also have a positive effect on life span. We found that two different Su(var)3–9 transgenic lines, SV2 and SV3, which constitutively increase Su(var)3–9 expression to 160–180% of the wild-type level in the absence of a driver (Fig. S4B), extend life span significantly compared with their genetically matched control (Fig. 4A). Similarly, increasing Dicer-2 expression in adult life in either all tissues (Fig. 4B) or selectively in adult neurons (Fig. 4C), using the conditional inducible GeneSwitch system, was able to extend life span, with flies fed the gene-activating drug RU486 outliving a genetically identical control cohort fed only the diluent control EtOH. Together with previously published data (21, 25), these observed life span extensions suggest a coordinated response integrating maintenance of heterochromatin structure, TE suppression, and longevity.
Fig. 4.
Increasing expression of Su(var)3–9 or Dicer-2 increases life span. (A) SV2 and SV3 lines [overexpressing Su(var)3–9] are long-lived compared with controls. Survivorship of adult female flies is shown (n = 250). WT mean life span = 40.6 d, median = 44 d; SV2 mean = 58.5 d, median = 60 d, 36% median extension, log-rank P < 10−10; SV3 mean = 58.3 d, median = 60 d, 36% median extension, P < 10−10. (B) Whole-adult animal Dicer-2 overexpressing flies are long-lived compared with controls. Survivorship of tubulin-GeneSwitch > UAS-Dicer-2 adult females (n = 250) is shown, either with EtOH (uninduced) or 200 µM RU486 (induced expression). EtOH mean = 52.3 d, median = 54 d; RU486 mean = 63.5 d, median = 66 d, 22% median extension, P < 10−10. (C) Flies expressing Dicer-2 in adult neurons are long-lived compared with controls. Survivorship of elav-GeneSwitch > UAS-Dicer-2 adult females is shown, for EtOH (uninduced) or 500 µM RU486 (induced) (n = 250). EtOH mean = 53.8 d, median = 56 d; RU486 mean = 61.7 d, median = 66 d, 18% median extension, P < 10−10. All life span experiments were repeated with similar results; representative experiments shown.
Pharmacological Suppression of Transposable Element Activity Delays Age-Related Transposition and Extends Life Span.
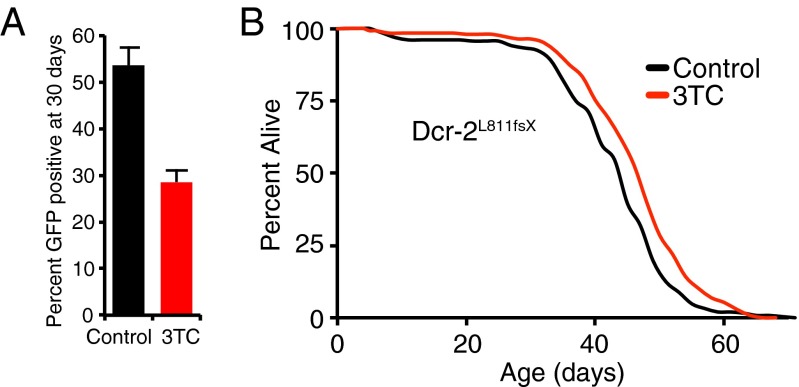
To test whether the age-related effects we observed were a result of increased TE activity, we sought to directly modulate transposition pharmacologically. We used the reverse transcriptase inhibitor lamivudine (3TC) to directly block TE transposition. Using the gypsy-TRAP reporter system in the fat body, we observed that flies treated with 3TC had a significant delay in age-related transposition compared with controls (Fig. 5A). This effect was observed whether the drug was administered constantly throughout life (Fig. 5A) or started during midadulthood at 20 d of age (Fig. S4C).
Fig. 5.
Administration of lamivudine (3TC) suppresses retrotranposition and extends Dicer-2 mutant life span. (A) Percentage of gypsy-TRAP flies with more than 30 fat body cells positive for GFP expression at 30 d of age for both untreated (control) and 10 µM 3TC treated fly cohorts. Fisher’s exact test P < 0.0001. (B) 10 µM 3TC treated flies are long-lived compared with untreated controls in the Dicer-2L811fsX genetic background. Survivorship of adult female flies is shown (n = 250). Control mean = 43.0 d, median = 45 d; 3TC mean = 46.4 d, median = 48 d, 6.6% median extension, log-rank P < 0.0001. Life span was repeated with similar results.
