Significance
Direct attack of target cell membranes by protein oligomerization is a powerful innate defense mechanism used widely throughout nature. Defensins are ubiquitous innate immunity mediators that are able to recognize certain phospholipids, and subsequently oligomerize to attack target cell membranes. We now show that the plant defensin NsD7 is able to bind the cellular phospholipid, phosphatidic acid (PA), which triggers defensin oligomerization in a unique manner. Our crystal structure of the NsD7–PA oligomer revealed a striking double-helical defensin–lipid oligomer that features a novel phospholipid-binding site mediating PA binding and membrane permeabilization. This demonstrates that defensins use their conserved small fold in a remarkably flexible way to specifically recognize a range of phospholipids during innate defense using different binding sites.
Keywords: antimicrobial peptides, defensins, phospholipids, innate defense, host–pathogen interactions
Abstract
Defensins are cationic antimicrobial peptides that serve as important components of host innate immune defenses, often by targeting cell membranes of pathogens. Oligomerization of defensins has been linked to their antimicrobial activity; however, the molecular basis underpinning this process remains largely unclear. Here we show that the plant defensin NsD7 targets the phospholipid phosphatidic acid (PA) to form oligomeric complexes that permeabilize PA-containing membranes. The crystal structure of the NsD7–PA complex reveals a striking double helix of two right-handed coiled oligomeric defensin fibrils, the assembly of which is dependent upon the interaction with PA at the interface between NsD7 dimers. Using site-directed mutagenesis, we demonstrate that key residues in this PA-binding site are required for PA-mediated NsD7 oligomerization and coil formation, as well as permeabilization of PA-containing liposomes. These data suggest that multiple lipids can be targeted to induce oligomerization of defensins during membrane permeabilization and demonstrate the existence of a “phospholipid code” that identifies target membranes for defensin-mediated attack as part of a first line of defense across multiple species.
Host defense peptides, which include cationic antimicrobial peptides (CAPs), are a group of innate immune molecules produced by essentially all plant and animal species that act as a first line of defense against microbial invasion. Numerous CAPs have been postulated to act by permeabilizing membranes using a range of mechanisms, including (i) carpet, (ii) barrel-stave, and (iii) toroidal-pore models (1). Although these models have been widely used to describe potential molecular mechanisms of action for various CAPs, the structural basis of interactions at the membrane remains to be determined.
Oligomerization of defensins has been shown to be a feature of their antimicrobial activity. The human α-defensin 6 (HD6) has been shown to oligomerize and form nanonets that trap bacteria and limit their motility (2). However, the structural basis of HD6 oligomerization, or indeed the molecular mechanism that triggers oligomerization, has not been well defined. Recently, the ability of defensins to lyse target cells has been linked to their capacity to recognize specific membrane phospholipids. The plant defensin NaD1 binds phosphatidylinositol 4,5-bisphosphate (PIP2), leading to the formation of large oligomeric NaD1:PIP2 assemblies. This formation of oligomeric complexes with PIP2 was shown to be critical for NaD1-mediated lysis of fungal cells (3). Studies on the plant defensin MtDef4 have indicated that binding of phosphatidic acid (PA), a key regulator of membrane curvature, membrane–cytoskeletal interactions, and phospholipid precursor molecule (4, 5), is important for its antifungal activity (6), suggesting that phospholipids other than PIP2 may be important targets for defensins. Furthermore, certain sphingolipids have been shown to be targets for various other plant defensins. For instance, glucosylceramide lipids, which are located in the cell walls and plasma membranes of filamentous fungi, are targeted by the RsAFP2 (7), Psd1 (8), and MsDef1 (9), whereas DmAMP1 interacts with the structurally related membrane lipid mannosyl–diinositolphospho–ceramide (10).
To understand the role of phosphatidic acid in defensin-mediated innate immunity, we investigated the molecular basis of PA binding to the Nicotiana suaveolens defensin NsD7 and its role in membrane permeabilization. Here we show that NsD7 binds and forms large oligomeric complexes with PA. Notably, NsD7:PA complexes adopt a radically different topology, compared with the previously described NaD1:PIP2 complex, to permeabilize target membranes containing PA. These findings suggest that multiple avenues exist to enable phospholipid recognition and sequestration by defensins in innate immunity and support the idea that different phospholipids in target cell membranes may act as recognition flags to trigger defensin-based innate immunity mechanisms.
