Significance
Zn2+ inhibition of voltage-gated proton (Hv1) channels has important physiological roles, such as quiescence of sperm in the male reproductive system. Here, we show that Zn2+ binds to different states of Hv1, and we propose a possible mechanism for Zn2+ inhibition of Hv1. Several residues are found to be involved in Zn2+ binding, and we provide detailed information about how these residues contribute to the functional effect of Zn2+ binding. This study provides valuable information for future drug development for Hv1 channels.
Keywords: Hv1, voltage-gated proton channel, Zn2+, inhibition, molecular model
Abstract
Voltage-gated proton (Hv1) channels are involved in many physiological processes, such as pH homeostasis and the innate immune response. Zn2+ is an important physiological inhibitor of Hv1. Sperm cells are quiescent in the male reproductive system due to Zn2+ inhibition of Hv1 channels, but become active once introduced into the low-Zn2+-concentration environment of the female reproductive tract. How Zn2+ inhibits Hv1 is not completely understood. In this study, we use the voltage clamp fluorometry technique to identify the molecular mechanism of Zn2+ inhibition of Hv1. We find that Zn2+ binds to both the activated closed and resting closed states of the Hv1 channel, thereby inhibiting both voltage sensor motion and gate opening. Mutations of some Hv1 residues affect only Zn2+ inhibition of the voltage sensor motion, whereas mutations of other residues also affect Zn2+ inhibition of gate opening. These effects are similar in monomeric and dimeric Hv1 channels, suggesting that the Zn2+-binding sites are localized within each subunit of the dimeric Hv1. We propose that Zn2+ binding has two major effects on Hv1: (i) at low concentrations, Zn2+ binds to one site and prevents the opening conformational change of the pore of Hv1, thereby inhibiting proton conduction; and (ii) at high concentrations, Zn2+, in addition, binds to a second site and inhibits the outward movement of the voltage sensor of Hv1. Elucidating the molecular mechanism of how Zn2+ inhibits Hv1 will further our understanding of Hv1 function and might provide valuable information for future drug development for Hv1 channels.
Voltage-gated proton (Hv1) channels are depolarization-activated channels that are highly selective for protons. Hv1 channel currents were first identified in snail neurons where they are thought to be important for acid extrusion to maintain physiological intracellular pH (1). In the immune system, proton currents through Hv1 channels compensate for the electrogenic currents and reduce the intracellular acidification caused by the activity of NADPH oxidase in human neutrophil (2–5) and lung airway epithelial cells (6). Hv1 channels in human microglia play an important role in NADPH oxidase-mediated brain damage in ischemic stroke (7). Hv1 channels are also found abundantly in human sperm cells, and the activation of Hv1 channels alkalizes the sperm cells during spermatozoa activation (8).
Hv1 channels belong to the superfamily of voltage-gated cation channels. However, in contrast to other voltage-gated cation channels, such as voltage-gated potassium (Kv) channels, which have six transmembrane (TM) segments per subunit and form tetramers, Hv1 has only four TM segments per subunit and forms dimers. The four TM segments in an Hv1 subunit are homologous to the first four TM segments of a Kv subunit that form the voltage-sensing domain in Kv channels. In contrast to Kv channels, which have a common pore formed by the assembly of the last two TM segments from all four subunits, each subunit of Hv1 contains a pore (9, 10).
Zn2+ is the most potent physiological inhibitor of Hv1 channels (11). Zn2+ is an essential mineral that is naturally present in our bodies. Zn2+ deficiency can cause severe mental retardation and immune dysfunction (12, 13), whereas excess Zn2+ accumulation leads to neurotoxicity and neurodegeneration (14). In addition to affecting a myriad of Zn2+-binding enzymes, Zn2+ inhibits Hv1, voltage-gated sodium (Nav), potassium (Kv), and calcium (Cav) channels (15–20). Sperm cells are quiescent in the male reproductive tract where high concentrations of Zn2+ inhibit Hv1. This helps the maturation of sperm cells in the male reproductive system. Sperm cells become active once introduced into the low-Zn2+ concentrations in the female reproductive tract by the removal of Zn2+ inhibition of Hv1 channels. Active Hv1 channels alkalize the cytosol, which activates CatSperm calcium channels and initiates sperm motility (8). Despite the importance of the inhibitory effect of Zn2+ on Hv1 channels, the molecular mechanism of how Zn2+ inhibits Hv1 is not completely understood.
The pH dependence of the Zn2+ inhibition of Hv1 suggests that Zn2+ is coordinated by several residues with a pKa similar to histidine residues (11). The Zn2+ sensitivity is profoundly attenuated when two extracellular histidines are mutated to alanines in human Hv1 channels (21). Musset et al. proposed that two Zn2+ ions are bound at the interface of two Hv1 subunits in the dimeric Hv1 channel and that each Zn2+ is coordinated by two histidines, one from each of the two subunits (22). Recently, an X-ray crystal structure of the voltage-gated proton channel was reported with a Zn2+ bound to a single subunit (23). Zn2+ is coordinated by several charged acidic residues of Hv1, in addition to the two previously proposed histidines. Therefore, the location of the Zn2+-binding site is still under debate. Here we compare the Zn2+ effect on both wild-type and mutant Hv1 channels in dimeric and monomeric forms to clarify the location of Zn2+ binding, the mechanism of Zn2+ inhibition, and the contribution of different amino acid residues to Zn2+ binding.
Our previous data showed that there are two different types of conformational changes involved in Hv1 channel opening (24). We proposed that, upon membrane depolarization, the first type of conformational change is the independent, outward movements of the two S4 segments and that the second conformational change is gate opening, which is most likely caused by a concerted conformational change in both subunits in the Hv1 dimer (24, 25). To determine the molecular mechanism of Zn2+ inhibition of Hv1, we characterize the effect of Zn2+ on the different conformational changes of the Hv1 channel using voltage clamp fluorometry (VCF). We find that Zn2+ inhibits Hv1 in two ways, most likely by binding to two different Zn2+-binding sites: at low concentration, Zn2+ decreases the proton current by preventing gate opening; and at high concentration, Zn2+ inhibits Hv1 further by inhibiting S4 movement. Mutating different residues in Hv1 channels affects the two inhibitory effects of Zn2+ differentially, suggesting that the two Zn2+-binding sites are made up of different residues. In addition, Zn2+ inhibits dimeric and monomeric Hv1 in a similar manner, suggesting that each Zn2+-binding site is made up of residues from within one Hv1 subunit.
Result
Zn2+ Shifts the Voltage Dependences of Channel Opening and Voltage Sensor Movement.
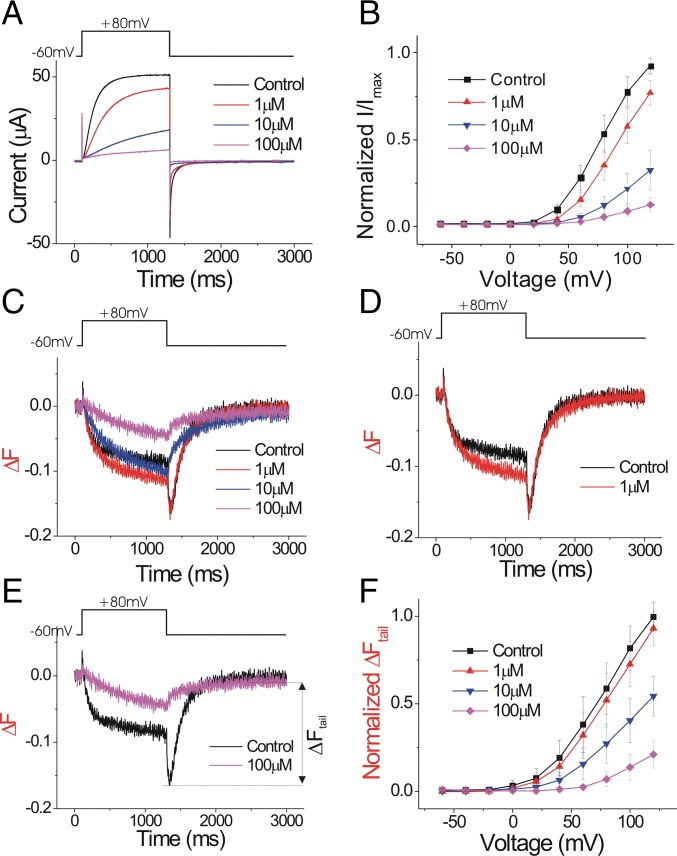
To measure the effect of Zn2+ on the conformational changes of the voltage sensor S4, we mutated S242 near the extracellular part of S4 in Ciona Hv1 (Ci-Hv1, also called Ci-VSOP) to cysteine and labeled it with the fluorophore Alexa488, as in our previous VCF experiments (24). We have previously shown that the voltage-dependent fluorescent change from Alexa488-labeled Ci-Hv1 S242C channels precedes channel opening, suggesting that the fluorescence reports on conformational changes of the voltage sensor S4 that triggers channel opening (24, 26). Zn2+ inhibits Hv1 current in a monotonic manner (Fig. 1 A and B). The introduced cysteine at S242C did not change the Zn2+ sensitivity of the Ci-Hv1 currents (Fig. 1 A and B and SI Appendix, Fig. S1A). Surprisingly, the Zn2+ inhibition of S4 movement is more complex. As we have shown in our previous study, the Hv1 channel activation involves two conformational changes in or near S4, which are reflected by a biphasic fluorescence change during channel opening and a fluorescence hook during channel closing (control trace in Fig. 1C and also see SI Appendix, Fig. S1C) (24). Upon membrane depolarization, the initial outward S4 movement generates a decrease in the fluorescence signal (SI Appendix, Fig. S1B). The subsequent gate-opening step increases the fluorescence signal to an intermediate level. During deactivation the process reverses: first there is a conformational change that closes the channel and decreases the fluorescence signal to a minimum, causing the formation of a hook in the tail fluorescence (SI Appendix, Fig. S1C). Then there is a second, charge-returning S4 movement that returns the fluorescence to baseline. Zn2+ has two effects on the fluorescence signal: at low Zn2+ concentration (≤1 μM), the amplitude of the fluorescence change increases during the depolarization, whereas the amplitude of the maximum fluorescence change remains the same during deactivation (Fig. 1D). This leads to an apparent decrease of the hook size. With higher Zn2+ concentration (≥10 μM), the amplitude of the whole fluorescence signal starts to decrease, and the hook completely disappears (Fig. 1E).