As our evidence suggests that maintenance of TE silencing is important for ensuring longevity, we tested whether attenuating TE activity directly could extend life span. In the Dicer-2L811fsX mutant background, which exhibits high levels of TE activity because of defective RNAi silencing (32), we found that administration of 3TC was able to extend life span significantly compared with controls (Fig. 5B). Together, these data suggest that heterochromatin-altering pathways that extend life span may do so at least in part by suppressing TE activity to ensure healthy longevity, and confirm that suppression of TE activation is likely to be an attractive target mechanism for pharmacological intervention.
Discussion
Aging is characterized by a breakdown in homeostasis throughout multiple cellular and organismal systems. Genomic and epigenomic stability is vital for somatic cells to maintain proper gene expression and silencing. A number of studies have shown changes in chromatin structure during organismal aging in various model systems, especially in heterochromatin (1, 2). Although heterochromatin regions contain some genes, a likely reason for the central importance of maintaining repressive heterochromatin is that heterochromatin is replete with TEs, and unchecked TE activation has particularly damaging ramifications for genomic stability. Indeed, the expression and mobilization of TEs is disruptive enough that cells have evolved RNAi pathways responsible for silencing and preventing TE activation. These cellular defenses against TE activation include siRNA-directed induction and maintenance of repressive heterochromatin to prevent TE expression, and degradation of target transcripts. siRNA pathway mutants exhibit both an activation of TEs and a shortening of life span (15, 24). Here we provide evidence that many heterochromatin genes and TEs do in fact increase expression with age in two different tissues. Increased transposition has been previously reported in the aging fly brain (15). Similarly, we also observe an increase in somatic TE transposition with age in the fat body. DNA double-strand breaks increase with age in the fat body (16), and we show this DNA damage also accumulates at a younger age in Dicer-2 mutant flies with high levels of TEs. The age-related increase in TE transcript levels as well as TE mobilization were suppressed by DR. Together, these data suggest that an increase in TE expression and transposition is an important hallmark of somatic aging, and preventing this increased TE activity is likely to preserve genomic stability and cellular homeostasis.
We demonstrate in a whole organism model that aging is associated with a loss of silencing of both TEs and genes natively located in heterochromatin regions, with a resultant increase in TE mobilization and an associated increase in DNA damage. It is highly likely that the pathways and processes of heterochromatin maintenance, RNAi, and gene and TE silencing represent potential homeostatic failure points that lead to the phenotypes of aging. Here we have also presented genetic evidence that these pathways are linked to aging, as manipulating the expression of several genes involved in maintaining heterochromatin structure, TE silencing, and RNAi efficacy directly suppresses age-related TE expression as well as extends life span. Furthermore, we show that DR, an environmental intervention that extends life span robustly and delays most age-related phenotypes, also opposes the loss of silencing we observe during normal aging and delays age-dependent transposition, further lending support to the idea that these processes are associated with and may be relevant to aging. We also demonstrate that inhibition of retrotransposon activity is strongly correlated with extension of life span. Our data support the retrotransposon theory of aging model, where proper maintenance of silent heterochromatin and prevention of TE activation are key to guaranteeing genomic stability and homeostasis during aging and ensuring healthy longevity. Furthermore, our demonstration that dietary (DR), genetic, and pharmacological interventions that reduce the age-related increases in TE activity can also extend life span suggests new and novel pathways for the development of interventions designed to extend healthy life span.
Materials and Methods
Gypsy-TRAP Assay.
Flies from the gypsy-TRAP line were aged for 5 d and initially examined for the number of GFP-positive cells, then examined every 7 d until age 49 d, using a fluorescent dissecting microscope. Flies were separated into three categories based on visual inspection, according to how many fat body cells were GFP positive: low (<20 cells), medium (20–100 cells), and high (>100 cells). The same cohort of flies was examined longitudinally throughout each of the experiments.
RNA-seq.
Total RNA from the indicated genotypes was collected from ∼100 fly heads or ∼50 dissected fat bodies, using the miRvana RNA prep kit (Ambion). Then, 100 ng total RNA was used as input for stranded RNA-seq library construction, using the Ovation Universal RNA-seq kit (Nugen) with Drosophila rRNA depletion module. Libraries were sequenced on an Illumina HiSEq. 2500 in 1 × 50 bp mode. All samples were prepared using at least three individually isolated biological replicates for each condition.
Bioinformatics.