Results
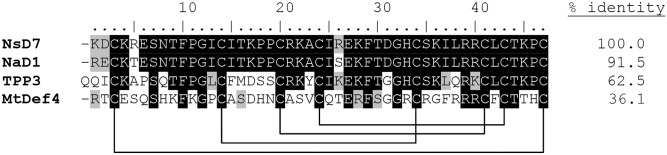
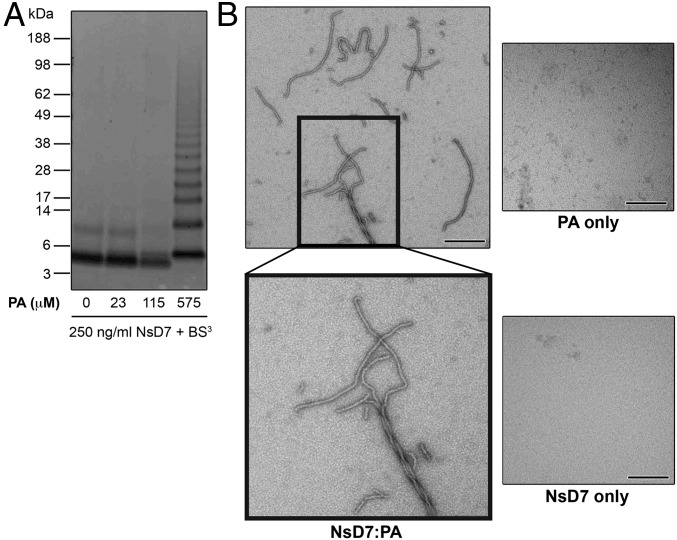
As PA has been proposed as an important target for plant defensins (6), we investigated the ability of NsD7, a homolog of NaD1 cloned from N. suaveolens (91.5% identical to NaD1) (Fig. S1), to interact with PA and form oligomers. NsD7 treated with the biochemical cross-linker bis[sulfosuccinimidyl] suberate (BS3) in the presence of PA resulted in the formation of covalently linked oligomers. In contrast, BS3 treatment of NsD7 on its own revealed only monomers and covalently linked dimers (Fig. 1A). We then investigated NsD7:PA complexes in solution using negative stain transmission electron microscopy (TEM). When PA was present, NsD7 formed elongated string-like structures that could assemble into multistranded helical fibrils. In contrast, no string-like structures or helical fibrils were observed for NsD7 or PA alone, indicating that these defensin assemblies are PA-dependent (Fig. 1B).
Fig. S1.
Sequence alignment of NsD7 with plant defensins NaD1 (AF509566, from ornamental tobacco), TPP3 (AAA80496, from tomato), and MtDef4 (2LR3, from barrel medic). The GenBank accession numbers and plant sources are given in parentheses. Residues that are identical to NsD7 are shaded in black, and similar residues are shaded in gray. The conserved cysteine residues involved in disulfide bonds are indicated by connecting lines, and the percent amino acid sequence identity relative to NsD7 is shown. The numbering above the sequences is relative to NsD7.
Fig. 1.
NsD7 binds PA and forms oligomers. (A) NsD7 forms oligomers in the presence of PA as determined by protein–protein cross-linking with BS3 followed by SDS/PAGE and Coomassie Brilliant Blue staining. (B) TEM of NsD7:PA complexes, NsD7 alone, or PA alone. (Scale bars, 100 nm.)
