Significance
The prototypical proprotein convertase furin proteolytically activates many precursor proteins and is essential for cellular homeostasis of the cell. Furin and other proprotein convertase are also associated with a number of diseases, including cancer, atherosclerosis, hypercholesterolaemia, and infectious diseases. A hallmark of furin and its physiologic function is its stringent specificity for polybasic substrates. Here we describe several structural states of the protein by X-ray crystallography and further characterize them by molecular dynamics simulations. The transition between these conformers is triggered by substrate binding and calcium ligation, respectively, and helps explain the stringent specificity of furin and its functional features. These studies may pave the way for novel strategies of structure-guided inhibitor development.
Keywords: serine-protease, activation, specificity, conformational transition
Abstract
Proprotein convertases (PCs) are highly specific proteases required for the proteolytic modification of many secreted proteins. An unbalanced activity of these enzymes is connected to pathologies like cancer, atherosclerosis, hypercholesterolaemia, and infectious diseases. Novel protein crystallographic structures of the prototypical PC family member furin in different functional states were determined to 1.8–2.0 Å. These, together with biochemical data and modeling by molecular dynamics calculations, suggest essential elements underlying its unusually high substrate specificity. Furin shows a complex activation mechanism and exists in at least four defined states: (i) the “off state,” incompatible with substrate binding as seen in the unliganded enzyme; (ii) the active “on state” seen in inhibitor-bound furin; and the respective (iii) calcium-free and (iv) calcium-bound forms. The transition from the off to the on state is triggered by ligand binding at subsites S1 to S4 and appears to underlie the preferential recognition of the four-residue sequence motif of furin. The molecular dynamics simulations of the four structural states reflect the experimental observations in general and provide approximations of the respective stabilities. Ligation by calcium at the PC-specific binding site II influences the active-site geometry and determines the rotamer state of the oxyanion hole-forming Asn295, and thus adds a second level of the activity modulation of furin. The described crystal forms and the observations of different defined functional states may foster the development of new tools and strategies for pharmacological intervention targeting furin.
Furin belongs to the family of highly specific, calcium-dependent proprotein/prohormone convertases (PCs) (1), endoproteinases that feature a catalytic domain of homology to subtilisin and activate a large number of secreted proteins by limited proteolysis. Furin is a type I transmembrane serine-protease that is ubiquitously expressed and cycles from the trans-Golgi network to the cell membrane, as well as through the endosomal system. In mammals, the PC family embraces seven members that cleave after multiple basic residues (furin, PC1, PC2, PC4, PACE4, PC5/6, and PC7) and at the general cleavage site (R/K)Xn(R/K)↓ (where “↓” represents the scissile peptide bond), with furin preferentially recognizing the motif R-X-K/R-R↓ (2). This highly sequence-specific cleavage is essential for the activation of numerous PC substrates (2). The PCs are also involved in a large number of pathologies, including bacterial and viral infections, as well as cancer and metastasis. Therefore, these enzymes are intensely investigated as drug targets (3), using for example, peptide-derived compounds (4–9), nonpeptidic small molecule compounds (10, 11), and protein-based inhibitors (12–15).
Different crystal structures of the inhibitor-bound murine (16) and human (6, 17) enzyme, together with structural studies of the yeast homolog Kex2p (18, 19) and modeling approaches (9, 20), gave hints of how substrates and substrate-derived inhibitors bind with high specificity to furin and to the PCs, but suffer from the lack of structural information of the unliganded enzyme. The commonly accepted notion is that the PCs bind their cognate substrates via recognition at several subsites, typically involving multiple tight contacts and hydrogen bonds. Catalytic efficiency, however, in addition to strong and fast binding of substrate, requires rapid chemical transformation and fast release of product.
The chemical reaction catalyzed by furin and other proteases—the hydrolysis of the scissile peptide bond—often involves limited structural changes of the active site residues, which are unlikely to perturb remote substrate-binding subsites of the enzyme. Furin has an extended substrate-binding area where typically all specificity-determining residues are located N-terminal to the scissile peptide bond, suggesting that the C-terminal segment is more loosely bound and may dissociate readily after hydrolysis. Potent noncovalent inhibitors are characterized by very slow off-rates, a feature that may apply to very tight binding substrates that are poorly cleaved as well, and can thus act as efficient inhibitors as those, for example, shown in other protease families for the slowly hydrolyzed dipeptidyl peptidase IV substrate diprotin A (21).
The high substrate selectivity and enzymatic activity of furin, and its strict Ca2+ dependence, demand a structure-based explanation. Although Ca2+ is not involved in the catalytic cycle per se, all PCs require calcium for their enzymatic activity (17, 22, 23).
One more distinctive feature of furin in the protease family is its very limited reactivity toward typical covalent inhibitors of serine proteases, like diisopropylfluorphosphat (DFP) and phenylmethylsulfonylfluorid (PMSF). Interestingly, the spatial restrictions at and around the catalytic serine residue 368 of furin are structurally reminiscent of typical trypsin-like proteases. This similarity is especially evident for the deep S1 pocket selecting for basic amino acid side chains at P1, the β-strand of the enzyme (Ser253-Gly255 in furin vs. Ser214-Gly216 in trypsin), displaying antiparallel β-sheet–like contacts with a peptidic substrate and a typical hydrophobic tryptophane (Trp254 in furin vs. Trp215 in trypsin) in its center. Therefore, one would expect a similar reactivity of furin against nonselective, covalent inhibitors that do not specifically interact with the different specificity pockets, but probe the nucleophilic catalytic site serine residue; however, this has not been observed. Furin is also completely inert against typical monobasic trypsin-substrates (our own observations and those, for example, in refs. 22 and 24). This is observed even though (i) the P1 requirements of the two enzymes are comparable, (ii) furin accepts different residues at its S2-pocket, and (iii) its S3-pocket is not very specific.
To address the above questions and to find out how the high negative-charge density within the active site cleft of furin is stabilized in its ligand-free state, we have studied the structure of the unliganded enzyme as well as of different Ca2+ and inhibitor-bound forms by X-ray crystallography at high resolution. Comparison of the different crystal structures together with solution studies and molecular dynamics (MD) simulations give hints of the enzymatic mechanism and the unique features of this central regulatory endoproteinase.
Results
Inhibitor-Induced Structural Changes of the Active-Site Cleft.
Human furin was crystallized in its unliganded state, its structure solved by molecular replacement (MR), and refined up to 1.8-Å resolution at high quality (Fig. 1A and Table S1). Structure alignments of unliganded furin with the inhibitor-bound human and mouse furin structures demonstrated high overall similarity (Cα-rmsd of 0.22 Å and 0.28 Å, respectively). A close inspection of the active-site residues and the substrate-binding cleft, however, revealed substantial differences.
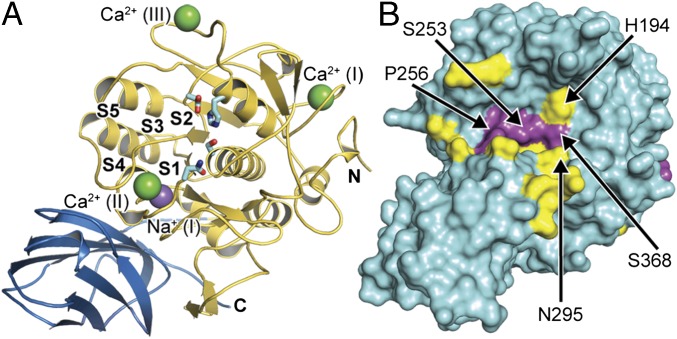
Fig. 1.
Analysis of substrate induced structural rearrangements of furin. (A) Overall structure of human unliganded furin (catalytic and the P-domains are in gold and blue, respectively). The catalytic residues (cyan stick model), the substrate binding pockets (labeled S1–S5), and bound ions (purple sphere, Na+; green spheres, Ca2+) are indicated. (B) The Cα-displacement values (intervals <0.3 Å, 0.3–0.8 Å, and >0.8 Å are highlighted in cyan, yellow, and magenta, respectively) were mapped to the molecular surface.
Table S1.
Data collection and refinement
| Dataset | Unliganded | Unliganded + inhibitor | EDTA | EDTA + inhibitor |
| Resolution range (Å) | 46.1–1.8 (1.91–1.80) | 43.1–2.0 (2.12–2.00) | 46.2–2.0 (2.12–2.00) | 46.1–2.0 (2.12–2.00) |
| Unit cell (P6522) (Å): a, c | 132.0, 155.6 | 131.8, 155.6 | 132.4, 155.7 | 132.1, 155.6 |
| Rsymm* (%) | 9.6 (62.3) | 10.8 (57.5) | 11.6 (59.5) | 13.6 (78.8) |
| <I/sig(I)> | 13.4 (2.8) | 13.5 (3.1) | 14.3 (3.0) | 11.9 (2.3) |
| Completeness (%) | 99.1 (98.3) | 99.2 (98.8) | 98.9 (99.1) | 98.9 (98.7) |
| Observations: total/unique | 364,023/73,616 | 266,429/53,872 | 268,857/54,336 | 266,967/53,880 |
| Rwork/Rfree | 15.4/17.3 | 15.7/18.6 | 15.8/18.5 | 16.7/18.9 |
| Nonhydrogen atoms | 4,263 | 4,204 | 4,126 | 4,130 |
| Protein/Inhibitor/ions/solvent | 3,608/–/12/643 | 3,608/54/8/534 | 3,597/–/9/520 | 3,608/54/8/460 |
| B-factors (Å2) | ||||
| Overall/Wilson plot | 20.9/24.2 | 23.5/26.4 | 22.1/24.6 | 24.5/27.0 |
| Protein/inhibitor/ions/solvent | 19.0/–/24.3/32.6 | 22.1/17.3/22.7/33.4 | 20.7/–/26.9/31.5 | 23.5/19.1/27.0/33.1 |
| Rmsd bond length (Å) | 0.007 | 0.007 | 0.007 | 0.007 |
| Rmds bonded B-factors (Å2) | 3.6 | 3.7 | 4.2 | 4.0 |
Values of the highest resolution shell are given in parentheses.