Fig. 1.
Zn2+ inhibits both current and fluorescence of Alexa488-maleimide–labeled Hv1 S242C. (A) Currents in response to a +80-mV voltage pulse in the presence of increasing extracellular Zn2+ concentrations on Alexa488-maleimide–labeled Hv1 S242C channels. (B) Normalized tail currents (Itail/Imax) versus voltage curves in the presence of increasing extracellular Zn2+ concentrations (n = 4). The curves are normalized to the maximum tail current (Imax) in ND96 control solution. (C) Fluorescence changes (ΔF) in response to a +80-mV voltage pulse in the presence of increasing extracellular Zn2+ concentrations. (D) One micromolar of Zn2+ increases the absolute change in the fluorescence signal during the +80-mV pulse. (E) One hundred micromolars of Zn2+ decreases the absolute change in the fluorescence signal. (F) Normalized fluorescence changes (ΔFtail/ΔFmax) versus voltage curves in the presence of increasing extracellular Zn2+ concentrations (n = 4). The ΔFtail is measured at the peak of the tail fluorescent signal, which previously has been shown to correlate with the outward movement of S4 (24). The curves are normalized to the maximum ΔFtail (ΔFmax) in ND96 control solution.
Zn2+ right-shifts the voltage dependence of both the G(V), i.e., channel opening (Fig. 1B), and the F(V), i.e., S4 movement, (Fig. 1F). Because S4 movement is thought to trigger the Hv1 channel opening, the right shift of the F(V) could by itself underlie the right shift of the G(V) and the decrease in the currents. However, the relative change in the amplitude of the fluorescence signal is slightly smaller than the decrease of the current, especially at low-Zn2+ concentrations (Fig. 1 B and F). In addition, the Zn2+ dose–responses of the fluorescence and currents clearly show that the current is affected at lower Zn2+ concentrations than the fluorescence (fivefold difference in affinities at +40 mV) (SI Appendix, Fig. S2). Therefore, an effect of Zn2+ on S4 movement cannot fully explain the current decrease, suggesting that, in addition to effecting S4 movement, Zn2+ has additional effects on Hv1 gate opening.
Zn2+ Binds Both the Activated Closed and the Resting Closed States of Hv1.
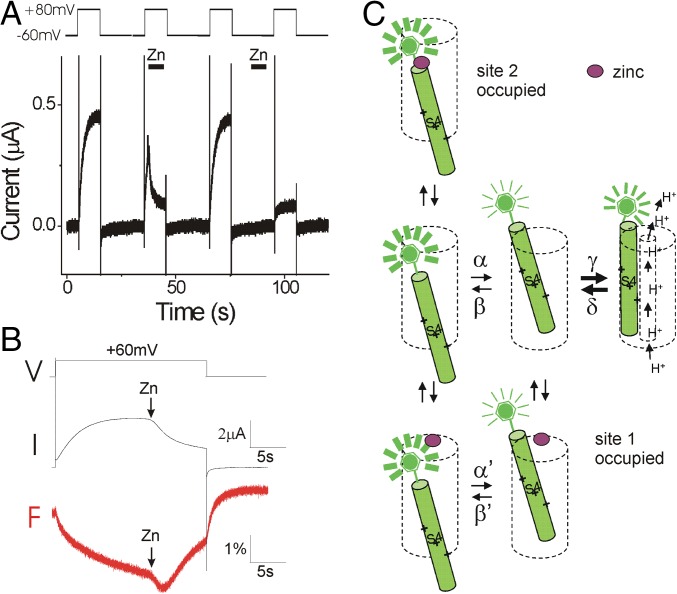
The previous experiments were all carried out under steady-state conditions, where Zn2+ was present throughout the whole recording. To identify which state Zn2+ binds to, we applied Zn2+ on open and closed channels separately (Fig. 2A). If we open the channel by stepping to +80 mV and then apply Zn2+ during the pulse, we observe a fast decrease of Hv1 currents (second voltage pulse in Fig. 2A), suggesting that Zn2+ binds to activated channels. The inhibitory effect is reversible and the currents recover after wash-out (third voltage pulse in Fig. 2A). We test Zn2+ inhibition of closed channel by perfusing Zn2+ at the holding potential between two depolarizations and washing Zn2+ away before the next depolarization, thereby exposing Zn2+ mainly to the resting closed state of Hv1. We wash the cell for several seconds to make sure Zn2+ is washed out of the recording chamber before the next depolarizing pulse. We find that the channel is prevented from opening by the previous exposure of Zn2+ at the holding potential (fourth voltage pulse in Fig. 2A), suggesting that Zn2+ also binds to the resting closed state of Hv1 channels.
Fig. 2.
Zn2+ stabilizes both the activated and the resting states of Hv1. (A) Currents in response to four +80-mV voltage pulses. The first voltage pulse is in control solutions. During the second voltage pulse, 100 μM Zn2+ is applied on open Hv1 channels. The third voltage pulse is again in control solutions. Before the fourth voltage pulse, 100 μM Zn2+ was applied on closed channels and then washed out. (B) Currents (I) and fluorescence (F) in response to a +60-mV voltage pulse during application of 100 μM Zn2+ on open Hv1 channels. (C) Proposed model for Zn2+ binding to the different states of the Hv1 channel (only one subunit is shown for simplicity). In response to a depolarizing voltage pulse in the absence of Zn2+ (Middle), Hv1 undergoes two different types of conformational changes (24): (i) first, a slow conformational change of S4 from the resting closed state to the activated closed state, which decreases the fluorescence; and (ii) a second conformational change opens the Hv1 channel and increases the fluorescence to an intermediate fluorescence. In the presence of low concentrations of Zn2+ (Bottom), Zn2+ binds to site 1, which prevents channel opening. At high concentrations of Zn2+ (Top), Zn2+ binds to site 2, which prevents outward S4 movement.
From the Zn2+-induced decrease in currents, we cannot differentiate between Zn2+ binding to the activated open state and physically blocking the H+ currents or Zn2+ binding to and stabilizing the resting closed state or the activated closed state, thereby preventing the opening conformational change of the channel. In all three cases, Zn2+ would inhibit the H+ currents. However, the fluorescent signals for these three states are different (SI Appendix, Fig. S1B): the activated closed state has a fluorescence that is lower than that of the open state, and the resting closed state has a fluorescence that is higher than that of the open state (24). Therefore, to find out to which state Zn2+ binds, we measured the effect of Zn2+ on the fluorescent signal during membrane depolarizations (Fig. 2B). If Zn2+ binds only to the resting closed state, we would expect to see an increase in the fluorescence signal upon Zn2+ binding. In contrast, if Zn2+ binds to the activated closed state, we would expect to see a decrease in the fluorescence signal upon Zn2+ binding. Finally, if Zn2+ binds only to the activated open state and blocks the proton currents, we would expect to see no change in the fluorescence signal upon Zn2+ binding. We open the channel by a voltage pulse to +60 mV and apply Zn2+ on the open channel during the +60 mV pulse (Fig. 2B). Upon Zn2+ application, we observed an initial decrease of the fluorescence signal followed by a subsequent increase of the fluorescence signal toward the baseline, as if Zn2+ initially stabilizes the channel in the activated closed state and then Zn2+ stabilizes the channel in the resting closed state (Fig. 2B, red line). This indicates that Zn2+ can bind to both the activated closed state and the resting closed state.
We therefore interpret the effect of different concentrations of Zn2+ on the fluorescence signal in Figs. 1 and 2 by the following mechanism (Fig. 2C). At low-Zn2+ concentrations, Zn2+ binds to a site (site 1) that does not interfere with the outward movement of S4 (the transition from resting closed state to activated closed state), whereas the second conformational change (the transition from the activated closed state to the open state) is inhibited. This will result in an increased amplitude of the fluorescence change during depolarizations, but decreased proton currents (Fig. 1D, red, and Fig. 2C, Bottom). At high-Zn2+ concentrations, Zn2+ also binds to a second site (site 2) and inhibits the outward S4 movement, which results in a smaller amplitude of the fluorescence change and a further decrease of the Hv1 current (Fig. 1E, purple, and Fig. 2C, Top).