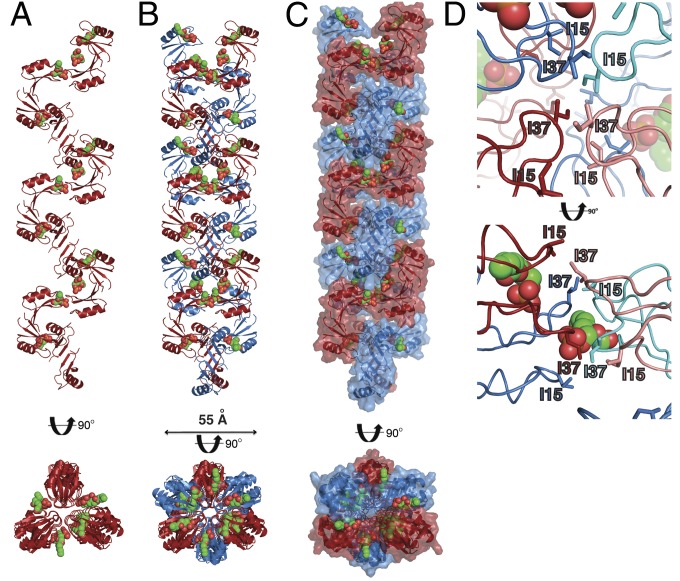
To understand the structural basis for PA-mediated oligomer formation of NsD7, we crystallized NsD7 in the presence of PA. Using monomeric NaD1 as a search model (11), the structure of a NsD7:PA complex was solved by molecular replacement and refined to a resolution of 1.70 Å with values of Rwork/Rfree of 0.183/0.216 (Table 1). In the crystal, NsD7 dimers form an extended continuous coil through the crystal lattice (Fig. 2A) that is held together by bound PA molecules. As a result, NsD7 forms a right-handed helical coil (Movie S1) whose fundamental building block comprises three NsD7 dimers bound to six PA molecules in the asymmetric unit. Furthermore, each helical coil was paired via crystallographic symmetry with a second coil, forming a double-stranded defensin–lipid helix that continues throughout the crystal (Fig. 2 B and C). Intriguingly, the interior of this double-helical protein–lipid complex is lined by a series of intercalating isoleucine residues (I15 and I37) arranged in a zipper-like manner (Fig. 2D).
Table 1.
Crystallographic data collection and refinement statistics
| Data collection and refinement | Statistics |
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 107.32, 62.32, 53.53 |
| α, β, γ (°) | 90.00, 118.6, 90.00 |
| Wavelength (Å) | 0.9537 |
| Resolution (Å)* | 31.16–1.7 (1.73–1.70) |
| Rsym or Rmerge* | 0.079 (0.778) |
| I/σI* | 8.9 (1.6) |
| CC(1/2) | 0.996 (0.593) |
| Completeness (%)* | 99.5 (98.2) |
| Redundancy* | 3.8 (3.8) |
| Refinement | |
| Resolution (Å) | 31.16–1.7 |
| No. reflections | 34,010 |
| Rwork/Rfree | 0.183/0.216 |
| No. nonhydrogen atoms | |
| Protein | 2,236 |
| Ligand/ion | 207 |
| Water | 263 |
| B-factors | |
| Protein | 23.70 |
| Ligand/ion | 48.90 |
| Water | 33.80 |
| Rmsd | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.615 |
| Molprobity score | 1.73 |
Values in parentheses are for highest-resolution shell.
Fig. 2.
The NsD7:PA complex forms a double-stranded helix. (A) The NsD7:PA complex adopts a right-handed coiled structure. The coil shown comprises the content of three asymmetric units. PA molecules are shown as green (carbon), orange (phosphate), and red spheres (oxygen). (B and C) Two neighboring NsD7:PA coils form a defensin double helix. (D) The interior of the NsD7:PA oligomer harbors a series of intercalating isoleucine residues (I15 and I37). Top view is down the center of the oligomer; Bottom view is the same view rotated 90° on the horizontal axis.
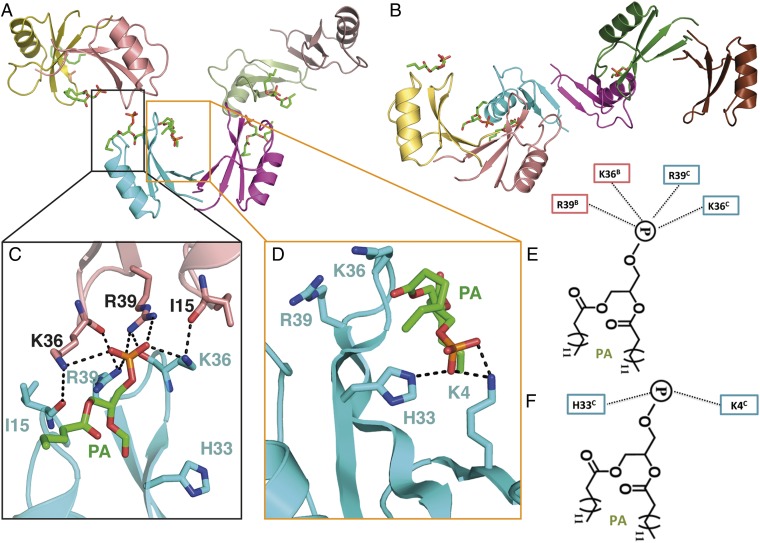
In the asymmetric unit, three NsD7 dimers are arranged in an extended configuration (Fig. 3A), bent in a twisted S-shape (Fig. 3B). In this configuration, two neighboring dimers contact each other via the tip of one arm of the dimer. With respect to the central dimer, the two neighboring dimers are arranged orthogonally (Fig. 3 A and B). A total of six PA molecules are bound by the three NsD7 dimers in the asymmetric unit, which are found in two different locations. A PA molecule is bound at the connecting point between two neighboring NsD7 dimers (designated a type I PA-binding site) (Fig. 3C) that appears critical for defensin oligomerization, with each connecting point between NsD7 dimers within the asymmetric unit containing one PA molecule. In contrast, a second PA is bound in a cationic grip formed by each NsD7 dimer (designated a type II PA-binding site) (Fig. 3D).
