Significance
Attachment to cellular glycans is a critical process in cell entry for several viruses. Antibodies that block this essential step can serve as neutralizing antibodies. Among human noroviruses (NoVs), serum antibodies that block histo-blood group antigen (HBGA) binding serve as correlates of protection. Escape from neutralization with evolving human NoVs (HuNoVs) through antigenic variation and differential HBGA binding is suggested to form a basis for the emergence of new strains. Currently, we are aware of no structural insights into antibody-mediated HBGA blockade or neutralization, or how emerging strains escape such neutralization. Our study reveals how a human IgA monoclonal antibody binds and blocks HBGA binding and indicates how other strains escape host immunity, laying the structural framework for understanding the immune correlates of protection against HuNoVs.
Keywords: norovirus, HBGA-blockade antibody, crystal structure, antibody neutralization, viral entry
Abstract
Human noroviruses (HuNoVs) cause sporadic and epidemic gastroenteritis worldwide. They are classified into two major genogroups (GI and GII), with each genogroup further divided into multiple genotypes. Susceptibility to these viruses is influenced by genetically determined histo-blood group antigen (HBGA) expression. HBGAs function as cell attachment factors by binding to a surface-exposed region in the protruding (P) domain of the capsid protein. Sequence variations in this region that result in differential HBGA binding patterns and antigenicity are suggested to form a basis for strain diversification. Recent studies show that serum antibodies that block HBGA binding correlate with protection against illness. Although genogroup-dependent variation in HBGA binding specificity is structurally well characterized, an understanding of how antibodies block HBGA binding and how genotypic variations affect such blockade is lacking. Our crystallographic studies of the GI.1 P domain in complex with the Fab fragment of a human IgA monoclonal antibody (IgA 5I2) with HBGA blocking activity show that the antibody recognizes a conformational epitope formed by two surface-exposed loop clusters in the P domain. The antibody engulfs the HBGA binding site but does not affect its structural integrity. An unusual feature of the antigen recognition by IgA 5I2 is the predominant involvement of the CDR light chain 1 in contrast to the commonly observed CDR heavy chain 3, providing a unique perspective into antibody diversity in antigen recognition. Identification of the antigenic site in the P domain shows how genotypic variations might allow escape from antibody neutralization and exemplifies the interplay between antigenicity and HBGA specificity in HuNoV evolution.
Human noroviruses (NoVs; HuNoVs) are the leading cause of viral gastroenteritis. They are associated with almost one fifth of all cases of acute gastroenteritis worldwide (1). It is estimated that ∼200,000 children under the age of 5 y die annually from HuNoV infections (2). Currently, there are no licensed vaccines or antiviral agents to treat the disease, although vaccine candidates are being investigated (3, 4). Development of efficient vaccines is limited by a lack of understanding of the immune correlates of protection and rapid evolution of NoVs based on antigenic variations and differential glycan binding.
NoVs are nonenveloped positive-strand RNA viruses belonging to the family Caliciviridae. They are phylogenetically classified into at least six genogroups (GI–GVI), with each genogroup divided into several genotypes. Genogroups GI, GII, and GIV contain human pathogens (5, 6). The prototype Norwalk virus (NV) is classified as genogroup I genotype 1 (i.e., GI.1). NoVs belonging to genotype GII.4 are the most prevalent and are associated with ∼70% of all HuNoV infections (7). HuNoVs recognize and bind to histo-blood group antigens (HBGAs) as receptors/coreceptors for cell entry. These glycoconjugates are also associated with susceptibility to HuNoV infection (8–10). HuNoVs bind HBGAs through their major capsid protein VP1, which, as 90 dimers, forms the T=3 icosahedral capsid (11). VP1, when expressed by itself in insect cells, self-assembles to form virus-like particles (VLPs) that are structurally and antigenically similar to the native virus. VP1 is composed of two principal domains, the shell domain, which is involved in the formation of the icosahedral shell, and the protruding (P) domain that projects out from the shell (11). The P domain is further divided into P1 and P2 subdomains, with the latter being an insertion in the P1 subdomain. Evolutionarily, the P2 subdomain is the least conserved and is implicated in strain diversity, differential HBGA binding, and antigenicity (12, 13).
HuNoVs are suggested to evolve through a coordinated interplay between differential HBGA binding specificities and antigenic variations that allow emerging strains to escape host immunity. Differential HBGA binding has been previously well characterized in GI and GII HuNoVs (14, 15). These studies show that both genogroups have evolved distinct HBGA binding sites localized on the outermost hypervariable P2 subdomain of VP1 (15–22). Human challenge studies show circulating serum antibodies that block HBGA binding correlate with protection from clinical disease and infection, and these antibodies have been proposed to serve as surrogate neutralizing Abs (NAbs) (4, 23–25). The presence of HBGA-blocking serum antibodies has also been associated with protection from infection in an i.v. challenge model in chimpanzees (26) and in the resolution of diarrhea in an immunocompromised patient with chronic gastroenteritis (27). Surrogate neutralization or HBGA-blockade assays have allowed identification of critical residues on VP1 that may be involved in NAb recognition (28). However, the lack of an efficient cell culture or small-animal model systems (29, 30) for HuNoVs has restricted the ability to define neutralization epitopes. This lack of information is in contrast to the fields of study of other viruses such as influenza virus, HIV, and dengue virus, in which immune correlates of protection and neutralization are better understood (31–33).