We here assume that Zn2+ binding to Hv1 does not directly affect Alexa488 or its environment, but only indirectly affects the fluorescence by preventing Hv1 conformational changes. Tetramethylrhodamine-methanethiosulfonate (TMR-MTS) is a fluorophore with a shorter linker between the cysteine reactive group and the fluorescent group than Alexa488-maleimide, and therefore TMR is expected to sense a microenvironment different from Alexa488 (SI Appendix, Fig. S3A). Hv1 S242C channels labeled with TMR-MTS display similar effects of Zn2+ on the fluorescence as Alexa488-maleimide–labeled Hv1 S242C channels (SI Appendix, Fig. S3B), supporting our assumption that Zn2+ affects Hv1 conformational changes and not directly the fluorophore or its environment. In addition, 100 μM Zn2+ did not quench the fluorescence from Alexa488-succinimidyl ester attached to a polylysine-coated coverslip, suggesting that these concentrations of Zn2+ do not alter the fluorescence directly. To more directly test whether Zn2+ inhibits S4 movement, we measured intracellular accessibility of N264C, located in the C-terminal end of S4, in the presence of 100 μM extracellular Zn2+. We have previously shown that, in the absence of Zn2+, intracellular 2-(trimethylammonium)ethyl methanethiosulfonate (MTSET) modifies N264C >20-fold faster at −60 mV than at +80 mV, as if S4 moves outward in response to depolarizations and that this outward S4 movement protects N264C from intracellular MTSET modification (SI Appendix, Fig. S4D, Left) (24, 26). However, intracellular MTSET modifies N264C at +80 mV in the presence of 100 μM extracellular Zn2+ (k = 247 ± 91 M−1⋅s−1, n = 4) at a similar rate as in the resting state at −60 mV in the absence of Zn2+ (k = 290 ± 32 M−1⋅s−1; ref. 26), suggesting that Zn2+ inhibits outward S4 movement (SI Appendix, Fig. S4). The increased intracellular cysteine accessibility at +80 mV in the presence of Zn2+ supports our interpretation of the fluorescence that Zn2+ inhibits the outward S4 movement.
E167 Is Involved in Zn2+ Binding to the Resting Closed State.
E115 in mouse Hv1 (mHv1) coordinates a Zn2+ ion in the mHv1 crystal structure (23). E167 (the homologous residue in Ci-Hv1) is localized at the external end of the proton permeation pathway and has been suggested to play an important role in stabilizing the open state of Hv1 by binding to the positively charged S4 residues (27). Here we tested how E167 contributes to Zn2+ binding in Ci-Hv1 and what effect Zn2+ has on the channel when binding to this residue.
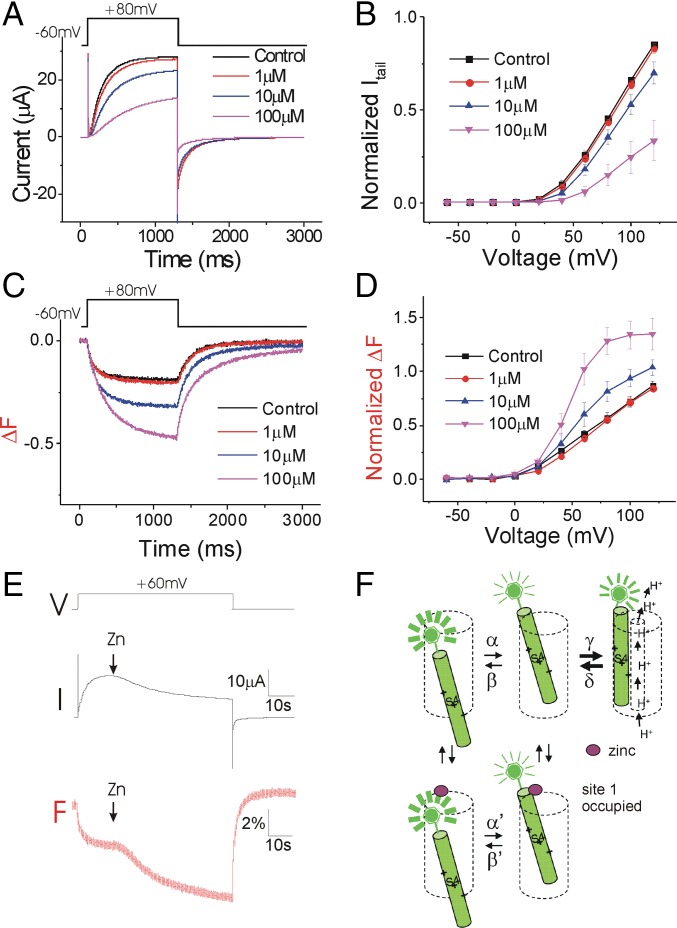
Neutralization of E167 decreases the apparent affinity of Zn2+ for inhibiting the currents in E167A/S242C (Fig. 3 A and B) and E167A (SI Appendix, Fig. S5) channels, suggesting that E167 is involved in Zn2+ binding. Surprisingly, Zn2+ increases the amplitude of the fluorescent change and shifts the voltage dependence of the total fluorescence signal to a more hyperpolarized voltage range in E167A/S242C (Fig. 3 C and D). The downward fluorescence change upon membrane depolarization is most likely due to the outward movement of S4, which is the transition from the resting closed state to the activated closed state (Fig. 2C and SI Appendix, S1 B and C) (24). The increased amplitude of the fluorescence and the left shift of the F(V) indicate that the activated S4 is stabilized by Zn2+. This suggests that in E167A/S242C, Zn2+ inhibits the currents without inhibiting the S4 movement. As we show above, Zn2+ binds to both the activated closed state and the resting closed state of WT Hv1. To test whether Zn2+ can bind to both of these states in the presence of the E167A mutation, we applied Zn2+ on E167A/S242C channels during depolarizations. In contrast to what occurs in WT Ci-Hv1 channels (Fig. 2B), Zn2+ decreased only the fluorescence (Fig. 3E), as if Zn2+ binds only to site 1 in E167A/S242C channels (Fig. 3F) and not to site 2.
Fig. 3.
E167A selectively removes the Zn2+ inhibition of the S4 movement. (A) Currents in response to a +80-mV voltage pulse in the presence of increasing extracellular Zn2+ concentrations on Alexa488-maleimide–labeled Hv1 E167A/S242C channels. (B) Normalized tail currents (Itail/Imax) versus voltage curves in the presence of increasing extracellular Zn2+ concentrations (n = 4). (C) Fluorescence changes (ΔF) in response to a +80-mV voltage pulse in the presence of increasing extracellular Zn2+ concentrations. (D) Normalized fluorescence changes (ΔFtail/ΔFmax) versus voltage curves in the presence of increasing extracellular Zn2+ concentrations (n = 4). (E) Currents and fluorescence in response to a +60-mV voltage pulse during application of 100 μM Zn2+ on open Hv1 channels. (F) Proposed model for Zn2+ binding to the E167A mutant channel. E167A decreases the affinity to site 2, but Zn2+ binding to site 1 is retained. Even in the presence of high concentrations of Zn2+, Zn2+ binds only to site 1.
We therefore hypothesize that E167 participates in Zn2+ binding to site 2 in the resting closed state and that Zn2+ binding to site 2 in the WT Hv1 inhibits S4 movement. However, at more depolarized voltages, the free energy of S4 movement is bigger than the free energy of Zn2+ binding. Therefore, S4 movement will still occur, but in a more depolarized voltage range. Zn2+ bound to site 2 will thus cause a right shift in the voltage dependence of S4 movement and channel opening (Fig. 1 B and F). If we mutate E167 (as in E167A), Zn2+ binds only to site 1. Zn2+ bound to site 1 prevents channel opening, but not outward S4 movement. This will result in an increased ΔF signal because channel opening normally reduces the ΔF. In addition, Zn2+ slows the return of the fluorescence signal. Therefore, Zn2+ binding to site 1 seems to stabilize the activated S4 while inhibiting channel opening (Fig. 3 A–D).
D233 Has a Similar Role as E167 in Zn2+ Inhibition.
D233 is a negatively charged residue at the extracellular end of S3. Similar to E167, this residue interacts with the positively charged S4 arginines in the open state and is important for the outward movement of S4 in Hv1 channels (27). The voltage dependence of S4 movement and channel opening is shifted to the right in D233N, and we observe currents only at very depolarized voltages. Zn2+ inhibits the currents of D233N channels, whereas the S4 movement is not affected (SI Appendix, Fig. S6 A and B). In D233N, the voltage dependence of channel opening is shifted so much that we cannot accurately determine the Zn2+-induced shifts of the G-V curve. However, Zn2+ does not shift the voltage dependence of S4 movement in D233N (SI Appendix, Fig. S6C). Zn2+ seems to decrease the current through D233N/S242C at +100 mV (SI Appendix, Fig. S6A). However, at this voltage (+100 mV), we cannot measure the proton current through Hv1 without activating some endogenous ion channels in Xenopus oocytes. To make sure that Zn2+ inhibits the proton current, we use 2′,7′-Bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein, acetoxymethyl ester (BCECF-AM) as an additional reporter for proton currents. BCECF is a pH-sensitive dye and therefore can detect pH changes caused by the Hv1 proton currents. Depolarizations induce BCECF fluorescence changes in oocytes expressing a WT Hv1 channel, but not in uninjected oocytes or oocytes injected with nonconducting Hv1 mutants (28). Zn2+ inhibits the BCECF signal from oocytes expressing D233N channels (SI Appendix, Fig. S6D), confirming that Zn2+ inhibits D233N currents.
Therefore, both D233 and E167 are necessary for the Zn2+ inhibition of S4 movement. We therefore propose that both D233 and E167 form part of site 2 that binds Zn2+ in the resting closed state in WT channels and prevents the outward S4 movement (Fig. 2C).
H188 Is Important for Zn2+ Binding in Both Activated and Resting Closed States.