Fig. 3.
Crystal structure of the NsD7:PA complex. (A) Three NsD7 dimers are arranged in a tip-to-tip configuration, with each neighboring dimer positioned orthogonally from its neighbor. A total of six PA molecules are bound by the three NsD7 dimers. (B) An identical view as in A, rotated 90° around the horizontal axis. The arrangement of the dimers leads to an S-shaped trimer of dimers. (C) Type I and (D) type II PA-binding sites. Hydrogen bonds or ionic interactions are marked as black dashed lines. Schematic representation of the (E) type I and (F) type II PA-binding sites.
For type I sites, K36 and R39 from two adjacent NsD7 monomers (which are each contributed by a different NsD7 dimer) form ionic interactions with the phosphate in the PA head group. In addition, K36 and R39 from one monomer form hydrogen bonds with the carbonyl groups of I15 and K36, respectively (Fig. 3 C and E). In contrast, for type II sites, K4 and H33 from a single chain form ionic interactions with the phosphate head group of PA (Fig. 3 D and F). K4 also forms a hydrogen bond with a carbonyl group in the acyl chain of PA.
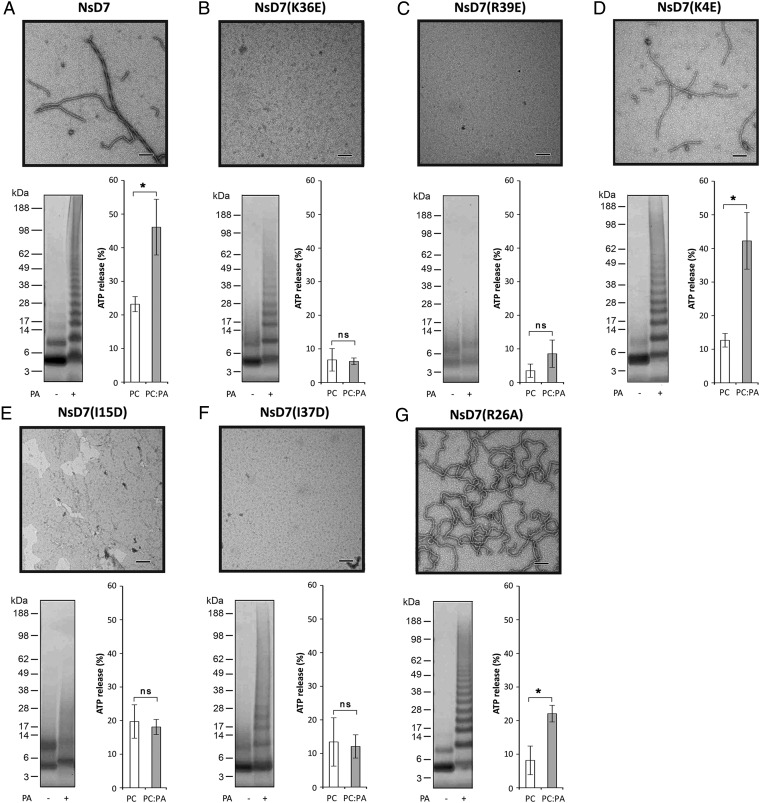
To further characterize the structural basis of the NsD7:PA oligomer–fibril formation and function in membrane permeabilization, we performed site-directed mutagenesis on NsD7 by generating mutants that targeted the: (i) type I PA-binding site, (ii) type II PA-binding site, (iii) isoleucine zipper, and (iv) interoligomer region. The ability of each mutant to form oligomers and multioligomer-containing fibrils was then assessed by biochemical cross-linking and TEM, as well as their capacity to lyse ATP-encapsulated liposomes reconstituted from phosphatidylcholines (PC) or PC:PA lipids (Fig. 4). NsD7 mutants K36E and R39E, which target the type I PA-binding site, showed complete loss of fibril formation under TEM (Fig. 4 B and C). NsD7(K36E) was still able to form some higher-order oligomers after BS3 chemical cross-linking, despite the inability to permeabilize PA-containing liposomes, whereas there was complete ablation of higher-order oligomer formation as well as loss of PA-containing liposome permeabilization by NsD7(R39E). In contrast, the K4E mutant that targets the type II PA-binding site in the cationic grip of each NsD7 dimer was comparable to wild-type NsD7 in all oligomerization and liposome permeabilization assays (Fig. 4 A and D). These results indicate that only the type I PA-binding site is important for PA-induced oligomerization. NsD7 mutants I15D and I37D, which both target the isoleucine zipper located on the inside of the oligomeric double helix, also displayed complete loss of fibril formation and lysis activity on PA-containing liposomes. Some higher-order oligomers were observed for NsD7(I37D), whereas this was absent for NsD7(I15D) after BS3 chemical cross-linking (Fig. 4 E and F). Together, these data suggest that the intercalation of I15 and I37 in the oligomer is critical for fibril formation. The final mutant we evaluated was NsD7(R26A), which is located on the side of the oligomeric fibril. We reasoned that the triple-stranded fibrils observed under TEM for wild-type NsD7:PA are formed due to either a charge-based interaction or PA-mediated interstrand contacts, and inspection of conspicuous charged residues protruding from the double-helical NsD7:PA oligomer identified R26 as the most prominent. This mutant displayed wild-type–like lysis activity on PA-containing liposomes and oligomerized in the presence of PA in the chemical cross-linking experiment, but TEM revealed a distinct lack of multistranded fibrils compared with wild-type NsD7, with the mutant forming only single-stranded fibrils that were bent and irregular (Fig. 4G).