In the absence of any structural studies of HuNoV in complex with HBGA blockade antibodies, many critical questions remain unanswered, including how NAbs recognize and bind to VP1 and what the structural determinants of such binding are, what the mechanism of HBGA blockade is, whether binding induces conformational changes, and how antigenic variation allows escape from host immunity. Understanding the molecular basis of HuNoV–antibody interactions is critical for the design and development of genotype-specific and broadly reactive immunotherapeutic agents in the form of antibody scaffolds, and can also facilitate the development of vaccine candidates that elicit blockade antibodies. In this study, we determined the crystal structure of the Fab fragment of a potentially neutralizing human monoclonal antibody, IgA 5I2, in complex with the P domain of VP1 from NV. Our studies reveal that Fab 5I2 binds to a conformational epitope on the P2 subdomain and elucidates the molecular determinants of NV P domain–Fab 5I2 interactions. The work further provides structural insights into the mechanism of HBGA blockade and how the sequence and structural variations among the different GI genotypes could allow escape from recognition by IgA 5I2.
Results
Interaction of Fab 5I2 with the P Domain of NV.
Among HuNoVs, the surface-exposed P2 subdomain in the P domain of the capsid protein VP1 is implicated in differential HBGA binding and antigenicity. Although HBGA binding to the P domain has been characterized extensively, information about antigenicity, neutralization, and how these HuNoVs evolve to escape host immunity remains limited. To understand the basis of antibody binding and neutralization among HuNoVs, we purified the P domain of GI.1 NV and the Fab fragment of IgA 5I2, which was selected from a panel of antibodies produced from a hybridoma generated with B cells isolated from a person previously challenged with GI.1 NV (34). By using ELISA-based assays, this monoclonal antibody is shown to be genotype-specific, to bind to the P domain of GI.1 NV, and to block hemagglutination of erythrocytes expressing H type HBGA when preincubated with NV VLPs (34).
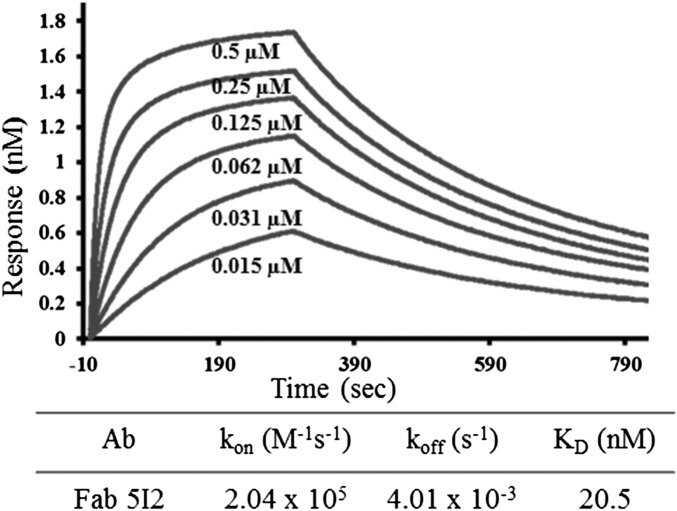
To assess the suitability of the Fab 5I2 and NV P domain complex for crystallographic studies, we first carried out binding studies using biolayer interferometry (BLI). In these studies, biotinylated P domain was immobilized on a streptavidin biosensor and titrated against serial dilutions of Fab 5I2. Data analysis showed that Fab 5I2 binds to the P domain with an affinity constant Kd of 20.5 nM and rate constants Kon of 2.04 × 105 M1·s−1 and Koff of 4.01 × 10−3 s−1 for association and dissociation, respectively (Fig. 1), indicating a tight interaction between Fab 5I2 and NV P domain.
Fig. 1.
Fab 5I2 binds tightly to NV P domain. BLI analysis of Fab 5I2 binding to NV P domain. P domain–Fab 5I2 association–dissociation curves were obtained through serial twofold dilutions of Fab 5I2 (0.5–0.015 µM) plus buffer controls by using Octet acquisition software. Sensograms for all concentrations are shown and labeled accordingly. The calculated KD, Kon, and Koff are shown in a tabular form.
Crystallographic Structure of Fab 5I2 in Complex with the NV P Domain.
