Significance
Lignin is the only renewable and abundant polymer on the earth with aromatic units as its building blocks; however, it remains as an untapped resource. The current approach to making the biofuel industry cost competitive with the petroleum industry is to derive more value from the lignin. In this study we combined the unique approaches of both chemical engineering and biology to gain a deeper understanding of the metabolism of a soil bacterium, Sphingobium sp. SYK-6, that enables it to survive on lignin-derived monomers and oligomers. Understanding the central metabolism of SYK-6 will enable researchers to redesign the metabolic pathways of Sphingobium sp. SYK-6 more effectively to provide a renewable route for the production of products currently sourced from petrochemicals.
Keywords: 13C-MFA, fingerprinting, gluconeogenesis, NADPH, lignin
Abstract
Sphingobium sp. SYK-6 is a soil bacterium boasting a well-studied ligninolytic pathway and the potential for development into a microbial chassis for lignin valorization. An improved understanding of its metabolism will help researchers in the engineering of SYK-6 for the production of value-added chemicals through lignin valorization. We used 13C-fingerprinting, 13C metabolic flux analysis (13C-MFA), and RNA-sequencing differential expression analysis to uncover the following metabolic traits: (i) SYK-6 prefers alkaline conditions, making it an efficient host for the consolidated bioprocessing of lignin, and it also lacks the ability to metabolize sugars or organic acids; (ii) the CO2 release (i.e., carbon loss) from the ligninolysis-based metabolism of SYK-6 is significantly greater than the CO2 release from the sugar-based metabolism of Escherichia coli; (iii) the vanillin catabolic pathway (which is the converging point of majority of the lignin catabolic pathways) is coupled with the tetrahydrofolate-dependent C1 pathway that is essential for the biosynthesis of serine, histidine, and methionine; (iv) catabolic end products of lignin (pyruvate and oxaloacetate) must enter the tricarboxylic acid (TCA) cycle first and then use phosphoenolpyruvate carboxykinase to initiate gluconeogenesis; and (v) 13C-MFA together with RNA-sequencing differential expression analysis establishes the vanillin catabolic pathway as the major contributor of NAD(P)H synthesis. Therefore, the vanillin catabolic pathway is essential for SYK-6 to obtain sufficient reducing equivalents for its healthy growth; cosubstrate experiments support this finding. This unique energy feature of SYK-6 is particularly interesting because most heterotrophs rely on the transhydrogenase, the TCA cycle, and the oxidative pentose phosphate pathway to obtain NADPH.
Lignocellulosic biomass with a worldwide annual production of 200 billion tons represents the most abundant renewable carbon source on the earth for the production of biofuels and chemicals (1, 2). Lignin is a heteropolymer that constitutes 25–35% of the woody biomass and is the second major component of the plant biomass. Lignin traditionally has been seen as a byproduct of limited usefulness and primarily has been burnt for its energy content. To make future potential biorefineries cost competitive with the petroleum industry, the current approach is to derive more value from the lignin. This approach has resulted in unprecedented research efforts in lignin deconstruction in recent years, with publications on lignin depolymerization in 2010–2013 increasing by two- to threefold in comparison with the previous decade (3). Lignin is the only renewable and abundant polymer with aromatic units as its building blocks. Therefore, lignin has the potential to serve as a cheap feedstock for the production of various value-added chemicals that currently are produced from crude oil (4). It has been estimated that using biomass waste for chemical production would yield at least 10 times more value than would be derived from burning it for electricity generation. For example, producing bulk chemicals from a ton of biomass would yield a value of $1,000 as against $60–$150 in value from electricity generation (5). Lignin valorization is one of the keys to realizing an affordable and scalable industrial biorefinery (6).
The lignin macromolecule is recalcitrant because of its structural complexity, hampering lignin valorization. Recent approaches toward lignin valorization can be generally grouped into the following categories: (i) lignin bioengineering to reduce the recalcitrance of the plant cell wall (7–9); (ii) chemical depolymerization (catalytic, ionic, and oxidative depolymerization) (10–13); and (iii) lignin valorization using microbes (14, 15). Microbial depolymerization of lignin can be performed at lower temperature and pressure than chemical depolymerization. In addition, product specificity can be improved through the use of microbes. However, a robust microbial chassis is required for effective lignin valorization. Fungal species are among the most efficient lignin degraders because they secrete some of the highly active ligninolytic enzymes, commonly referred to as “ligninases” or “lignases.” The dominant ligninases produced by fungi include lignin peroxidase, versatile peroxidase, manganese peroxidase, and laccase (16). Although fungi are effective lignin degraders, fungal genomes are difficult to engineer, thereby limiting the development of a ligninolytic technology based on fungi. Thus, attention has now shifted toward bacterial lignin degraders (17–19).
In the last few years, several bacteria with lignin-degrading capability have been discovered, and some of them were found to have unique pathways for the cleavage of lignin linkages (20, 21). Sphingobium SYK-6, a soil bacterium discovered in the ponds of a kraft pulp mill, has been one of the most studied bacterial lignin degraders, and its genome was sequenced by Eiji Masai and others in 2011 (22). SYK-6 can grow on a wide variety of lignin-derived monoaryls and biaryls, and most of the research thus far has focused on understanding its lignin catabolic pathways (23, 24). SYK-6 is also amenable to genetic engineering, as demonstrated in an earlier work by the construction of a ligV (vanillin dehydrogenase) mutant in SYK-6 through insertional inactivation (25). Inactivating the expression of ligV in SYK-6 is an intuitive technique for the accumulation of vanillin in SYK-6 with lignin as a feedstock, but the mutant strain lost its normal ability to grow with vanillin as the sole carbon source. These results underscore the obvious and growing need for understanding the central metabolism of SYK-6 in association with lignin degradation to establish a robust molecular and cellular biology toolbox for further manipulation and engineering of the organism for lignin valorization.
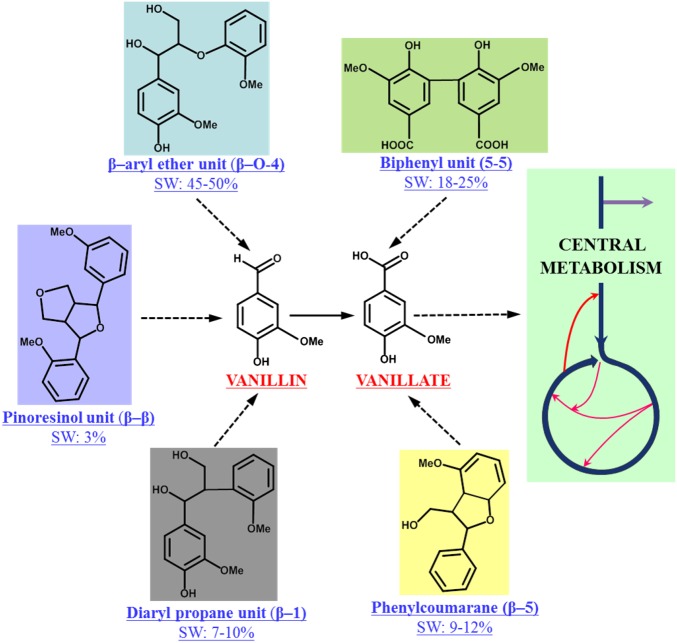
13C metabolic flux analysis (13C-MFA) is a powerful technique for determining steady- state in vivo fluxes of central metabolic pathways in living cells (26–29). 13C-MFA also has been conducted to understand the metabolism of biofuel- and biochemical-producing microbial strains (30–32). However, nonmodel organisms often possess uncommon metabolic pathways that could lead to inaccuracies in flux calculations. Therefore, a priori work using 13C-fingerprinting to understand the central metabolic pathways in nonmodel species is essential before a 13C-MFA study is performed. Recently, several studies have used multiple 13C substrates to identify unique metabolic pathways (evaluations also called “13C-fingerprinting “or “13C-pathway analysis”) and to determine product yields accurately (33, 34). In this work we used 13C-fingerprinting, 13C-MFA, and RNA-sequencing (RNA-seq) data analysis to unravel the unique pathways SYK-6 has evolved with and to elucidate its central metabolism with the goal of identifying the unique metabolism that lets it survive on lignin-derived oligomers. The insights to be gained from this study will enable the optimal engineering of this organism for the purpose of lignin valorization. The 13C-MFA to study the ligninolysis-based central metabolism of SYK-6 was performed using 13C vanillin for several reasons. (i) Vanillin and vanillic acid are the monomers resulting from the enzymatic cleavage of the major linkages present in lignin (Fig. 1). (ii) The presence of the lignin-derived oligomers would alter the concentration of vanillin depending on the rate of bond cleavage, and this alteration in turn would change the absolute fluxes of the central metabolism. However, the relative fluxes would remain independent of the vanillin concentration present in the medium. Therefore, the relative fluxes to be measured with vanillin as a substrate are hypothesized to be approximate enough to the fluxes that would otherwise be observed in the presence of lignin oligomers. (iii) 13C-labeled lignin-derived oligomers are not commercially available. (iv) Tracking the carbon transitions resulting from a mixture of labeled aromatic substrates is still challenging, thus preventing 13C-MFA from computing the flux of the metabolism accurately. For example, assume that 13C-labeled oligomers are available for all the major linkages present in lignin, as shown in Fig. 1. The cleavage of all these units will result in a mixture of vanillic acid isotopes that are labeled at multiple positions and at varying ratios. Identifying the composition of this isotopic mixture and tracking carbon from the mixture is complicated and can result in huge inaccuracies in the flux calculation. Therefore, using 13C-vanillin as a representative carbon substrate to study the ligninolysis-based central metabolism of SYK-6 is a reasonable approximation.