Fig. 4.
Structure–function analysis of the interaction between NsD7 and PA. (A) Wild-type NsD7, type I PA-binding site mutants (B) NsD7(K36E) and (C) NsD7(R39E), type II PA-binding site mutant (D) NsD7(K4E), isoleucine zipper mutants (E) NsD7(I15D) and (F) NsD7(I37D), and interoligomer fibril formation mutant (G) NsD7(R26A). Each subpanel displays data for defensin oligomerization in the presence of PA as assessed by TEM (Top) or biochemical cross-linking using BS3 followed by SDS/PAGE and Coomassie Brilliant Blue staining (Bottom Left) and liposome permeabilization as measured by percentage of ATP release (relative to a Triton-X100 control) from liposomes comprising PC or PC:PA (Bottom Right). Graphs are representative of at least three independent experiments. Error bars, SEM. *P < 0.05; ns, not significant. (Scale bars, 100 nm.)
Discussion
Oligomerization of defensins has recently been shown as a critical mechanistic step in their antimicrobial functions (2, 3). For example, human α-defensin 6 (HD6) oligomerizes to form nanonets that act as a physical barrier to trap and reduce the movement of bacteria (2). It was postulated that the interaction of HD6 with various surface proteins encountered on intruding bacteria is the trigger for dynamic self-assembly into these nanonets. However, the structural basis for triggering self-assembly, the role of specific ligands to induce nanonets or the molecular structure of such nanonets, remains undefined. Furthermore, we have shown that the plant defensins NaD1 and TPP3 oligomerize upon interaction with the phospholipid PIP2 to mediate membrane permeabilization of target cells (3, 12). Using X-ray crystallography, we defined the structure of an NaD1–PIP2 oligomeric complex, revealing that dimers of NaD1 bind PIP2 molecules in a highly cooperative manner that mediates oligomerization and the formation of long fibrils (3). These observations prompted the question of whether formation of large oligomeric complexes by defensins is a conserved feature for their biological activity.
In this study, we show that the plant defensin NsD7 specifically recognizes a different type of phospholipid, PA, to form oligomers that mediate permeabilization of PA-containing liposomal membranes. These data indicate that defensin oligomerization is mediated by interaction with different specific phospholipids and highlight an important mechanism underlying defensin antimicrobial activity. Our structural analysis of the NsD7–PA oligomeric complex indicates that the basic building block is an NsD7–dimer. Indeed, a NsD7 monomer–dimer equilibrium is suggested on the basis of the biochemical cross-linking experiment. We have previously defined similar dimeric configurations for the plant defensins NaD1 and TPP3 (3, 12), suggesting that defensin dimers are an important functional unit.
