Significance
Cold stress is an influential environmental factor that affects plant distribution and can strongly limit crop productivity. Plants have evolved sophisticated signaling cascades that enable them to withstand chilling or even freezing temperatures. These cascades alter the biochemical composition of cells for protection from damage caused by low-temperature stress. In addition, cold stress has a profound impact on plant morphologies, causing growth repression and reduced yields. In this work we reveal that the brassinosteroids, a class of steroid hormones that is known for its role in growth control, also confers freezing tolerance in plants and describe regulatory circuits that contribute to this activity. Implications for the breeding of cold-resistant plants are discussed.
Keywords: brassinosteroid, CESTA, cold responses, CBFs, CBF1
Abstract
Brassinosteroids (BRs) are growth-promoting plant hormones that play a role in abiotic stress responses, but molecular modes that enable this activity remain largely unknown. Here we show that BRs participate in the regulation of freezing tolerance. BR signaling-defective mutants of Arabidopsis thaliana were hypersensitive to freezing before and after cold acclimation. The constitutive activation of BR signaling, in contrast, enhanced freezing resistance. Evidence is provided that the BR-controlled basic helix–loop–helix transcription factor CESTA (CES) can contribute to the constitutive expression of the C-REPEAT/DEHYDRATION-RESPONSIVE ELEMENT BINDING FACTOR (CBF) transcriptional regulators that control cold responsive (COR) gene expression. In addition, CBF-independent classes of BR-regulated COR genes are identified that are regulated in a BR- and CES-dependent manner during cold acclimation. A model is presented in which BRs govern different cold-responsive transcriptional cascades through the posttranslational modification of CES and redundantly acting factors. This contributes to the basal resistance against freezing stress, but also to the further improvement of this resistance through cold acclimation.
Seasonal and diurnal temperature changes are influential environmental factors that affect plant distribution and can strongly limit crop productivity. Whereas chilling-sensitive plants from tropical or subtropical regions suffer damage already even above freezing temperatures, plants from temperate geographical zones, such as Arabidopsis thaliana, commonly display a certain level of constitutive (intrinsic or basal) freezing tolerance. This basal tolerance can be further enhanced by exposure to low but nonfreezing temperatures in a process termed cold acclimation that leads to transcriptome reprogramming (1) and induces biochemical, physiological, and morphological changes, including growth repression, which allows plants to increase their freezing resistance (2).
A group of genes induced by cold and other types of abiotic stress such as drought or osmotic stress is the COLD-RESPONSIVE (COR) genes. The promoters of certain COR genes contain a cis-acting element responsible for drought and low-temperature responsiveness, the C-REPEAT/DEHYDRATION-RESPONSIVE ELEMENT (CRT/DRE) (3), which is bound by the CRT/DRE BINDING FACTOR (CBF/DREB) family of APETALA 2 (AP2) domain transcription factors (TFs) (4). CBF activity is controlled by upstream components of which the best characterized is INDUCER OF CBF EXPRESSION 1 (ICE1), a basic helix–loop–helix (bHLH) transcription factor (5), which, in response to cold, is modified by SUMOylation and promotes CBF3 expression (6).
Like most physiological traits in plants, chilling and freezing tolerance is controlled by phytohormones (7), and evidence suggests that the brassinosteroids (BRs) are involved. When externally applied, BRs increased chilling tolerance (8, 9). However, the molecular modes of this control remain unclear (10). BRs are steroid hormones that regulate vegetative and reproductive development by promoting cell division and cell elongation (11). A plasma membrane-localized receptor complex containing the receptor-like kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1) perceives the hormones and initiates signaling to control the activity of BR-regulated transcription factors (11). The best-studied members of BR-controlled TFs are BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE-RESISTANT 1 (BZR1) (12, 13). They are substrates of BR-repressed ARABIDOPSIS GSK3/SHAGGY-LIKE KINASES (ASKs) including BRASSINOSTEROID INSENSITIVE 2 (BIN2), which are negatively regulated by BRs and repress BES1/BZR1 activity through phosphorylation (11).
In addition, bHLH TFs such as CESTA/HALF FILLED (CES/HAF) are also targets of BIN2 (14). CES is the closest homolog of BRASSINOSTEROID ENHANCED EXPRESSION 1 (BEE1) and BEE3 (15) and is required for elongation growth in both vegetative (14) and reproductive tissues (16). BRs control CES protein activity, abundance, and subnuclear localization via BIN2-mediated phosphorylation and phosphorylation-repressed SUMOylation (17).
Here we show that BRs promote the freezing resistance of plants and elucidate molecular modes that contribute to this activity. BR-deficient mutants were hypersensitive to freezing stress, whereas an activation of BR signaling increased freezing tolerance both before and after cold acclimation. We provide evidence that CES can directly bind to CBF promoters and contribute to the regulation of constitutive CBF expression, in particular of CBF1 and CBF3. In addition, non-CBF regulon types of COR genes are identified that are regulated by the BR-CES/BEE pathway during cold acclimation. A model for the role of BRs in basal and acquired freezing tolerance is presented and discussed.
Results
BR Signaling Contributes to Basal Freezing Tolerance of Plants.
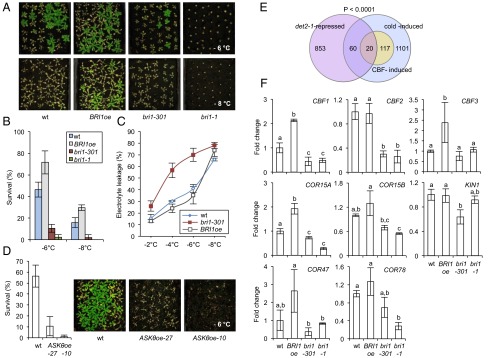
Previously, it was shown that BR treatment improves the chilling tolerance of Arabidopsis plants (8–10). In addition, we found that application of the BR epibrassinolide (epiBL) improved Arabidopsis survival following subzero temperature exposure (SI Appendix, Fig. S1A). To investigate if BRs are involved in basic freezing tolerance, BR-signaling mutant plants were treated at either −6 °C or −8 °C for 4 h, and survival was assessed as the ability to form new leaves after 2 wk of recovery at 21 °C. WT Columbia-0 (Col-0) showed survival rates of ∼45% following −6 °C exposure and of ∼15% following −8 °C exposure (Fig. 1 A and B). These rates were increased in BRI1oe (with ∼70% and 30%, respectively) and strongly reduced in bri1-1 and bri1-301, with <10% survival (Fig. 1 A and B). The phenotype was associated with a decrease in electrolyte leakage, an indicator of damage to cellular membranes, in BRI1oe and with an increase in bri1-301 (Fig. 1C).
Fig. 1.