Fig. 1.
Vanillin and vanillic acid are the monomers generated from the enzymatic cleavage of some of the major linkages present in lignin. Vanillin and vanillic acid undergo further chemical transformation through a linear set of reactions before cleaving into pyruvate and oxaloacetate. Pyruvate and oxaloacetate then enter the central metabolic pathway of SYK-6 for the generation of its cellular building blocks and energy. SW, softwood.
Results
Carbon Substrate Utilization.
To understand carbon substrate preference by Sphingobium sp. SYK-6, cultures of SYK-6 were grown in minimal medium containing different carbon substrates (Table 1). The mass distributions of amino acids or the mass isotopomer distribution (MID) obtained from cells fed with different labeling substrates are presented in SI Appendix, Table S1. (The abbreviations “m0,” “m1,” and “m2,” and so forth, present in the MID stand for the mass isotopomer fraction of unlabeled, singly-labeled, doubly-labeled, and multi-labeled amino acids, respectively.) Cells grew well in the presence of vanillin (the preferred substrate for SYK-6) and reached a maximum OD600 of ∼0.7 in 6 d. The resulting proteinogenic amino acids from 13C-vanillin tracer experiments were highly labeled. SYK-6 grew very poorly in the glucose medium with yeast extract, reaching an OD600 of about 0.16 in 6 d after inoculation. From Fig. 2, it can be seen that the amino acids obtained from cultures fed with 13C-glucose were highly unlabeled (m0 >90%), clearly indicating that SYK-6 lacks essential enzymes for glucose catabolism. Indeed, annotation of the genes encoding enzymes of the phosphotransferase system (PTS), which mediates the transport and phosphorylation of extracellular glucose into glucose-6-phosphate, is missing in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Thus SYK-6 cannot obtain reducing cofactors, NAD(P)H, or building blocks from glycolysis of sugar substrates. We also tested cell growth using only organic acids (common substrates for bacterial species) for 6 d. In the cultures fed with acetate and in the absence of yeast extract, the cell OD600 reached only 0.1. Cell OD600 was also low (0.16) in cultures fed with pyruvate, a product of the vanillin catabolic pathway. The incorporation of carbons from 13C-acetate or 13C-pyruvate into SYK-6 biomass can be verified by a decrease in m0 for several of the amino acids (Fig. 2). A high m0 (>0.90) for valine, methionine, phenylalanine, lysine, and histidine (SI Appendix, Table S1) from 13C-acetate– or 13C-pyruvate–fed cultures also indicates that SYK-6 was not able to synthesize those amino acids from 13C-pyruvate or 13C-acetate. Therefore, the cells had to depend on the amino acids that were carried over during the inoculation, and the growth may have come to a halt as soon as the culture ran out of those particular amino acids.
Table 1.
Carbon substrates used for the 13C-fingerprinting experiments in SYK-6
| Primary carbon substrate | Primary substrate concentration, g/L | Secondary carbon source |
| Vanillin (phenyl-13C) | 0.75 | — |
| Vanillin (methoxy-13C) | 0.75 | — |
| Vanillin (aldehyde-13C) | 0.75 | — |
| Acetate (1-13C) | 1 | — |
| Acetate (1,2 -13C) | 1 | — |
| d-glucose (U-13C) | 1 | Yeast extract, 0.2 g/L |
| Xylose (1,2-13C) | 1 | Yeast extract, 0.2 g/L |
| Pyruvate (U-13C) | 1 | Malate, 1 g/L |
| Pyruvate (U-13C) | 1 | Glyoxylate, 1 g/L |
| Pyruvate (U-13C) | 1 | Oxaloacetate, 1g/L |
| Pyruvate (unlabeled) | 1 | Formate (13C), 1 g/L |
| Vanillin (unlabeled) | 0.75 | Formate (13C), 0.75 g/L |
| Vanillin (phenyl-13C) | 0.75 | Methanol, 0.75 g/L |
| Vanillin (phenyl-13C) | 0.75 | Glyoxylate, 0.75 g/L |
Fig. 2.
Incorporation of 13C-carbon into the amino acids ([M-57]+) of SYK-6 after feeding with the following labeled carbon substrates: (A) (1,2-13C) glucose and yeast extract; (B) (phenyl-13C) vanillin; (C) (1,2-13C) acetate; and (D) (U-13C) pyruvate*. The asterisk indicates that unlabeled glyoxylate was added to the labeled pyruvate culture to explore the presence of glyoxylate shunt in SYK-6. The SD represents the 2% technical error of the instrument. The Embden–Meyerhof–Parnas pathway is shown in purple; the TCA is shown in green; the PPP is shown in red; and the amino acid synthesis pathway is shown in black.
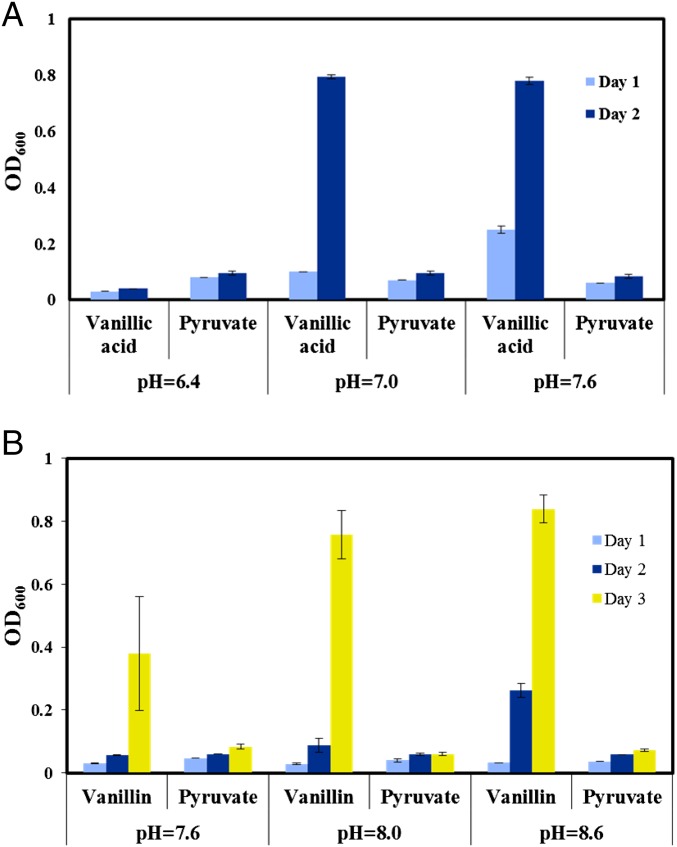
Last, we tested the effect of pH on cell growth. Growth in the presence of pyruvate remained unaltered for all the tested pHs, but higher pH (7.0–8.6) improved the growth of SYK-6 in the presence of vanillin or vanillic acid (Fig. 3). A lower pH of 6.4 was found to reduce the growth of SYK-6 drastically with vanillic acid as a carbon source. The OD600 obtained with vanillic acid as a carbon substrate was found to be less than that obtained with pyruvate as a carbon substrate at a pH of 6.4. These results indicating that SYK-6 prefers alkaline growth conditions are not surprising, given that the bacterium was initially isolated from high-pH waste streams from a pulp mill. Therefore, SYK-6 can serve as an efficient host for the consolidated bioprocessing of lignin.
Fig. 3.
Growth of SYK-6 under varying pH conditions. (A) With vanillic acid plus pyruvate as the carbon source. (B) With vanillin plus pyruvate as the carbon source.
Tetrahydrofolate-Dependent C1 Metabolism of SYK-6.