NsD7 forms extended coiled oligomers in complex with the phospholipid PA, with PA lodged at the connecting points between individual NsD7 dimer tips, leading to the formation of a right-handed defensin–lipid double helix. It is likely that several of these defensin double helices associate to form the multistranded fibrils observed using TEM. Strikingly, the overall topology of the NsD7:PA oligomer is very different compared with the NaD1:PIP2 oligomer, which is due to the distinct manner of how neighboring dimers of NsD7 or NaD1 are connected via PA (Fig. 5 A and C) or PIP2 (Fig. 5 B and D) molecules, respectively. In NsD7, the critical residues that enable assembly of the dimers into an oligomer are K36 and R39, which are contributed from each monomer in two adjacent dimers to form crucial interactions with a single bridging PA molecule (Fig. 5A), resulting in a tip-to-tip arrangement of NsD7 dimers (Fig. 5C). In contrast, the critical residues in NaD1 are K36 and R40 (Fig. 5B). These residues mediate the binding of two PIP2 molecules per dimer. Crucially, K36 and R40 are intercalated between neighboring PIP2 molecules within the grip-like structure of an individual NaD1 dimer, and the lateral association of two NaD1 dimers allows continuous intercalation of K36 and R40 in an alternating manner across multiple PIP2 molecules, resulting in a side-to-side arrangement of neighboring NaD1 dimers bridged by two PIP2 molecules (Fig. 5D).
Fig. 5.
Oligomeric NsD7:PA and NaD1:PIP2 complexes have distinct assembly mechanisms. (A) Cartoon diagram of NsD7:PA complex illustrating the tip-to-tip dimer–dimer arrangement in the oligomer mediated by interaction with PA (dimer 1 in cyan and magenta and dimer 2 in sand and green). The key residues comprising the PA-binding site are shown as sticks; PA is shown as green sticks. (B) Cartoon diagram of NaD1:PIP2 complex illustrating the side-to-side dimer–dimer arrangement in the oligomer mediated by interaction with PIP2 (dimer 1 in cyan and magenta and dimer 2 in sand and green). The view of the cyan and magenta dimer 1 is conserved from A. Key residues comprising the PIP2 binding sites are shown as sticks, and PIP2 molecules are shown as sticks. (C) Schematic space-fill diagram of NsD7–PA oligomer assembly. NsD7 dimers are colored as in A with one dimer–dimer unit in the assembly highlighted by a red boundary; PA molecules are shown as black circles. (D) Schematic space-fill diagram of NaD1-PIP2 oligomer assembly. NaD1 dimers are colored as in B with one dimer–dimer unit in the-assembly highlighted by a red boundary; PIP2 molecules are shown as black circles.
Consequently, lipid engagement and dimer:dimer contacts are achieved using different sets of defensin residues, highlighting the functional flexibility inherent in defensins despite their small overall size and rigid architecture. Inspection of the NsD7 type I PA-binding site suggests that it is likely to be selective for PA over PIP2, due to the small pocket of the type I site only being able to accommodate the single-phosphate head group of PA but not the substantially larger head group of PIP2 that contains three phosphate moieties. In contrast, the much larger type II site of NsD7 is likely to accommodate both PA and PIP2, with the critical K4, K36, and R40 PIP2-binding residues in NaD1 being conserved in NsD7.
The site-directed mutagenesis of NsD7 indicates that PA binding and oligomerization is important for permeabilization and demonstrates that an alternative phospholipid-binding site is crucial for PA-mediated defensin oligomerization compared with the previously reported PIP2-induced oligomers. Taken together, with the observation that PA binding is critical for the antimicrobial activity of the legume defensin MtDef4 (6), these data provide strong evidence that PA-induced defensin oligomerization is functionally significant. Considering our findings on PA-mediated oligomerization of NsD7 and membrane permeabilization, we propose that the previously suggested mechanisms of action for innate defense peptides based on the carpet, barrel-stave, or toroidal-pore models (1) are unlikely to be applicable to NsD7. Our data support a model of irrecoverable membrane disruption through a direct destabilization mechanism involving sequestration of PA by interaction with and oligomerization of NsD7.