BR signaling promotes basal freezing tolerance. (A and B) Freezing tolerance of nonacclimated BRI1oe, bri1-301, and bri1-1 plants compared with WT. Plants were grown in LD growth conditions at 21 °C. After 3 wk they were treated at −6 °C or −8 °C for 4 h. Survival was assessed after 2 wk of recovery at 21 °C. (A) Representative plants of each line and (B) the quantified results. Error bars show the SD of three biological replicates. (C) Electrolyte leakage in leaves of nonacclimated plants of the WT, BRI1oe, and bri1-301 plants grown under the same conditions as described in A and treated with the indicated temperatures. Error bars show SD of three biological replicates. (D) Freezing tolerance of nonacclimated ASKθoe-27 and ASKθoe-10 plants compared with WT. Plants were grown in the same conditions as in A and treated at −6 °C for 4 h. Quantified results (Left) and representative plants (Right) are shown. (E) Venn diagram showing the overlap of cold-induced genes [as defined by Park et al. (25); in purple] with genes repressed in det2-1 (determined from dataset TAIR ME00335; in lavender). The significance of representation of the CBF regulon of COR genes [as defined by Park et al. (25); in yellow] in the overlay was calculated with a χ2 test giving the P value shown. (F) Transcript levels of CBFs and COR genes were determined in 3-wk-old plants of WT, 35S:BRI1-GFP (BRI1oe), bri1-301, and bri1-1 by qPCRs. Error bars show SD of three biological replicates. The letters indicate significant differences between genotypes (P < 0.05; ANOVA).
To investigate at which step in the BR-signaling pathway the regulatory effects may occur, the survival rates of two independent lines overexpressing ASKθ, a BIN2 homolog (18), were assessed following −6 °C treatment. In ASKθoe plants, BR signaling is constitutively repressed, which results in BR-deficient phenotypes even severer than those found in bin2-1 (18). Importantly, both lines were hypersensitive to freezing (Fig. 1D), providing evidence that BR function in constitutive freezing tolerance is conferred downstream of ASKs.
In cold and freezing stress responses, COR gene regulation occurs (1). To investigate if BRs may participate, we used a bioinformatic analysis and determined on a genome-wide scale whether COR genes may be constitutively repressed in a BR-deficient situation. This analysis showed that ∼6% of all cold-induced genes were repressed in the BR-deficient mutant det2-1 (de-etiolated 2) (18). Among them CBF-induced genes were significantly enriched (Fig. 1E and SI Appendix, Table S1). To investigate whether BRs are required for the basal expression of CBFs and downstream COR genes, transcript levels were measured by quantitative real-time PCR (qPCR) in the BR-hypersignaling line 35S:BRI1-GFP (BRI1oe) (19), the two BR-signaling defective mutants bri1-1 (20) and bri1-301 (21), and the BR-biosynthetic mutant constitutive photomorphogenic dwarf (cpd) (22), which has severer phenotypes than det2-1 (11). The results showed that BR-signaling deficiency repressed CBF3 transcription in seedlings (SI Appendix, Fig. S1B), and CBF1 and CBF2 transcription in adult plants (Fig. 1F). Moreover, the downstream targets COR15A, COR15B, COR47, COR78, and KIN1 were all repressed in BR-signaling deficient mutant backgrounds. In particular COR15A and COR15B levels were reduced (Fig. 1F and SI Appendix, Fig. S1B). In BRI1oe plants, complementary changes occurred, but they were subtler (Fig. 1F and SI Appendix, Fig. S1B). Growth on media containing BR or the BR-biosynthesis inhibitor brassinazole (Brz) (23) produced corresponding changes in CBF expression (SI Appendix, Fig. S2 A and B), confirming previous studies, which had shown BR induction of CBF1 and COR gene expression following BL (brassinolide) treatment (8, 24). At early time points after BR treatment, CBFs were not significantly induced in the conditions we tested (SI Appendix, Fig. S2C).
BR Signaling Contributes to Cold Acclimation.
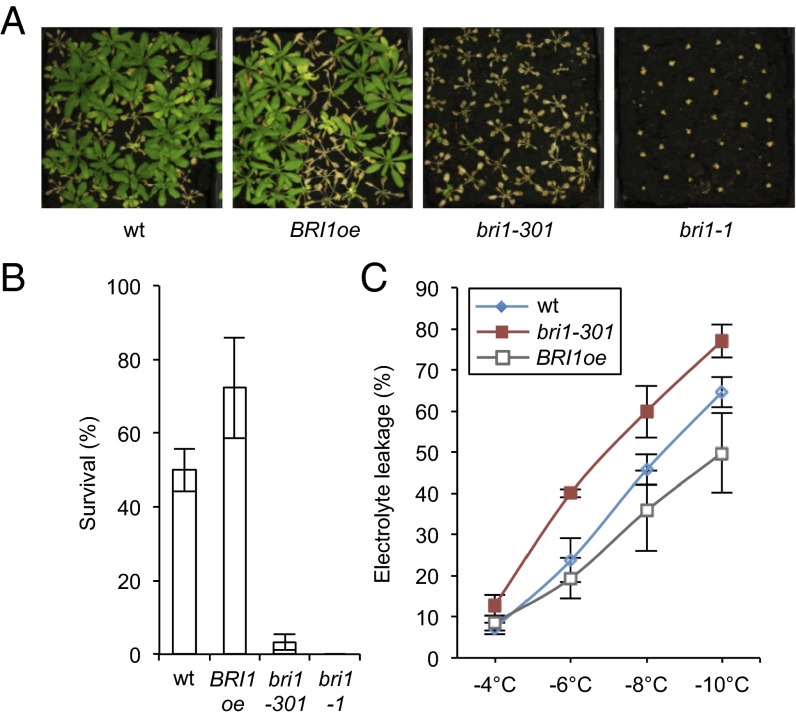
Given the evidence that BR signaling promotes freezing tolerance of plants under nonacclimated conditions, we investigated whether BRs also affect freezing tolerance following cold acclimation. Freezing tolerance assays of cold-acclimated BR-signaling deficient mutants showed that the survival rates of bri1-1 and bri1-301 were strongly reduced, whereas BRI1oe survival was increased in comparison with that of the WT (Fig. 2 A and B). In agreement, electrolyte leakage of acclimated plants was increased in bri1-301 and decreased in BRI1oe (Fig. 2C).
Fig. 2.
BR signaling contributes to cold acclimation. (A and B) Freezing tolerance of BRI1oe, bri1-301, and bri1-1 plants compared with WT after cold acclimation. Three-week-old plants grown in LDs at 21 °C were acclimated for 3 d at 4 °C and then treated at −10 °C for 6 h. Survival was scored after 2 wk of recovery at 21 °C. Pictures of representative plants (A) and the quantified results (B) are shown. Error bars show SD of three biological replicates. (C) Electrolyte leakage in acclimated plants of the WT, BRI1oe, and bri1-301. Plants were grown and acclimated as described in B, and ion leakage was measured in detached leaves following exposure to the indicated subzero temperatures. Error bars show SD of three biological replicates.
Because BRs can impact on basal CBF expression levels, it was of interest to analyze if BRs may also contribute to the induction of CBF transcription in response to cold stress. Therefore, bri1-1 and bri1-301 were treated at 4 °C, and mRNA levels of CBFs, COR15A and COR15B were quantified in a time-course manner by qPCR. The result showed that in response to cold stress, the transient induction of CBF1 expression was slightly reduced in BR-signaling defective mutants, whereas CBF2 and CBF3 induction were unaffected (SI Appendix, Fig. S3). Consistently, the levels of COR15A and COR15B were only slightly decreased, with the most prominent effects 24 h after treatment (SI Appendix, Fig. S3).
CES and Homologs Promote Basal Freezing Tolerance.