Rsymm=ΣhΣl|Ihl-<Ih>|/ ΣhΣl|<Ih>|.
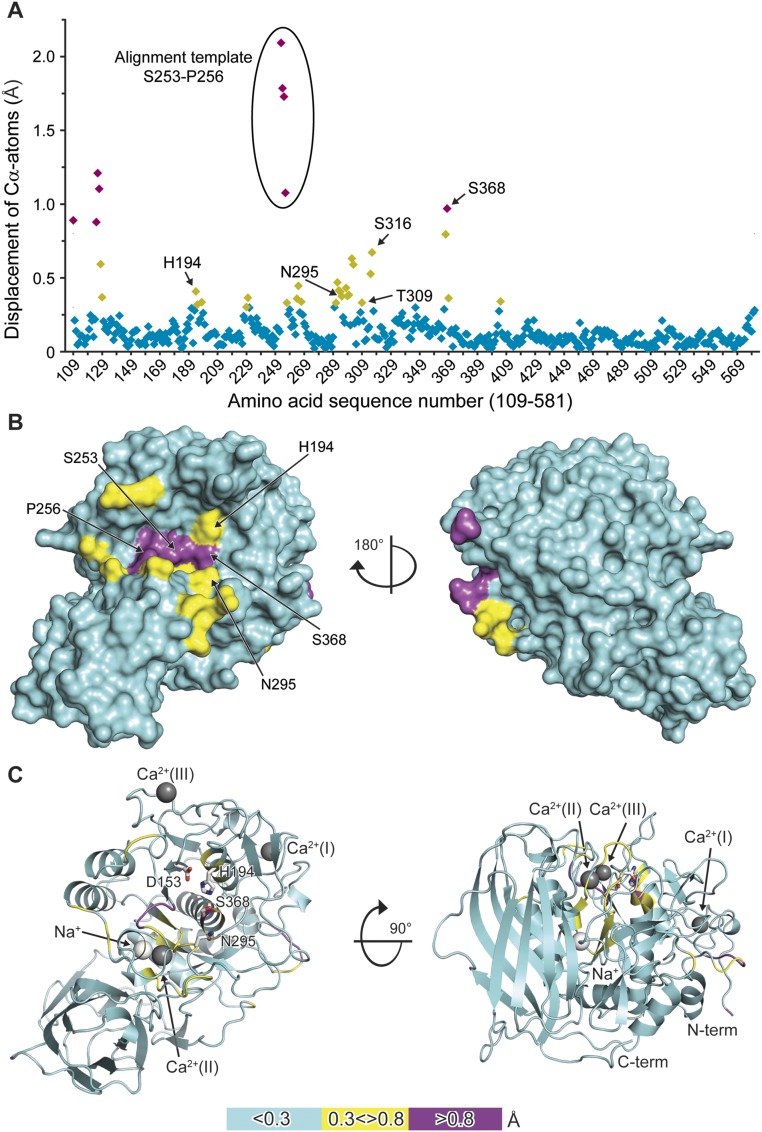
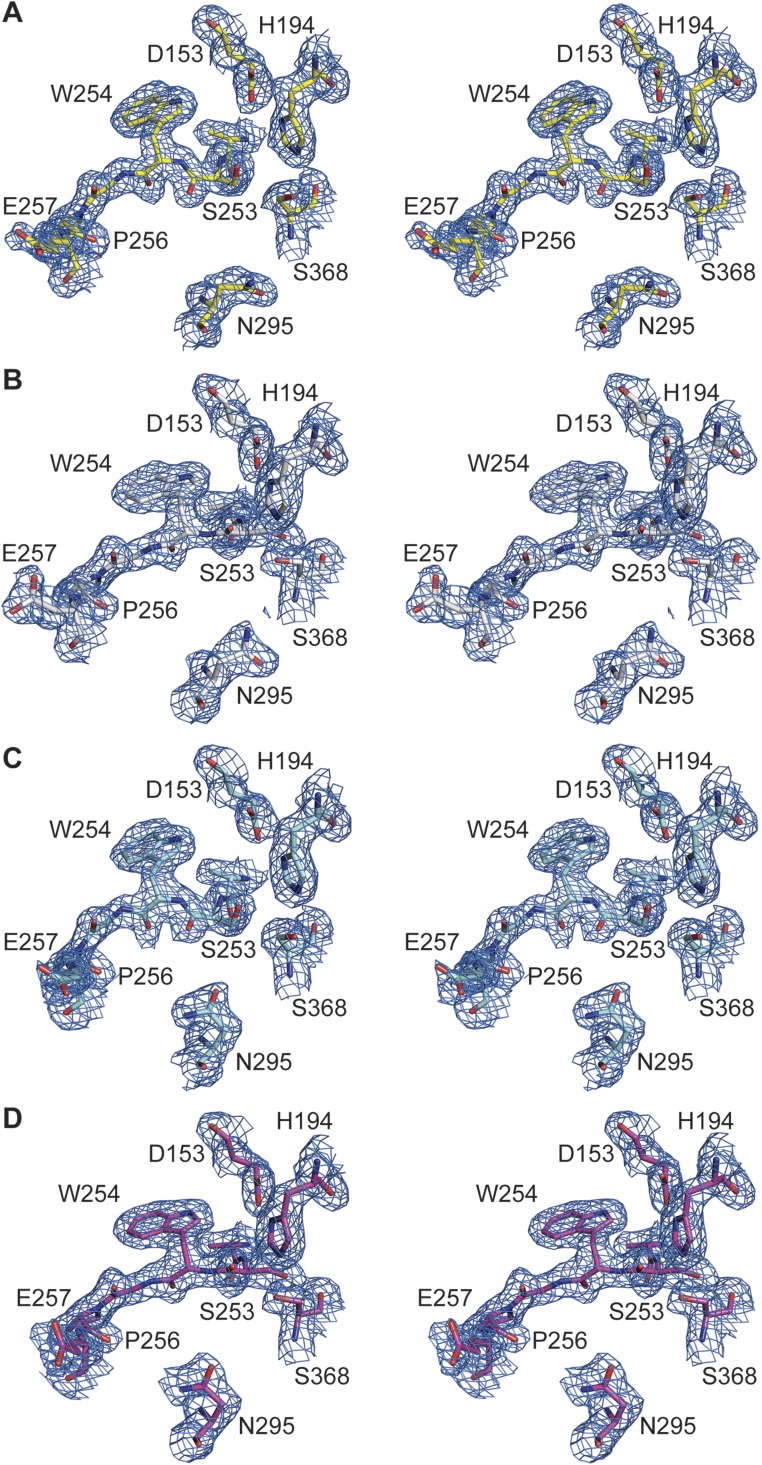
To directly investigate structural changes occurring upon substrate binding in identical crystal forms, we soaked the unliganded crystals with the substrate analogous, noncovalent inhibitor m-guanidinomethyl-phenylacetyl-Arg-Val-Arg-(4-amidomethyl)-benzamidine (MI-52, #26 in ref. 4). The active-site cleft of furin is oriented toward large solvent channels in these crystals, enabling the binding of large peptidic compounds to the protease unperturbed by crystal packing. The crystal structure was refined at a resolution of 2.0 Å (Table S1). A residue-wise comparison of unliganded and inhibitor-bound forms revealed a number of significant structural changes (Fig. S1A). Hotspots of conformational changes include the catalytic residues His194, Ser368, Asn295 of the oxyanion hole, the sodium binding site (Thr309 and Ser316), and residues in direct contact with the inhibitor peptide (e.g., the region Ser253–Pro256, the alignment template). Mapping of the Cα displacement to the surface of the structure induced by inhibitor binding revealed a concerted local rearrangement at the substrate-binding cleft (Fig. 1B and Fig. S1B). These displacements propagate with diminishing amplitude to the core of the protease (Fig. S1C). The catalytic residues and the alignment template are well defined in the electron-density maps (Fig. S2 A and B) and characterized by low flexibility.
Fig. S1.
Structural analysis of furin in its unliganded vs. its MI-52–bound states. (A) Plot of the Cα-displacement values of the Cα atoms covering all amino acids of furin (intervals <0.3 Å, 0.3–0.8 Å, and >0.8 Å are highlighted in cyan, yellow, and magenta, respectively). (B) Mapping of the respective values to the molecular surface and colored according to A. (C) Cartoon representation colored according to A. Bound ions are shown as light gray (Na+) and dark gray (Ca2+) spheres.
Fig. S2.
Electron-density maps observed for the active-site residues and the alignment template of human furin. Stereo panels show the structures in stick representation. The 2Fo–Fc simulated annealing composite-omit electron density maps are given as blue-colored mesh, which is contoured at 1.0 σ. (A) Unliganded furin. (B) Furin complexed with MI-52. (C) Unliganded furin in presence of EDTA. (D) Furin in presence of EDTA and complexed with MI-52.