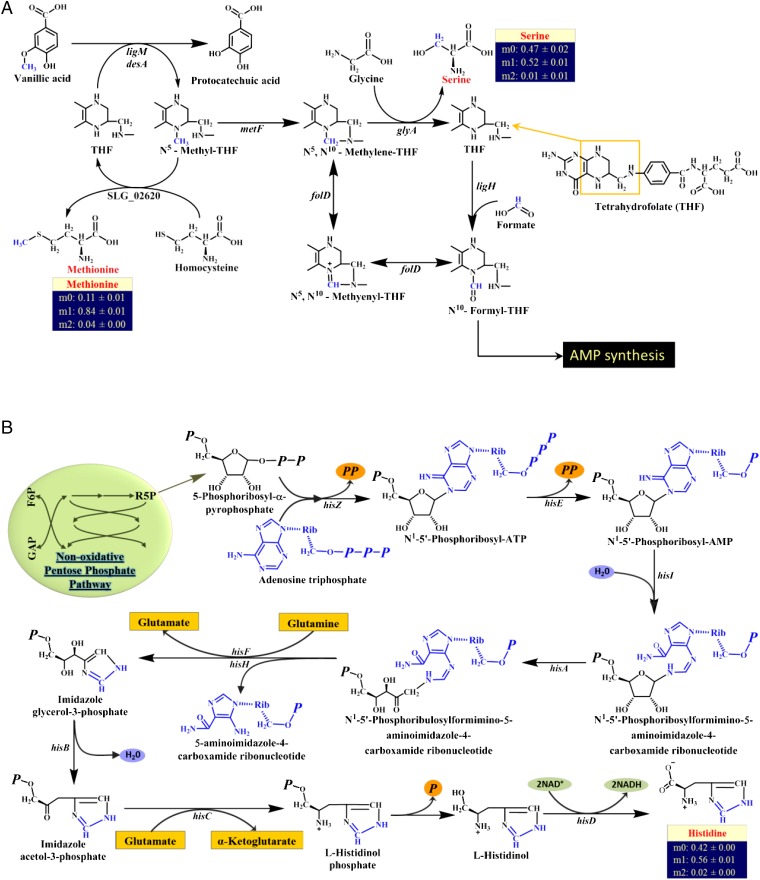
Demethylation is an essential step involved in the conversion of some of the important lignin-derived monomers into the key central metabolites pyruvate and oxaloacetate (35, 36). Earlier works have reported the presence of demethylation in various organisms (e.g., Pseudomonas sp. Strain ATCC 19151 and Acetobacterium dehalogenans) that use vanillate as the carbon substrate (37, 38). In the ligninolytic pathways of SYK-6, demethylation occurs during three major reactions: (i) during the conversion of vanillic acid into protocatechuic acid, (ii) during the conversion of syringate into 3-O-methylgallate, and (iii) during the conversion of 3-O-methylgallate into gallate. Demethylation in SYK-6 has been shown to occur in the presence of the products of either syringate O-demethylase gene (desA) or tetrahydrofolate-dependent O-demethylase gene (ligM) (35, 36). Earlier works have shown that the methyl group cleaved from the three metabolites (vanillate, syringate, and 3-O-methylgallate) participates in a tetrahydrofolate (THF)-dependent metabolic pathway (C1-THF) for amino acid and nucleic acid synthesis (36, 39, 40).
To identify the fate of the methyl group in the amino acid pool precisely, SYK-6 was cultivated with a vanillin substrate in which the 13C label was in the methoxy position. Detailed mass isotopomer analysis indicated that only methionine, serine, and histidine were labeled (Fig. 4). These data clearly indicate that the methyl group liberated from vanillin was being used for the biosynthesis of these three amino acids. Also, methionine was the most highly labeled of the three amino acids, indicating a high flux toward methionine synthesis from the C1-THF– dependent pathway. In addition, our labeling data suggest that serine and histidine are synthesized in almost equal proportions by other competing pathways. Moreover, genome annotation confirmed the presence of the following genes belonging to the C1-THF pathway: SLG_ 02620 (cobalamin-independent methionine synthase); SLG_12750 (5,10-methylenetetrahydrofolate reductase, metF); SLG_29740 (glycine hydroxymethyltransferase, glyA); SLG_12760 (10-formyltetrahydrofolate synthetase, ligH); SLG_03300 and SLG_20320 (methenyltetrahydrofolate cyclohydrolase, folD); and SLG_03300 and SLG_20320 (5,10-methylenetetrahydrofolate:NADP oxidoreductase). The C1 pathway deduced from our study for the synthesis of methionine and serine in SYK-6 is shown in Fig. 4A.
Fig. 4.
C1 metabolism in SYK-6. (A) The THF-dependent one-carbon metabolic pathway in SYK-6 for the synthesis of methionine, serine, and AMP. The demethylation step is shown for vanillic acid as an example. The carbon derived from the methyl group is shown in blue in each molecule to allow the tracking of this carbon in the formation of amino acids. The gene symbols in the figure are based on KEGG annotations. A detailed pathway for the synthesis of AMP is presented in SI Appendix, Fig. S1. (B) Pathway for the biosynthesis of histidine in SYK-6. Carbon from the C1-THF pathway enters into the histidine backbone through adenosine triphosphate that was synthesized from N10-formyl THF. The remaining five carbons for histidine synthesis come from R5P, an intermediate of the PPP. ATP is shown in blue to track the carbons that take part in histidine synthesis. The gene symbols in the figure are based on KEGG annotations. F6P, fructose-6-phosphate; GAP, glyceraldehyde-3-phosphate; P, phosphite (PO3) ion; R5P, ribose-5-phosphate.
Histidine obtained its labeled carbon from the C1-THF pathway via a more indirect route. SI Appendix, Fig. S1 shows that the labeled carbon of N10-formyl THF produced from the C1-THF pathway takes part in the synthesis of AMP in a pathway that has ribose-5-phosphate (R5P) as its primary precursor. AMP in turn takes part in the synthesis of ATP, thereby transferring its labeled carbon to ATP. The labeled ATPs contribute some of their labeled carbons to histidine biosynthesis as they take part in a nine-step metabolic pathway (Fig. 4B). However, the KEGG map for SYK-6 has a missing annotation for one gene (for the conversion of l-histidinol phosphate to l-histidinol) of the 10 genes that are required for the conversion of R5P to histidine, and the loci of this gene need to be identified. The labeled carbon in histidine that was derived from ATP is shown in blue in Fig. 4B.
Formic acid is a well-known C1 source for the C1-THF pathway (41, 42). To probe the C1 metabolism further, SYK-6 was fed with (i) pyruvate plus 13C formate and (ii) vanillin plus 13C formate. In the cells fed with 13C formate in the presence of pyruvate, only serine was labeled among the three amino acids that were found to be synthesized from C1-THF pathway. When SYK-6 was fed vanillin plus 13C formate, both serine and histidine were labeled, leaving methionine unlabeled (Table 2). These observations indicate that the carbon from formate was not used for methionine synthesis. The absence of methionine synthesis from formate could be caused either by the irreversibility of the reaction N5-methyl-THF → N5,N10-methylene-THF (catalyzed by 5,10-methylenetetrahydrofolate reductase) or by the very low activity of 5,10-methylenetetrahydrofolate reductase in the reverse direction, because it is NADH dependent (43). Also, when SYK-6 was fed with pyruvate plus 13C formate, few amino acids other than serine were discovered to be labeled and this pathway needs to be elucidated through future studies (Table 2 and SI Appendix, Table S1).
Table 2.
Mass isotopomer distribution of few key amino acids ([M-57]+) from 13C formate SYK-6 cultures
| Amino acid | 13C formate | ||||||
| Pyruvate | Vanillin | ||||||
| m0 | m1 | m2 | m0 | m1 | m2 | m3 | |
| Alanine | 0.96 ± 0.00 | 0.04 ± 0.00 | 0.97 ± 0.01 | 0.03 ± 0.01 | |||
| Glycine | 0.98 ± 0.01 | 0.02 ± 0.01 | 0.97 ± 0.01 | 0.03 ± 0.01 | |||
| Methionine | 0.94 ± 0.03 | 0.05 ± 0.01 | 0.01 ± 0.01 | 0.95 ± 0.01 | 0.04 ± 0.02 | 0.01 ± 0.01 | |
| Serine | 0.49 ± 0.01 | 0.50 ± 0.01 | 0.01 ± 0.01 | 0.73 ± 0.01 | 0.26 ± 0.00 | 0.01 ± 0.01 | |
| Glutamate | 0.88 ± 0.02 | 0.11 ± 0.02 | 0.01 ± 0.01 | 0.94 ± 0.02 | 0.05 ± 0.01 | 0.01 ± 0.01 | |
| Histidine | 0.95 ± 0.01 | 0.05 ± 0.01 | 0.57 ± 0.01 | 0.40 ± 0.01 | 0.02 ± 0.00 | 0.01 ± 0.01 | |
Cataplerotic Pathways of the Tricarboxylic Acid Cycle.