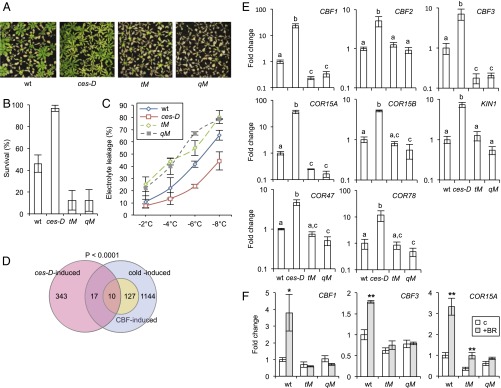
To identify BR-controlled TFs that promote freezing tolerance downstream of ASKs, a candidate gene approach was used. The dominant bes1-D (12), bzr1-1D (13), and ces-D (14) mutants were tested for their responses to freezing stress. Subzero temperature treatments of nonacclimated adult plants showed that, whereas the bzr1-1D mutation did not confer an effect, the bes1-D mutation slightly suppressed freezing tolerance in this experimental setting (SI Appendix, Fig. S4 A–C). On the contrary, the ces-D mutation clearly increased survival rates, conferring a high level of resistance to −6 °C treatment (Fig. 3 A and B) and reducing electrolyte leakage in the treated plants (Fig. 3C).
Fig. 3.
CES and homologs confer basal freezing tolerance. (A and B) Freezing tolerance of nonacclimated ces-D, tM, and qM lines compared with that of the WT. Plants were grown in soil in LD growth conditions at 21 °C for 3 wk and were treated at −6 °C for 4 h. Survival was assessed following 2 wk of recovery at 21 °C. Shown are representative plants of each line (A) and a quantification of the results (B). Error bars show SD of three biological replicates. (C) Electrolyte leakage in leaves of nonacclimated plants of the WT, ces-D, tM, and qM plants grown under the same conditions as described in C and treated at the indicated temperatures. Error bars show SD of at least two biological replicates. (D) Venn diagram showing the overlap of cold-induced genes [as defined by Park et al. (25); in purple] with genes induced in ces-D [as defined by Poppenberger et al. (14); in pink]. The significance of representation of the CBF regulon of cold-induced genes [as defined by Park et al. (25); in yellow] in the overlay was calculated with a χ2 test giving the P value shown. (E) mRNA levels of CBFs and COR genes in 3-wk-old, nonacclimated soil-grown ces-D, tM, and qM plants determined by qPCRs. Error bars show SD of at least two biological replicates. The letters indicate significant differences between genotypes (P < 0.05; ANOVA). (F) Transcript levels of CBF1, CBF3, and COR15A were assayed in 10-d-old WT seedlings grown on 1/2 MS media supplemented with 250 nM epi-BL. Error bars show SD of at least two biological replicates. Asterisks indicate significant differences (*P < 0.05, **P < 0.01; Student’s t test).
In the Col-0 background, CES acts redundantly with the BEEs in floral organ development (16) and it therefore seemed likely that the BEEs would also compensate for a loss of CES function in freezing tolerance. Accordingly, to evaluate the effects of CES loss of function on freezing tolerance, a ces-2 bee1 bee2 bee3 quadruple mutant (qM) was generated by introducing a newly identified CES knock-out mutant allele, ces-2 (whose characterization is shown in SI Appendix, Fig. S5), into the bee1-3 mutant background (15). In addition, the haf/ces-3 bee1 bee3 triple mutant (SI Appendix, Fig. S5 A and B) (16) was included in the analysis (termed tM from here forward). The results of freezing tolerance assays showed that the tM and qM were hypersensitive to freezing and showed increased electrolyte leakage (Fig. 3 A–C).
To address the molecular basis of the ces mutants freezing tolerance phenotypes, a bioinformatic analysis of available ces-D microarray data (14) was carried out. It was determined to which extent the dominant ces-D mutation can induce the expression of COR genes on a genome-wide scale. This analysis revealed that 7.5% of ces-D–induced genes were also cold inducible and that the CBF regulon, as defined by Park et al. (25), was significantly enriched among them (Fig. 3D and SI Appendix, Table S2). qPCR confirmed that CBFs were significantly induced in ces-D. However, the degree of regulation varied between developmental stages with only CBF1 induced in seedlings (SI Appendix, Fig. S6), but all CBFs up-regulated in adult plants (Fig. 3E). This induction was correlated with an increased expression of all downstream COR genes tested. In the CES tM and qM, CBF1 and CBF3 were reduced by approximately fivefold in adult plants (Fig. 3E), whereas in seedlings, a significant reduction on a whole-plant scale was detectable only for CBF3 (SI Appendix, Fig. S6). This finding was correlated with a reduction of downstream COR genes, in particular of COR15A, in both developmental settings (Fig. 3E and SI Appendix, Fig. S6). Moreover, in tM and qM plants, BR treatment was ineffective in inducing CBF1, CBF3, and COR15A expression (Fig. 3F). Therefore, in summary, there is evidence that the CES/BEE bHLH subfamily participates in basal freezing tolerance and the BR induction of CBF expression.
CES and Homologs Participate in Cold Acclimation.
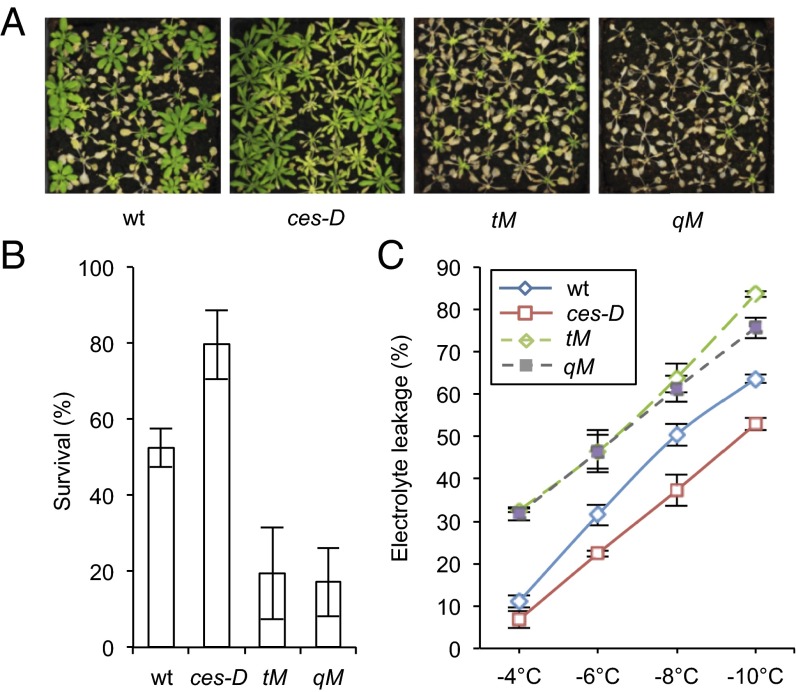
To test whether CES and the BEEs may also affect freezing tolerance following cold adaptation, ces mutant plants were cold acclimated before exposure to subzero temperatures. Also in these assays, ces-D was more resistant to freezing stress than WT, whereas the tM and qM lines were clearly hypersensitive (Fig. 4 A and B). Electrolyte leakage assays confirmed that ces-D was less affected, whereas tM and qM plants were more affected by the treatment (Fig. 4C).
Fig. 4.
CES and homologs promote acquired freezing tolerance. (A and B) Freezing tolerance of ces-D, tM, and qM plants. Three-week-old plants grown in LD growth conditions at 21 °C were acclimated for 3 d at 4 °C and then treated at −10 °C for 6 h. Survival was scored after 2 wk of recovery at 21 °C. Images of representative plants (A) and quantified results (B) are shown. Error bars show SD of three biological replicates. (C) Electrolyte leakage in detached leaves of acclimated plants of the WT, ces-D, tM, and qM. Plants were grown and acclimated as described in B before electrolyte leakage assays were performed. Error bars show SD of three biological replicates.