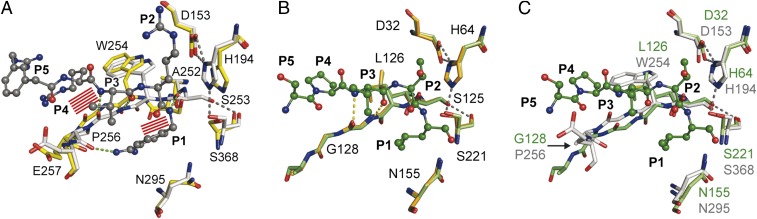
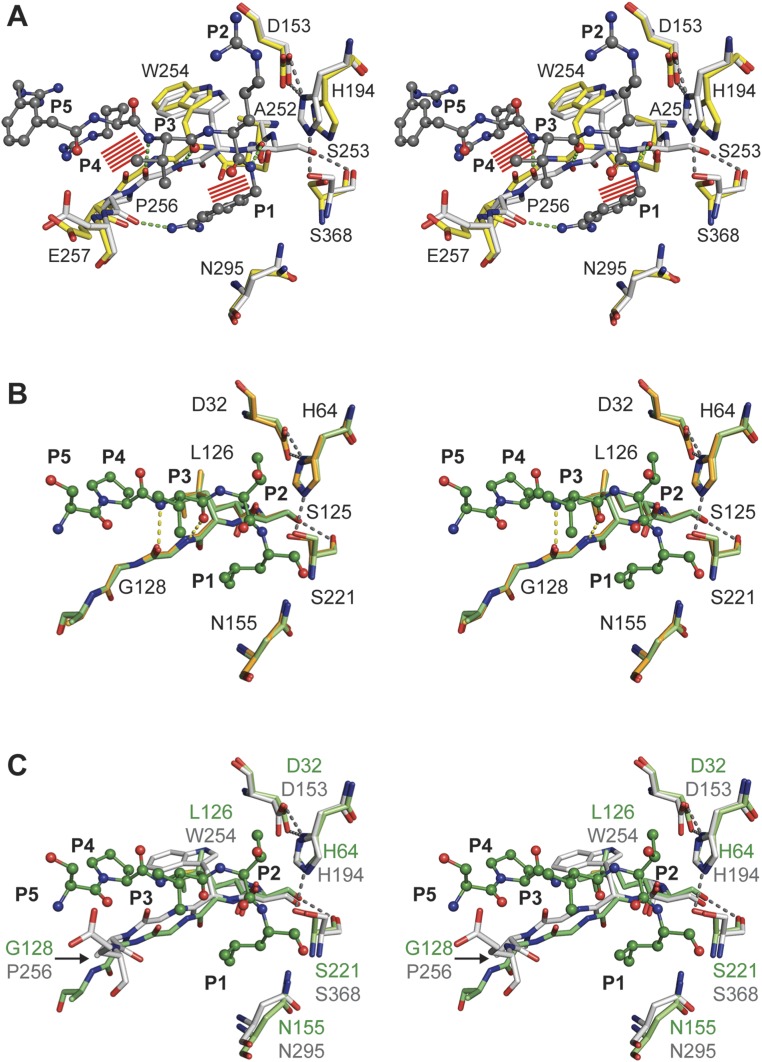
A structural comparison of the substrate-binding region indeed revealed two distinct conformational states for furin (Fig. 2A and Fig. S3A). In contrast, the homologous unspecific protease subtilisin showed a high overall structural similarity (Cα rmsd of 0.24 Å) of the unliganded structure [PDB ID code 3UNX (25)] and the subtilisin–eglin-c complex [PDB ID code 1CSE (26)] as well as identical conformations of the active site cleft (Fig. 2B and Fig. S3B).
Fig. 2.
Structural comparison of substrates binding to the active sites of furin and to subtilisin Carlsberg. (A) Structural alignment of selected residues of unliganded furin (yellow carbons) and inhibitor-bound furin (gray carbons; inhibitor: ball-and-stick). Steric clashes between bound inhibitor/substrate and unliganded furin are highlighted as red line patterns. (B) Structural alignment of unliganded subtilisin (orange carbons) and inhibitor-bound subtilisin (green carbons). (C) Structural alignment of inhibitor-bound subtilisin (green carbons) and furin (gray-colored stick model). Important interactions are always highlighted by dashes.
Fig. S3.
Stereo representation of the structural comparison of substrate binding by furin and subtilisin Carlsberg. Detailed view of the alignment template and the active-site residues of inhibitor-bound furin (protein: gray-colored stick model; inhibitor: gray-colored ball-and-stick model), unliganded furin (yellow-colored stick model), unliganded subtilisin (light orange-colored stick model), and eglin-c (shown P5–P1 segment Ser-Pro-Val-Thr-Leu) bound subtilisin (protein: light green-colored stick model; inhibitor: dark green-colored ball-and-stick model). (A) Structural alignment of selected residues of unliganded furin and inhibitor-bound furin. Steric clashes between bound inhibitor/substrate and unliganded furin are highlighted as red line patterns around the respective atoms. (B) Structural alignment of unliganded subtilisin and inhibitor-bound subtilisin. Important interactions of the active-site residues and between the alignment template and the inhibitor are highlighted as gray and yellow dashes, respectively. (C) Structural alignment of inhibitor-bound subtilisin and furin. Important homologous amino acids of the proteases are labeled in green (subtilisin) and gray (furin).
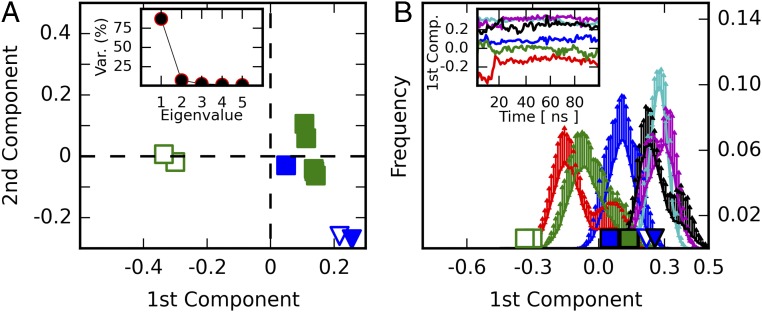
A principal component analysis (PCA) was carried out to gain further insight into the structural modifications induced by inhibitor binding and their relationship with furin’s homologous proteins. To allow a direct comparison between furin and the closely related protease subtilisin Carlsberg, a common core of 191 residues was selected to perform the PCA on their Cα atoms. Projecting the crystallographic structures on the first principal component, which captures more than 80% of the variance, shows a clear separation among the furin structures with and without ligand-bound conformations (Fig. 3A), but not among the subtilisin Carlsberg structures, where both unliganded and inhibitor-bound conformations exhibit closer similarity to the furin inhibitor-bound structures. These observations prompted us to consider the first principal component as an appropriate metric to monitor the unliganded and inhibitor-bound states. A detailed inspection revealed that the conformation of residues proximal to the alignment template and the active site of subtilisin and furin are similar in the ligand-bound state (overall Cα rmsd of 0.38 Å) (Fig. 2C and Fig. S3C): specifically, the rotamers of Ser221 of subtilisin and Ser368 of furin, respectively. Conversely, the unliganded conformations of both enzymes differ and reflect the structural transition in furin absent in subtilisin. Interestingly, the PCA covariance matrix unveils a concerted movement between the alignment template (Ser253–Pro256), residues neighboring the sodium binding site (314–317), and the oxyanion hole (290–297 and 365–369) again.
Fig. 3.
PCA. (A) Projections of furin (squares) and subtilisin Carlsberg (triangles) on the first two principal components. Only human furin structures (green) were used to compute the covariance matrix. Murine furin structures (blue) are shown for comparison only. Open and closed markers represent unliganded and inhibitor-bound structures, respectively. (Inset) The proportion of the structural variation encoded within each principal component. (B) Histograms of MD-simulation frames projected on the first principal component for each simulated system: furin unliganded state (red), furin inhibitor-bound (blue), furin inhibitor-removed (green), subtilisin unliganded state (black), substilisin inhibitor-bound (cyan), and subsitilisn inhibitor-removed (magenta). Projections of the crystallographic structures are shown on the x axis as in A. (Inset) Examples of MD trajectories.
The unliganded structure of furin has the side chain of Ser368 rotated 180° away from the S1 pocket and the scissile peptide bond, as defined in the dec–RVKR–cmk complex structure (16), altering the interaction geometry of the catalytic triad and disrupting the essential hydrogen bond to His194. However, a nucleophilic attack at the scissile peptide bond requires a conformation, as observed in the inhibitor-bound structure. The rotamer of Ser368 as observed in the unliganded structure can thus not enable substrate processing. Interestingly, the rotameric state of Asn295 in the unliganded and the ligand-bound forms is largely identical as long as the Ca2+-(II) site is occupied (see Calcium-Dependent Activity Regulation of Furin, below).
The loss of the hydrogen bond between Ser253-OH and the carbonyl-O of Ser368 appears to facilitate the reorientation of Ser368 in unliganded furin (Fig. 2A and Fig. S3A). In addition, binding of peptide chains to the active-site cleft of furin requires a specific orientation of the alignment template, enabling characteristic backbone interactions and a P1–side-chain recognition. These interactions are incompatible with the unliganded structure (Fig. 2A and Fig. S3A). Furthermore, steric clashes between ligand and the enzyme would occur, especially at P1 and P4. In conclusion, the catalytic residues and the substrate-binding region adopt an inactive conformation in the unliganded furin structure, representing an “off” state. By binding of the substrate-analog inhibitor MI-52, the enzyme switches to a catalytically active “on” state.
MD Calculations Show Switching Between the On and Off States.
Next we assessed the stability of the unliganded and inhibitor-bound conformations by performing MD simulations. For this purpose, furin and subtilisin were simulated in three different conditions: (i) an unliganded state, (ii) an inhibitor-bound state, and (iii) an inhibitor-removed state (protein in its inhibitor-bound conformation with the inhibitor molecule deleted). Histograms collected from projecting MD trajectories on the first principal component show that for furin, the inhibitor-removed state drifts away from its initial inhibitor-bound conformation. It relaxes toward a conformation that resembles the crystallographic unliganded conformation and approaches the relaxation state reached by simulations of furin’s unliganded state (Fig. 3B). In contrast the inhibitor-bound simulations show that the furin–MI-52 complex is highly stable, sampling conformations around its crystallographic pose. Notably, furin simulations in its unliganded state eventually visit the ligand-bound conformation, thereby suggesting that the latter is a thermally accessible state. In subtilisin, all three systems retain their crystallographic conformations and sample inhibitor-bound–like structures (Figs. 2B and 3B and Fig. S3B). These results suggest that in furin, but not in subtilisin, a fraction of the ligand binding energy is spent in transforming the enzyme from an off state to an on state.
Prompted by these observations, we reanalyzed the melting temperatures (Tm) of furin–inhibitor complexes and their dependence on the Ki-values (6). In a semi-logarithmic representation of the data, an apparent linear relationship between pKi (range 6.4–11.3) and Tm can be found. Interestingly, by extrapolation to pKi-values < 6, Tm falls below the value observed for unliganded furin. We reason that the dependence of melting temperature and stability on ligand affinity rests on two opposing factors: a gain in bonding interactions and energy with increasing affinity and a loss by the conformational switch with all ligands, such that the latter prevails with weakly binding ligands.