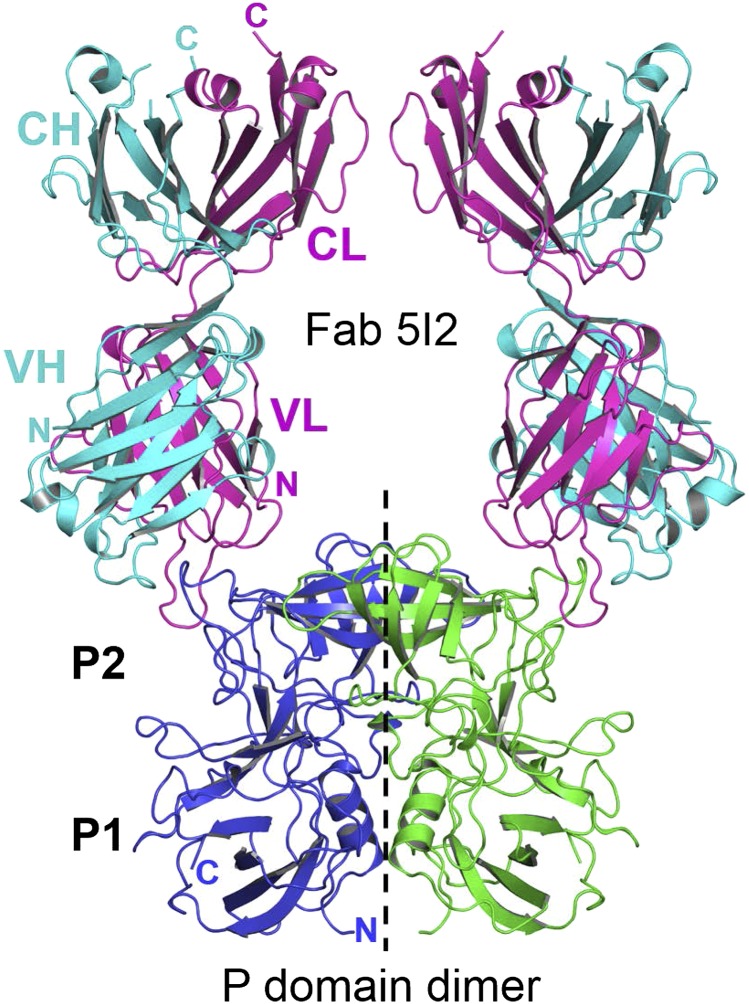
To understand the molecular details of how Fab 5I2 recognizes the NV P domain and what the mechanism of HBGA blockade is, we carried out crystallographic studies of the recombinant NV P domain (amino acids 229–519) in complex with the Fab 5I2. The P domain–Fab 5I2 complex crystals diffracted to ∼2.3 Å, and the structure was determined in the space group P6522, with one P domain–Fab complex in the crystallographic asymmetric unit. The structure of the complex was determined by using molecular replacement techniques and refined with final Rwork and Rfree values of 18% and 21%, respectively (Table 1). The P domains related by crystallographic twofold symmetry associate to form a dimer, as typically found in the NV capsid and other NoV P domain structures, with each of the dimeric subunits interacting separately with a Fab 5I2 molecule (Fig. 2). The Fab recognizes and interacts with a conformational epitope in the P2 subdomain. Superposition of the unbound and the Fab 5I2-bound P-domain structures showed that Fab binding does not alter the overall structure of the P domain (rmsd of 0.5 Å), but Fab binding does induce local conformation changes in some of the loop regions.
Table 1.
Data processing and refinement statistics for P domain–Fab 5I2 complex
| Space group | P6522 |
| Cell dimensions, Å | 162, 162, 146.81 |
| α,β,γ, ° | 90, 90, 120 |
| Resolution, Å | 81–2.3 (2.42–2.3) |
| Wavelength, Å | 0.97935 |
| No. of reflections | 563,010 |
| Unique reflections | 50,863 |
| R merge, % | 12.1 (82.9) |
| I/σI | 15.1 (3.00) |
| Completeness, % | 100 (100) |
| Redundancy | 11.1 (10.7) |
| R work, %c | 18.63 |
| R free, % | 21.23 |
| rmsd | |
| Bond lengths, Å | 0.003 |
| Bond angles, ° | 0.678 |
Numbers in parentheses correspond to highest-resolution shell.
Fig. 2.
Fab 5I2 recognizes a conformational epitope on top of the NV P domain. Cartoon representation of the overall structure of the Fab 5I2–P domain complex showing one Fab 5I2 molecule bound to each subunit of the P domain dimer. The individual P dimer subunits are shown in blue and green with the P1 and P2 subdomains labeled. The dashed black line indicates the twofold symmetry axis. Fab 5I2 is depicted with the heavy and light chains shown in cyan and magenta, respectively. The variable and constant domains of the Fab 5I2 light chain and heavy chain are labeled VL-CL and VH-CH respectively. The N- and C-terminal ends are also indicated for the P domain and Fab 5I2.
The overall structure of the bound Fab 5I2 is similar to those of other structurally characterized Fabs. The constant (CH and CL) and variable (VH and VL) domains of the heavy and light chains exhibit a typical Ig fold. The CH and CL interact closely with one another, as do the VL and VH. As expected, the three hypervariable complementarity determining regions (CDRs) from each heavy (CDR-H1, -H2, and -H3) and light chain (CDR-L1, -L2, -L3) in the variable domains are oriented facing the P2 subdomain. The CDR loops of Fab 5I2 are of varying lengths, with CDRH3 and CDRL1 being the longest, each consisting of 17 residues. Although the length of CDRH3 with 17 residues is typical, the 17-residue length of CDRL1 is unusual, and analyses of the interfacial interactions between the P domain and Fab show that CDRL1 plays a dominant role in antigen recognition.
Molecular Determinants of Antigen Recognition.