When in the ces mutant lines the expression of CBFs was assessed following cold treatment, it was found that in ces-D the induction of CBF1 was more pronounced than in the WT (SI Appendix, Fig. S7). This was correlated with a more pronounced increase in the mRNA levels of COR15A and COR15B (SI Appendix, Fig. S7). In qM plants, no significant differences from WT were detectable.
CES Directly Binds to CBF Promoters in Planta.
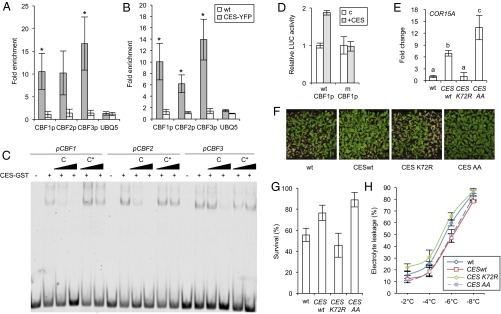
In view of the evidence that CES has the ability to promote CBF expression, it was investigated whether the CBFs are direct CES targets. The promoters were searched for CES binding sites and all three promoters were found to contain G-box motifs. To investigate whether CES can bind to these regulatory elements, chromatin immunoprecipitation (ChIP) assays were performed with 35S:CES-YFP–expressing plants before and after cold treatment. The result showed that CES was significantly enriched on the G-box containing promoter regions of all CBFs, both in untreated conditions and following cold treatment (Fig. 5 A and B). In vitro DNA binding studies with recombinant protein confirmed that CES directly bound to the G-box motifs in the CBF promoters (Fig. 5C). LUCIFERASE (LUC) reporter assays in Arabidopsis protoplasts from the ces-3/haf bee1 bee3 tM with a fragment of the CBF1 promoter containing the G-box and one in which the G-box was mutated showed that the G-box was necessary for CES transcriptional activity in vivo (Fig. 5D).
Fig. 5.
CES directly binds to the promoters of CBFs and activates their expression. (A and B) ChIP of CES-YFP followed by qPCR of DNA fragments containing the G-box motifs in the CBF1 (G box: −106 from the transcriptional start), CBF2 (G box: −111 from the transcriptional start), and CBF3 (G box: −2,151 from the transcriptional start) promoter. Leaves of 3-wk-old CES-YFPoe plants and WT grown in soil in LDs, either untreated (A) or treated at 4 °C for 3 h (B), were used. The 5sr RNA gene was used for normalization. UBQ5 was measured as a control. Values are fold enrichment of CES-bound DNA containing the G-box motif in immunoprecipitated samples relative to the total input DNA. Error bars show the SD of three biological replicates. Asterisks indicate significant differences (*P < 0.05, **P < 0.01; Student’s t test). (C) CES binds to G-box motifs in the promoters of CBF1, CBF2, and CBF3 in vitro. Fluorescent-labeled probes representing parts of the CBF1, CBF2, or CBF3 promoter that contain a G box were incubated with CES-GST. Competitor (C) or mutated competitor oligos with the G box mutated (C*) were added in 10× and 100× molar excess to analyze the specificity of binding. (D) Luciferase transactivation assays in Arabidopsis protoplasts from the ces-3/haf bee1 bee3 tM. The LUC reporter constructs (wtCBF1p: WT CBF1 promoter and mCBF1p: CBF1 promoter with mutated G box) were transiently expressed in protoplasts either alone (control) or with CES as an effector. Error bars show SD of three biological replicates. Asterisks indicate significant differences (*P < 0.05, Student’s t test). (E) Transcript level of COR15A in plants overexpressing either WT CES (CESwt), a mutant impaired in SUMOylation (CESK72R) or a mutant constitutively SUMOylated (CESS75A+S77A: CESCESAA). Error bars show SD of three biological replicates. The letters indicate significant differences between genotypes (P < 0.05; ANOVA). (F and G) Freezing tolerance of CESwt-, CESK72R-, and CESAA-expressing plants. Three-week-old plants grown in LDs at 21 °C were acclimated for 3 d at 4 °C and then treated at −10 °C for 6 h. Survival was scored after 2 wk of recovery at 21 °C. Pictures of representative plants (F) and the quantified results (G) are shown. Error bars show SD of three biological replicates. (H) Electrolyte leakage in leaves of nonacclimated plants of WT and CESwt-, CESK72R-, and CESAA- expressing plants grown in the same conditions as in B and treated at the indicated temperatures. Error bars show SD of three biological replicates.
CES activity is altered by SUMOylation, which is induced in response to an activation of BR signaling (17). Because protein SUMOylation plays a central role in cold stress responses (6), we addressed whether the SUMOylation state impacts on CES activity in COR15A expression and freezing tolerance. COR15A expression levels were determined in plants expressing CES WT (35S:CESwt-YFP, line 32) (14) or CES mutant versions, with either impaired (35S:CESK72R-YFP, line 411) or enhanced SUMOylation (35S:CESS75A+S77A-YFP, line 310) (17). The result showed that, compared with CESwt-expressing plants, plants expressing non-SUMOylated CESK72R had decreased, whereas plants expressing constitutively SUMOylated CESS75A+S77A had increased COR15A levels (Fig. 5E). Also, plants expressing CESK72R suffered more, and plants expressing CESS75A+S77A suffered slightly less damage by freezing than the CESwt-expressing control (Fig. 5 F–H). Therefore, there is evidence that SUMOylation promotes CES activity in freezing tolerance.
BRs and CES Control Common Non–CBF-Regulon Genes in Response to Cold.
Although there was strong evidence that CES can regulate CBF expression, the subtle changes in CBF and downstream COR gene expression in BR and CES loss-of-function mutants, in particular during cold acclimation, compared with their clear freezing hypersensitivity, indicated that, in addition to the CBF regulon, other types of cold-responsive genes are regulated by BRs and CES as well. To identify these genes, we assessed global changes in gene expression in 3-wk-old bri1-301, ces-D, the qM, and WT in response to cold stress using the new Affymetrix Arabidopsis Gene 1.1 ST Array.
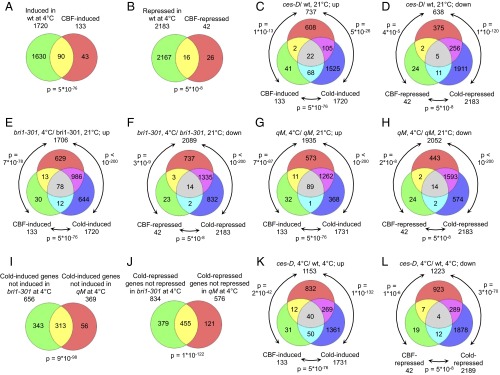
Comparison of cold-treated (4 °C) and untreated (21 °C) WT plants showed that 1,720 genes were significantly induced [>1.5-fold change (FC); false discovery rate (FDR) <0.05] and 2,183 were significantly repressed in response to cold stress (<−1.5-fold change; FDR <0.05) (Dataset S1, Table S3). Among the cold-induced genes, a highly significant share (hypergeometric test P value: 5 × 10−76; Fig. 6A) was previously identified as CBF-induced genes (25). Also, a significant share of genes repressed (Fig. 6B), were previously identified as CBF repressed (25), although less significantly (P value: 5 × 10−8). A relatively large number of CBF regulon genes, as defined by Park et al. (25) escaped detection in cold-treated WT plants (Fig. 6 A and B), which may be due to differences in ecotypes, experimental settings, and array types used. The Arabidopsis Gene 1.1 ST Array differs from the ATH1, which had been used to define the CBF regulon (25), because it employs a random primer for reverse transcription of RNA and a different design of probe sets, resulting in differences in signal strength between the two array types (26). This is important to consider, in particular when the mutant data are interpreted.