Specific Entities Define the Structural Difference Between the Two Conformational States.
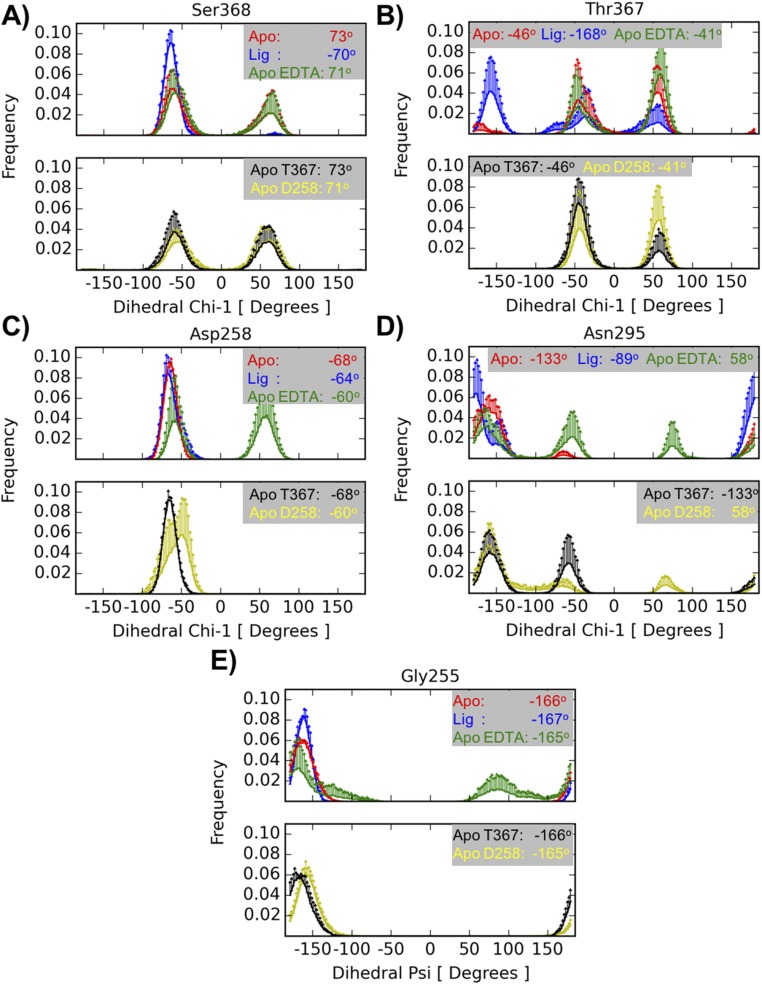
Comparisons of the unliganded and inhibitor-bound crystallographic structures provide insight regarding to the key structural features that characterize the on state. Different alignment template configurations allow hydrogen bond formation between Ser253γOH and the carbonyl oxygen of Ser368 in the on state, whereas this interaction is absent in the off state. This interaction prompts a Ser368 side-chain rotation of 180° away from the S1 pocket, thereby altering the interaction geometry of the catalytic triad. The MD simulations support this mechanism because, in the furin–inhibitor complex, the Ser253γOH–Ser368O hydrogen bond is observed 88% of the time, considering all repetitions (SI Materials and Methods), as a part of a dynamic process, and the rotameric state of Ser368 is stable in the experimentally observed conformation (Fig. S4A). In contrast, in the furin unliganded system this hydrogen bond is practically never observed (1% of the time) and the distribution of Ser368 χ-angle is bimodal, suggesting a dynamic equilibrium between the two orientations. Interestingly, the dynamic behavior of Thr367 appears to be also related to the on and off states. The Thr367 χ1 dihedral angle (Fig. S4B) displays a single orientation in the liganded state corresponding to the crystal structure, whereas the unliganded form shows, as for Ser368, two stable orientations. However, only one of these orientations corresponds to that observed in the crystals. The MD data suggest that Thr367 χ1 also samples an orientation where its hydroxyl group approaches the S1 pocket and the Asn295’s side chain.
Fig. S4.
Rotameric states from MD simulations. Histograms are computed from five independent runs of each simulated system: unliganded furin (red), furin with inhibitor MI-52 (blue), unliganded furin in the presence of EDTA (green), unliganded furin with Thr367’s χ1 initially set at 60° (black), unliganded furin in the presence of EDTA and with Asp258 protonated (yellow). For residues Ser368 (A), Thr367 (B), Asp258 (C), and Asn295 (D) the measured quantity is the χ1 dihedral-angle, whereas for Gly255 (E) it is the ψ dihedral-angle. The information is split into two panels for clarity. The Upper panel contains plots from the simulations of unliganded furin, furin with inhibitor, and unliganded furin in the presence of EDTA structures; the Lower panel contains plots corresponding to simulations of unliganded furin with Thr367's χ1 initially set at 60° and unliganded furin in the presence of EDTA with Asp258 protonated. Each curve represents an average histogram (solid lines) with 1 SD as upper limit (error bar). The averages were computed from five independent runs. The values in each inset correspond to the values found in the crystallographic structures.
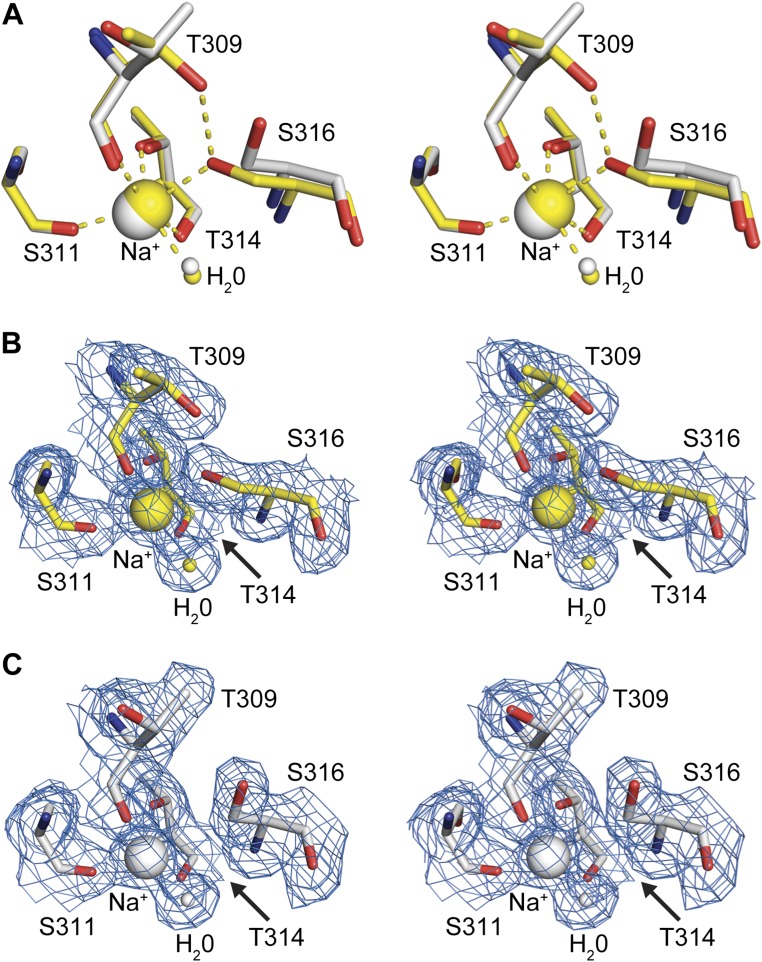
Conformational changes are also transduced to the single sodium binding site of the protease (Fig. S5). In the unliganded structure the sodium ion is coordinated by the carbonyl oxygens of Thr309, Ser311, and Thr314, the hydroxyls of Thr314 and Ser316, as well as one water molecule, in the preferred octahedral coordination. Upon inhibitor binding, the coordination changes to a tetragonal-pyramidal geometry that is less frequently observed. The side-chain rotations of Ser316 and Thr309 result in breakage of the coordinative bond to the sodium ion and loss of the hydrogen bond between these residues, indicating a switch from a lower (unliganded form) to a higher (liganded form) energy state. Accordingly, in the MD simulations of the unliganded system the rotameric state of Ser316 is highly stable and the Ser316γOH–Thr309γOH interaction is relatively well conserved, being present 50% time in a dynamic fashion considering all repetitions.
Fig. S5.
Substrate-induced conformational changes of the sodium site in furin. Stereo panels show a superposition of selected residues of unliganded furin (yellow, protein: stick model; nonbonded atoms: spheres) and inhibitor-bound furin (gray, protein: stick model; inhibitor: ball-and-stick; nonbonded atoms: spheres). (A) Superposition of the unliganded inhibitor-bound states. (B and C) Electron-density maps observed for the sodium binding site. Sodium ions and water molecules are given as big and small spheres, respectively. The 2Fo–Fc simulated annealing composite-omit electron-density maps are given as blue-colored mesh and are contoured at 1.0 σ. (B) Unliganded furin. (C) Furin complexed with MI-52.
Reaction of Furin with Nonspecific Inhibitors.
Previous studies had demonstrated a lack of reactivity of furin for nonspecific covalent serine protease inhibitors (22). We have reanalyzed here the inihibitory potency of DFP, 4-(2-aminoethyl)benzensulfonylfluorid (AEBSF), and PMSF. All three inhibitors readily inhibited the activity of the two prototypical serine proteases, trypsin and subtilisin, to below 3% of their original activity, but did not significantly affect the activity of human furin (SI Materials and Methods). As modeling excluded steric reasons, the failure of these inhibitors to bind may reflect the electronic properties and activation state of furin’s active site, and thus represent an electrophilic probe and measure for the reactivity of the protease.
Calcium-Dependent Activity Regulation of Furin.