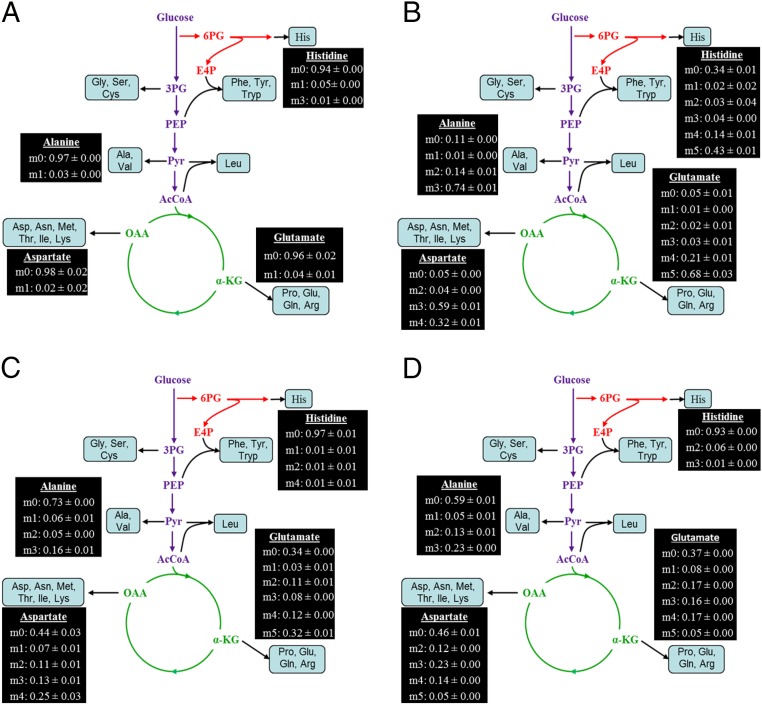
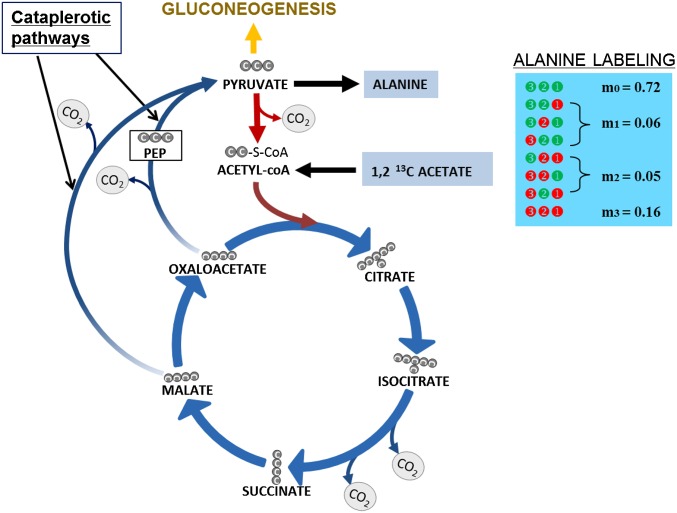
The anaplerotic/cataplerotic pathways play a significant role in regulating the flux of the tricarboxylic acid (TCA) cycle (44). The cataplerotic pathways convert the four-carbon intermediates of the TCA cycle, malate and oxaloacetate, to pyruvate and phosphoenolpyruvate (PEP), via two parallel routes. The first step in this path is a single-reaction pathway catalyzed by the malic enzyme that converts malate directly to pyruvate. The second cataplerotic pathway converts oxaloacetate into PEP. To confirm the cataplerotic pathway, SYK-6 cultures were grown in the presence of either 1-13C acetate or 1,2-13C acetate. There are two routes to convert acetate into pyruvate: The enzyme pyruvate synthase can drive the reductive carboxylation of acetyl-CoA to pyruvate (45) or acetate can enter the TCA cycle, and then cataplerotic pathways can pull the carbons from the TCA cycle into the pyruvate pool. If only pyruvate synthase is active, alanine cannot be labeled in all three of its carbons, because the first position of pyruvate would be an unlabeled carbon from CO2. However, cells fed with 1,2-13C acetate had an m3 of 0.16 for [M-57] alanine (Fig. 5). A more probable way for all of the three carbons of pyruvate (the alanine precursor) to be labeled is through the cataplerotic pathway. Furthermore, the MID data from 1-13C acetate–fed SYK-6 cultures provide additional proof of the presence of cataplerotic pathways in SYK-6 (Table 3). An m1 of 0.13 was present for [M-57] but dropped to 0.03 for [M-159], suggesting that about 10% of the pyruvate in SYK-6 had a carbon in the first position labeled via cataplerotic pathways.
Fig. 5.
Alanine labeling reveals the activity of cataplerotic pathways in SYK-6. The MID shown was obtained from SYK-6 cells fed with 1,2-13C acetate.
Table 3.
Mass isotopomer distribution of amino acids from cultures designed to explore the presence and activity of anaplerotic pathways in SYK-6
| Amino acid | TBDMS fragment | 1-13C acetate | 1,2-13C acetate | |||||||
| m0 | m1 | m2 | m0 | m1 | m2 | m3 | m4 | m5 | ||
| Glutamate | [M-57]+ | 0.40 ± 0.03 | 0.37 ± 0.01 | 0.23 ± 0.02 | 0.34 ± 0.00 | 0.03 ± 0.01 | 0.11 ± 0.01 | 0.08 ± 0.00 | 0.13 ± 0.01 | 0.32 ± 0.01 |
| [M-159]+ | 0.42 ± 0.01 | 0.57 ± 0.01 | 0.01 ± 0.01 | 0.35 ± 0.01 | 0.06 ± 0.01 | 0.12 ± 0.00 | 0.12 ± 0.00 | 0.35 ± 0.00 | ||
| Aspartate | [M-57]+ | 0.51 ± 0.01 | 0.47 ± 0.00 | 0.02 ± 0.01 | 0.44 ± 0.03 | 0.07 ± 0.01 | 0.11 ± 0.00 | 0.13 ± 0.01 | 0.25 ± 0.03 | |
| [M-159]+ | 0.74 ± 0.01 | 0.26 ± 0.01 | 0.43 ± 0.01 | 0.10 ± 0.01 | 0.17 ± 0.00 | 0.30 ± 0.01 | ||||
| Alanine | [M-57]+ | 0.87 ± 0.00 | 0.13 ± 0.00 | 0.73 ± 0.00 | 0.06 ± 0.01 | 0.05 ± 0.00 | 0.16 ± 0.01 | |||
| [M-159]+ | 0.95 ± 0.02 | 0.03 ± 0.00 | 0.02 ± 0.02 | 0.73 ± 0.00 | 0.08 ± 0.00 | 0.19 ± 0.00 | ||||
13C-MFA of Ligninolysis-Based Metabolism.
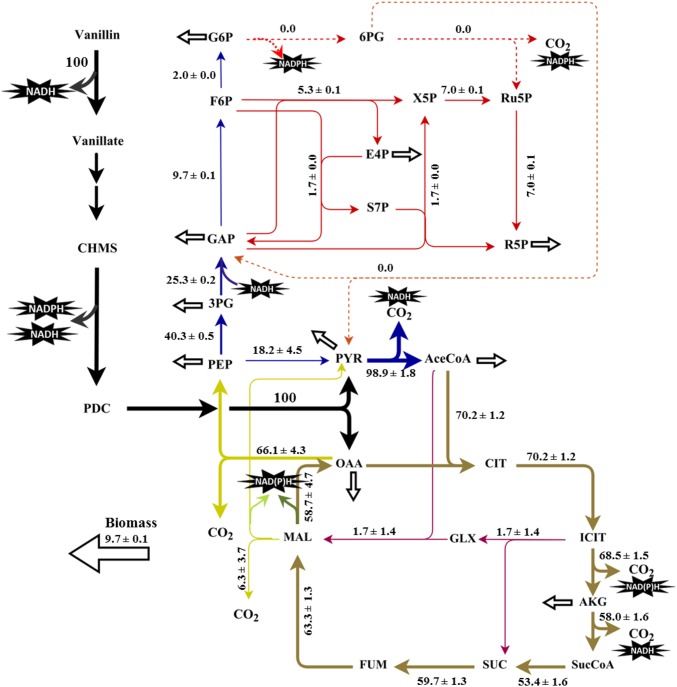
13C-MFA using parallel labeling experiments with vanillin was conducted to shed light on the central metabolism of Sphingobium sp. SYK-6. The flux map is presented in Fig. 6. In Sphingobium sp. SYK-6, a vanillin molecule is broken down to a pyruvate, an oxaloacetate, and a C1 unit. Pyruvate and oxaloacetate are both key intermediates in the central carbon metabolism and are precursors to building blocks, such as aspartate and alanine. As a consequence, vanillin degradation leads to strong fluxes in the TCA cycle (>70 units) and gluconeogenesis pathways, especially in the oxaloacetate→PEP reaction. Because of the highly negative Gibbs free energy (ΔGΘ = −31.7kJ/mol) of the PEP→pyruvate reaction, pyruvate cannot be favorably converted to PEP directly (46). Therefore, the ligninolysis coproduct pyruvate must be rerouted through the TCA cycle to start the gluconeogenesis from oxaloacetate. As shown in the flux map, more than 66 flux units are observed for oxaloacetate conversion via PEP carboxykinase (i.e., the rate-controlling step of gluconeogenesis). Moreover, the glyoxylate shunt had a minimal flux that cannot reduce carbon loss from the TCA cycle. Additionally, the flux (approximately five units) through the nonoxidative pentose phosphate pathway (PPP) is indispensable and provides building blocks such as erythose-4-phosphate and R5P for biomass synthesis. Last, Sphingobium sp. SYK-6 grown with vanillin shows ∼60% carbon loss as CO2 compared with the ∼30% carbon loss observed in glucose-fed Escherichia coli (31). The greater CO2 loss indicates that the energy requirement is more demanding, and therefore biomass synthesis is less efficient in SYK-6 than in E. coli.