Fig. 6.
Whole transcriptome changes in BR and CES mutants in response to cold. (A and B) Overlap of genes induced (A) or repressed (B) in WT in response to 4 °C, WT grown at 21 °C, and CBF-induced or -repressed genes. P values were calculated with the hypergeometric test. (C and D) Overlap of genes induced (C) or repressed (D) in ces-D compared with WT at 21 °C, cold-induced genes of WT and CBF-induced genes. (E and F) Overlap of genes induced (E) or repressed (F) in bri1-301 in response to 4 °C treatment compared with qM grown at 21 °C and CBF- and cold-induced or -repressed genes. (G and H) Overlap of genes induced (G) or repressed (H) in qM in response to 4 °C treatment compared with qM grown at 21 °C and CBF- and cold-induced or -repressed genes. (I) Overlap of cold-induced genes that failed to be induced in bri1-301 and qM plants treated with 4 °C. (J) Overlap of cold-repressed genes that failed to be repressed in bri1-301 and qM plants treated with 4 °C for 3 d. (K) Overlap of genes up-regulated in ces-D in response to 4 °C treatment compared with WT, CBF-induced genes, and cold-induced genes. (L) Same as K but for down-regulated genes.
Comparison of WT and ces-D at 21 °C revealed that of 737 ces-D-induced genes (Dataset S1, Table S4), 24 were also CBF induced (P = 1 × 10−13). A further 105 identified cold-induced genes are not members of the CBF regulon (P = 5 × 10−26) (Fig. 6C). Among those, a significant enrichment of annotations associated with membrane was found (SI Appendix, Fig. S8A). Interestingly, among the 638 ces-D-repressed genes a highly significant share of 261 cold-repressed genes was present (P = 1 × 10−120); of those 7 were CBF-repressed genes, which again is a significant share (P = 5 × 10−8; Fig. 6D). Gene Ontology (GO) analysis of the 256 CBF-independently up- down-regulated genes revealed a number of enriched terms, many associated with lipid and fatty acid biosynthesis or metabolism (SI Appendix, Fig. S8B and Dataset S1, Tables S5 and S6).
With the microarray analysis, relatively few genes were found to be misregulated in bri1-301 and the qM at 21 °C (15 up and 41 down in bri1-301; 3 up and 19 down in the qM; SI Appendix, Fig. S9). Among the genes induced in bri1-301 were the BR biosynthesis genes BR6ox2 and ROT3 (Dataset S1, Table S4), which are feedback-induced in BR-deficient mutants (23, 27). No significant share between genes induced in bri1-301 or the qM and CBF- or cold-induced genes was found (SI Appendix, Fig. S9 A–C). Comparison of genes repressed in bri1-301 and the qM with CBF- and cold-induced genes revealed that only a few CBF-induced genes were also constitutively repressed in these mutants. Although this share was not significant, the microarray analysis did identify COR15A as repressed in both bri1-301 and the qM (Dataset S1, Table S4). Given that the clear reduction in CBF1 expression in bri1-301 and the qM and the extent of increase in ces-D at 21 °C (Figs. 1B and 3B), escaped detection with the arrays, it is evident that the arrays were less sensitive than the qPCR analysis we applied. In support, COR15A, which, according to qPCRs, was approximately 80-fold increased in ces-D, was determined as being increased only approximately 8-fold with the arrays. This reduced sensitivity will have masked changes in gene expression that prevail in the knockout lines. However, the high stringency also bears benefits, because identified changes can be seen with stronger confidence.
In bri1-301, in response to 4 °C, 656 genes failed to be induced and 834 genes failed to be repressed compared with WT (Fig. 6 E and F and Dataset S1, Tables S7 and S8), indicating that they rely on BRI1 function for cold regulation. Among those, CBF regulon members were represented (12 and 2, respectively), but also many additional types of COR genes, which are shown in Dataset S1, Tables S8 and S9). In the qM, 369 genes failed to be induced and 576 genes failed to be repressed in response to 4 °C compared with WT. Almost all of these genes were not CBF regulon genes (Fig. 6 G and H and Dataset S1, Tables S10 and S11) providing evidence that, during cold adaptation, CES largely impacts CBF-independent routes of COR gene regulation. Interestingly, in both bri1-301 and the qM, several CBF-induced genes were more strongly up-regulated by cold than in WT (Dataset S1, Tables S8 and S10), which may result from the lower basal levels of CBF regulon expression that prevail in these mutants (Figs. 1B and 3B).
Importantly, there was a strikingly large overlap of 313 COR genes (P < 9 × 10−98) that failed to be induced in both bri1-301 and the qM mutant (Fig. 6I and Dataset S1, Table S12). GO enrichment analysis revealed terms related to aromatic compound and phenylpropanoid biosynthesis and metabolism, but also terms associated with defense and immune responses (SI Appendix, Fig. S10A and Dataset S1, Table S13). Moreover, there was a highly significant overlap of 455 COR genes (P < 1 × 10−122) that failed to be repressed in both bri1-301 and the qM (Fig. 6J and Dataset S1, Table S12). Very interestingly, in addition to GO terms associated with lipids and fatty acids as in ces-D, genes involved in cell cycle regulation, cell skeleton, and microtubule activity were highly overrepresented (SI Appendix, Fig. S10B and Dataset S1, Table S14). Also, a number of cyclins were present (Dataset S1, Table S13).
In ces-D, the expression of a multitude of genes was altered in response to 4 °C (Dataset S1, Table S3). A total of 1,153 genes showed enhanced expression, whereas 1,223 had reduced expression compared with WT. Notably, the expression of 12 CBF-regulated genes was stronger in ces-D than in WT (Fig. 6K), confirming that ces-D has a larger capacity to activate the CBF regulon during cold acclimation. Also, 7 CBF-repressed genes were more strongly repressed in ces-D than in WT (Fig. 6L). In addition, 269 non–CBF-regulon genes were more strongly induced and 298 more strongly repressed in ces-D. GO analysis again revealed terms associated with stress responses and lipid biosynthesis, respectively, but also ribosome biogenesis and rRNA metabolism (SI Appendix, Figs. S8 C and D and Dataset S1, Tables S5 and S6). Thus, in addition to a clear impact on the CBF regulon, ces-D also affects additional types of COR genes. Particularly fatty acid and lipid biosynthesis, metabolism, and localization appear to be targets.
Discussion
BRs are steroid hormones with versatile roles throughout plant development (11). In the early days of BR research, it became apparent that in addition to promoting growth, BR application increases plant resistance against different types of abiotic stress, including chilling stress (9). In this study we provide evidence that BRs control freezing tolerance and that this ability is conferred by an effect of BRs on the expression of different classes of COR genes, including the CBF regulon.