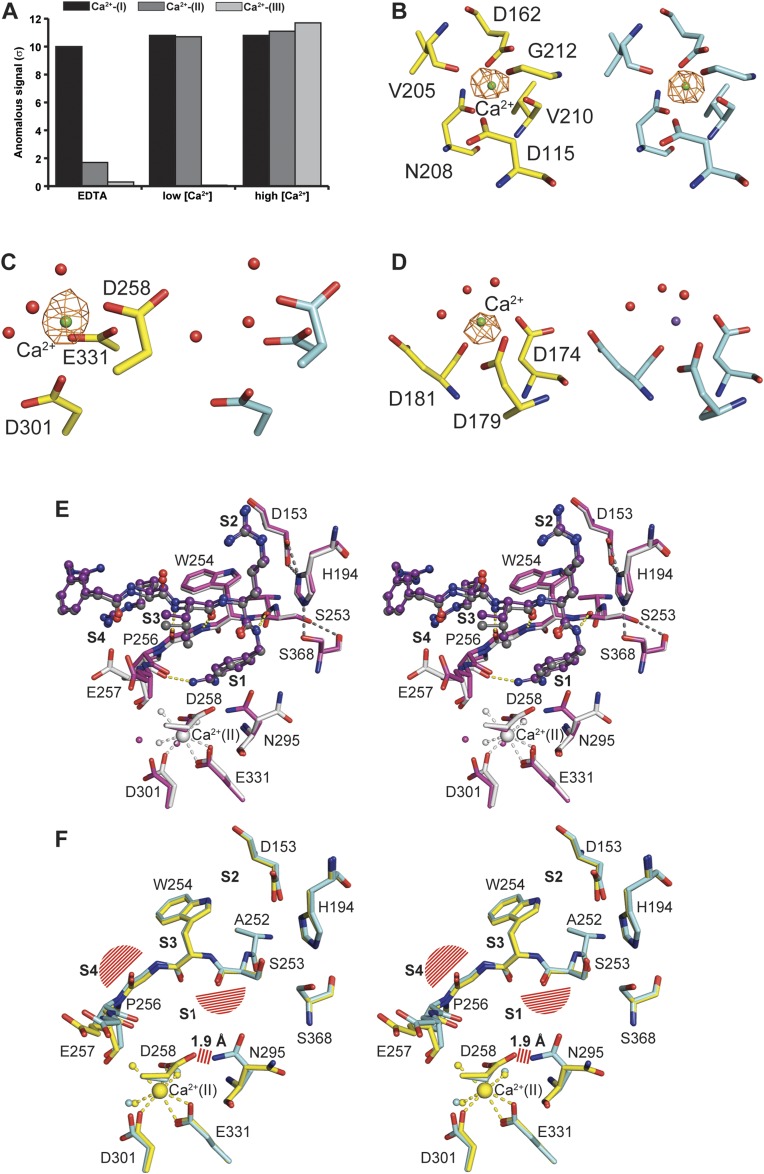
Previous studies have shown that the activity of furin is dependent on the presence of calcium (22). To investigate its role in activity regulation and substrate binding, we soaked unliganded furin crystals with EDTA, and analyzed and refined the structure to 2.0-Å resolution (Table S1). To our surprise, only the Ca2+-(II) and Ca2+-(III) ions (Fig. 1A) were removed by addition of the strong calcium chelator, suggesting different affinities of the three calcium binding sites. We further analyzed their anomalous diffraction at defined concentrations of Ca2+ (Table S2), finding decreasing affinities in the order Ca2+-(I) > Ca2+-(II) > Ca2+-(III) (Fig. S6 A–D).
Table S2.
Statistics for diffraction datasets collected at low energy to analyze Ca2+-binding
| Dataset | Unliganded low energy | Low Ca2+ low energy | EDTA low energy |
| Wavelength (Å)/energy (eV) | 2.07/6,000 | 2.07/6,000 | 2.07/6,000 |
| Resolution range (Å) | 47.2–2.4 (2.58–2.44) | 47.2–2.6 (2.73–2.58) | 47.2–2.6 (2.73–2.58) |
| Unit cell (P6522) (Å): a, c | 132.1, 155.4 | 132.4, 155.4 | 132.1, 155.3 |
| Rsymm* (%) | 11.0 (58.0) | 10.6 (55.3) | 9.9 (47.7) |
| <I/sig(I)> | 10.1 (2.1) | 11.3 (2.2) | 12.0 (2.6) |
| Anomalous completeness (%) | 99.1 (97.4) | 98.4 (96.5) | 98.2 (96.0) |
| Observations: total/unique | 186,624/56,144 | 159,820/47,320 | 159,072/47,096 |
| Initial model refinement | |||
| Rwork/Rfree | 18.6/21.8 | 17.0/20.5 | 17.3/20.7 |
Values of the highest resolution shell are given in parentheses.
Rsymm=ΣhΣl|Ihl-<Ih>|/ ΣhΣl|<Ih>|.
Fig. S6.
Binding of calcium to furin and its structural effects. (A) Bar diagram of the anomalous signal observed for the calcium binding sites at high (1 mM) and low (formally 1 nM, but see SI Materials and Methods for details) Ca2+ concentrations as well as in the presence of EDTA. (B–D) The coordinating amino acids are given for Ca binding sites Ca-I (B), Ca-II (C), and Ca-III (D) as stick model (yellow carbons correspond to high calcium concentration, cyan carbons correspond to the presence of EDTA) together with selected water molecules (red spheres). The Ca2+ ions (green spheres) are shown together with the anomalous electron-density map (countoured at 6.0 σ and given as orange mesh) at high calcium concentration or in presence of EDTA. At low Ca2+-concentration, Ca2+-(III) is displaced by Na+ (purple sphere). (E) Stereo representation of the superposition of EDTA and MI-52 soaked furin (purple carbons, purple inhibitor) with non-EDTA–treated furin (gray). Selected hydrogen bonds are highlighted as dashes according to Fig. 2. Coordinative bonds of Ca2+-(II) are highlighted as gray dashes. Calcium ions and water molecules are represented by large and small spheres, respectively (coloring according to the carbon atoms of the respective structures). (F) Superposition of unliganded furin (yellow) and additionally EDTA-treated unliganded furin (cyan). Basically, identical structural alterations as shown in Fig. 2A are observed. Steric clashes with the superposed inhibitor (according to Fig. 2A and Fig. S4) and with N295 in its inhibitor-bound conformation are highlighted as red line pattern. Coordinative bonds of Ca2+-(II) are highlighted as yellow dashes. Calcium ions and water molecules are represented by large and small spheres, respectively (coloring according to the carbon atoms of the respective structures).
A structural comparison of unliganded furin crystals soaked with either EDTA or grown in the presence of 1 mM Ca2+ revealed almost an identical result in the PCA (Fig. 3A) and in the Cα overlay (rmsd 0.09 Å) (Fig. S7). In addition, the distribution of B-factors of surface-exposed residues is highly similar. However, a detailed inspection of the active-site residues revealed differences at the Ca2+-(II) binding site. Although the calcium ligands Asp258, Asp301, and Glu331 undergo only slight changes upon removal of Ca2+-(II), the side chain of Asn295 displays a different rotamer and loses its role as central component of the oxyanion hole, where it forms a hydrogen bond to and thus polarizes the carbonyl oxygen of the scissile peptide (Fig. S6 E and F). In the Ca2+-(II)–free state the side chain of Asp258 may be protonated. It moves slightly toward the position of the previously bound calcium ion such that it now forms a hydrogen bond with the flipped side-chain amide of Asn295. Charging of the Ca2+ site pushes Asp258 away and causes a reorientation of the Asn295 side chain to the catalytically competent conformation. Distortion of the oxyanion hole explains the calcium requirement for catalysis. EDTA-soaked crystals were also subjected to a second soaking step with MI-52 to study the impact of ligand binding (Table S1). Interestingly, the observed changes triggered by the inhibitor are nearly identical to those observed in the Ca2+-(II)–bound forms (Fig. S6E). The conformation of the Ca2+-(III) ligands are identical in presence and absence of Ca2+ (Fig. S6D).
Fig. S7.
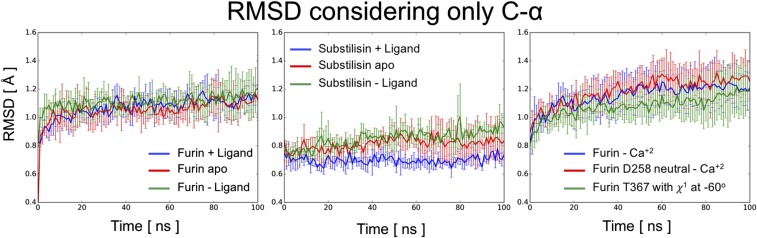
Rmsd curves for the simulated systems. In the panels are shown the average rmsd curves (solid lines) with 1 SD as upper and lower limits (error bars). The averages were computed out of five and four independent runs for furin and substilisin systems, respectively. In all cases, the starting protein-fold is preserved.
A series of five independent MD simulations was performed on the EDTA-soaked unliganded furin structure to evaluate the consequences of the empty Ca2+-(II) site on dynamics. These simulations were carried out considering two Asp258 protonation states. Interestingly, in simulations where Asp258 is protonated, its side-chain orientation corresponds to that observed in the crystal, but showed a second conformation in its charged state (Fig. S4C). The experimentally observed orientation of the Asn295 side chain in Ca2+-(II)–free unliganded furin crystals was only seen as a minor species in MD; it mainly sampled conformations more similar to those observed in the Ca2+-(II)–bound state (Fig. S4D). Nevertheless, prominent differences were observed in Gly255 and in the catalytic triad geometry. The empty ion pocket allows more rotational freedom to Gly255’s ψ angle (Fig. S4E). Because Gly255 is a key element of the alignment template, its structural alteration impairs substrate recognition. Although in this case the MD simulations and the crystallographic observations do not reinforce each other in detail, they both suggest that Ca2+-(II) stabilizes substrate binding and processing.
SI Materials and Methods
Protein Expression and Purification.