Fig. 6.
Metabolic flux distribution of Sphingobium sp. SYK-6 growing on vanillin in W medium. The flux values are all relative to the vanillin uptake rate. 3PG, 3-phosphoglycerate; 6PG, 6-phosphogluconate; AceCoA, acetyl-CoA; AKG, α-ketoglutarate; CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; CIT, citrate; E4P, erythrose 4-phosphate; F6P, fructose 6-phosphate; FUM, fumarate; G6P, glucose 6-phosphate; GAP, glyceraldehyde 3-phosphate; GLX, glyoxylate; ICT, isocitrate; MAL, malate; OAA, oxaloacetate; PDC, 2-pyrone-4,6-dicarboxylate; PEP, phosphoenolpyruvate; PYR, pyruvate; R5P, ribose 5-phosphate; Ru5P, ribulose-5-phosphate; S7P, sedoheptulose-7-phosphate; SER, serine; SUC, succinate; SucCoA, succinyl-CoA; X5P, xylulose-5-phosphate.
For energy metabolism, the TCA cycle provides rich sources of reducing power via NADH, which is necessary for powering gluconeogenesis and oxidative respirations for ATP generation. However, key enzymes of the OPP pathway are not annotated, and the absence of the OPP pathway suggests that cell metabolism must find an alternative route for NADPH production. Consequently, the ligninolysis pathway becomes indispensable, because it produces both NADH and NADPH. Its NADPH production derives mainly from the dehydrogenation reaction of vanillin to vanillate (catalyzed by vanillin dehydrogenase, LigV) and the enzymatic conversion of 4-carboxy-2-hydroxymuconate-6-semialdehyde (CHMS) to 2-pyrone-4,6-dicarboxylate (PDC) (catalyzed by CHMS dehydrogenase, LigC) (25, 47). The essential role of the vanillin-degrading pathway explains the inability of SYK-6 to grow well using pyruvate or acetate. Unlike glycolysis, which supplies both ATP and NADH, a strong gluconeogenesis activity results in the consumption of ATP and NADH. If we assume that SYK-6 and E. coli require the same amount of ATP for biomass synthesis (31), the net ATP requirement from respirations for Sphingobium sp. SYK-6 is even higher. The ATP gap is compensated by oxidizing NADH, which is generated mainly from the TCA cycle and vanillin degradation.
RNA-Seq Analysis of the Differential Expression of Genes Involved in NAD(P)H Metabolism.
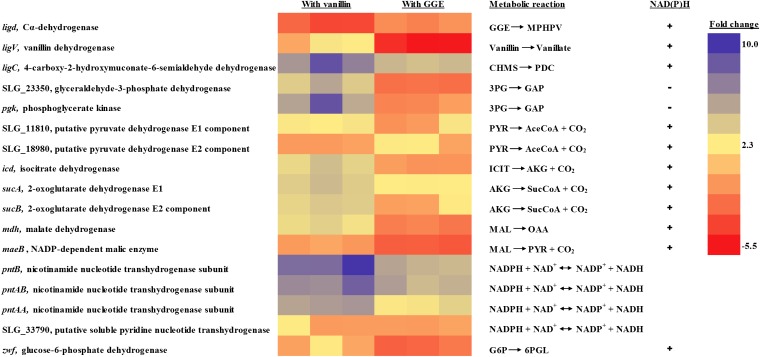
RNA-seq is a highly sensitive and accurate technique for measuring the expression levels of various genes at the transcriptome level and can be used to validate our findings from the fluxomics study. Therefore, RNA-seq data analysis was performed with SYK-6 cells grown in the presence of one of the following substrates: vanillin, guaiacylglycerol-β-guaiacyl ether (GGE), or pyruvate. The dimeric compound GGE was chosen over other dimers for this analysis because the β–O-4 linkage represents the most abundant bond present in lignin (Fig. 1). The heat map for the differential expression of genes involved in NAD(P)H metabolism is shown in Fig. 7. The fold changes in the expression of genes are reported for cells grown in the presence of vanillin (a monomeric product of lignin) or GGE (a dimeric product of lignin) vs. cells grown in the presence of pyruvate. In comparison with the genes of the central metabolic pathway involved in the generation of NAD(P)H, the CHMS dehydrogenase gene ligC, belonging to the vanillin catabolic pathway, was expressed at the highest level in the presence of both vanillin and GGE. However, the expression levels of the TCA cycle genes (icd, sucA, sucB, and mdh) were less than 50% of ligC. In addition, the expression level of the cataplerotic pathway gene maeB (malic enzyme) was lower than that of mdh (malate dehydrogenase). This observation is in agreement with the results of the 13C-MFA showing that the conversion of TCA cycle metabolites into gluconeogenesis intermediates happens largely via oxaloacetate (OAA) and not directly from malate. The orphan gene of the OPP pathway, zwf, had some level of expression in the presence of vanillin; therefore it is unclear whether this lone gene of the OPP pathway is involved in NADPH production. The nicotinamide nucleotide transhydrogenases play a crucial role in the energy metabolism of a cell through the interconversion of NADH and NADPH. With the absence of a complete OPP pathway in SYK-6, the transhydrogenases must play a more important role in SYK-6 in the generation of NADPH from NADH. The RNA-seq data confirm this hypothesis with the detection of high levels of expression for three of the transhydrogenases—pntB, pntAB, and pntAA—in the presence of vanillin and GGE.
Fig. 7.
Heat map of differentially expressed genes involved in NAD(P)H metabolism of Sphingobium SYK-6 based on RNA-seq data analysis. The fold changes are reported for cells grown in the presence of vanillin (a monomeric product of lignin) or GGE (a dimeric product of lignin) vs. cells grown in the presence of pyruvate (a carbon source lacking the vanillin catabolic pathway). A plus or minus sign indicates that the corresponding gene is involved in the production or consumption of NAD(P)H, respectively. The results are shown in triplicate for each carbon source, and each column represents the results from one of the biological replicates. 3PG, 3-phosphoglycerate; 6PGL, 6-phosphogluconolactone; CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; PDC, 2-pyrone-4,6-dicarboxylate; AceCoA, acetyl-CoA; AKG, α-ketoglutarate; G6P, glucose 6-phosphate; GAP, glyceraldehyde 3-phosphate; GGE, Guaiacylglycerol-β-guaiacyl ether; ICIT, isocitrate; MAL, malate; MPHPV, α-(2-methoxyphenoxy)-β-hydroxypropiovanillone; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PYR, pyruvate; SucCoA, succinyl-CoA.
The mRNA for the phosphoglycerate kinase gene (pgk) of the gluconeogenesis pathway that uses an NADH for the conversion of 3-phosphoglycerate (3PG) into glyceraldehyde 3-phosphate (GAP) was also found at high levels (shown in dark blue in Fig. 7) in the presence of vanillin. With a highly active gluconeogenesis pathway, the cells need an extra source for NADH generation apart from the TCA cycle, and therefore the lignin catabolic pathway is indispensable for the healthy survival of SYK-6. The RNA-seq analysis provides further strong evidence that the lignin catabolic pathway plays a major role in supplying reducing equivalents for SYK-6, taking over the TCA cycle that is commonly the major supplier of NADH in most microbes.