BR-deficient Arabidopsis mutants were impaired in the basal expression of CBFs and downstream targets, whereas an activation of BR signaling through BRI1 overexpression increased the basal levels of CBFs, in particular of CBF1 and CBF3. This result supports earlier findings of increased basal CBF expression in BRI1oe plants (8–10) and increased COR15A levels in plants overexpressing the BR biosynthesis gene DWF4 (DWARF4) (24). In contrast, a semiquantitative analysis had indicated that in the BRI1 allele bri1-9 CBF expression is increased (9). Our study has now quantitatively assessed CBF and downstream COR gene expression in several BR mutants and provides evidence that BR signaling promotes basal expression of CBFs. The fact that BRs did not induce CBF expression on a whole-plant level at early time points after BR treatment, but only when plants were exposed to BR for longer periods of time, supports the notion that the BR status affects basal CBF expression, but does not significantly impact CBF expression in early responses to BR or cold.
There is evidence that the activity of BRs in basal CBF transcription is mediated by the BR-regulated bHLH TF CES and its homologs BEE1 and BEE3. In the dominant ces-D mutant basal expression levels of CBFs and downstream COR genes were strongly increased. In ces bee knockout plants, basal CBF1 and CBF3 levels were decreased and CBF1 and CBF3 induction by BRs was compromised. Moreover, in response to cold, the ces-D mutant activated the CBF regulon to a larger extend than WT, which could be due to the elevated basal CBF expression levels that exist in this mutant.
Although the constitutive changes in CBF and COR gene expression revealed in BR and CES loss-of-function plants, particularly compared with their clear freezing hypersensitivity, may be considered mild, there are several things to be considered. First, BR responses at the transcriptional level are small, usually only two- to threefold (28, 29). Second, tissue-specific differences in CBF expression, that may be instrumental for basal freezing tolerance, could have been disguised when whole plants were used for qPCR and microarray analyses. Third, in loss-of-function mutants, functional redundancy can mask regulatory effects and it is clear that additional factors and pathways can also regulate CBF expression. Therefore, the relatively mild changes in basal CBF expression levels in the loss-of-function plants do not exclude a role of the BR-CES/BEE-CBF1/3-COR pathways in basal freezing tolerance. However, because we here show that additional pathways participate, it will be important to determine the relative contribution of the CBF regulon to BR-CES/BEE conferred basal freezing tolerance.
In addition to their role in basal freezing tolerance, BRs and CES are also of importance for freezing stress resistance that requires cold adaptation, because BR and CES/BEE mutants have clear defects in acquired freezing tolerance. However, in this process, BRs appear to act largely in a CBF-independent manner, because in BR and ces bee knockout mutants, the CBF regulon is activated to similar extends like in WT. A whole-transcriptome analysis identified non–CBF-regulon types of COR genes that depend on BRI1 and CES for activation or repression in response to cold. Importantly, there is a highly significant overlap of genes misregulated in both bri1-301 and qM plants following cold exposure, providing evidence that the role of BRs in cold adaptation is conferred to a significant degree by CES and the BEEs. Within the COR genes that failed to be repressed in BR and CES knockout mutants, annotations for cell cycle regulation, cell skeleton, and microtubule activity were overrepresented. This finding indicates that without a functional BR-CES/BEE module, plants are unable to decrease cell division and metabolism during cold adaptation, which could, at least in part, explain the freezing hypersensitivity of the mutants. Also, annotations for fatty acid and lipid synthesis, metabolism, and transport were overrepresented. Because it is well established that in response to cold stress changes in the structure and composition of membranes occur, which are dedicated to reduce damage caused by freezing (2, 7), it is possible that a role of CES/BEEs in the synthesis of fatty acid and lipids, which are essential membrane building blocks, contributes to the freezing hypersensitivity of the mutants. In the future it will be important to investigate which factors are directly regulated by CES/BEE during cold acclimation and to determine their relative contribution to BR-CES/BEE-conferred cold acclimation-dependent freezing tolerance.
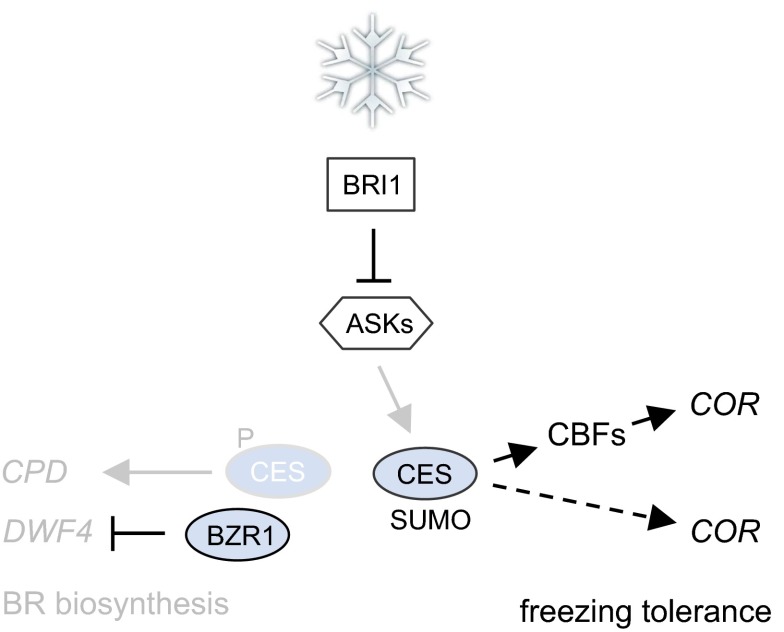
Unlike the BEEs, whose expression is BR induced (15), CES is not BR regulated at the transcriptional level but is subject to BR-induced posttranslational modification. The current postulation is that in response to BR, CES phosphorylation by BIN2 is inhibited, CES accumulates in an unphosphorylated state, promoting CES SUMOylation and nuclear compartmentalization (17). This may restrict CES activity on one type of promoters, for example those of BR biosynthesis genes (14), which are repressed when BR levels become high (23, 27), but may facilitate activity on other types such as CBFs. In support of this hypothesis, we have evidence that SUMOylation promotes CES activity in COR15A expression and freezing tolerance, suggesting a model in which BRs induce CES SUMOylation to alter COR gene expression.
In cold responses, protein SUMOylation plays a key role. The overall SUMOylation of proteins drastically increases in response to cold stress (6), and the SUMOylation of ICE1 enhances its activity in CBF3 transcription. The manner by which ICE1 SUMOylation is induced is currently unknown, although a phosphodeficient mutant of ICE1 is more readily SUMOylated (30), and also in CES, dephosphorylation promotes SUMOylation (17). It is thus possible that in response to cold, BR signaling is activated to alter the phosphorylation state of TFs, inducing SUMOylation and activating them in COR gene expression. This, in a secondary response, would reduce BR biosynthesis, given that when BR signaling is activated, BR-biosynthetic gene expression is repressed (23, 27). In support of this model (illustrated in Fig. 7), the BR biosynthesis gene CPD, a direct CES target (14), is down-regulated by cold both in Arabidopsis (31) and in mungbean (32), and also in our hands, CPD, as well as DFW4 and BRASSINOSTEROID-6-oxidase 2 (BR6ox2) mRNA abundance were reduced in response to cold (SI Appendix, Fig. S11). However, importantly, in BR-deficient mutants, unlike in other mutants with growth defects (33, 34), growth repression is not correlated with increased stress tolerance. This lack of correlation supports the notion that the function of BRs in freezing tolerance is not principally to cause morphological changes that promote plant survival as a secondary consequence, but that BRs directly participate in signaling events required for freezing tolerance.