Details regarding the plasmids, expression system, and purification procedure used for production of human furin were described previously (17). In short, the protein was expressed by transient transfection of human embryonic kidney cells, comprised of the catalytic and P-domains, and was purified by a three-step chromatographic procedure consisting of immobilized metal-affinity chromatography, immobilized inhibitor affinity chromatography, and gel-permeation chromatography. To increase the yield of recombinant protein, a modified expression and harvesting procedure was applied. For this purpose, transfection was carried out in 150 × 25-mm tissue culture dishes (Falcon) with 0.17 mL/cm2 DMEM supplemented with 2% (vol/vol) FBS, and 5 mM CaCl2. Twenty-four hours postinduction the medium was exchanged by DMEM without FBS but supplemented with 2 mM CaCl2. Finally, conditioned medium was harvested after 72 h of cultivation. Medium containing the recombinant protease was centrifuged at 4,500 × g for 20 min, filtered with a Durapore membrane (Merck-Millipore) of 0.45-µm pore size, and concentrated ∼25-fold in a stirred pressure-based concentration cell (Merck-Millipore) with an Omega 10K ultrafiltration membrane (Pall). Concentrated medium was flash-frozen in liquid nitrogen and stored at −80 °C. Immediately before purification, concentrated medium was thawed in a water bath at 25 °C and centrifuged at 4,500 × g for 20 min. For purification by immobilized metal-affinity chromatography, the supernatant was diluted 1:4 in 100 mM Tris/HCl, pH 8.0, 500 mM NaCl, and 5 mM CaCl2.
Inhibition Assays.
To biochemically probe for the inhibitory potency of typical, nonspecific covalent inhibitors of serine proteases against different proteases, DFP, AEBSF, and PMSF were incubated at 1, 2.5, and 0.5 mM, respectively, with subtilisin (250 µg/mL; ∼9.3 µM), trypsin (5 µg/mL; ∼0.2 µM), and furin (170 µg/mL; ∼3.3 µM) in 100 mM Hepes, 5 mM CaCl, 0.5% Triton X-100, pH 7.0 for 1 h at 37 °C. Subsequently, 1 µL was transferred into an activity test in 100 mM Hepes/NaOH, 5 mM CaCl2, 0.5% Triton X-100, pH 7.0 at 37 °C using 0.2 mM of the substrates Z-Gly-Gly-Leu-AMC (subtilisin), H-Gly-Arg-AMC (trypsin), and pGlu-Arg-Thr-Lys-Arg-AMC (furin). The increase in fluorescence was analyzed by linear regression (Tecan Infinity M1000, excitation 380 nm, emission 460 nm).
The resulting activity data are given in nanomoles of AMC per hour: for DFP: subtilisin: 4.64 ± 0.08 vs. <0.04, trypsin: 23.82 ± 0.71 vs. 0.35 ± 0.02, furin: 7.33 ± 0.47 vs. 7.67 ± 0.32; for AEBSF: subtilisin: 5.37 ± 0.30 vs. <0.04, trypsin 23.40 ± 0.71 vs. 0.29 ± 0.02, furin: 7.52 ± 0.63 vs. 6.65 ± 0.33; for PMSF: subtilisin: 4.85 ± 0.21 vs. <0.04, trypsin: 23.83 ± 0.85 vs. 0.48 ± 0.02, furin: 7.19 ± 0.60 vs. 7.62 ± 0.18.
Crystallization, Crystal Soaking, and Structure Determination.
Crystals of human furin were grown in sitting drops mixing equal volumes of the protein solution (∼9 mg/mL) and crystallization solution [100 mM Mes, 200 mM K/NaH2PO4, pH 5.5–6.0, 3–4 M NaCl and 3% (vol/vol) DMSO]. The reservoir contained 3–4 M NaCl. To study inhibitor binding in situ, the crystals were soaked overnight with 100 mM Mes, 200 mM K/NaH2PO4, pH 6.0, 3.0 M NaCl, 1 mM CaCl2, and 3% (vol/vol) DMSO supplemented with 0.2 mM inhibitor MI-52 (#26 in ref. 4). To study the structure of furin in the presence of EDTA, crystals were washed with 100 mM Mes, 200 mM K/NaH2PO4, pH 6.0, 3.0 M NaCl, and 3% (vol/vol) DMSO and soaked for 1 h with the same solution supplemented with 5 mM EDTA. To study inhibitor binding to furin in the presence of EDTA, crystals were washed with 100 mM Mes, 200 mM K/NaH2PO4, pH 5.0, 3.16 M NaCl, and 3% (vol/vol) DMSO, soaked for 1 h with the same solution supplemented with 5 mM EDTA, and finally overnight supplemented with 5 mM EDTA and 0.2 mM MI-52. To study calcium binding at defined concentrations in the absence of phosphate by anomalous diffraction, crystals were washed in 100 mM Mes, pH 6.0, 100 mM KCl, 3.16 mM NaCl, and 3% (vol/vol) DMSO and soaked for 1 h with the same solution supplemented with 2 mM EDTA. To complex the crystals with Ca2+, they were washed with 100 mM Mes, pH 6.0, 100 mM KCl, 3.16 mM NaCl, and 3% (vol/vol) DMSO supplemented with the respective concentration of CaCl2 and soaked in the same solution. Two Ca2+ concentrations were applied: (i) in the presence of 1 mM CaCl2 (high Ca2+) the crystals were washed 1× and soaked 2× for 0.5 h and 1× for 1 h; (ii) in the presence of normally 1 nM CaCl2 (low Ca2+) the crystals were washed 2× and soaked 4× for 0.5 h and 1× for 1 h. Because of the high concentrations, the inherent calcium content of the used chemicals contributes mainly to the overall calcium concentration of the soaking solution with very low Ca2+ supplementation. The maximum calcium contribution as a result of contamination from the used chemicals was calculated to be 142 µM, according to the specifications of the manufacturers. The drop size for washing and soaking of the crystals was always ∼1 µL. To measure diffraction data at 100 K, the crystals were cryo-protected by supplementation of the respective reservoir or soaking solutions with 13.1% (vol/vol) ethylene glycol and flash-cooled in liquid nitrogen. Diffraction data were collected at the BESSY-II beamline 14.1 of the Helmholtz-Zentrum Berlin (35). Data processing was performed with XDS (36) (v10/2014) via the XDSAPP interface (v1.0) and programs of the CCP4-suite (37) (CCP4 v6.3.0, CCP4 interface v2.2.0). The Phase Problem was solved by MR using the structure of human furin [PDB ID code 4RYD (6)] as a search model in PHASER (38). Model building was carried out in COOT (39) (v0.6.2). PHENIX (40) (v1.9-1692) was used for refinement. Refinement parameters of the MI-52 were generated with the PRODRG-server. Simulated annealing composite-omit electron-density maps were calculated in PHENIX, whereby the omitted region comprised 2.5% of the model. PYMOL (www.pymol.org) was used for structure alignments and molecular graphics.
Principal Component Analysis.
A PCA was carried out considering furin’s crystallographic structures reported in this publication: (i) furin unliganded state (PDB ID code 5JXG), (ii) furin unliganded state soaked with EDTA (PDB ID code 5JXI), (iii) furin with MI-52 bound (PDB ID code 5JXH), and (iv) furin soaked with EDTA and with MI-52 bound (PDB ID code 5JXJ); together with previously reported structures of the human enzyme [PDB ID codes 4RYD (6), 4OMC (17), and 4OMD (17)]. The analysis was performed considering a subset of 191 Cα atoms to allow a direct comparison with subtilisin Carlsberg, a homologous bacterial protease. Furin residues considered were: 121–124, 133–162, 193–206, 213–226, 235–242, 248–256, 286–297, 308–343, 350–361, 364–401, and 423–436; which correspond to the following subtilisin Carlsberg residues: 5–8, 12–41, 63–76, 82–95, 106–113, 120–128, 146–157, 168–203, 204–215, 217–254, and 262–275. The projections of the human furin structures (mentioned above), the murine furin structure [PDB ID code 1P8J (16)], and two subtilisin Carlsberg structures [PDB ID codes 1CSE (26) and 3UNX (25)] are shown in Fig. 3. The PCA analysis was conducted using the GROMACS (41) subroutines g_covar and g_anaeig.
MD Simulations.
The starting coordinates of the modeled systems were taken from the refined crystallographic structures (PDB IDs codes 5JXG and 5JXH), as well as from the comparable subtilisin structures [PDB ID codes 1CSE (26) and 3UNX (25)]. The furin unliganded EDTA-soaked (PDB ID code 5JXI) structure was used as a model to evaluate the influence of an empty Ca2+-(II) ion pocket. Four additional systems were established from the experimental data. Two of these systems were after removing the coordinates of the bound ligand from furin and substilisin-complexed structures (PDB ID codes 5JXG and 1CSE), a third one was to evaluate the influence of protonating Asp258 in a furin unliganded EDTA-soaked (PDB ID code 5JXI) structure, and the last one was to assess the influence of setting Thr367’s χ1 angle at 60° as the initial conformation in the furin unliganded structure (PDB ID code 5JXH). Crystallographic waters and ions located at the ion binding sites were included in the simulated models of furin and substilisin. All systems were simulated with GROMACS 5.0.3 (41) using the AMBER99SB-ILDN all-atom force field in an explicit water-solvent scheme in periodic boundary conditions. The topology and force-field parameter files for the ligand MI-52 were generated with ACPYPE (42). The protein protonation was performed with Molprobity (43). The water molecule model used was that based on the Transferable Intermolecular Potential with three sites, TIP3P. The solvent also included sodium chloride (NaCl) at 0.15 mM plus a small surplus of ions for electric charge neutralization. The long-range electrostatic interactions were computed using the fast particle-mesh Ewald setting, using a grid spacing of 1.2 Å. The van der Waals and short-range electrostatic interactions were computed using a cut-off of 1.1 Å for both parameters. The neighbor list was updated every 20 steps using the Verlet cut-off scheme. All bonds were constrained using LINCS, enabling a simulation time step of 2 fs.