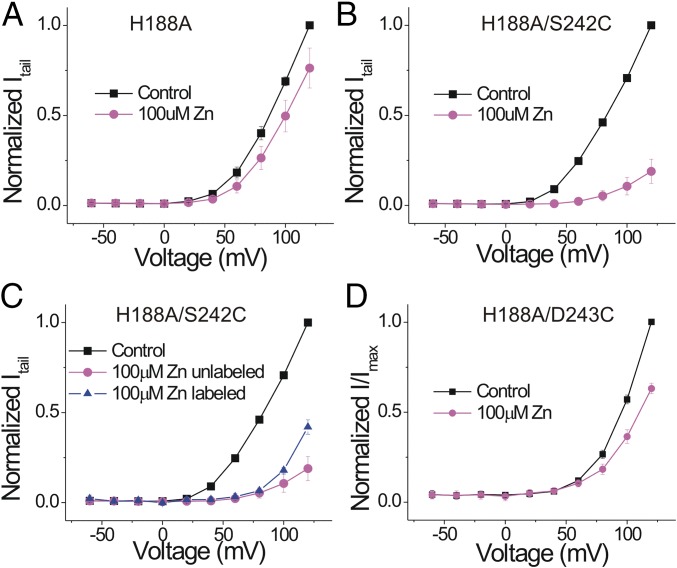
Two histidines—one at the extracellular end of S2 and the other one in the S3-S4 loop—in human Hv1 (hHv1) and mHv1 have been suggested to participate in Zn2+ coordination during channel inhibition (11, 21–23). Mutation of each individual histidine decreased the inhibitory effect of Zn2+ moderately, and mutation of both histidines abolished the Zn2+ inhibition (22). In contrast to hHv1, Ci-Hv1 lacks the histidine in the S3-S4 loop and has only one histidine: H188 at the beginning of S2. Mutating H188 to alanine almost completely abolished the Zn2+ inhibition of Ci-Hv1 (Fig. 4A), similar to the double-histidine mutation in hHv1. So, in contrast to E167A and D223N, H188A removes the inhibitory effect of Zn2+ on the proton currents. This suggests that H188 contributes to Zn2+ binding sites 1 and 2 because binding to either one of these sites should inhibit the currents.
Fig. 4.
H188A removes the Zn2+ inhibition of both the S4 movement and the currents. Normalized tail currents (Itail/Imax) versus voltage curves in the absence and presence of 100 μM Zn2+ (n = 4) for (A) H188A, (B) unlabeled H188A/S242C, (C) Alexa488-maleimide–labeled H188A/S242C, and (D) Alexa488-maleimide–labeled H188A/D243C Hv1 channels.
Zn2+ Effect on H188A Can Be Rescued by S242C.
To test whether Zn2+ has any effect on S4 movement in H188A, we introduced S242C in H188A to be able to label S4 with a fluorophore. Surprisingly, the H188A/S242C double mutant is sensitive to Zn2+ (Fig. 4B). S242 is very close to the homologous position of the second histidine that has been implicated in Zn2+ binding in hHv1 and mHv1. In addition, there are two negatively charged residues, E241 and E243, near S242. Introduction of a cysteine at S242 could do two things: S242C could participate in the Zn2+ binding directly or the S242C mutation could introduce a conformational change so that the nearby acidic residues can coordinate Zn2+. To test the direct involvement of S242C, we labeled S242C with the fluorophore Alexa488 and measured the effect of Zn2+ after labeling. The Zn2+ response after fluorophore labeling is decreased compared with unlabeled H188A/S242C channels (Fig. 4C), suggesting the possible direct involvement of S242C in Zn2+ binding. Because Zn2+ inhibition is not completely removed by fluorophore labeling, we cannot rule out a possible contribution of the nearby negative charges to Zn2+ binding. As a control, we introduced a cysteine at E243 in the H188A mutant. The H188A/E243C mutant has a similar decreased Zn2+ response as H188A (Fig. 4D), showing that E243C cannot rescue the Zn2+ effect in H188A. This suggests that the cysteine introduced at S242 is specific for Zn2+ binding in the H188A/S242C mutant and that S242C can compensate for the impaired Zn2+ binding in H188A. The effect of Zn2+ on the fluorescence from Alexa488-labeled H188A/S242C channels is similar to the effect of Zn2+ on the fluorescence from Alexa488-labeled S242C channels (SI Appendix, Fig. S7; cf. Fig. 1): low concentrations of Zn2+ alter the fluorescence during the voltage pulse but have no effect on the tail fluorescence, whereas higher concentrations of Zn2+ decrease the tail fluorescence. This suggests that the S242C mutation restores Zn2+ binding to both sites 1 and 2 in H188A channels, although with a lower affinity than in WT Hv1 channels.
D171 Is Not Part of the Zn2+-Binding Sites.
There are three conserved negatively charged residues in or near S1 (D160, E167, and D171). The X-ray crystal structure of mHv1 suggested that E115 and D119 (which are equivalent to E167 and D171 in Ci-Hv1) are part of the binding site for Zn2+. We therefore tested the role of D171 in Zn2+ binding by mutating it to alanine and measuring the Zn2+ response of D171A. We found that both the currents and the S4 movement in the D171A/S242C mutant are inhibited by Zn2+ in a similar way as S242C, suggesting that D171 does not contribute to Zn2+ binding in Ci-Hv1 (SI Appendix, Fig. S8 A and B).
D160 Mutation Changes Zn2+ Binding.
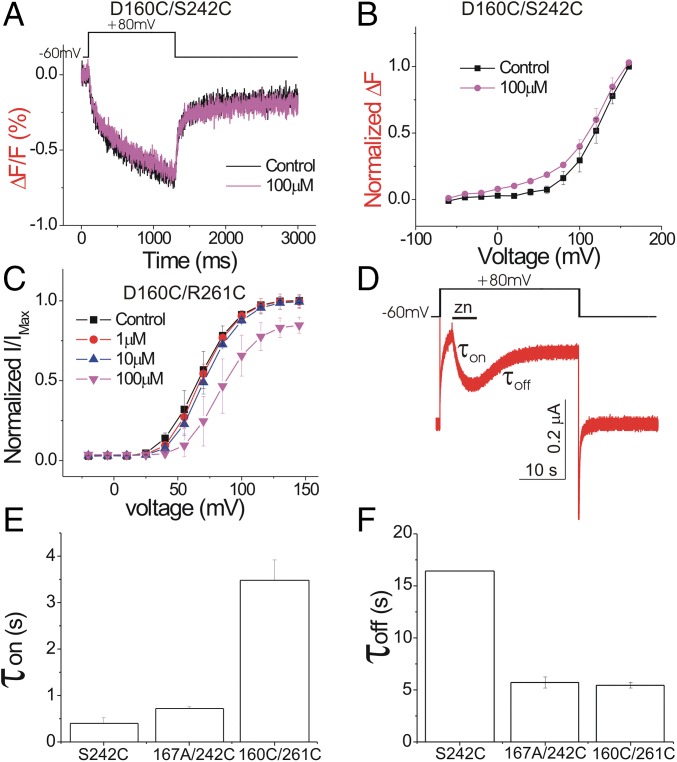
D112 in hHv1 (equivalent to D160 in Ci-Hv1) has been proposed to be the selectivity filter of Hv1 channels (29). Cysteine mutation of D112 in hHv1 changes the selectivity of the channel so that Cl− is also permeable (29). However, cysteine mutation at the homologous residue D160 in Ci-Hv1 results in a nonconducting channel (28). The absence of current in D160C is not due to an impaired plasma membrane expression or an absence of voltage-dependent S4 movement because we observed a fluorescence signal caused by the S4 movement in D160C/S242C (Fig. 5A). This suggests that D160C impairs proton conduction, but not S4 movement. Here we use the fluorescent signal as a reporter to measure the Zn2+ effect on D160C and test whether D160 is involved in Zn2+ binding.
Fig. 5.
D160C removes most of the Zn2+ effect. (A) Fluorescence changes (ΔF/F) from D160C/S242C in response to a +80-mV voltage pulse in the presence and absence of 100 μM Zn2+. (B) Normalized fluorescence changes (ΔFtail/ΔFmax) versus voltage curves from D160C/S242C in the absence and presence of 100 μM Zn2+ (n = 4). (C) Normalized tail currents (Itail/Imax) versus voltage curves for D160C/R261C in the presence of increasing extracellular Zn2+ concentrations (n = 4). (D) Currents in response to a +80-mV voltage pulse from S242C Hv1 channels. One hundred micromolars of Zn2+ is perfused briefly (black bar) during the 25-s-long depolarization to measure the time constants for Zn2+ inhibition (τon) and recovery from Zn2+ inhibition (τoff), using a fast perfusion system with an exchange time of <1 s (SI Appendix, Fig. S9). (E and F) Time constants for (E) Zn2+ inhibition and (F) recovery from Zn2+ inhibition measured as in D for S242C, D160C/R261C, and E167A/S242 channels.
The Zn2+ inhibition of the S4 movement is impaired in D160C/S242C because 100 μM Zn2+ has no effect on the fluorescent signal (Fig. 5 A and B). To test whether this impaired inhibition of S4 movement is correlated with impaired inhibition of channel opening, we test the Zn2+ effect on the D160C/R261C double mutant. Combining R261C with D160C rescues channel conduction, but proton selectivity is disrupted (28). The effect of Zn2+ on the current in the D160C/R261C double mutant is largely decreased compared with WT Hv1 (Fig. 5C). A total of 10 μM Zn2+ has no effect on D160C/R261C (compared with an almost 50% decrease of current in WT Hv1), whereas 100 μM Zn2+ decreases the current only moderately. Therefore, it seems that the mutation D160C impairs the Zn2+ inhibition of both the currents and the S4 movement.
To further test the role of D160 and E167 in Zn2+ binding, we measure the kinetics of Zn2+ binding and unbinding in E167A/S242C and D160C/R261C and compare them to the kinetics of Zn2+ binding/unbinding in the WT (i.e., S242C) (Fig. 5D). The Zn2+ on-rate is impaired in D160C/R261C (Fig. 5E), whereas the Zn2+ off-rate is much increased in the two mutants E160A/S242C and D160C/R261C compared with S242C (Fig. 5F; for perfusion exchange see SI Appendix, Fig. S9). The increased off-rate in the D160 and E167 mutants compared with S242C suggests a decreased strength of Zn2+ binding in these two mutations. The different on-rates of Zn2+ inhibition in the D160 and E167 mutants suggest that these two residues play different roles in Zn2+ binding.