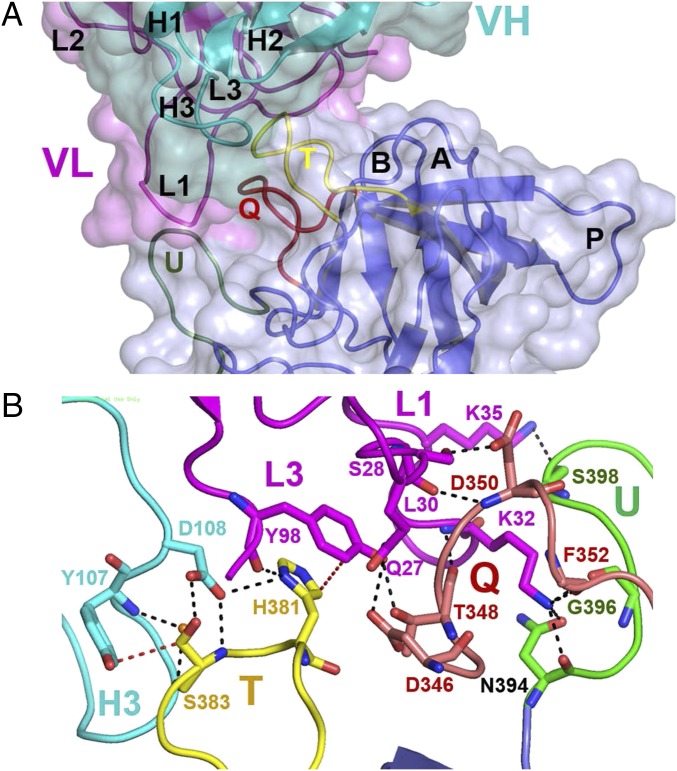
The epitope on the P2 subdomain that is recognized by Fab 5I2 is formed by residues from three surface-exposed loops: T (377–386), U (394–405), and Q (345–354; Fig. 3A). Some of these loop regions have been observed to have sequence and structural changes in other genogroups and within genotypes contributing to variations of HBGA binding specificities (13, 15, 21). The paratope of Fab 5I2 comprises three of the six CDRs, including CDRL1 (residues 24–40) and CDRL3 (residues 96–103) in the light chain and CDRH3 (residues 97–113) in the heavy chain (Fig. 3A). The P domain–Fab 5I2 interaction is achieved through several hydrogen bonding, electrostatic, and hydrophobic interactions involving a total buried surface area of ∼735 Å2 (Fig. 3B), consistent with the low nanomolar affinity indicated by binding assays (Fig. 1).
Fig. 3.
Detailed view of Fab 5I2–P domain interactions. (A) Close-up view of P domain (blue) bound to Fab 5I2 (heavy chain in cyan; light chain in magenta) depicted in surface and cartoon representation. All six CDRs, three from the light chain (L1–L3) and three from the heavy chain (H1–H3), were identified and labeled respectively. Similarly, six loop regions were identified on the P2 subdomain. Five of these loops have been previously identified in other genotypes and labeled according to the convention (loops A, B, P, T, and U). The sixth loop, labeled loop Q, was identified in this study. Three of these loops, loops T (yellow), Q (red), and U (green), form the conformational epitope recognized by the Fab 5I2. (B) Molecular details of Fab 5I2–P domain interactions. Fab 5I2 binds P domain through CDRs L1, L2, and H3 that make a network of hydrogen bonding (black dashed lines) and hydrophobic interactions (red dotted line) with the loops U, Q, and T of the P domain. CDRL1 makes the predominant interactions. All interacting loops are shown in cartoon representation with interacting residues shown as stick model per the aforementioned color convention, with nitrogen and oxygen atoms in blue and red, respectively.
Of the three CDRs, CDRL1 makes the most extensive interaction with the P domain. Its residues Q27, S28, L30, K32, and K35 contribute to eight hydrogen bonding interactions with the P domain residues N346, T348, D350, and F352 in loop Q and residues N394, G396, and S398 in loop U. CDRL1 also contributes to several water-mediated hydrogen bonding interactions. CDRL3 participates in the epitope recognition through its residues Y98 and I100. Y98 makes two hydrogen bonds, one with residue T348 in loop U and another with H381 in loop T. I100 is involved in a water-mediated hydrogen-bonding interaction with residue S380 in loop T. CDRH3 is the lone CDR from the heavy chain of Fab 5I2 that interacts with the P domain. Interactions involve its residues, Y107, and D108. Y107 is involved in hydrophobic and water-mediated hydrogen bond interactions with residue S383 in loop T of P domain, and D108 makes two direct hydrogen bonds with residue S383 on the T loop of the P domain (Fig. 3B).
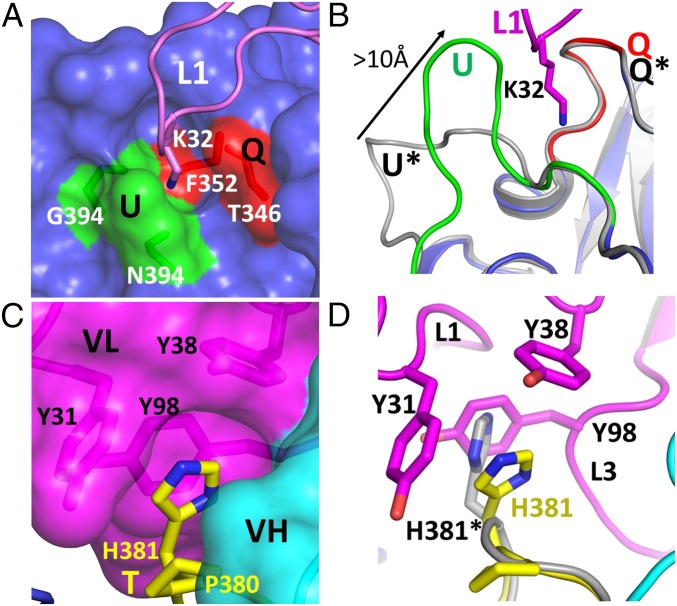
The P domain–Fab complex structure also exhibits interactions that contribute significantly to surface complementarity. First, the side chain of K32 from CDRL1 buries itself into a narrow, ∼8-Å-deep pocket on the surface of P domain, contributing to a network of hydrogen bonding interactions. Whereas the main-chain amide group of K32 hydrogen bonds with the main chain carbonyl group of T348 on the rim of the pocket, its side-chain hydrogen bonds with the main-chain carbonyl groups of F352, N394, and G396 lying at the bottom of the pocket (Fig. 4A). Second, the side chain of H381 from the P domain buries itself in a hydrophobic pocket on the surface of Fab 5I2 (Fig. 4C) making π–π stacking and cation–π interactions with the aromatic side chains of residues Y31 (CDRL1), Y38 (CDRL1), and Y98 (CDRL3) and hydrogen-bonding interaction with the main chain carbonyl group of Y98. This surface complementarity is enhanced by subtle conformational alterations in the P2 subdomain in response to Fab 5I2 binding. A comparison of the Fab 5I2-bound P domain structure with the P domain structure of the NV VLP [Protein Data Bank (PDB) ID 1IHM] (11) reveals that Fab binding induces conformational changes in loop U. This loop shifts by as much as 10 Å (at maximum Cα divergence) to make favorable interactions with CDRL1, including hydrogen bonding interactions described earlier involving K32 of CDRL1 (Fig. 4B). Similarly, the orientation of the side chain of H381 is flipped compared with its orientation in the unbound P domain structure (Fig. 4D). This flipping ensures that the H381 side chain does not sterically hinder the binding of Fab 5I2, and allows it to participate in favorable intramolecular stacking interactions with the side chain of P382 of the P domain.