Discussion
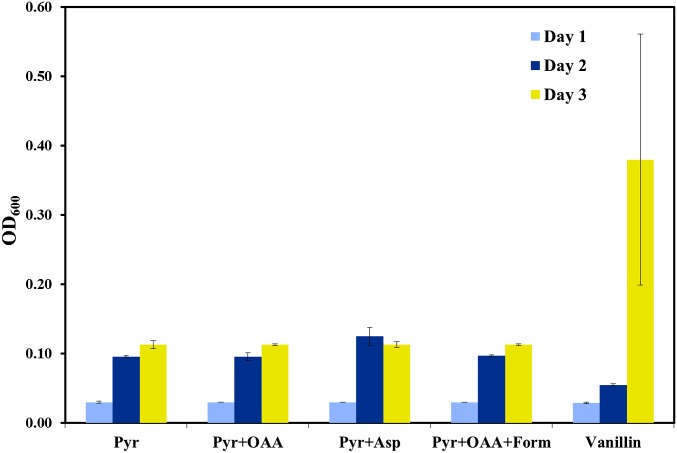
In this study, we have discovered several traits of SYK-6 metabolism by using a rapid 13C-based technique, 13C-fingerprinting. The experiments proved that SYK-6 lacks the ability to use glucose and xylose as its carbon source even with the supplementation of yeast extract. Vanillin was found to be the most preferred carbon substrate by SYK-6 among the different substrates that were tried in this study. Pyruvate, acetate, and formate were also tested with SYK-6 and were found to be metabolized to a small extent. Experiments conducted to verify the influence of initial medium pH on the growth of SYK-6 in the presence of pyruvate, vanillin, and vanillic acid suggested that SYK-6 is highly tolerant of high pH conditions (Fig. 3). To verify if cosubstrate metabolism is required for the growth of SYK-6, experiments were conducted that included oxaloacetate (the other catabolic product of vanillin), aspartate (the amino acid precursor to oxaloacetate), and formic acid (C1 carbon) along with pyruvate. However, none of these cosubstrates encouraged SYK-6 to grow at levels similar to those observed with vanillin as the carbon source (Fig. 8). This result further confirms that ligninolysis is an essential pathway for the cell to obtain its reducing equivalents.
Fig. 8.
Growth of SYK-6 in the presence of multiple substrates. This experiment was designed to verify the cosubstrate metabolism of SYK-6. However, the addition of other carbon intermediates to cultures containing pyruvate did not improve the growth of SYK-6. This experiment supports the need for the vanillin catabolic pathway to provide the extra NAD(P)H to boost the growth of SYK-6. Asp, aspartate; Form, formate; OAA, oxaloacetate; Pyr, pyruvate.
The ligninolysis pathways, such as the β-aryl ether cleavage pathway, the biphenyl cleavage pathway, and others, result in the formation of either vanillin or vanillic acid (23). Therefore, vanillin is at a pivotal location in the lignin catabolic pathways of SYK-6, and a study based on vanillin metabolism is important step toward lignin valorization. The labeling experiments revealed that the C1-THF pathway contributes C1 units for the synthesis of methionine, serine, and histidine. Methionine was synthesized from homocysteine and N5-methyl-THF catalyzed by a cobalamin-independent methionine synthase. Serine can be synthesized either from 3-phosphoglycerate or from glycine and N5,N10-methylene-THF. Furthermore, RNA-seq data obtained for cells grown with vanillin reveal that the genes of the C1-THF pathway are well expressed in relation to ligC, a critical gene of the vanillin catabolic pathway (SI Appendix, Fig. S2). The presence of carbon from the C1-THF pathway in histidine confirmed the presence of an active nonoxidative PPP. The oxidative portion of the PPP in SYK-6 is incomplete, and only the first gene responsible for the conversion of glucose-6-phosphate into 6-phosphoglucono-lactone is known for SYK-6. There are a few other organisms that lack the OPP pathway. For example, Methanocaldococcus jannaschii is a methanogenic archaea that lacks an active PPP but uses a ribulose monophosphate pathway for the synthesis of ribulose-5-phosphate and R5P (48). Because the PPP is the major source of NADPH in bacteria, this finding is important for understanding SYK-6 metabolism. Also, the labeling experiments show no evidence for a functional Entner–Doudoroff (ED) pathway in SYK-6. The genome, however, confirms the presence of the two genes of the ED pathway, edd and eda, coding for phosphogluconate dehydratase and 2-keto-3-deoxygluconate 6-phosphate aldolase, respectively. These two genes are required for the conversion of 6-phosphogluconate into pyruvate and glyceraldehyde 3-phosphate. However, the OPP gene required for the formation of 6-phosphogluconate from 6-phosphogluconolactone is unannotated in SYK-6. Therefore, the ED pathway is essentially like an island in the metabolic pathway map of SYK-6. Also, in comparison with some of the essential genes in SYK-6, the transcript levels of both edd and eda were very low (SI Appendix, Fig. S2). Last, cataplerotic pathways and gluconeogenesis that act as a liaison between the intermediates of the TCA cycle and the OPP pathway were found to be active.
The reduced form of NADH is a crucial cofactor for the survival of a cell because it drives several metabolic reactions, maintains the normal redox status of the cell, and is involved in energy metabolism and production (49, 50). Therefore, a cell with active gluconeogenesis but lacking a glycolysis pathway must be under huge stress because of the heavy burden placed on its NADH pool. The 13C-MFA results in our study also affirm that the vanillin catabolism and the central pathways produces excessive NAD(P)H, which can be oxidized for ATP synthesis to satisfy cell growth, maintenance, and ligninolytic enzyme production. From this perspective, the presence of ligninolytic pathways in SYK-6 not only may have provided SYK-6 with a pathway for the utilization of a carbon source that would be rejected by other microbes but also might have relieved the stress experienced by the cell from cofactor imbalance by producing two NAD(P)H from vanillin. In addition, some of the catabolic pathways from the breakdown of lignin oligomers to vanillin formation also produce NADH, adding to the NADH pool. As an example, the formation of α-(2-methoxyphenoxy)-β-hydroxypropiovanillone from GGE (a β-aryl ether model compound) in the ligninolytic pathway involves the formation of one NADH (23). A low cell density observed in SYK-6 fed with either pyruvate or acetate also may be caused by the absence or inactivity of ligninolytic pathway that provides NAD(+)/NADH balance.
Last, this study provides insights into the energy metabolism of SYK-6. In common heterotrophic microbial species, these catabolic pathways (EMP, OPP, ED, and the TCA cycle) are major sources of NAD(P)H that tightly regulate cell functions and biosynthesis. However, SYK-6 employs an entirely different strategy by using the ligninolysis pathway as a major route to obtain NADH and NADPH, replacing the OPP pathway and glycolysis routes. Such unique energy generation from a recalcitrant substrate not only may play a key role for in the cell’s survival but also may offer guidelines for SYK-6 engineering, which must carefully account for cofactor balances during the reorganization of cell fluxomes. We hypothesize that vanillin has the following advantages as a carbon source for SYK-6: (i) It is a carbon source that is least preferred by other microbes and thus SYK-6 encounters less competition in the environment; (ii) it provides intermediates (pyruvate and oxaloacetate) of a central metabolic pathway without any carbon loss; and (iii) it generates reducing equivalents to compensate for the absence of glycolysis and the presence of gluconeogenesis.
Conclusion
Gene annotations do not always reflect the actual functions of the corresponding pathways. To decipher the ligninolysis-based metabolism in Sphingobium sp. SYK-6, this study aimed to unravel its fluxome for catabolizing oligomers derived from recalcitrant heterogeneous polymeric lignin. Using 13C-fingerprinting, 13C-MFA, and RNA-seq data analysis, we have deciphered both carbon and energy metabolisms in SYK-6. This work sheds light on the unique physiologies of a ligninolytic bacterium using an uncommon aromatic substrate, which is particularly innovative because none of the previous fluxomics studies have used a lignin-derived carbon substrate. Based on the fluxome results, we determine that the lignin catabolic pathway plays a critical role in maintaining the NAD(+)/NADH balance in SYK-6 along with ATP production. Therefore, any efforts to engineer SYK-6 to synthesize high-value product from lignin must ensure that the redox state of the cell remains unaffected. For example, knocking out genes of the vanillin catabolic pathway may not be a straightforward way to accumulate vanillin or vanillic acid, because doing so would interfere severely with the cell’s energy metabolism. Instead, knocking down the vanillin catabolic pathway or combining the knockout of the vanillin catabolic pathway with overexpression of the OPP pathway genes to balance the cofactors may prove to be productive approaches for lignin valorization but needs to be validated through further research. The pioneering research work reported here provides an unprecedented understanding of the physiology of SYK-6 and will be of immense value for future research on engineering SYK-6 as a microbial chassis for lignin valorization.
Materials and Methods
Chemicals, Culture Medium, and Growth Conditions.
All chemicals and 13C-labeled carbon substrates were purchased from Sigma-Aldrich or Cambridge Isotope Laboratories as required. Sphingobium sp. strain SYK-6 (NBRC 103272) was grown in LB or W minimal salt medium at 30 °C in 125-mL or 250-mL glass flasks and the agitation of the shaker was maintained at 200 rpm. One liter of W medium consisted of 0.85 g KH2PO4, 4.9 g Na2HPO4⋅7H2O, 0.5 g (NH4)2SO4, 0.1 g MgSO4⋅7H2O, 9.5 mg FeSO4⋅7H2O, 10.75 mg MgO, 2 mg CaCO3, 1.44 mg ZnSO4·7H2O, 1.12 mg MnSO4⋅4H2O, 0.25 mg CuSO4⋅5H2O, 0.28 mg CoSO4⋅7H2O, and 0.06 mg H3BO4 (51). Growth of SYK-6 was monitored by measuring OD600 using a UV spectrophotometer. 16S ribosomal DNA (rDNA) sequencing of the strain was confirmed by sequencing the bacterial pellet by Genewiz Inc.