Monomeric Hv1 Channel Has a Similar Response to Zn2+ Inhibition as Dimeric Hv1.
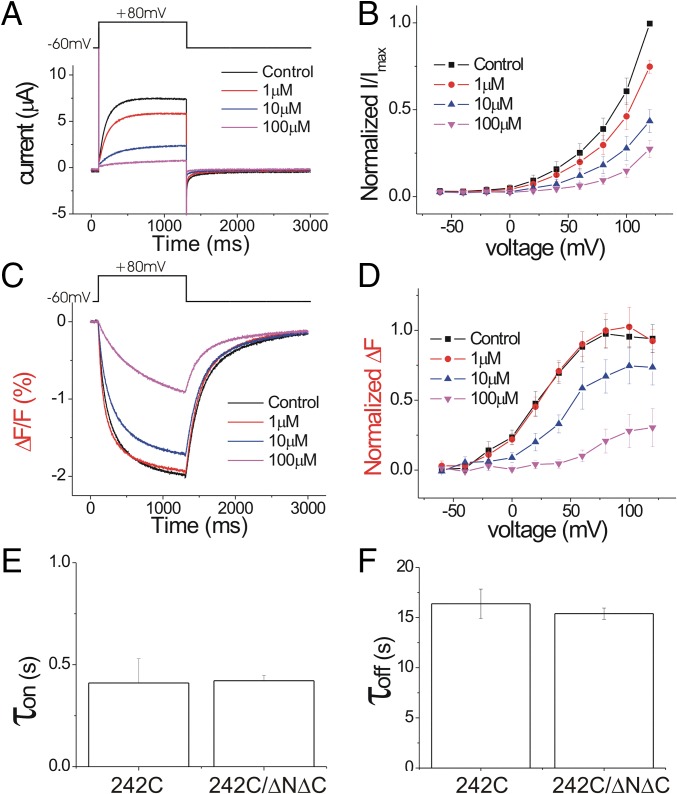
To test whether the Zn2+-binding sites are localized between the two subunits [as suggested earlier (22)] or within one subunit [as suggested by the mHv1 crystal structure (23)], we measured the Zn2+ inhibition of the C- and N-terminus–deleted mutant Ci-Hv1 ΔNΔC. The Ci-Hv1 ΔNΔC channel functions as a monomeric voltage-gated proton channel (9, 26). The apparent Zn2+ affinity is slightly decreased in the monomeric Hv1 compared with the dimeric Hv1, but the overall effect of Zn2+ on Hv1 ΔNΔC is very similar to its effect on WT Hv1 (cf. Figs. 6 and 1). Zn2+ shifts both the voltage dependence of channel opening, G(V) (Fig. 6 A and B), and the voltage sensor movement, F(V) (Fig. 6 C and D), in a dose-dependent manner similar to WT Hv1. We also measured the kinetics of Zn2+ binding and unbinding in the dimer and monomer Hv1 (Fig. 6 E and F). Both the Zn2+ on-rates (Fig. 6E) and off-rates (Fig. 6F) are similar for dimeric S242C and monomeric S242C ΔNΔC, suggesting that Zn2+ is coordinated mainly by residues within one subunit. These results are consistent with the findings of Takeshita et al. for their experiments on the monomeric/dimeric Zn2+ affinities in mammalian Hv1 channels (23).
Fig. 6.
Zn2+ inhibits similarly monomeric and dimeric Hv1 channels. (A) Currents in response to a +80-mV voltage pulse in the presence of increasing extracellular Zn2+ concentrations on Alexa488-maleimide–labeled Hv1 ΔNΔC channels. (B) Normalized tail currents (Itail/Imax) versus voltage curves in the presence of increasing extracellular Zn2+ concentrations (n = 4). (C) Fluorescence changes (ΔF) in response to a +80-mV voltage pulse in the presence of increasing extracellular Zn2+ concentrations. (D) Normalized fluorescence changes (ΔFtail/ΔFmax) versus voltage curves in the presence of increasing extracellular Zn2+ concentrations (n = 4). (E and F) Time constants for (E) Zn2+ inhibition and (F) recovery from Zn2+ inhibition for dimeric S242C and monomeric S242/ΔNΔC channels.
We also tested the Zn2+ effect in the monomeric version of the E167A mutant and H188A mutant. Similar to what we found in the E167A dimer (Fig. 3), Zn2+ facilitates the S4 movement by shifting the voltage dependence of the fluorescence signal in E167A/S242C ΔNΔC to the left (SI Appendix, Fig. S10A), whereas Zn2+ still inhibits the currents (SI Appendix, Fig. S10B). Similar to the H188A dimer, Zn2+ has very little effect in H188A ΔNΔC, whereas introducing S242C in this mutant rescues the Zn2+ inhibition (SI Appendix, Fig. S10 C and D). The overall effects of Zn2+ on dimeric Hv1 and monomeric Hv1 are very similar, suggesting that Zn2+ binding occurs inside each subunit.
Mapping Zn2+-Binding Sites in Ci-Hv1 and mHv1 Channel Models.
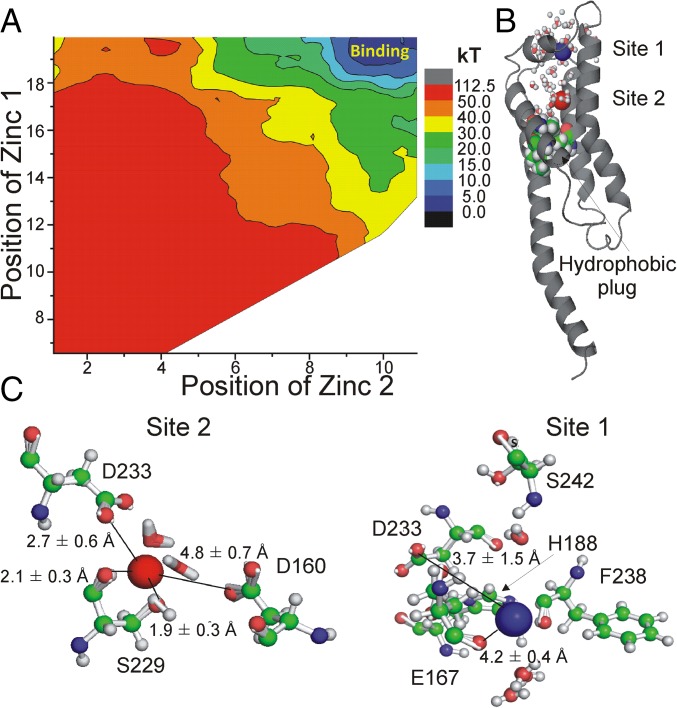
To further identify how and where Zn2+ interacts with the Ci-Hv1 channel, we investigate the binding of Zn2+ to Hv1 channels by molecular dynamics (MD) and free-energy simulations with additive and polarizable force fields applied to the available X-ray structure of mHv1 (23) and to a previously developed model of the Ci-Hv1 channel in the state that most closely corresponds to the X-ray structure (28). The molecular representation of the simulation box is shown in SI Appendix, Fig. S11. The root-mean-square difference for the heavy atoms forming the TM segments in the refined model of the Ci-Hv1 channel and X-ray coordinates is 2.5 Å for helices and 3.8 Å for flexible elements, such as loops (29). The X-ray structure of mHv1 and the Ci-Hv1 model are thought to be in the closed state with S4 in an intermediate position (28), which we here will interpret as the activated closed state (SI Appendix, Fig. S1B). SI Appendix, Fig. S11, shows the 1D potential of mean force (PMF) calculation for Zn2+ unbinding from the X-ray structure of mHv1 (purple line) and the model of Ci-Hv1 channel (green line). The reaction coordinate was defined as a normal to the bilayer, and the use of cylindrical boundaries ensuring sampling across the permeation pathway was identified in previous studies (27, 28). The crystallographic site for Zn2+ in mHv1 is formed by E115, D119, and H136 (23). Placing Zn2+ into the proposed binding site in a relaxed mHv1 channel based on the X-ray crystal structure leads to very rapid unbinding of Zn2+ because the site is disassembled and the residues are fully exposed to the bulk solution, allowing for rapid exchanges (SI Appendix, Fig. S11, Left, red residues). Despite the topological similarities between these two models of Hv1 channels (Ci-Hv1 and mHv1), there are notable differences in the location and organization of Zn2+-binding sites in these two proteins.
The 1D PMF shows a relatively large shallow binding region in the activated-closed state of the Ci-Hv1 model, corresponding closely to the binding site reported in the mHv1 crystal structure (25). One of the tentative binding sites identified by PMF evaluation (site 1) is formed by E115 and E192 in mHv1 and by E167, H188, and partially by S242 in the Ci-Hv1 channel. The bound Zn2+ at site 1 retains two to three water molecules at all times in both structures. The analysis of the Zn2+ coordination in the Ci-Hv1 channel suggests that the side-chain of S242 is involved in hydrogen bonding to one of the nitrogen atoms of the imidazole ring of H188 (20% of time) or direct water-mediated coordination of bound Zn2+ in site 1. These results are consistent with the experimental observations reported above for the role played by H188 and S242 in Zn2+ binding in the activated closed state of Ci-Hv1. Mutations of E167 also changed the affinity of the Zn2+ inhibition of the currents, suggesting that E167 also contributes to site 1 in the activated closed state of Ci-Hv1.