Fig. 4.
Complementary surface residues involved in the Fab 5I2–P domain interaction. Presentation of complementary surfaces is important to antigen–antibody recognition and binding; two pockets, one on the P domain surface (A) and one on the Fab surface (C), were identified in the study and are shown to accommodate a complementary residue from the other molecule. (A) A pocket on the P domain surface (blue) buries a lysine 32 residue (pink stick model) contributed by the CDRL1 of Fab 5I2. The P domain residues, shown as sticks and labeled N394, G396 from loop U (green) and T346, F352 from loop Q (red), contribute to hydrogen bonding interactions with K32. (C) A similar pocket on the surface of Fab 5I2 (magenta) is shown to accommodate a histidine residue (H381; yellow stick model). H381 makes hydrophobic and stacking interactions with three tyrosine residues labeled Y31, Y38, and Y98 contributed by CDRs L1 and L3 of Fab 5I2. (B and D) Interestingly, superposition of the Fab 5I2-bound P domain structure and native NV VLP structure (PDB ID 1IHM; gray) shows that Fab binding induces local conformational changes on the P domain to make favorable interactions. (B) Loop U moves approximately 10 Å to make favorable interactions with CDRL1 and forms one side of the pocket that buries residue K32. Loop U from VLP is labeled in gray and marked with an asterisk. The movement of loop U is indicated by an arrow. (D) Similarly, Fab binding induces a flip in the orientation of the side chain of an H381, allowing it to make favorable hydrophobic and stacking interactions. In the native VLP structure, the side chain of H381 (gray; asterisk) would sterically clash with the Y98 residue of CDRL3.
Structural Basis for How IgA 5I2 Blocks HBGA Binding.
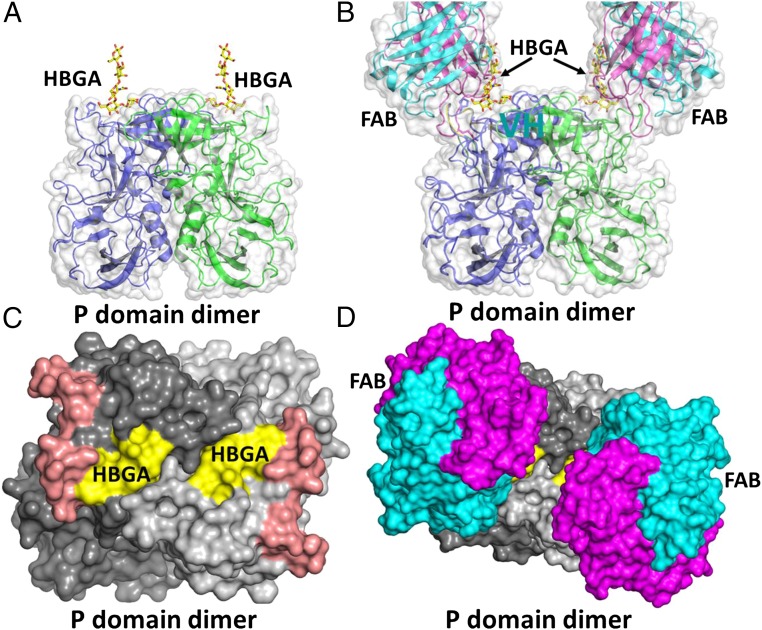
To gain insight into the mechanism of HBGA blockade or neutralization by IgA 5I2, we superimposed our Fab 5I2–P domain complex structure with an HBGA-bound NV P-domain structure (Fig. 5 A and B). This comparison shows that Fab 5I2 binds in close proximity to the primary HBGA binding site (Fig. 5B), but does not alter the structural integrity of the HBGA binding site, including the side chain orientations of the residues that participate in HBGA binding. It is clear that binding of Fab 5I2 sterically blocks HBGA from accessing the binding site (Fig. 5 C and D). Thus, HBGA blockade or neutralization by IgA 5I2 antibody is principally through steric hindrance as opposed to direct competition or disruption of the HBGA binding site.
Fig. 5.