13C Cultures for Fingerprinting and Parallel Labeling Experiments.
A seed culture of Sphingobium SYK-6 was started in LB medium from the frozen glycerol stock. After 2 d of growth in the LB medium, the seed culture was transferred to W medium containing appropriate 13C carbon substrates (Table 1) at an inoculation ratio of 1% (vol/vol). The biomasses for the fingerprinting experiments were harvested 6 d after the culture was started in minimal medium. The parallel labeling experiment for the flux work was conducted as follows. SYK-6 cultures were grown in parallel in W medium containing three different 13C isotopes of vanillin: (i) 13C phenyl vanillin; (ii) 13C-aldehyde vanillin; and (iii) 50% 13C ring vanillin and also 50% 13C aldehyde vanillin. The exponentially growing cultures from the labeled medium were transferred to fresh 13C-labeled medium to minimize the influence of carryover from the LB medium. The biomass samples for amino acid analysis were collected during the exponential phase (SI Appendix, Fig. S3). The biomass samples were stored at −20 °C until further analysis.
Proteinogenic Amino Acid Analysis for Mass Isotopomer Distributions.
The proteinogenic amino acids from the biomass were hydrolyzed using 6-M HCl for 24 h at 100 °C. The amino acids were derivatized with N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide (TBDMS) with tetrahydrofuran as the solvent (52). The derivatized amino acids were analyzed for their 13C mass fraction by GC-MS using a Hewlett Packard 7890A gas chromatograph and an Agilent Technologies 5975C GS-MS system equipped with a DB5-MS column (J&W Scientific) (53). The m/z ions [M-57]+ or [M-15]+ containing information of the entire amino acid, [M-159]+ or [M-85]+ containing information of the amino acid after loss of its first carbon, and f(302)+ containing the first and second carbons of the amino acid backbone were used for isotopic tracing (M: the molecular mass of the derivatized amino acids). The fragment [M-15]+ was used only for leucine, because its [M-57]+ overlaps with other mass peaks (54). The presence of natural isotopes such as 13C (1.13%), 18O (0.20%), 29Si (4.70%), and 30Si (3.09%) contributes background noise to the mass isotopomer spectrum. Therefore the MIDs were corrected for the natural stable isotopes using a published algorithm; the detailed protocol can be found in ref. 55.
13C-MFA.
The 13C-MFA model was constructed in MATLAB R2015b, using the elementary metabolite units (EMU) algorithm (56) to simulate the labeling patterns of proteinogenic amino acids. Our model also accounted for the reversibility of reactions (57). The metabolic reactions are listed in SI Appendix, Table S2. The built-in function of MATLAB, fmincon, was used to minimize the differences between simulation and experiment results, i.e., the value of the objective function. To avoid local minima, more than 100 random initial guesses were tested, and the best fit was chosen as the minimum value of the objective function. SI Appendix, Fig. S4 shows the comparison between experimentally obtained MIDs and the computationally obtained MIDs. The confidence interval was calculated using the Monte Carlo method (58) in which the MID data were perturbed with normally distributed noise (µ = 0, δ = 0.01) 1,000 times, and a new set of flux predictions was generated each time. Our model was constructed to find the best fit for the MIDs obtained from the SYK-6 cells grown simultaneously on three different labeled vanillin substrates. The MATLAB software for flux analysis (WUFlux) can be downloaded from the website (www.13cmfa.org/).
RNA-Seq and Differential Expression Analysis.
A seed culture of Sphingobium SYK-6 was started in fresh LB medium from a frozen glycerol stock. One milliliter of the overnight culture was transferred into 20 mL of fresh LB medium and cultured at 30 °C, 220 rpm until it reached an OD600 of 1.0–1.2. The cultures were collected and spun down at 5,000 rpm at 4 °C for 5 min. The cell pellets were rinsed three times with W medium and were resuspended in 20 mL of W medium containing one of the following carbon substrates: vanillin, GGE, or pyruvate. Cultures with each carbon substrate were grown in triplicate. After 6 h of growth in the minimal medium, the cultures were centrifuged to remove the supernatant. The cell pellets were frozen immediately in liquid nitrogen for 20 min and stored at −80 °C.
RNA library preparation, sequencing, and data analysis were performed by SeqWright Genomic Services (GE Healthcare). In brief, the frozen pellets were ground by a mortar and pestle containing liquid nitrogen, and the total RNA was extracted using TRIzol (Invitrogen) reagent as previously described (59, 60). The extracted total RNA was purified using the RNeasy mini kit (Qiagen). The quality and integrity of the total RNA were examined using an Agilent 2100 Bioanalyzer. The RNA library was constructed using the TruSeq RNA Library Preparation Kit (Illumina) and sequenced using a NextGen Sequencing machine (Illumina). The adapter sequences were trimmed from the sequence data using Trim galore v0.3.3. The sequences then were aligned to the reference genome of SYK-6 (downloaded from Joint Genome Institute, https://gold.jgi.doe.gov/) using Tophat 2.0.8b, and gene-expression differential analysis was performed using Partek Genomics Suite 6.6.
Supplementary Material
Acknowledgments
We thank Joint BioEnergy Institute for the SYK6 strain, Yu Yi for the technical discussions, and Whitney Hollinshead for her help with the GCMS. This project was supported by Laboratory Directed Research and Development Program 16-0758 of Sandia National Laboratories to Principal Investigator S.S. Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the US Department of Energy's National Nuclear Security Administration under Contract DE-AC04-94AL85000. L.H. was supported by NSF Grant DBI 1356669.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606043113/-/DCSupplemental.
References
- 1.Szczodrak J, Fiedurek J. Technology for conversion of lignocellulosic biomass to ethanol. Biomass Bioenergy. 1996;10(5–6):367–375. [Google Scholar]
- 2.Simmons BA, Loque D, Blanch HW. Next-generation biomass feedstocks for biofuel production. Genome Biol. 2008;9(12):242. doi: 10.1186/gb-2008-9-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu C, Arancon RAD, Labidi J, Luque R. Lignin depolymerisation strategies: Towards valuable chemicals and fuels. Chem Soc Rev. 2014;43(22):7485–7500. doi: 10.1039/c4cs00235k. [DOI] [PubMed] [Google Scholar]
- 4.Varanasi P, et al. Survey of renewable chemicals produced from lignocellulosic biomass during ionic liquid pretreatment. Biotechnol Biofuels. 2013;6(1):14. doi: 10.1186/1754-6834-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuck CO, Pérez E, Horváth IT, Sheldon RA, Poliakoff M. Valorization of biomass: Deriving more value from waste. Science. 2012;337(6095):695–699. doi: 10.1126/science.1218930. [DOI] [PubMed] [Google Scholar]
- 6.Ragauskas AJ, et al. Lignin valorization: Improving lignin processing in the biorefinery. Science. 2014;344(6185):1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- 7.Wilkerson CG, et al. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science. 2014;344(6179):90–93. doi: 10.1126/science.1250161. [DOI] [PubMed] [Google Scholar]
- 8.Chapple C, Ladisch M, Meilan R. Loosening lignin’s grip on biofuel production. Nat Biotechnol. 2007;25(7):746–748. doi: 10.1038/nbt0707-746. [DOI] [PubMed] [Google Scholar]
- 9.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25(7):759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 10.Zakzeski J, Jongerius AL, Bruijnincx PC, Weckhuysen BM. Catalytic lignin valorization process for the production of aromatic chemicals and hydrogen. ChemSusChem. 2012;5(8):1602–1609. doi: 10.1002/cssc.201100699. [DOI] [PubMed] [Google Scholar]
- 11.Socha AM, et al. Efficient biomass pretreatment using ionic liquids derived from lignin and hemicellulose. Proc Natl Acad Sci USA. 2014;111(35):E3587–E3595. doi: 10.1073/pnas.1405685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruijnincx PCA, Weckhuysen BM. Biomass conversion: Lignin up for break-down. Nat Chem. 2014;6(12):1035–1036. doi: 10.1038/nchem.2120. [DOI] [PubMed] [Google Scholar]
- 13.Rahimi A, Ulbrich A, Coon JJ, Stahl SS. Formic-acid-induced depolymerization of oxidized lignin to aromatics. Nature. 2014;515(7526):249–252. doi: 10.1038/nature13867. [DOI] [PubMed] [Google Scholar]
- 14.Linger JG, et al. Lignin valorization through integrated biological funneling and chemical catalysis. Proc Natl Acad Sci USA. 2014;111(33):12013–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picart P, et al. From gene towards selective biomass valorization: Bacterial β-etherases with catalytic activity on lignin-like polymers. ChemSusChem. 2014;7(11):3164–3171. doi: 10.1002/cssc.201402465. [DOI] [PubMed] [Google Scholar]
- 16.Dashtban M, Schraft H, Syed TA, Qin W. Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol. 2010;1(1):36–50. [PMC free article] [PubMed] [Google Scholar]