The second site (site 2) identified in PMF computations performed for mHv1 is formed by D108/D181 (D160/D233 in Ci-Hv1). The absence of this site in the Ci-Hv1 model can be rationalized by comparing differences in water accessibility between two channels. We used the Grand-Canonical Monte-Carlo (GCMC) protocol (Methods) to equilibrate the number of water molecules in the permeation pathway. The modeled Ci-Hv1 is considerably “drier” than the crystal structure or the equilibrated mHv1 model. We observed that hydrophobic interaction between Val112, Val144, and Leu185 was rapidly disrupted in all simulations of mHv1 with either an additive or a polarizable force field allowing for formation of water pockets connecting the channel interior to the extracellular solution. In contrast, Ci-Hv1 shows very little hydration in the vicinity of D160/D233 for the modeled activated closed state. The water access in this state is controlled by the secondary constriction region centered at V164, which represents a barrier to water permeation. Our prior studies of the tentative gating pathways suggest that the region around D160/D233 is likely to be more accessible to external solution (and Zn2+) in the resting closed states of the channel, in which S4 has moved further toward the intracellular solution. These results are consistent with the experimental observations reported above for the role played by D160 and D233 in Zn2+ binding in the resting closed state of Ci-Hv1.
The limiting factor in the 1D PMF analysis is in the use of only one, and apparently the intermediate, state of Hv1. To circumvent these limitations, we opted to perform 2D PMF computations to unravel a partial (with one Zn2+ ion already in the channel) energetic landscape of two-Zn2+ binding to the resting closed state of Ci-Hv1. The resulting 2D PMF is shown in Fig. 7A. Interestingly, the presence of the second Zn2+ ion bound to the channel appears to result in an energy minima relative to the situation where the second Zn2+ is still in the bulk solution. The result is consistent with our experimental data that suggest the existence of two Zn2+-binding sites and indirectly validates the Ci-Hv1 resting closed state. The resting closed state of the channel is fully hydrated in the region above the hydrophobic plug and thus allows effective screening of unfavorable electrostatic repulsion between the two bound divalent Zn2+ ions. Analysis of the 2D PMFs (Fig. 7 B and C) allows us to understand in-depth organization of these two binding pockets in the resting closed state of Ci-Hv1 labeled as “site 1” and “site 2.” It is important to note that in both cases the coordination environment is highly dynamic with two to three water molecules involved in direct coordination of each bound Zn2+ ion. Site 2 in the resting closed state of Ci-Hv1 mapped from 2D PMF computations is defined by D233 and S229 in the first coordination sphere of bound Zn2+, whereas D160 participates in water-mediated interactions with bound ion. This finding is consistent with our experimental data that D160 and D233 play direct roles in Zn2+ coordination for site 2 in the resting closed state of Hv1. Site 1 is open to the extracellular solution and is composed of the main chain of F238, the side-chain of H188, the highly mobile side-chain of D233, and the side-chain of E167 involved in water-mediated interactions. This finding is consistent with our experimental data that H188 and E167 play direct roles in Zn2+ coordination for site 1. Similarly to the site mapped from 1D PMF computations, S242 is pivotal for orientation of H188. The sites mapped from 2D PMF are in general agreement with a large shallow binding “pocket” formed by a constellation of negatively charged residues, particularly D160, D233, and E167. The mobility of D233 in the presence of two bound Zn2+ ions allows for direct ligation of one Zn2+ ion at a time, but it bifurcates between coordination site 2 (∼60% of all structures) and site 1. Interestingly, the composition of the binding site labeled site 1 in our 2D PMF computations is reminiscent of one of the most common coordination modes found in Protein Data Bank (PDB) for Zn2+ buried sites (30, 31). Dudev and Lim (30) noted that, in about 80% of all cases where Zn2+-bound His is present, the second coordination sphere contains an Asp or Glu carboxylate. Dudev and Lim (30) proposed that the presence of a carboxylate group in the second coordination sphere plays a substantial role in the protonation/deprotonation dynamics, the orientation of the imidazolium ring for optimal ion coordination, and the hydration pattern of the ion binding pocket. The use of a polarizable Drude force field allowed us to partially capture polarization effects (32), but quantum mechanics (QM) computations will be required to account for charge and proton transfer dynamics in future.
Fig. 7.
Potential of Mean Force Profiles (PMFs) for two-ion channel blockade. (A) Two-dimensional PMF plots for the single-file movement of two ions relative to the center of the bilayer. (B) Location of the binding sites (ZN1 and ZN2) in the lowest energy state for closed state of Ci-Hv1 with hydration sphere shown. (C) Average structures and coordinating residues (within 3.5 Å) from 2D PMF computations.
The simulation studies are in line with most of the experimental data, supporting the existence of two Zn2+-binding sites and the involvement of different amino acid residues in the two binding sites. However, the match between the experimental and simulation data is not 100% (e.g., E167 is not directly part of site 2 in the simulations), which we attribute to the flexible nature of the side-chains proposed to be part of the binding sites (e.g., E167 and D233 are very mobile in simulations) and the possible need for QM computations to capture proton dynamics and deprotonation events (of, e.g., H188 during Zn2+ binding). Clearly more simulations are needed to further test the proposed Zn2+-binding sites. Despite these limitations, the simulations still strengthen our conclusions that D233 and D160 participate in site 2 and that H188 (and S242C), D233, and E167 form part of site 1.
Kinetic Model of Hv1 with Two Zn2+-Binding Sites Reproduces Experimental Data.
We previously developed a kinetic model of Ci-Hv1 that can reproduce the fluorescence and currents in response to different voltage pulses (24). Here we incorporated two different Zn2+-binding sites, with different affinities, in this model (SI Appendix, Fig. S12). Zn2+ binding to the high-affinity site 1 prevents channel opening, but not outward S4 movement. Zn2+ binding to the low-affinity site 2, in addition, prevents outward S4 movement. This model can reproduce the inhibitory effects of Zn2+ on the currents and fluorescence of WT Ci-Hv1 channels (SI Appendix, Fig. S13 A and B). At low concentrations of Zn2+, Zn2+ binds to site 1, which reduces the currents by inhibiting the opening conformational change. Zn2+ inhibition of the opening conformational change also increases the amplitude of fluorescence change during the voltage pulse because normally the opening conformational change reduces the amplitude of the fluorescence change (SI Appendix, Fig. S1C). However, the amplitude of the fluorescence tail is not affected. At higher Zn2+ concentrations, Zn2+ binds to site 2, which further reduces the currents. Zn2+ binding to site 2 also reduces the fluorescence signal because Zn2+ binding to site 2 inhibits the outward S4 movement. The model can also reproduce the effects of Zn2+ on the current and fluorescence of Ci-Hv1 E167A and D233N channels (SI Appendix, Fig. S13 C–F), if we assume that the E167A and D233N mutations mainly prevent Zn2+ binding to site 2 (but also reduce the affinity to site 1) (SI Appendix, Table S1).
Discussion
By using VCF, we show that Zn2+ binding inhibits two different conformational changes of the Ci-Hv1 channel—outward S4 movement and channel opening—with different affinities. This suggests that there are two binding sites for Zn2+, site 1 and site 2. Zn2+ binding to site 1 inhibits Hv1 channel opening, but not outward S4 movement. Zn2+ binding to site 2 in the resting closed state of Hv1, in addition, inhibits the outward S4 movement. By mutating putative binding residues, we show that different Hv1 residues contribute to the two sites. For example, the D233N and E167A mutations remove only the Zn2+ inhibition of the outward S4 movement, but not the Zn2+ inhibition of channel opening. This suggests that D233 and E167 contribute to binding site 2 in the resting closed state of Hv1. However, Zn2+ inhibition of the Hv1 channel opening is preserved in E167A and D233N. Therefore, in these two mutations, Zn2+ binding to site 1 still occurs, which inhibits the transition from the activated closed state to the open state. Our results suggest that D160, E167, D233, and H188 contribute to site 2 and that E167, D233, and H188 contribute to site 1.
We have previously shown that E167 in S1 and D233 in S3 are important for the stabilization of the S4 segment in the open state of Hv1 by binding to the positively charged S4 residues in the open state (27). We propose that Zn2+ binding to E167 and D233 in WT Hv1 prevents the outward S4 movement by competing with the S4 charges in binding to these two residues, thereby shifting the voltage dependence of both S4 movement and channel opening.
In E167A, Zn2+ causes a left shift of the voltage dependence of S4 movement [F(V)] and an increase in amplitude of the fluorescence signal, in contrast to the right shift of the F(V) curve in WT Ci-HV1 channels. We propose that this left shift and increase in amplitude of the F(V) in the E167A mutant is because Zn2+ preferably binds to site 1. Zn2+ thereby stabilizes the activated closed state in the E167A mutant channels, which left-shifts the F(V) and, at the same time, inhibits the transition to the open state, which prevents the decrease in the fluorescence that normally accompanies the transition to the open state in WT Ci-Hv1.
Note that in E167A (and D233N) there is no fluorescence hook, even in the absence of Zn2+. In WT Hv1 channels, fast channel closing in combination with slow S4 deactivation gives rise to the fluorescence hook because of a transient accumulation of channels in the activated closed state during hyperpolarizations. However, in E167A, the closing is very slow, so there is no accumulation of activated closed-state channels and therefore no fluorescence hook in E167A. Our simulations of E167A with a slower closing rate recapitulate this behavior (SI Appendix, Fig. S13C). From our data from E167A we interpret that the E167A channel undergoes the same conformational changes as the WT channel, but the difference in rates masks the fluorescence hook. Indeed, one can actually see the equivalent of the hook in E167A channels when we apply Zn2+. Zn2+ decreases the fluorescence during the voltage pulse while inhibiting the currents (Fig. 3E). We interpret this to mean that Zn2+ accumulates E167A channels in the activated closed state, thereby unmasking the low fluorescence of the activated closed state and the hook. In contrast, the absence of a fluorescence hook in D233N is most likely due to a different reason. Opening is shifted to very depolarized voltages in D233N, so that only a small percentage of the channels open even at +100 mV. Because not many D233N channels reach the open state, there will not be a large fluorescence hook. Our simulations of D233N with a shifted opening voltage dependence recapitulate this behavior (SI Appendix, Fig. S13 E).