Fig. 7.
Working model for the contribution of BRs to freezing tolerance. Cold stress stimulates BR signaling to induce dephosphorylation and SUMOylation of CES and activate it in both CBF-dependent and CBF-independent modes of COR gene regulation. As a consequence of an activation of BR signaling, BR biosynthesis is feedback repressed through positive and negative regulation of activators (including CES) and repressors (including BZR1) of BR-biosynthetic gene transcription.
Although tolerance to different types of abiotic stress can be enhanced by CBF overexpression (35), this is not an agronomically feasible strategy, given that overexpressing CBFs impairs plant growth by inhibiting gibberellin (GA) metabolism and signaling (33). In contrast, the overexpression of CES induces CBF transcription and enhances freezing tolerance, but does not also produce dwarf plants (14), uncoupling freezing tolerance from growth repression. Different, but not mutually exclusive mechanisms may account for this. First, CBF mRNA levels in ces-D may be sufficient to increase COR expression and freezing tolerance, but not to repress growth. Second, the growth-repressive activity of CBFs may be released by the positive regulatory role of ces-D in BR biosynthesis (14), and/or third, ces-D may directly interfere with the effects of CBFs on GA biosynthesis and/or signaling.
In conclusion, our study reveals that BRs, in addition to their growth-promoting capacities, enhance plant resistance against freezing stress. We present evidence that this function is conferred by the activity of the CES/BEE proteins in controlling COR gene expression. Given that BRI1- and CES overexpression increases freezing tolerance, but does not also repress growth, this approach may be promising for enhancing crop resistance against freezing, and possibly also other types of abiotic stress. Such a strategy would be of high relevance for plant production in agriculture and horticulture.
Materials and Methods
Plant Material.
The transfer DNA (T-DNA) insertional mutant ces-2 was obtained from the Nottingham Arabidopsis Stock Center (NASC) (SALK_124840), and the site of insertion was mapped by PCR (primers CES KO fwd and CES KO rev in combination with LBb1; all primers used in this study are listed in Table S15) to the location 1380 downstream of the ATG (SI Appendix, Fig. S5A). qPCR analysis showed that CES expression was reduced in ces-2 (SI Appendix, Fig. S5C).
ces-2 was crossed with bee1 bee2 bee3 (15), and the F2 offspring were genotyped using the primer pairs CES KO fwd/CES KO rev, BEE1 KO fwd/BEE1 KO rev, BEE2 KO fwd/BEE2 KO rev, and BEE3 KO fwd/BEE3 KO rev. These primers, for the WT, and plants heterozygous for the corresponding gene, yielded amplicons of 484 bp, 1077 bp, 458 bp, and 709 bp, respectively, whereas they did not yield amplicons for homozygous plants.
For phenotypic analysis of silique filling, 8-wk-old, soil-grown ces mutant plants were used. Siliques were harvested from a single branch of at least five plants of the indicated genotypes. The siliques were incubated in a clearing solution (20 g of chloral hydrate, 4.6 mL of water, 2 mL of glycerol, 87%) for 24 h at room temperature and assessed by optical bright field microscopy.
Plant Freezing Assay.
Plant freezing assays were performed as described previously (6, 36) with modifications. Plants were grown in soil in Bright Boy growth chambers (CLF Plant Climatics) at 21 °C under long-day (LD) conditions (16 h light at 80 µmol m−2⋅s−1/ 8 h dark) for 3 wk before the treatments were performed. For treatments without cold acclimation, plants were incubated in a controlled temperature chamber (Panasonic MIR-154, Panasonic Biomedical) for 30 min at 4 °C, then for 1 h at 0 °C, before the temperature was decreased by 2 °C per hour. The final desired subzero temperature was maintained for the indicated period before the temperature was again increased at the same rate to 4 °C. The plants were then kept at 4 °C for 1 d before they were returned to 21 °C. Survival was scored 2 wk later, with only those plants able to develop new leaves counted as survivors.
For cold acclimation experiments, 3-wk-old plants were acclimated for 3 d at 4 °C in the light. Freezing treatment was then performed in the same manner as for nonacclimated plants with the final freezing temperature of −10 °C maintained for 6 h.
Electrolyte Leakage Assays.
Electrolyte leakage from fully expanded rosette leaves of 3-wk-old plants was measured as described previously (37) with modifications. Plants were grown in soil at 21 °C in LD conditions, and the fifth and sixth leaf pairs were used. The leaves were placed in tubes containing 100 µL of deionized water. An ice chip was added to facilitate nucleation, and the tubes were kept at −2 °C for 2 h followed by a temperature decrease at 2 °C per hour. Samples were removed at the indicated temperature points and immediately placed on ice for gradual thawing overnight. On the next day, 6 mL of deionized water was added to each tube, and the samples were incubated for 5 h at 21 °C with gentle shaking, after which the conductivity of the solution was determined with a conductivity meter (GMH 3430; Greisinger Electronic). The tubes were then incubated at 95 °C for 3 h, and the conductivity of the solution was measured again. Electrolyte leakage was quantified as a percentage of the conductivity after treatment relative to total conductivity.
qPCRs.
For qPCRs, plants were grown vertically on agar plates or in soil depending on the experiment. Total mRNA was extracted with a Plant RNA Kit (Omega Bio-Tek) and treated with DNaseI to digest traces of DNA. First strand cDNA was synthesized from 1 µg of RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions. qPCRs were performed with the SensiFAST SYBR Lo-ROX Kit (Bioline) using the Mastercycler Realplex (Eppendorf). Ubiquitin-conjugating enzyme 21 (UBC21) (38) was used for the normalization of the results. qPCRs were typically performed with at least two independent biological samples, each measured in at least three technical repeats. For qPCR analysis of cold-treated plants, seedlings were grown vertically on agar plates. Twelve-day-old seedlings were treated at 4 °C in the light, and plant material was collected in a time-course manner. Statistical significance of differences between two samples was assessed using Student’s t test. For quantitative analyses among multiple samples, statistical significance was assessed using analysis of variance (ANOVA) followed by the Tukey’s post hoc test.
ChIPs.
For ChIPs, 3-wk-old, soil-grown 35S:CES-YFP plants (line 32) (14) and WT were treated for 3 h with 4 °C or left at 21 °C. Approximately 0.6 g of plant material was harvested for each biological replicate. ChIP was performed in three biological repeats according to ref. 39.
Protoplast Transformation and Luciferase Transactivation Assays.
For transactivation assays, a 35S:CES construct was used (14). For reporter plasmid generation, a CBF1 promoter fragment containing the G-box was PCR amplified with the primer pair CBF1-luc-FW/RV. The primer pair CBF1-luc-FW1/RV was used to mutate the G-box sequence, yielding the mCBF1 promoter fragment. The resulting PCR fragments were cloned into the pGreenII-0800-LUC vector (40).
Protoplasts were isolated from haf bee1 bee3 plants (16) and transiently transformed using a PEG-mediated transformation protocol. Luciferase assays were performed using a Dual-Luciferase Reporter Assay System (Promega) and a Lumat LB9501 luminometer (Berthold) as described previously (17).
Whole Transcriptome Analysis.