Standard RNA-seq analysis was performed using Tophat 2.0 for sequence alignment, followed by gene-based counting using the easyRNASeq R package from Bioconductor. Finally, trimmed mean of M-values (TMM) normalization and fold change calculation was performed using edgeR, with a generalized linear model (GLM) correcting for batch effects, as described in the edgeR vignette. To account for repetitive sequencing reads and perform TE quantification, RepEnrich (33) was used to generate a count table of reads for each TE, and edgeR was used to normalize, calculate fold changes, and perform differential expression testing. The Repbase annotation was used as input for RepEnrich.
Heat maps were generated by ranking the TEs or genes in order of log2 fold change of expression with age (old/young) for the reference (control/HC) condition and then plotting log2 fold change with age for all conditions. To show consistent trends in our data and include genes/TEs that behaved consistently but did not meet the threshold of statistical significance, we displayed genes or TEs where log2 fold change was greater than 0.322 (1.25× fold; TEs) or 0.263 (1.2x fold; heterochromatin genes). Plots were generated using the heatmap.2 function in the gplots2 R package.
See SI Materials and Methods for additional details on fly stocks and methods.
SI Materials and Methods
Fly Stocks and Culturing.
Drosophila stocks were maintained at 25 °C with a 12 h light/dark cycle at 60% relative humidity. Lab stocks of Canton S and w1118 were used for wild-type experiments. The Adar hypomorph (dAdarhyp), as well as its genetically matched control (dAdarloxP), were a gift from Robert Reenan and described in ref. 21. The LacI-Su(var)3–9 lines (SV2 and SV3) were a gift from Kristen Johansen and described in ref. 34. SV2 and SV3 were backcrossed for 20 generations to w1118. Flies expressing Dicer-2 were generated by crossing Bloomington stock #25750 with tubulin-GeneSwitch or elav-GeneSwitch, and homozygosing both chromosomes. Experimental flies were generated by crossing these tub-GS/UAS-Dcr-2 or elav-GS/UAS-Dcr-2 stocks with w1118 such that both the GeneSwitch driver and UAS-Dcr-2 were heterozygous. Sir2-expressing EP2300 flies were obtained from Bloomington stock center (#24859). The fat body gypsy-TRAP reporter line was generated from the gypsy-TRAP line generously provided to us by Josh Dubnau (15), r4-GAL4 (Bloomington #33832), and UAS-GFP (Bloomington #1522). Briefly, the fat body specific r4-GAL4 driver line was first recombined with a UAS-GFP line generating a new stable line that expresses GFP exclusively in the fat body and balanced on the second chromosome with If/CyO. This line was then crossed with the gypsy-TRAP reporter line, which contains a Tubulin-ovo-GAL80 that suppresses GFP expression until TE transposition disrupts reporter function (15). For the line shown in Fig. 2, If F1 flies from this cross were taken (w1118; If/+; r4-GAL4, UAS-GFP/tub-ovo-GAL80). CyO F1 flies were taken to generate CyO line #1 in Fig. S2 (w1118; CyO/+; r4-GAL4, UAS-GFP/tub-ovo-GAL80). For CyO line #2 in Fig. S2, piwi2/CyO flies were crossed to the gypsy-TRAP reporter line, and CyO F1 flies were taken (w1118; CyO/+; r4-GAL4, UAS-GFP/tub-ovo-GAL80). The mutant ovo gypsy-TRAP line was constructed as described earlier, except using a gypsy-TRAP line with mutated ovo sites that were no longer hot spots for gypsy transposition.
Life Span Assays.
Food composition for life span experiments was as follows: for normal/HC food, 15% dextrose, 15% autolyzed yeast, 2% cornmeal, and 2% agar; for DR/low calorie food, 5% dextrose, 5% autolyzed yeast, 2% cornmeal, and 2% agar (all wt/vol). For experiments using GeneSwitch drivers, either RU486 dissolved in EtOH at the indicated concentration or EtOH alone (for controls) was added to the food at 20 mL/L. Upon eclosion, flies were sorted under CO2 anesthesia and placed in food vials at a density of 25 males and 25 females per vial, with a total of at least 10 vials (n = 250) for each condition. Flies were passed to fresh food every other day, and dead flies scored and counted. Life span statistics and log-rank P values were determined using the web-based OASIS tool (35).
Double-Strand Break Assay.
Sectioned female flies were stained with α-γH2Av antibody and imaged with a Zeiss Axiovision epifluorescence scope at a constant exposure time of 500 ms (AlexaFluor 568) or 100 ms (DAPI). Images from slices containing fat body nuclei were used to quantify fluorescence levels using the technique described in ref. 16 (n = 50 nuclei).