These different effects of the mutations of E167 and D233 on Hv1 channel behavior also explain the differences in the F(V) response to Zn2+ for these two mutants. Because not many D233N channels reach the open state, the amplitude of the fluorescence F(V) in D233N is mainly a measure of the difference in fluorescence between the resting closed state and the activated closed state. If we now apply Zn2+ to prevent opening, not much is expected to change in the fluorescence because most channels never reach the open state even in the absence of Zn2+. This is in contrast to E167A, for which most channels reach the open state and the fluorescence F(V) amplitude is mainly a measure of the difference of fluorescence between the resting closed state and the open state. If we now apply Zn2+ to prevent opening in E167A, the fluorescence amplitude of the F(V) is expected to increase in this case because now most channels will transit only between the resting closed state and the activated closed state, and therefore the F(V) will measure the difference between the resting closed state and the activated closed state.
Ci-Hv1 has a lower affinity for Zn2+ than the mammalian Hv1 channels. The lower affinity might be due to the fact that Ci-Hv1 lacks the proposed Zn2+-binding histidine in the S3-S4 loop, corresponding to H193 in hHv1 and H189 in mHv1 (22, 23). However, introducing the E241H mutation in the corresponding position in the S3-S4 loop in Ci-Hv1 did not improve the affinity in Ci-Hv1 (SI Appendix, Fig. S14). However, the S3-S4 loops are slightly different in lengths between mammalian Hv1 and Ci-Hv1, which makes it hard to identify equivalent positions in the S3-S4 loop between Ci and mammalian Hv1 channels and could also induce changes in the local secondary structure of the S3-S4 loop. In contrast, the conserved histidine in S2 (H140 in hHv1, H136 in mHv1, and H188 in Ci-Hv1) is important for Zn2+ binding in all of the Hv1 channels. Removal of this histidine, H188A, has a more dramatic effect on Zn2+ binding in Ci-Hv1 than in human or mouse Hv1. The effect of Zn2+ on Ci-Hv1 H188A is comparable to the effect of Zn2+ on the double-histidine mutant in human Hv1. The facts that S242C can rescue Zn2+ binding in H188A and that single histidine mutations in mammalian Hv1 still bind Zn2+ suggest that the two histidine residues in human or mouse Hv1 might be redundant and functionally compensate for each other in the case of mutations, thereby safeguarding the Zn2+ sensitivity in mammalian Hv1. The H188A mutation removes almost all effects of Zn2+ on Ci-HV1, suggesting that H188 is part of, or essential for, both site 1 and site 2.
The overall similarity of the Zn2+ data in dimeric and monomeric Hv1 suggests that the Zn2+-binding site is mainly localized within each subunit. A previous study suggested that Zn2+ binds at the interface between two Hv1 subunits because mutations in only one of the two Hv1 subunits in a linked Hv1 dimer had specific effects on the Zn2+ inhibition of the Hv1 currents (22). However, the two subunits in the dimeric Hv1 activate cooperatively (10, 24, 26). Therefore, Zn2+ binding in one subunit of the Hv1 dimer could affect cooperative channel opening in both subunits. Conversely, if Zn2+ binding is impaired in one subunit, this might impact Zn2+ binding in the other subunit, as has been seen with other Hv1 blockers (33), or the functional effect of Zn2+ binding in the other subunit on the cooperative channel opening transition. More studies are clearly needed to understand Zn2+ inhibition and cooperative opening. The Zn2+ inhibitory effect in the monomer is slightly lower than in the dimer, especially for the inhibition of S4 movement (cf. Figs. 1 and 6). One possible explanation for this is that the dimerization alters slightly the structure of each subunit, so that there is a slightly higher affinity of Zn2+ in the dimeric Hv1 than in the monomeric Hv1.
In conclusion, our study showed that Zn2+ is coordinated mainly by residues within each subunit of Ci-Hv1. The negatively charged residues D160, E167, D233, and H188 in the middle and at the extracellular end of S1 and S3 are important for Zn2+ binding to site 2 in the resting closed state and prevent outward S4 movement upon membrane depolarization. H188, D233, and E167, in addition, seem to contribute to Zn2+ binding to site 1 that prevents channel opening. We have shown here that Zn2+ can inhibit proton currents by binding to either the activated closed or resting closed state of the Hv1 channel, providing several conformational states that can be used as platforms for designing inhibitors for Hv1.
Methods
Mutagenesis and Expression of Ci-Hv1 Channels.
We performed site-directed mutagenesis, in vitro transcription of cRNA, and injection of cRNA encoding the Ci-VSOP (here called Ci-Hv1) into Xenopus laevis oocytes as described previously (26). The ΔNΔC Ci-Hv1 was constructed with a stop codon at Val270 and initiator methionine replacing Glu129 (26).
Two-Electrode Voltage Clamp and VCF Recordings.
We performed VCF experiments as described previously (26). Briefly, we labeled oocytes for 30 min with 100 μM Alexa-488 maleimide (Molecular Probes) in Na+ Ringer’s solution. Fluorescence was monitored through a FITC filter cube: exciter, HQ480/40; dichroic, Q505LP; and emitter, HQ535/50. Fluorescence intensities were low-pass-filtered at 200–500 Hz and digitized at 1 kHz. We performed two-electrode voltage clamp (TEVC) recordings as described earlier. Solutions for TEVC contained 88 mM NaCl, 1 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 100 mM Hepes (pH = 7.4). We injected oocytes with 50 nL of 1 M Hepes (pH = 7.0) to minimize pH changes due to the proton currents. This results in ∼100 mM Hepes in the cytosol. We also added 100 mM Hepes (pH = 7.4) to the external solutions for these recordings. Currents were leak subtracted off-line, assuming ohmic leak and using currents from potentials between –80 and –40 mV. Solutions were applied with a gravity-driven perfusion system with a solution exchange time of <2 s.
BCECF Measurements.
We incubated the oocytes with 50 μM BCECF dye (Life Technologies) in ND96 solutions for 30 min and washed before recording. Fluorescence was monitored through a FITC filter cube: exciter, HQ480/40; dichroic, Q505LP; and emitter, HQ535/50. Fluorescence intensities were low-pass-filtered at 200–500 Hz and digitized at 1 kHz.
MD Simulations and Potential of Mean Force Computations.
The simulation system was constructed initially using CHARMM-GUI protocol and CHARMM27 (34, 35) for proteins with NBFIX (36) and CHARMM36 (37) for lipids. A crystal structure of hHv1 (PDB: 3WKV) was embedded into lipid bilayer 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) constructed with a protein-membrane builder from the CHARMM-GUI project (38). The homology model of Ci-Hv1 was developed previously (27, 28). To ensure correct hydration of the permeation pathway, we used the GCMC protocol for water insertion on two protein structures. The details of the protocol were similar to that of Deng and Roux (39, 40). The permeation pathway or water insertion-deletion region for the GCMC simulations was defined as a box with boundaries of 30 Å × 30 Å × 60 Å. The two constructed protein-lipid systems were equilibrated for 50 ns with additive force fields. The average representative structures were used to create fully polarizable systems with a Drude force field developed by the groups of MacKerell and Roux (41–43). The positions of auxiliary Drude particles attached only to heavy atoms were propagated via an extended Lagrangian formalism through the assignment of a small mass (0.4 amu) at low temperature (1 K) using a separate thermostat. The Velocity-Verlet (VV2) integrator and the Langevin thermostat were used for all simulations involving polarizable models (44). The 1D PMF computations with umbrella sampling were carried out by applying a total of 80 windows (Zn2+ was positioned between 0 Å and 20 Å from the center of the membrane, with a force constant of 25 kcal/mol/Å2) along a reaction coordinate normal to the membrane (z axis). We used Zn2+ parameters published by Riahi et al. (45) Each PMF window was pre-equilibrated for 250 ps before a production run of 1 ns. The data were analyzed using weighted histogram analysis method (WHAM) to yield a converged PMF (tolerance was set to 0.0001 kcal/mol). 2D PMF computations were performed for the resting closed-state model of Ci-Hv1 developed previously (27, 28). To unravel energetics of two Zn2+-binding/permeation, we used multidimensional umbrella sampling methods, a powerful computational technique used with a considerable success in the past for various ion channels (46). Umbrella sampling simulations were performed with harmonic biasing potentials with a force constant of 20 kcal/(mol·Å2) along the z axis. The sampling windows for 2D PMF computations were spaced every 0.5 Å from 0 to 10 Å for the first Zn2+ ion and from 0 to 19 Å from the center of the lipid bilayer for the second Zn2+ ion. During the simulations used to calculate the single-ion and two-ion PMFs, other ions were excluded from the sphere of radius of 20 Å defining the tentative blockade pathway, using a repulsive flat-bottom spherical harmonic restraint with force constant 5 kcal/mol. To limit the lateral displacement of the Zn2+ ion, and thus ensure good sampling in a well-defined region of configurational space, we used a flat-bottom cylindrical restraint with radius 12 Å (relative to the center of mass of the monomer) and force constant 10 kcal/mol/Å (47). The 2D PMF setup resulted in 760 windows total. The simulation time per window was set to 1 ns with total simulation time for the 2D PMF map of ∼0.76 μs. The energy surfaces were rebuilt with WHAM (48, 49). The tolerance for WHAM was set to 0.001.