Whole transcriptome analysis was performed from 3-wk-old, soil-grown plants, treated with 4 °C in the light for 3 d. Controls were kept at 21 °C for 3 d. RNA was prepared from aerial tissues of plants; sample concentration and purity was determined by spectrophotometry; and RNA integrity was confirmed using an Agilent 2100 Bioanalyzer with the RNA 6000 Nano Kit (Agilent Technologies). Whole transcriptome analysis was conducted by hybridizing total RNA of four independent biological replicates for each line and treatment (32 biological samples in total) to Affymetrix Arabidopsis Gene 1.1 ST Array Strips (Affymetrix).
Gene expression data were analyzed using Partek Genomics Suite 6.6 software (Partek). The raw CEL files were normalized using the robust multiarray average (RMA) background correction with quantile normalization, log base 2 transformation, and mean probe-set summarization with adjustment for GC content. One 4 °C-treated WT sample was identified as an outlier using Partek and was thus excluded from further analysis.
Bioinformatic Analyses.
The det2-1 mutant comparison against the WT was performed using the AtGenExpress dataset “Brassinolide time course in wild-type and det2-1 mutant seedlings” (TAIR ME00335). The CEL data were normalized using the RMA algorithm as provided by the justRMA method in the Bioconductor Affy package (41). For each time point (30 min, 1 h, and 3 h) significantly regulated genes (FDR-adjusted P value <0.05) were called with the limma package (42). The union of these sets was considered to represent the det2-1-associated genes. COR and CBF regulon-related genes were assigned as described previously (25). The significance of the overlap of COR genes and genes related to the CBF regulon was determined with a χ2 test of independence.
For bioinformatics analysis of the microarray data differentially expressed genes (DEGs) were identified by a two-way ANOVA, and P values were adjusted using the FDR method to correct for multiple comparisons. DEG were considered significant if P value was ≤0.05 at a FC of ≥2 with a FDR of <0.05. The raw data were included in the Gene Expression Omnibus (GEO) database. Venn diagrams were constructed using the Venn diagram generator (bioinformatics.lu/venn.php). Significance of overlaps was calculated with the hypergeometric test. Analysis for enrichment of GO terms was performed with agriGO (bioinfo.cau.edu.cn/agriGO/analysis.php).
EMSAs.
HEX-labeled probes for EMSAs were prepared by PCR using the primer pairs CBF1-ChIP-FW/HEX-CBF1-ChIP-RV, HEX-CBF2-ChIP-FW/CBF2-ChIP-RV, and CBF3-ChIP-FW/HEX-CBF3-ChIP-RV. The amplicons were purified using an E.Z.N.A. Cycle Pure kit (Omega Bio-Tek). The probes (30 ng per reaction) were incubated with purified CES-GST protein and subsequently separated on 6% PAGE gels as described previously (14). The bands were detected using a Molecular Imager FX Pro (Biorad) equipped with a 532-nm laser for excitation and a 555-nm-long pass emission filter.
Supplementary Material
Acknowledgments
We thank Joanne Chory for seeds of the bee1 bee2 bee3, bes1-D, and bzr1-1D mutants and the 35S:BRI1-GFP line; Irene Ziegler, Clarissa Fahrig, and Renata Milcevicova for technical assistance; and the horticultural staff of the TUMs Gewächshauslaborzentrum Dürnast for plant care. This work was supported by funds from the Austrian Science Fund (Project P22734 to B.P.), the Deutsche Forschungsgemeinschaft (Project PO1640/4 to B.P. and SFB924 to B.P. and K.F.X.M.), and a TUM doctoral fellowship (to M.E.). M.E. and S.J.U. were members of the TUM graduate school.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE86605).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611477113/-/DCSupplemental.
References
- 1.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14(8):1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA. 2004;101(42):15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6(2):251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmour SJ, et al. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16(4):433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 5.Chinnusamy V, et al. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17(8):1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura K, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19(4):1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eremina M, Rozhon W, Poppenberger B. Hormonal control of cold stress responses in plants. Cell Mol Life Sci. 2016;73(4):797–810. doi: 10.1007/s00018-015-2089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta. 2007;225(2):353–364. doi: 10.1007/s00425-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 9.Kim SY, Kim BH, Lim CJ, Lim CO, Nam KH. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiol Plant. 2010;138(2):191–204. doi: 10.1111/j.1399-3054.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- 10.Divi UK, Krishna P. Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. N Biotechnol. 2009;26(3-4):131–136. doi: 10.1016/j.nbt.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Clouse SD. Brassinosteroids. Arabidopsis Book. 2011;9:e0151. doi: 10.1199/tab.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 13.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2(4):505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 14.Poppenberger B, et al. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 2011;30(6):1149–1161. doi: 10.1038/emboj.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrichsen DM, et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162(3):1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford BC, Yanofsky MF. HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development. 2011;138(14):2999–3009. doi: 10.1242/dev.067793. [DOI] [PubMed] [Google Scholar]
- 17.Khan M, et al. Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat Commun. 2014;5:4687. doi: 10.1038/ncomms5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozhon W, Mayerhofer J, Petutschnig E, Fujioka S, Jonak C. ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 2010;62(2):215–223. doi: 10.1111/j.1365-313X.2010.04145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410(6826):380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 20.Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123(4):1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, Huang J, Li B, Li J, Wang Y. Is kinase activity essential for biological functions of BRI1? Cell Res. 2008;18(4):472–478. doi: 10.1038/cr.2008.36. [DOI] [PubMed] [Google Scholar]
- 22.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85(2):171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 23.Asami T, et al. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123(1):93–100. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Divi UK, Rahman T, Krishna P. Gene expression and functional analyses in brassinosteroid-mediated stress tolerance. Plant Biotechnol J. 2016;14(1):419–432. doi: 10.1111/pbi.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S, et al. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015;82(2):193–207. doi: 10.1111/tpj.12796. [DOI] [PubMed] [Google Scholar]
- 26.Kakei Y, Shimada Y. AtCAST3.0 update: A web-based tool for analysis of transcriptome data by searching similarities in gene expression profiles. Plant Cell Physiol. 2015;56(1):e7. doi: 10.1093/pcp/pcu174. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka K, et al. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 2005;138(2):1117–1125. doi: 10.1104/pp.104.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130(3):1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müssig C, Fischer S, Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129(3):1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura K, Ohta M, Nakazawa M, Ono M, Hasegawa PM. ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J. 2011;67(2):269–279. doi: 10.1111/j.1365-313X.2011.04589.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17(11):3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang B, Chu CH, Chen SL, Juan HF, Chen YM. A proteomics study of the mung bean epicotyl regulated by brassinosteroids under conditions of chilling stress. Cell Mol Biol Lett. 2006;11(2):264–278. doi: 10.2478/s11658-006-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Achard P, et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20(8):2117–2129. doi: 10.1105/tpc.108.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54(5):767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- 35.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280(5360):104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 36.Eremina M, Rozhon W, Yang S, Poppenberger B. ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1. Plant J. 2015;81(6):895–906. doi: 10.1111/tpj.12775. [DOI] [PubMed] [Google Scholar]
- 37.Gilmour SJ, Hajela RK, Thomashow MF. Cold acclimation in Arabidopsis thaliana. Plant Physiol. 1988;87(3):745–750. doi: 10.1104/pp.87.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unterholzner SJ, et al. Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell. 2015;27(8):2261–2272. doi: 10.1105/tpc.15.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellens RP, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 42.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.