The following start-up protocol was applied to all modeled systems: First, two consecutive minimization processes, with (i) 1,000 steps under the steepest descendant algorithm and (ii) 500 steps using conjugate gradient algorithm. Subsequently, the following two simulations were carried out to equilibrate the system at 300 K: (i) 500 ps in the canonical ensemble at 300 K using a V-rescale thermostat (τ = 0.1 ps) and (ii) 500 ps in the equilibrium isobaric (NTP) ensemble using Berendsen pressure coupling at 1 bar (τ = 2.0 ps). During these processes, a restraint constant of 1,000 kJ⋅mol−1⋅nm−2 was applied to all protein heavy atoms and to all main-chain heavy atoms for the equilibrium isochoric (NVT) and NPT equilibrations, respectively. An additional 500 ps were simulated within NTP conditions without restraints. Finally, 100 ns of MD simulations were performed in the NTP ensemble using the Parrinello–Rahman barostat with no restraints. The protocol was repeated five and four times, starting from the minimized structures with a new assignment of velocities for the furin and subtilisin systems, respectively. The rmsd analysis (Fig. S7) indicates equilibrated systems with stabilizations at around 1.2 Å and 0.8 Å for furin and substilisin simulations, respectively. To perform all of the reported analysis, the first 20 ns of each simulation were discarded.
Discussion
The herein described crystals of unliganded furin allowed us to investigate conformational rearrangements triggered by substrate and calcium binding to PCs. These studies revealed two activity-switches affecting the functionality of the active site dependent upon substrate and calcium, respectively. Comparing the structures of unliganded and inhibitor-bound furin, we observe a significant structural rearrangement of the substrate-binding cleft without alteration of the global fold of the protease, which provides a structural framework and should be highly conserved in orthologs of this enzyme as well (6, 16, 17). The active-site cleft shuttles between two structurally well-defined states of the enzyme, the inactive off and the active on states.
Furin and the bacterial subtilisin show highly conserved catalytic domains in their inhibitor-bound states, but differ significantly in their unliganded forms. In addition, inhibitor-free subtilisin displays a relatively high structural flexibility at the S1 and S4 pockets (27), indicating plasticity and hence facilitating the binding of different sequence motives. In furin, the substrate-binding area is characterized by low B-factors, indicating a rigid framework in both the unliganded and the ligand-bound forms. Ligand binding and rearrangements of the S1 and S4 sites and the main chain are cooperatively coupled with the active-site residues, in particular Ser368. The observed inactivity of furin with nonspecific covalent inhibitors that probe the nucleophilicity of the active site Ser368 but lack S1 to S4 substituents reflects this cooperativity.
We suppose that the switch between two rigid conformations requires precise bonding interactions and restricts substrate binding to a well-defined sequence motif. Exceptional substrate specificity is a hallmark of furin and other PCs, and hence a fundamental requirement of a functional secretory system and for cellular homeostasis (2).
The MD simulations demonstrate relaxation of the inhibitor-bound on state to the unliganded conformation following ligand dissociation (Fig. 3B). This finding shows that furin can switch between these two states and that the active on state of furin is less stable and requires the presence of a bound substrate or substrate-like inhibitor. This finding is in line with our extrapolation of the Tm to weak Ki-values that also suggests a lower structural stability of an inhibitor-free on state of furin. The MD simulations indicate that the on state is already visited in the ligand-free condition, thus suggesting that the substrate may bind directly to this state without energy expenditure (Fig. 3B). However, in such a scenario a better performance would be expected from the nonspecific covalent inhibitors of the serine protease family (22) (SI Materials and Methods). This evidence argues in favor of a mechanism, where the energy cost of the activation and stabilization of the protease in its on state is paid by ligand binding. Further support to this mechanism comes from the observed change in the coordination sphere of the sodium binding site in furin that is linked to inhibitor binding. The previously noted rather atypically bent or kinked conformation of the Arg-side chain at P4 (16, 20), which also corresponds to an energetically unfavorable yet well-defined conformation, may also consume some of the binding energy. In addition, a general stabilization of furin by inhibitor binding was demonstrated previously (6) and is recapitulated in the MD simulations, showing a substantially narrower distribution of conformers of the complex compared with the unliganded forms (Fig. 3B).
These considerations are relevant for the catalytic properties of furin. The requirement of energy to convert the protease from the off to the on state affects substrate specificity, because only ligands with sufficient binding energy may activate the enzyme. It also nicely explains the efficient cleavage of Arg-X-X-Arg↓ substrates that occupy the S1 and S4 sites. These are highly preferred to X-X-(Lys/Arg)-Arg↓ substrates. Monobasic peptides are even completely inert against proteolysis by furin (24, 28). A substrate-induced conformational shift might also influence the catalytic efficacy of the protease. Extensive protease–substrate interactions as described for furin usually result in large binding energies, and hence slow off-kinetics of the formed products and, in essence, in slow turnover of the substrate. A structural rearrangement could absorb some of the energy of substrate binding; it might reduce the net binding energy and thus allow for the observed fast kinetics of the PCs (28) that is comparable, for example, to the much less-specific subtilisin.
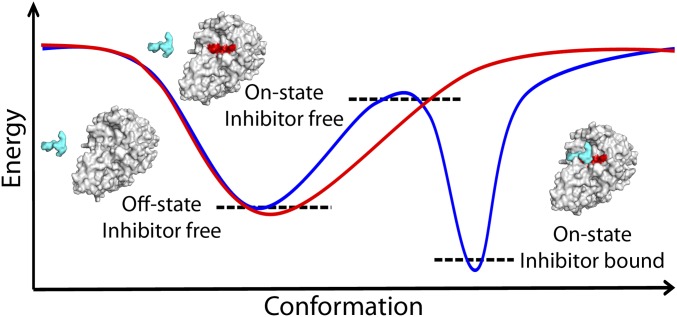
Two principal mechanisms may govern the transition from the off to the on state of furin: that is, induced fit triggered by ligand binding or conformational selection (Fig. 4). In the absence of experimental kinetic data, the MD simulations may provide a hint for the latter, as the ligand-removed form relaxes toward the unliganded structure within the first 20 ns and the unliganded form shows a relatively broad distribution of conformers (Fig. 3B). In addition, the χ1 angle histograms of the amino acid side chains with alternative torsions in unliganded and liganded forms show mostly bimodal distributions in the former, in accord with conformational equilibria (Fig. S4).
Fig. 4.
Schematic energy landscapes according to different scenarios. Furin, in an inhibitor-free condition (red solid line), resides most of the time at the bottom of an energy well adopting an off-state conformation, whereas its catalytically active on state is not energetically stable. In the presence of an inhibitor (blue solid line), the energy landscape is modified and the furin–ligand interaction stabilizes the on-state conformation. Furin and the ligand are represented as continuous gray/red and cyan surfaces, respectively. Black dashed lines represent the corresponding energy states.
Ligand-induced structural changes in proteases have been found and intensely studied by structural and MD studies in the proteasome, where only precisely defined enzyme main-chain peptidic ligand interactions induce opening and closing of the substrate-binding cleft, whereas other ligands do not have this effect. In addition, a variant of the proteasome, the immune proteasome, does not shuttle between different conformers and remains in the enzymatically competent conformation in the ligand-free form. This finding was interpreted as an important factor, contributing to the superior activity of the immune proteasome (29).
Another example of a protease shown to switch the conformation upon interaction with substrate (or inhibitor) is the αI-tryptase variant rHTαGI, which exhibits equilibria between the two states (30). Comparing furin to the 3C protease family, which embraces cysteine-type proteases with trypsin-like fold—including for example, tobacco etch virus protease or human rhinovirus 14 3C protease [HRV14, PreScission protease (31)]—shows that the conformation of the catalytic residues is here unaffected by substrate binding, pointing to a different mechanism to achieve the required high specificity. Additional reports on substrate-induced activation of proteases regard, for example, the caspase family (32, 33), where a more complex mechanism (also depending on dimerization, proteolytic cleavage, and conformational change) regulates their cleavage activity, and the oligopeptidase family (34), where [in essence similar to furin’s regulation by Ca2+-(II)] structural alterations in the vicinity of the oxyanion hole of the protease are described.
We also studied the three binding sites of calcium observed for human furin (17). Whereas Ca2+-(I) and Ca2+-(III) are conserved in bacterial subtilases, Ca2+-(II) is specific for furin and the other PCs (17, 23). Ca2+-(I) is tightly bound and fulfils a stabilizing, structural role in subtilisin and in furin. Ca2+-(II) and Ca2+-(III) show a less-tight interaction with binding strengths of the three sites in the order Ca2+-(I) > Ca2+-(II) > Ca2+-(III). The crystal structures without Ca2+-(II) do not indicate a significant effect on the geometry of inhibitor binding, except for an alternative rotamer of Asn295 and an inoperable oxyanion hole. Thus, Ca2+-(II) is mainly responsible for stabilizing the catalytically competent state of furin (22).
The dynamic data of the MD simulations suggest, however, that with an empty Ca2+-(II) pocket substrate binding has to pay an extra energetic penalty because of the destabilization of the Gly255 backbone and the side chain of Asp258 (Fig. S4 C–E). Interestingly, when Asp258 was protonated, together with an empty Ca2+-(II) pocket, its crystallographic pose was retained and the effect on Gly255 disappeared (Fig. S4 C–E). Here the MD simulations do not reflect the crystallographic data in detail, demonstrating only a partial reorientation of the Asn295 side chain in the Ca2+-(II)–free form (Figs. S3 and S4D). A closer inspection of furin and subtilisin offers a possible explanation for this apparent failure. In subtilisin Asn155’s orientation is reinforced by a hydrogen bond with Thr220Oγ, equivalent to Thr367 in furin. In the subtilisin simulations, the Asn155Nδ–Thr220Oγ hydrogen bond was indeed observed over 95% of the whole simulated time.
In accord with subtilisin and the murine furin structure [PDB ID code 1P8J (16)], crystallized in the presence of a covalent inhibitor, the Asn295 orientation is reinforced by establishing a hydrogen bond with Thr367. In both proteins, murine furin and subtilisin, the Asn295–Thr367 interaction is achieved by positioning the Thr367 side chain with a χ1 dihedral angle at ∼60°. Strikingly, this apparently crucial orientation is very well observed in the human furin simulations (Fig. S4B). The failure to observe the Asn295 conformation in our simulations may be because of the fact that Thr367 was not initially oriented to support this interaction. Remarkably, further stabilization of Asn295’s crystallographic pose was achieved in MD simulations of the furin unliganded structure, when the interaction between Thr367 and Asn295 was artificially set as a starting condition (Fig. S4D).