These findings suggest that despite their small size, the defensin fold is highly adaptable for phospholipid recognition and sequestration to repel microbial attacks and can form distinct oligomeric assemblies with unique topologies. The observation that defensin oligomers form by interaction with different cellular phospholipids supports the concept that multiple phospholipids in target cell membranes may act as recognition flags to trigger defensin-based innate immunity mechanisms.
Methods
Cloning and Recombinant Expression of NsD7 and Its Mutants.
Oligonucleotide primers specific to the 5′ and 3′ regions of the DNA corresponding to the endoplasmic reticulum signal sequence and C-terminal propeptide of the Nicotiana alata defensin NaD1 protein (GenBank accession no. AF509566) were used in a PCR with genomic DNA extracted from the leaves of Nicotiana suaveolens. A homologous genomic clone, named NsD7, was identified following DNA sequencing. Subsequently, the mature defensin domain of NsD7 was amplified by PCR and subcloned into the pPIC9 vector for recombinant protein production in the methylotrophic yeast Pichia pastoris as described previously (13). NsD7 point mutants were prepared using the QuikChange mutagenesis kit (Stratagene) as per manufacturer’s instructions.
Crystallization and Structure Determination.
The NsD7:PA complexes were generated by mixing NsD7 at 10 mg/mL and PA at a molar ratio of 1:1.2. Crystals were grown in sitting drops at 20 °C in 0.2 M ammonium sulfate, 25% (wt/vol) PEG MME 2000, and 0.1 M sodium acetate pH 4.9. Diffraction data were collected from crystals flash-cooled in mother liquor supplemented with 10% (vol/vol) ethylene glycol at 100 K at the Australian Synchrotron (beamline MX2) and processed with Xds (14). The structure was solved by molecular replacement with PHASER (15) using the structure of NaD1 (11) as a search model. The final model was built with Coot (16) and refined with Phenix (17) to a resolution of 1.70 Å. All data collection and refinement statistics are summarized in Table 1. Refinement yielded Rwork and Rfree values of 18.3% and 21.6%, respectively. Analysis of the final refined structure using MolProbity (18) revealed a MolProbity score of 1.73, whereas a Ramachandran plot classified 91.1% of the residues in the core region, 8.1% in the allowed region, and 0.8% in the generously allowed region, with none in the disallowed region. All programs were accessed via the SBGrid suite (19). The coordinates have been deposited in the Protein Data Bank (PDB ID code 5KK4). Figures were prepared using PyMol.
Chemical Cross-Linking.
NsD7 at 1 mg/mL (5 μL) were incubated with 0, 0.092, 0.46, or 2.3 mM PA (5 μL) at room temperature for 30 min. Protein complexes were cross-linked through primary amino groups by the addition of 12.5 mM bis[sulfosuccinimidyl] suberate (BS3; 10 μL) in a buffer containing 20 mM sodium phosphate and 150 mM NaCl, pH 7.1, at room temperature for 30 min. Samples were reduced and denatured and subjected to SDS/PAGE before Coomassie Brilliant Blue staining. For the comparative cross-linking experiments with NsD7 and the NsD7 mutants, 2.3 mM PA was used.
Transmission Electron Microscopy.
TEM imaging was performed according to the procedure described previously (20). In brief, samples were prepared by mixing 1 mg/mL protein with 2.3 mM PA at a 9:1 ratio. The samples (10 μL) were applied to 400-mesh copper grids coated with a thin layer of carbon for 2 min. Excess material was removed by blotting, and samples were negatively stained twice with 10 μL of a 2% (wt/vol) uranyl acetate solution (Electron Microscopy Services). The grids were air-dried and viewed using a JEOL JEM-2010 transmission electron microscope operated at 80 kV.
ATP Release Assay with ATP-Encapsulated Liposomes.