- 17.Deangelis KM, et al. Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1. Front Microbiol. 2013;4:280. doi: 10.3389/fmicb.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abd-Elsalam HE, El-Hanafy AA. Lignin biodegradation with ligninolytic bacterial strain and comparison of Bacillus subtilis and Bacillus sp. isolated from Egyptian soil. Am Eurasian J Agric Environ Sci. 2009;5:39–44. [Google Scholar]
- 19.Bandounas L, Wierckx NJP, de Winde JH, Ruijssenaars HJ. Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol. 2011;11:94. doi: 10.1186/1472-6750-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol. 2011;22(3):394–400. doi: 10.1016/j.copbio.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Brown ME, Chang MCY. Exploring bacterial lignin degradation. Curr Opin Chem Biol. 2014;19:1–7. doi: 10.1016/j.cbpa.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Masai E, et al. Complete genome sequence of Sphingobium sp. strain SYK-6, a degrader of lignin-derived biaryls and monoaryls. J Bacteriol. 2012;194(2):534–535. doi: 10.1128/JB.06254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem. 2007;71(1):1–15. doi: 10.1271/bbb.60437. [DOI] [PubMed] [Google Scholar]
- 24.Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep. 2011;28(12):1883–1896. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- 25.Masai E, et al. Characterization of ligV essential for catabolism of vanillin by Sphingomonas paucimobilis SYK-6. Biosci Biotechnol Biochem. 2007;71(10):2487–2492. doi: 10.1271/bbb.70267. [DOI] [PubMed] [Google Scholar]
- 26.Cordova LT, Antoniewicz MR. 13C metabolic flux analysis of the extremely thermophilic, fast growing, xylose-utilizing Geobacillus strain LC300. Metab Eng. 2015;33:148–157. doi: 10.1016/j.ymben.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Au J, Choi J, Jones SW, Venkataramanan KP, Antoniewicz MR. Parallel labeling experiments validate Clostridium acetobutylicum metabolic network model for (13)C metabolic flux analysis. Metab Eng. 2014;26:23–33. doi: 10.1016/j.ymben.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Ma F, Jazmin LJ, Young JD, Allen DK. Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc Natl Acad Sci USA. 2014;111(47):16967–16972. doi: 10.1073/pnas.1319485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goudar C, et al. Metabolic flux analysis of CHO cells in perfusion culture by metabolite balancing and 2D [13C, 1H] COSY NMR spectroscopy. Metab Eng. 2010;12(2):138–149. doi: 10.1016/j.ymben.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Yim H, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7(7):445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 31.He L, et al. Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol Bioeng. 2014;111(3):575–585. doi: 10.1002/bit.25124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker J, Zelder O, Häfner S, Schröder H, Wittmann C. From zero to hero--design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab Eng. 2011;13(2):159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Varman AM, He L, You L, Hollinshead W, Tang YJ. Elucidation of intrinsic biosynthesis yields using 13C-based metabolism analysis. Microb Cell Fact. 2014;13(1):42. doi: 10.1186/1475-2859-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang JK, You L, Blankenship RE, Tang YJ. Recent advances in mapping environmental microbial metabolisms through 13C isotopic fingerprints. J R Soc Interface. 2012;9(76):2767–2780. doi: 10.1098/rsif.2012.0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masai E, et al. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol. 2004;186(9):2757–2765. doi: 10.1128/JB.186.9.2757-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol. 2005;187(6):2030–2037. doi: 10.1128/JB.187.6.2030-2037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunel F, Davison J. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol. 1988;170(10):4924–4930. doi: 10.1128/jb.170.10.4924-4930.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaufmann F, Wohlfarth G, Diekert G. O-demethylase from Acetobacterium dehalogenans--substrate specificity and function of the participating proteins. Eur J Biochem. 1998;253(3):706–711. doi: 10.1046/j.1432-1327.1998.2530706.x. [DOI] [PubMed] [Google Scholar]
- 39.Sonoki T, et al. Tetrahydrofolate-dependent vanillate and syringate O-demethylation links tightly to one-carbon metabolic pathway associated with amino acid synthesis and DNA methylation in the lignin metabolism of Sphingomonas paucimobilis SYK-6. J Wood Sci. 2002;48(5):434–439. [Google Scholar]
- 40.Bailey LB, Gregory JF., 3rd Folate metabolism and requirements. J Nutr. 1999;129(4):779–782. doi: 10.1093/jn/129.4.779. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Moore M, King J. Investigating the regulation of one-carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2003;44(3):233–241. doi: 10.1093/pcp/pcg029. [DOI] [PubMed] [Google Scholar]
- 42.Zhuang W-Q, et al. Incomplete Wood-Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc Natl Acad Sci USA. 2014;111(17):6419–6424. doi: 10.1073/pnas.1321542111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson DL, Nelson DL, Lehninger AL, Cox MM. Lehninger Principles of Biochemistry. W.H. Freeman; New York: 2008. [Google Scholar]
- 44.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277(34):30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 45.Furdui C, Ragsdale SW. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem. 2000;275(37):28494–28499. doi: 10.1074/jbc.M003291200. [DOI] [PubMed] [Google Scholar]
- 46.Zhao J, Shimizu K. Metabolic flux analysis of Escherichia coli K12 grown on 13C-labeled acetate and glucose using GC-MS and powerful flux calculation method. J Biotechnol. 2003;101(2):101–117. doi: 10.1016/s0168-1656(02)00316-4. [DOI] [PubMed] [Google Scholar]
- 47.Masai E, et al. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J Bacteriol. 2000;182(23):6651–6658. doi: 10.1128/jb.182.23.6651-6658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grochowski LL, Xu H, White RH. Ribose-5-phosphate biosynthesis in Methanocaldococcus jannaschii occurs in the absence of a pentose-phosphate pathway. J Bacteriol. 2005;187(21):7382–7389. doi: 10.1128/JB.187.21.7382-7389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teodoro JS, Rolo AP, Palmeira CM. The NAD ratio redox paradox: Why does too much reductive power cause oxidative stress? Toxicol Mech Methods. 2013;23(5):297–302. doi: 10.3109/15376516.2012.759305. [DOI] [PubMed] [Google Scholar]
- 50.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: An old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31(2):194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng X, et al. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl Environ Microbiol. 1998;64(7):2520–2527. doi: 10.1128/aem.64.7.2520-2527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varman AM, Yu Y, You L, Tang YJ. Photoautotrophic production of D-lactic acid in an engineered cyanobacterium. Microb Cell Fact. 2013;12(1):117. doi: 10.1186/1475-2859-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You L, et al. Metabolic pathway confirmation and discovery through (13)C-labeling of proteinogenic amino acids. J Vis Exp. 2012;(59):e3583. doi: 10.3791/3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng X, Tang KH, Blankenship RE, Tang YJ. Metabolic flux analysis of the mixotrophic metabolisms in the green sulfur bacterium Chlorobaculum tepidum. J Biol Chem. 2010;285(50):39544–39550. doi: 10.1074/jbc.M110.162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wahl SA, Dauner M, Wiechert W. New tools for mass isotopomer data evaluation in (13)C flux analysis: Mass isotope correction, data consistency checking, and precursor relationships. Biotechnol Bioeng. 2004;85(3):259–268. doi: 10.1002/bit.10909. [DOI] [PubMed] [Google Scholar]
- 56.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): A novel framework for modeling isotopic distributions. Metab Eng. 2007;9(1):68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiechert W, Siefke C, de Graaf AA, Marx A. Bidirectional reaction steps in metabolic networks: II. Flux estimation and statistical analysis. Biotechnol Bioeng. 1997;55(1):118–135. doi: 10.1002/(SICI)1097-0290(19970705)55:1<118::AID-BIT13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 58.Wiback SJ, Famili I, Greenberg HJ, Palsson BO. Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space. J Theor Biol. 2004;228(4):437–447. doi: 10.1016/j.jtbi.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Wu W, Hildebrand A, Kasuga T, Xiong X, Fan Z. Direct cellobiose production from cellulose using sextuple beta-glucosidase gene deletion Neurospora crassa mutants. Enzyme Microb Technol. 2013;52(3):184–189. doi: 10.1016/j.enzmictec.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 60.Wu W, Kasuga T, Xiong X, Ma D, Fan Z. Location and contribution of individual β-glucosidase from Neurospora crassa to total β-glucosidase activity. Arch Microbiol. 2013;195(12):823–829. doi: 10.1007/s00203-013-0931-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.