Fab 5I2 blocks HBGA binding to P domain through steric hindrance. Superposition of HBGA-bound P domain (PDB ID 2ZL6) and our Fab 5I2-bound P domain structure reveals steric hindrance as the mechanism of HBGA blockade. (A) Surface representation (gray) and cartoon representation of the P domain dimer (side view) bound to H-type HBGA (yellow sticks). (B) Superposition of Fab- and HBGA-bound P domain structures clearly shows that Fab 5I2 will sterically hinder binding of HBGA. (C) Surface representation of the P-domain dimer (top view) highlighting the footprint of the HBGA binding site (yellow; labeled HBGA) and the Fab 5I2 epitope (brick red). (D) Superposition of the Fab-bound structure onto this P-domain structure shows that Fab binding limits access to the HBGA binding site, indicated by masking of the HBGA footprint (yellow).
Structural Basis for Why IgA 5I2 Is Genotype-Specific.
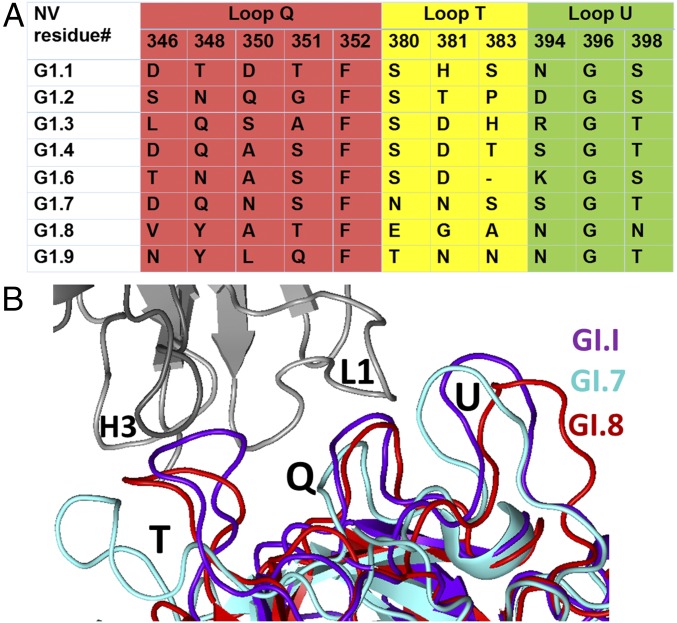
To understand the basis of the genotype specificity exhibited by IgA 5I2 and how other genotypes escape neutralization by IgA 5I2, we aligned the P-domain sequences of various GI genotypes, focusing on the residues that are involved in Fab 5I2 binding (Fig. 6A). Sequence alignment of these residues clearly shows that these residues are poorly conserved and might not be able to participate in the observed hydrogen bonding and hydrophobic interactions with Fab 5I2. Particularly interesting is residue H381 that, in our structure, is involved in highly stabilizing stacking interactions with residues from Fab 5I2. This histidine residue is not conserved and varies significantly between other GI genotypes (Fig. 6A). Superposition of our Fab-bound GI.1 P-domain structure with other available P-domain structures in the GI genogroup, including GI.7 and GI.8, shows that, in addition to sequence changes, the loop regions involved in Fab 5I2 binding are susceptible to significant conformational changes (Fig. 6B). These structural alterations among the other GI genotypes lead to disruption of the conformational epitope recognized by Fab 5I2 on the GI.1 P domain. Thus, sequence and structural changes allow other genotypes to escape Fab 5I2-mediated HBGA blockade.
Fig. 6.
Sequence and structural changes mediate escape from Fab 5I2 neutralization in other genotypes. (A) Amino acid sequence alignment of representative GI variants showing the poor conservation of residues at positions that correspond to residues from the three loop regions in GI.1 involved in interaction with Fab 512. The residues in loops Q, T, and U are colored in red, yellow, and green, respectively. (B) Superposition of P-domain structures from GI genotypes GI.7 (green; PDB ID 4P12), GI.8 (orange; PDB ID 4RDJ), and GI.1 (purple; present study) show that the loop regions (Q, T, and U) involved in interacting with the Fab 5I2 (gray) in GI.I undergo structural alterations that would disrupt the conformational epitope recognized by Fab 5I2 on GI.1 NV. The loops and CDRs from Fab 5I2 are respectively labeled.
Discussion
HuNoVs are unique among viral pathogens in exploiting the genetically controlled polymorphic nature of the HBGAs among host populations for their sustained evolution (14, 16). In response to adaptive immunity, the distally located P2 subdomain can evolve to escape neutralization and differentially interact with HBGAs, as underscored by recent studies that show that HBGA-blocking antibodies confer protection against HuNoV illness or infection (23, 35). Although there are numerous studies delineating HuNoV interactions with HBGAs, the understanding of the mechanism by which a human antibody blocks HBGA binding is limited. Our crystallographic structure of the NV (GI.1) P domain in complex with the Fab of a human IgA 5I2 monoclonal antibody addresses key questions such as how a blockade antibody recognizes HuNoV, what the mechanism of HBGA blockade is, and how sequence alterations allow other genotypes to escape neutralization. The IgA 5I2 was selected from a panel of HBGA blocking antibodies obtained by generating hybridomas from B cells isolated from an individual challenged with GI.1 NV. IgG and IgA antibodies were identified (34). We chose an IgA antibody for our structural studies because of the important role of IgA compared with IgG in conferring mucosal immunity. The subnanomolar binding affinity of this antibody, together with its HBGA blockade activity in vitro, is suggestive of its high potency in virus neutralization.