Liposomes were prepared as described in ref. 21 using l-α-PC (chicken egg) and l-α-phosphatidic acid (PA, chicken) purchased from Avanti Polar Lipids. Lipids dissolved in chloroform were combined with the desired molar ratio of lipid components (pure PC or PC:PA, 95:5). The lipid mixture was dried under a stream of nitrogen gas followed by further drying under a vacuum for 16 h. The lipid films were rehydrated to 5 mg/mL in 20 mM Hepes (pH 7.4) containing 5 mg/mL ATP for 20 min at 45 °C with periodic gentle agitation. Lipid mixtures were then freeze-thawed three times before nine rounds of extrusion at 45 °C through an Avanti Polar Lipids Mini-Extruder with 0.2 μM membrane. Liposomes were washed three times in 20 mM Hepes (pH 7.4) by centrifugation at 8,000 × g to remove nonencapsulated ATP. Liposomes were used within 24 h. The release of ATP from liposomes treated with NsD7 or NsD7 mutants was determined by measuring the bioluminescence signal using a SpectraMax M5e microtitre plate reader. ATP-encapsulated liposomes in 20 mM Hepes (pH 7.4) equivalent to 200 μg dried lipid (40 μL) were combined with 50 μL luciferase reagent (Roche) and added to 10 μL protein prepared in buffer (final concentration of 30 μM) immediately before recording luminescence over a 30-min time course. Peak luminescence corrected by subtracting background luminescence (buffer only) was then determined for each reading. One-hundred percent lysis was determined as a 5-min treatment with 1% Triton X-100. Defensin-mediated membrane lysis was then expressed as a percentage of total lysis.
Supplementary Material
Acknowledgments
We thank the MX2 beamline staff at the Australian Synchrotron for help with X-ray data collection, and Janet Newman and Shane Seabrook at the Commonwealth Scientific and Industrial Research Organization C3 Collaborative Crystallization Centre for assistance with crystallization. This work was supported by Australian Research Council Fellowship FT130101349 (to M.K.), Balmoral Australia Pty Ltd., and Hexima Ltd. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Footnotes
Conflict of interest statement: M.D.H. is a former Vice President of Research in Hexima Ltd.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5KK4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607855113/-/DCSupplemental.
References
- 1.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 2.Chu H, et al. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337(6093):477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon IKh, et al. Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. eLife. 2014;3:e01808. doi: 10.7554/eLife.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athenstaedt K, Daum G. Phosphatidic acid, a key intermediate in lipid metabolism. Eur J Biochem. 1999;266(1):1–16. doi: 10.1046/j.1432-1327.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- 5.Testerink C, Munnik T. Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005;10(8):368–375. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Sagaram US, et al. Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: Identification of an RGFRRR motif governing fungal cell entry. PLoS One. 2013;8(12):e82485. doi: 10.1371/journal.pone.0082485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thevissen K, et al. Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem. 2004;279(6):3900–3905. doi: 10.1074/jbc.M311165200. [DOI] [PubMed] [Google Scholar]
- 8.Gonçalves S, et al. Evaluation of the membrane lipid selectivity of the pea defensin Psd1. Biochim Biophys Acta. 2012;1818(5):1420–1426. doi: 10.1016/j.bbamem.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Ramamoorthy V, et al. Glucosylceramide synthase is essential for alfalfa defensin-mediated growth inhibition but not for pathogenicity of Fusarium graminearum. Mol Microbiol. 2007;66(3):771–786. doi: 10.1111/j.1365-2958.2007.05955.x. [DOI] [PubMed] [Google Scholar]
- 10.Thevissen K, et al. A gene encoding a sphingolipid biosynthesis enzyme determines the sensitivity of Saccharomyces cerevisiae to an antifungal plant defensin from dahlia (Dahlia merckii) Proc Natl Acad Sci USA. 2000;97(17):9531–9536. doi: 10.1073/pnas.160077797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lay FT, et al. Dimerization of plant defensin NaD1 enhances its antifungal activity. J Biol Chem. 2012;287(24):19961–19972. doi: 10.1074/jbc.M111.331009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baxter AA, et al. The tomato defensin TPP3 binds phosphatidylinositol (4,5)-bisphosphate via a conserved dimeric cationic grip conformation to mediate cell lysis. Mol Cell Biol. 2015;35(11):1964–1978. doi: 10.1128/MCB.00282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lay FT, Veneer PK, Hulett MD, Kvansakul M. Recombinant expression and purification of the tomato defensin TPP3 and its preliminary X-ray crystallographic analysis. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 3):314–316. doi: 10.1107/S1744309112001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 3):432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adda CG, et al. Plasmodium falciparum merozoite surface protein 2 is unstructured and forms amyloid-like fibrils. Mol Biochem Parasitol. 2009;166(2):159–171. doi: 10.1016/j.molbiopara.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan TK, et al. Human β-defensin 3 contains an oncolytic motif that binds PI(4,5)P2 to mediate tumour cell permeabilisation. Oncotarget. 2016;7(2):2054–2069. doi: 10.18632/oncotarget.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.