The activity of furin seems to be controlled by two intertwined mechanisms, including (i) a calcium-triggered switch mediated by the Ca2+-II site and (ii) a conformational on/off switch, both connected to substrate specificity and catalytic efficiency. This rather elaborate activation mode is probably essential for the unusually high substrate selectivity seen of furin and the other highly homologous PC-family members (20). The described twofold activation mechanism and the underlying structural basis offer opportunities for activity modulation of either individual PCs or multiple family members.
Materials and Methods
Expression, Crystallization, and Structure Determination.
Details regarding the plasmids, the expression system, purification procedure, and functional analysis used for human furin were described previously (17) and are also given in SI Materials and Methods. Crystals of human furin were grown in sitting drops mixing equal volumes of the protein solution (∼9 mg/mL) and crystallization solution [100 mM Mes, 200 mM K/NaH2PO4, pH 5.5–6.0, 3–4 M NaCl, and 3% (vol/vol) DMSO]. The reservoir contained 3–4 M NaCl. For inhibitor binding studies, the crystals were soaked with MI-52 (4) and for the calcium-studies the crystals were soaked with EDTA and Ca2+. Diffraction data were collected at beamline 14.1 of the Helmholtz-Zentrum Berlin (35), processed with XDS (36) and programs of the CCP4-suite (37). Phasing was performed by MR using the human furin–inhibitor complex structure [PDB ID code 4RYD (6)] and PHASER (38); the model was built in COOT (39) and refined in PHENIX (40). Structural alignments and graphics were prepared in PYMOL (www.pymol.org) (see SI Materials and Methods for further details).
Principal Component Analysis.
A PCA was carried out on Furin’s crystallographic structures (SI Materials and Methods).
MD Simulations.
The MD simulations were performed using the GROMACS (41) (v5.0.3) molecular simulation package (see SI Materials and Methods for further details).
Note Added
During preparation and revision of our report, PDB ID structure 4Z2A (10.2210/pdb4z2a/pdb; www.rcsb.org/pdb/explore.do?structureId=4Z2A) became available, which also shows a structure of human unliganded furin.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5JXG, 5JXH, 5JXI, and 5JXJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613630113/-/DCSupplemental.
References
- 1.Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artenstein AW, Opal SM. Proprotein convertases in health and disease. N Engl J Med. 2011;365(26):2507–2518. doi: 10.1056/NEJMra1106700. [DOI] [PubMed] [Google Scholar]
- 3.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11(5):367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 4.Becker GL, et al. Highly potent inhibitors of proprotein convertase furin as potential drugs for treatment of infectious diseases. J Biol Chem. 2012;287(26):21992–22003. doi: 10.1074/jbc.M111.332643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garten W, et al. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76(3-4):217–225. doi: 10.1016/0300-9084(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 6.Hardes K, et al. Novel furin inhibitors with potent anti-infectious activity. ChemMedChem. 2015;10(7):1218–1231. doi: 10.1002/cmdc.201500103. [DOI] [PubMed] [Google Scholar]
- 7.Kacprzak MM, et al. Inhibition of furin by polyarginine-containing peptides: Nanomolar inhibition by nona-D-arginine. J Biol Chem. 2004;279(35):36788–36794. doi: 10.1074/jbc.M400484200. [DOI] [PubMed] [Google Scholar]
- 8.Levesque C, et al. PACE4 inhibitors and their peptidomimetic analogs block prostate cancer tumor progression through quiescence induction, increased apoptosis and impaired neovascularisation. Oncotarget. 2015;6(6):3680–3693. doi: 10.18632/oncotarget.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, et al. Peptidomimetic furin inhibitor MI-701 in combination with oseltamivir and ribavirin efficiently blocks propagation of highly pathogenic avian influenza viruses and delays high level oseltamivir resistance in MDCK cells. Antiviral Res. 2015;120:89–100. doi: 10.1016/j.antiviral.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Jiao GS, et al. Synthetic small molecule furin inhibitors derived from 2,5-dideoxystreptamine. Proc Natl Acad Sci USA. 2006;103(52):19707–19712. doi: 10.1073/pnas.0606555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komiyama T, et al. Inhibition of furin/proprotein convertase-catalyzed surface and intracellular processing by small molecules. J Biol Chem. 2009;284(23):15729–15738. doi: 10.1074/jbc.M901540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jean F, et al. alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: Application as an antipathogenic agent. Proc Natl Acad Sci USA. 1998;95(13):7293–7298. doi: 10.1073/pnas.95.13.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komiyama T, Fuller RS. Engineered eglin c variants inhibit yeast and human proprotein processing proteases, Kex2 and furin. Biochemistry. 2000;39(49):15156–15165. doi: 10.1021/bi001907c. [DOI] [PubMed] [Google Scholar]
- 14.Scamuffa N, et al. Prodomain of the proprotein convertase subtilisin/kexin Furin (ppFurin) protects from tumor progression and metastasis. Carcinogenesis. 2014;35(3):528–536. doi: 10.1093/carcin/bgt345. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J, et al. Generation and characterization of non-competitive furin-inhibiting nanobodies. Biochem J. 2012;448(1):73–82. doi: 10.1042/BJ20120537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrich S, et al. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10(7):520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 17.Dahms SO, et al. X-ray structures of human furin in complex with competitive inhibitors. ACS Chem Biol. 2014;9(5):1113–1118. doi: 10.1021/cb500087x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holyoak T, Kettner CA, Petsko GA, Fuller RS, Ringe D. Structural basis for differences in substrate selectivity in Kex2 and furin protein convertases. Biochemistry. 2004;43(9):2412–2421. doi: 10.1021/bi035849h. [DOI] [PubMed] [Google Scholar]
- 19.Wheatley JL, Holyoak T. Differential P1 arginine and lysine recognition in the prototypical proprotein convertase Kex2. Proc Natl Acad Sci USA. 2007;104(16):6626–6631. doi: 10.1073/pnas.0701983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: Explanation of their specificity. J Mol Biol. 2005;345(2):211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 21.Rahfeld J, Schierhorn M, Hartrodt B, Neubert K, Heins J. Are diprotin A (Ile-Pro-Ile) and diprotin B (Val-Pro-Leu) inhibitors or substrates of dipeptidyl peptidase IV? Biochim Biophys Acta. 1991;1076(2):314–316. doi: 10.1016/0167-4838(91)90284-7. [DOI] [PubMed] [Google Scholar]
- 22.Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem. 1992;267(23):16396–16402. [PubMed] [Google Scholar]
- 23.Than ME, et al. The endoproteinase furin contains two essential Ca2+ ions stabilizing its N-terminus and the unique S1 specificity pocket. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 5):505–512. doi: 10.1107/S0907444905002556. [DOI] [PubMed] [Google Scholar]
- 24.Hatsuzawa K, et al. Purification and characterization of furin, a Kex2-like processing endoprotease, produced in Chinese hamster ovary cells. J Biol Chem. 1992;267(23):16094–16099. [PubMed] [Google Scholar]
- 25.Fisher SJ, Blakeley MP, Cianci M, McSweeney S, Helliwell JR. Protonation-state determination in proteins using high-resolution X-ray crystallography: Effects of resolution and completeness. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 7):800–809. doi: 10.1107/S0907444912012589. [DOI] [PubMed] [Google Scholar]
- 26.Bode W, Papamokos E, Musil D. The high-resolution X-ray crystal structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis. Structural analysis, subtilisin structure and interface geometry. Eur J Biochem. 1987;166(3):673–692. doi: 10.1111/j.1432-1033.1987.tb13566.x. [DOI] [PubMed] [Google Scholar]
- 27.Martin JR, et al. The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site. Structure. 1997;5(4):521–532. doi: 10.1016/s0969-2126(97)00208-6. [DOI] [PubMed] [Google Scholar]
- 28.Krysan DJ, Rockwell NC, Fuller RS. Quantitative characterization of furin specificity. Energetics of substrate discrimination using an internally consistent set of hexapeptidyl methylcoumarinamides. J Biol Chem. 1999;274(33):23229–23234. doi: 10.1074/jbc.274.33.23229. [DOI] [PubMed] [Google Scholar]
- 29.Arciniega M, Beck P, Lange OF, Groll M, Huber R. Differential global structural changes in the core particle of yeast and mouse proteasome induced by ligand binding. Proc Natl Acad Sci USA. 2014;111(26):9479–9484. doi: 10.1073/pnas.1408018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohr KB, et al. X-ray structures of free and leupeptin-complexed human alphaI-tryptase mutants: indication for an alpha→beta-tryptase transition. J Mol Biol. 2006;357(1):195–209. doi: 10.1016/j.jmb.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 31.Bjorndahl TC, Andrew LC, Semenchenko V, Wishart DS. NMR solution structures of the apo and peptide-inhibited human rhinovirus 3C protease (Serotype 14): Structural and dynamic comparison. Biochemistry. 2007;46(45):12945–12958. doi: 10.1021/bi7010866. [DOI] [PubMed] [Google Scholar]
- 32.Datta D, McClendon CL, Jacobson MP, Wells JA. Substrate and inhibitor-induced dimerization and cooperativity in caspase-1 but not caspase-3. J Biol Chem. 2013;288(14):9971–9981. doi: 10.1074/jbc.M112.426460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pop C, Salvesen GS. Human caspases: Activation, specificity, and regulation. J Biol Chem. 2009;284(33):21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szeltner Z, Renner V, Polgár L. Substrate- and pH-dependent contribution of oxyanion binding site to the catalysis of prolyl oligopeptidase, a paradigm of the serine oligopeptidase family. Protein Sci. 2000;9(2):353–360. doi: 10.1110/ps.9.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19(Pt 3):442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sousa da Silva AW, Vranken WF. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]