IgA 5I2 Recognizes a Conformational Epitope Formed by the P2 Subdomain Loops.
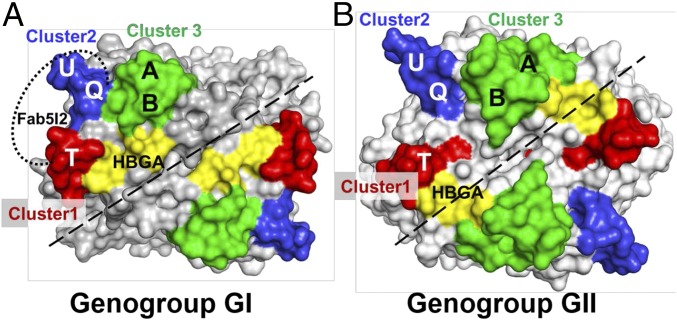
Our crystal structure of the Fab 5I2 in complex with the NV P domain shows that the Fab recognizes a conformational epitope composed of residues from the solvent-exposed loops in the distal portion of the P2 subdomain. Involvement of the surface loops in antibody recognition is a common feature as observed in many antigen–antibody structures. The distal surface of the P2 subdomain consists of six loops that project out into the solvent, which can be grouped into three clusters, numbered 1–3 (Fig. 7A). Despite sequence changes, differences in their lengths and orientations, these loops are similarly clustered in GII.4 (Fig. 7B) as well as in murine NoVs. Residues from clusters 1 and 2 in GI.1 constitute the antigenic site recognized by IgA 5I2 (Fig. 7A). Although IgA 5I2 specifically recognizes clusters 1 and 2, it is possible that other blockade antibodies can recognize residues in other clusters, as suggested by previous biochemical studies characterizing such antibodies in GI and GII.4. In GI, residues in the A and B loops in cluster 3 have been identified as important for binding blockade antibodies (26). In GII.4, three blockade epitopes (A, D, and E) have been suggested; residues in epitope A map to loops A and B (cluster 3), whereas epitopes D and E map to residues in T and U loops (clusters 1 and 2), respectively (36–39). In murine NoVs and rabbit hemorrhagic disease virus (an animal calicivirus), neutralization epitopes have been mapped to A and B loops (40–42). Together with our structural studies and available epitope mapping on other NoVs, the data indicate that these loops allow for differential antigenic presentations contributing to serotypic differences in HuNoVs.
Fig. 7.
Mapping of neutralizing epitopes on NoVs. Shown are surface representations of the P domains from GI (A) and GII (B) NoV genogroups. Based on this study and other biochemical studies, we hypothesize that the majority of the neutralizing epitopes on the surface of the NoV capsid protein lie in one of the three clusters identified in the present study. Cluster 1 (red) comprises the evolving residues in the T loop. Cluster 2 (blue) comprises residues in the Q and U loops. Cluster 3 (green) comprises residues in the A and B loops. The identified clusters are in close proximity to the HBGA binding site (yellow). NAbs can either bind to individual clusters or use a combination of these clusters to bind and neutralize NoVs. The epitope of Fab 5I2 is located in clusters 1 and 2 and is indicated by a dotted line and labeled respectively.
CDRL1 Plays a Dominant Role in Antigen Recognition.
A rather unusual feature of IgA 5I2 is the dominant involvement of CDRL1 in antigen recognition, providing a unique perspective into antibody diversity and antigen interactions. Typically, in an antibody–antigen interaction, including those involving antiviral antibodies, CDRH3 encoded by the highly diverse D-JH joining genes plays a dominant role because of the inherent sequence diversity and consequent conformational variability. The H3 loop is also a common site for somatic hypermutations, allowing affinity maturation of the antibodies (43, 44). In the case of IgA 5I2, five of eight residues in the CDRs that interact with P domain are from CDRL1. In k-chain human antibodies, the length of the CDRL1 varies between 10 and 17 residues, with the majority of the antibodies exhibiting a loop length of 11 residues. In IgA 5I2, CDRL1 is 17 residues long. Despite its unusual length, it exhibits the expected canonical conformation. In IgA 5I2, the CDRH3 is also 17 residues long and is within the expected range of 10–30 residues. The general expectation is that H3 loops with longer lengths (>14) play a predominant role in antigen recognition, as illustrated by the analysis of several antibody–antigen crystal structures (43–46), whereas, in those with shorter lengths, antigen interactions involve other loops. In IgA 5I2, the H3 loop is positioned slightly away from the P2 subdomain, with just two of its residues interacting with the P2 subdomain. The other non-H3 CDRs are of normal lengths with canonical conformations as observed in other antibody structures. With the exception of two residues in the CDRL3, residues from other CDRs do not participate in antigen recognition. CDRL1, together with L3 and H3 residues, provide the complementary residues for optimal hydrogen bond and hydrophobic interactions with the loop residues of the P2 subdomain, as well as appropriate topographical features to enhance the surface complementarity with the P2 subdomain, consistent with the observed binding affinity in the low nanomolar range. It remains to be seen whether the dominant role of CDRL1 observed with IgA 5I2 is a common feature in HuNoV blockade antibodies.
HBGA Blockade by Fab 5I2 Is by Steric Hindrance.