Supplementary Material
Acknowledgments
We thank Drs. Van Ngo and Yibo Wang for useful discussion regarding free-energy simulations. This work was supported by Grants National Heart, Lung, and Blood Institute (NHLBI) R01-HL095920 (to H.P.L.) and Fondecyt 1160261 (to C.G.). Work in the S.Y.N. laboratory was supported by grants from the Natural Sciences and Engineering Research Council (Canada) to S.Y.N. (RGPIN-315019) and the Alberta Innovates Technical Futures Strategic Chair in (Bio)Molecular Simulations. All of the computations were performed at the West-Grid/Compute Canada facilities, and the University of Calgary TNK cluster was acquired with direct support from the Canada Foundation for Innovation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604082113/-/DCSupplemental.
References
- 1.Thomas RC, Meech RW. Hydrogen ion currents and intracellular pH in depolarized voltage-clamped snail neurones. Nature. 1982;299(5886):826–828. doi: 10.1038/299826a0. [DOI] [PubMed] [Google Scholar]
- 2.Henderson LM, Chappell JB, Jones OT. The superoxide-generating NADPH oxidase of human neutrophils is electrogenic and associated with an H+ channel. Biochem J. 1987;246(2):325–329. doi: 10.1042/bj2460325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson LM, Chappell JB, Jones OT. Internal pH changes associated with the activity of NADPH oxidase of human neutrophils. Further evidence for the presence of an H+ conducting channel. Biochem J. 1988;251(2):563–567. doi: 10.1042/bj2510563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson LM, Chappell JB, Jones OT. Superoxide generation by the electrogenic NADPH oxidase of human neutrophils is limited by the movement of a compensating charge. Biochem J. 1988;255(1):285–290. [PMC free article] [PubMed] [Google Scholar]
- 5.DeCoursey TE, Morgan D, Cherny VV. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 2003;422(6931):531–534. doi: 10.1038/nature01523. [DOI] [PubMed] [Google Scholar]
- 6.DeCoursey TE. Hydrogen ion currents in rat alveolar epithelial cells. Biophys J. 1991;60(5):1243–1253. doi: 10.1016/S0006-3495(91)82158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu LJ, et al. The voltage-gated proton channel Hv1 enhances brain damage from ischemic stroke. Nat Neurosci. 2012;15(4):565–573. doi: 10.1038/nn.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lishko PV, Botchkina IL, Fedorenko A, Kirichok Y. Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell. 2010;140(3):327–337. doi: 10.1016/j.cell.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 9.Koch HP, et al. Multimeric nature of voltage-gated proton channels. Proc Natl Acad Sci USA. 2008;105(26):9111–9116. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tombola F, Ulbrich MH, Isacoff EY. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58(4):546–556. doi: 10.1016/j.neuron.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherny VV, DeCoursey TE. pH-dependent inhibition of voltage-gated H(+) currents in rat alveolar epithelial cells by Zn(2+) and other divalent cations. J Gen Physiol. 1999;114(6):819–838. doi: 10.1085/jgp.114.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad AS. Discovery of human zinc deficiency: Impact on human health. Nutrition. 2001;17(7-8):685–687. doi: 10.1016/s0899-9007(01)00598-6. [DOI] [PubMed] [Google Scholar]
- 13.Keen CL, Gershwin ME. Zinc deficiency and immune function. Annu Rev Nutr. 1990;10:415–431. doi: 10.1146/annurev.nu.10.070190.002215. [DOI] [PubMed] [Google Scholar]
- 14.Morris DR, Levenson CW. Ion channels and zinc: Mechanisms of neurotoxicity and neurodegeneration. J Toxicol. 2012;2012:785647. doi: 10.1155/2012/785647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilly WF, Armstrong CM. Divalent cations and the activation kinetics of potassium channels in squid giant axons. J Gen Physiol. 1982;79(6):965–996. doi: 10.1085/jgp.79.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison NL, Gibbons SJ. Zn2+: An endogenous modulator of ligand- and voltage-gated ion channels. Neuropharmacology. 1994;33(8):935–952. doi: 10.1016/0028-3908(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 17.Horning MS, Trombley PQ. Zinc and copper influence excitability of rat olfactory bulb neurons by multiple mechanisms. J Neurophysiol. 2001;86(4):1652–1660. doi: 10.1152/jn.2001.86.4.1652. [DOI] [PubMed] [Google Scholar]
- 18.Magistretti J, Castelli L, Taglietti V, Tanzi F. Dual effect of Zn2+ on multiple types of voltage-dependent Ca2+ currents in rat palaeocortical neurons. Neuroscience. 2003;117(2):249–264. doi: 10.1016/s0306-4522(02)00865-5. [DOI] [PubMed] [Google Scholar]
- 19.Gruss M, Mathie A, Lieb WR, Franks NP. The two-pore-domain K(+) channels TREK-1 and TASK-3 are differentially modulated by copper and zinc. Mol Pharmacol. 2004;66(3):530–537. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 20.Teisseyre A, Mozrzymas JW. Inhibition of the activity of T lymphocyte Kv1.3 channels by extracellular zinc. Biochem Pharmacol. 2002;64(4):595–607. doi: 10.1016/s0006-2952(02)01227-3. [DOI] [PubMed] [Google Scholar]
- 21.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440(7088):1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musset B, et al. Oligomerization of the voltage-gated proton channel. Channels (Austin) 2010;4(4):260–265. doi: 10.4161/chan.4.4.12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeshita K, et al. X-ray crystal structure of voltage-gated proton channel. Nat Struct Mol Biol. 2014;21(4):352–357. doi: 10.1038/nsmb.2783. [DOI] [PubMed] [Google Scholar]
- 24.Qiu F, Rebolledo S, Gonzalez C, Larsson HP. Subunit interactions during cooperative opening of voltage-gated proton channels. Neuron. 2013;77(2):288–298. doi: 10.1016/j.neuron.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mony L, Berger TK, Isacoff EY. A specialized molecular motion opens the Hv1 voltage-gated proton channel. Nat Struct Mol Biol. 2015;22(4):283–290. doi: 10.1038/nsmb.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez C, Koch HP, Drum BM, Larsson HP. Strong cooperativity between subunits in voltage-gated proton channels. Nat Struct Mol Biol. 2010;17(1):51–56. doi: 10.1038/nsmb.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chamberlin A, et al. Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc Natl Acad Sci USA. 2014;111(2):E273–E282. doi: 10.1073/pnas.1318018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chamberlin A, Qiu F, Wang Y, Noskov SY, Larsson HP. Mapping the gating and permeation pathways in the voltage-gated proton channel Hv1. J Mol Biol. 2015;427(1):131–145. doi: 10.1016/j.jmb.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musset B, et al. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 2011;480(7376):273–277. doi: 10.1038/nature10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev. 2003;103(3):773–788. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 31.Dudev T, Lin YL, Dudev M, Lim C. First-second shell interactions in metal binding sites in proteins: A PDB survey and DFT/CDM calculations. J Am Chem Soc. 2003;125(10):3168–3180. doi: 10.1021/ja0209722. [DOI] [PubMed] [Google Scholar]
- 32.Ngo V, et al. Quantum effects in cation interactions with first and second coordination shell ligands in metalloproteins. J Chem Theory Comput. 2015;11(10):4992–5001. doi: 10.1021/acs.jctc.5b00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong L, Singh V, Wulff H, Tombola F. Interrogation of the intersubunit interface of the open Hv1 proton channel with a probe of allosteric coupling. Sci Rep. 2015;5:14077. doi: 10.1038/srep14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 35.Mackerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 36.Noskov SY, Bernèche S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431(7010):830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 37.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu EL, et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem. 2014;35(27):1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Y, Roux B. Computation of binding free energy with molecular dynamics and grand canonical Monte Carlo simulations. J Chem Phys. 2008;128(11):115103. doi: 10.1063/1.2842080. [DOI] [PubMed] [Google Scholar]
- 40.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J Phys Chem B. 2009;113(8):2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Lopes PEM, Roux B, MacKerell AD., Jr Recent advances in polarizable force fields for macromolecules: Microsecond simulations of proteins using the classical Drude oscillator model. J Phys Chem Lett. 2014;5(18):3144–3150. doi: 10.1021/jz501315h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamoureux G, MacKerell AD, Roux B. A simple polarizable model of water based on classical Drude oscillators. J Chem Phys. 2003;119(10):5185–5197. [Google Scholar]
- 43.Li H, et al. Representation of ion-protein interactions using the Drude polarizable force-field. J Phys Chem B. 2015;119(29):9401–9416. doi: 10.1021/jp510560k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H, et al. Simulating monovalent and divalent ions in aqueous solution using a Drude polarizable force field. J Chem Theory Comput. 2010;6(3):774–786. doi: 10.1021/ct900576a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riahi S, Roux B, Rowley CN. QM/MM molecular dynamics simulations of the hydration of Mg(II) and Zn(II) ions. Can J Chem. 2013;91(7):552–558. [Google Scholar]
- 46.Finol-Urdaneta RK, et al. Sodium channel selectivity and conduction: Prokaryotes have devised their own molecular strategy. J Gen Physiol. 2014;143(2):157–171. doi: 10.1085/jgp.201311037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allen TW, Andersen OS, Roux B. Energetics of ion conduction through the gramicidin channel. Proc Natl Acad Sci USA. 2004;101(1):117–122. doi: 10.1073/pnas.2635314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J Comput Chem. 1992;13(8):1011–1021. [Google Scholar]
- 49.Grossfield A. 2012 WHAM: The Weighted Histogram Analysis Method. Version 2.0.6. http://membrane.urmc.rochester.edu/content/wham.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.