Supplementary Material
Acknowledgments
We thank Will Lightfoot and Gillian Horwitz for technical assistance and Christoph Schorl and the Brown University Genomics Core Facility for sequencing. We also thank Joshua Dubnau, Robert Reenan, Kristen Johansen, and the Bloomington Drosophila Stock center for fly stocks and Robert Reenan and John Sedivy for helpful discussions. This work was supported in part by National Institute on Aging (NIA) T32 Training Grant AG41688 and Fellowship Award AG47736 (to B.C.J.); NIA Grants AG16667 and AG24353, a Glenn/American Federation for Aging Research (AFAR) Breakthroughs in Gerontology award, NIH Program Project Grant AG51449, and Grant P30AI042853 from the Providence/Boston Center for AIDS Research (to S.L.H.); Ellison/AFAR Postdoctoral Fellowship awards (to J.G.W. and N.J.); a Nathan Shock Center Pilot Grant (to J.G.W.); and NIH Grants AG28753 and AG50582 (to N.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11069.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604621113/-/DCSupplemental.
References
- 1.Wood JG, Helfand SL. Chromatin structure and transposable elements in organismal aging. Front Genet. 2013;4(December):274. doi: 10.3389/fgene.2013.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585(13):2041–2048. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16(10):593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood JG, et al. Chromatin remodeling in the aging genome of Drosophila. Aging Cell. 2010;9(6):971–978. doi: 10.1111/j.1474-9726.2010.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang N, et al. Dietary and genetic effects on age-related loss of gene silencing reveal epigenetic plasticity of chromatin repression during aging. Aging (Albany, NY) 2013;5(11):813–824. doi: 10.18632/aging.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Cecco M, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12(2):247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Cecco M, et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany, NY) 2013;5(12):867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, et al. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 2011;10(17):3016–3030. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75(1):26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Meter M, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis S, Sheth U, Feldman JL, English KA, Priess JR. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 2012;8(3):e1002591. doi: 10.1371/journal.ppat.1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell PH, Burhans WC, Curcio MJ. Retrotransposition is associated with genome instability during chronological aging. Proc Natl Acad Sci USA. 2011;108(51):20376–20381. doi: 10.1073/pnas.1100271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16(5):529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Zheng X, Xiao D, Zheng Y. Age-associated de-repression of retrotransposons in the Drosophila fat body, its potential cause and consequence. Aging Cell. 2016;15(3):542–552. doi: 10.1111/acel.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schotta G, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21(5):1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castel SE, Martienssen RA. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14(2):100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savva YA, et al. RNA editing regulates transposon-mediated heterochromatic gene silencing. Nat Commun. 2013;4:2745. doi: 10.1038/ncomms3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg MI, Parkhurst SM. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell. 2002;109(4):447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- 23.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim DH, et al. The endogenous siRNA pathway in Drosophila impacts stress resistance and lifespan by regulating metabolic homeostasis. FEBS Lett. 2011;585(19):3079–3085. doi: 10.1016/j.febslet.2011.08.034. [DOI] [PubMed] [Google Scholar]
- 25.Whitaker R, et al. Increased expression of Drosophila Sir2 extends life span in a dose-dependent manner. Aging (Albany, NY) 2013;5(9):682–691. doi: 10.18632/aging.100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477(7365):E1–E2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 27.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477(7365):482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann J, Romey R, Fink C, Yong L, Roeder T. Overexpression of Sir2 in the adult fat body is sufficient to extend lifespan of male and female Drosophila. Aging (Albany, NY) 2013;5(4):315–327. doi: 10.18632/aging.100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riddle NC, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21(2):147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charlesworth B, Langley CH. The population genetics of Drosophila transposable elements. Annu Rev Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. [DOI] [PubMed] [Google Scholar]
- 31.Dej KJ, Gerasimova T, Corces VG, Boeke JD. A hotspot for the Drosophila gypsy retroelement in the ovo locus. Nucleic Acids Res. 1998;26(17):4019–4025. doi: 10.1093/nar/26.17.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320(5879):1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 2014;15(1):583. doi: 10.1186/1471-2164-15-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boeke J, et al. Phosphorylation of SU(VAR)3-9 by the chromosomal kinase JIL-1. PLoS One. 2010;5(4):e10042. doi: 10.1371/journal.pone.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang JS, et al. OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS One. 2011;6(8):e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.