HBGA blockade by an antibody potentially can occur in a number of ways, including directly competing for the HBGA binding site, allosterically disrupting the HBGA binding site by inducing conformational changes in the P domain, or sterically masking the HBGA binding site. Our crystallographic studies show that, in the case of IgA 5I2, the mechanism of HBGA blockade is principally through steric hindrance. The Fab 5I2 binds to the NV P domain without affecting the dimeric conformation of the P domain or the structural integrity of the HBGA binding site. In NV, and generally in other HuNoVs, the HBGA binding site is located in a shallow depression on the distal surface of the P2 subdomain surrounded by clusters of loop regions. Given the considerably larger size of the Fab compared with HBGA, its direct access to the HBGA binding site for optimal interactions is perhaps restricted. This restriction is evident from the structure of the complex; that, despite the Fab entirely engulfing the primary HBGA binding site, none of the residues in the HBGA binding site make contact with the Fab. Their side-chain orientation also remains unaltered by antibody binding. It is interesting to hypothesize that steric hindrance could be a common mechanism used by blockade antibodies. Several observations support such a hypothesis. First, as noted, the HBGA binding sites in GI and GII are surrounded by loop regions. Second, the majority of the residues mapped by biochemical studies as being critical for blockade antibody binding are outside of the primary HBGA binding site, whether it is the β-gal binding site in the case of GI or the αFuc site, as seen in GII.4 HuNoVs. Third, most of the HBGA blocking mAbs characterized thus far are genotype-specific and do not cross-react, even within the same genogroup, similar to IgA 5I2, suggesting that these mAbs also primarily interact with the loop regions that are prone to genotypic alterations. Although HBGA blocking polyclonal antibodies from HuNoV-infected individuals have shown cross-reactivity (47), many derived mAbs, such as IgA 5I2, are genotype-specific (48, 49). Inaccessibility of the HBGA binding site, which represents the most conserved region of the P2 subdomain and is highly genogroup-specific, is consistent with the observation that IgA 5I2 is highly specific for GI.1 HuNoV (34). Although HBGA-blocking, and thus potentially neutralizing, antibodies recognize epitopes in the surface-exposed P2 subdomain, broadly reactive antibodies, which can be used as diagnostic agents, likely recognize other regions in the NoV P domain, as demonstrated by recent crystallographic studies of the GII P domain in complex with a broadly reactive mAb (5B18) (50). In contrast to HBGA-blocking and GI.1-specific 5I2, 5B18 recognizes multiple GII genotypes and binds to a highly conserved region in the P1 subdomain that is close to the shell domain in GII NoVs.
Sequence and Structural Changes Allow Other GI Genotypes to Escape Neutralization.
Our structural studies show that the observed genotype specificity of IgA 5I2 is mainly because the conformational epitope it recognizes is formed by residues in the loop regions that are prone to sequence and structural alterations. The residues in GI.1 P domain that interact with the Fab 5I2 are poorly conserved, and some of the loop regions involved in antibody binding show significant conformational variations, including changes in their orientations. Although it has not been well documented in the case of GI genotypes, in the case of GII.4, several studies have suggested that some of the residues in the corresponding loop regions represent evolutionary hotspots contributing to epochal strain diversity (48, 51, 52). These changes, likely in response to adaptive immunity, alter HBGA binding profiles of these epochal variants. Our structural studies have previously indicated how a small change in the T loop of GII.4 2004 alters the HBGA binding profile (15). The present studies suggest a similar phenomenon in the case of GI genotypes. The H381 residue in the T loop of NV P domain is critical for IgA 5I2 binding, as it is involved in multiple stabilizing interactions with the antibody and is not conserved in other GI genotypes. Although this residue in GI.1 is far removed from the HBGA binding site, because of the conformational changes, the structurally corresponding residue S391 in GI.7 becomes a part of the primary HBGA binding site (13), clearly illustrating a coordinated interplay between antigenic variation and HBGA binding in the evolution of NoVs.
In conclusion, by determining the crystal structure of a human HBGA blockade antibody in complex with the NV P domain, we have provided atomic details of how a potentially neutralizing HBGA blockade antibody recognizes and binds to HuNoV. Our structural study shows that the mechanism by which 5I2 antibody neutralizes the NV is by sterically blocking the HBGA binding. Further, the structural basis for the genotype specificity is also clearly evident from our crystallographic studies. Based on the observation from our structural studies, as well as from epitope mapping studies by others, that the antigenic sites in HuNoVs are mainly composed of residues from loop regions that are hypervariable raises a question whether it will be possible to obtain broadly reactive HBGA blocking antibodies for therapeutic intervention. Although the naturally occurring human HBGA blockade mAbs tend to be genotype-specific, one possibility is to use a mixture of such antibodies or to design antibody scaffolds with a smaller footprint such as single-chain antibodies or even small-molecule mimics that specifically target highly conserved HBGA binding sites. Further studies are clearly required to explore such a possibility.
Materials and Methods
Expression and Purification of the P Domain and Fab 512.
The P domain was expressed and purified as described previously (15). In brief, we cloned the P-domain construct (amino acids 216–519) of GI.1 NV into the pMal-C2E expression vector, overexpressed the protein in Escherichia coli cells, and purified the P domain by using basic chromatography techniques. The purified P domain was concentrated to ∼10 mg/mL in a buffer containing 25 mM Tris·HCl, pH 7.5, 150 mM NaCl, and 5 mM MgCl2 and stored at −80 °C until further use.
Determination of variable-domain sequences of IgA 5I2 and synthesis of expression-optimized genes was done as described previously (34). The VH domain was cloned as an EcoRI/HindIII fragment into a pHC-huCg1Fab expression vector. The VL domain was cloned as a BglII/NotI fragment into pML-huCk κ-expression vector (53). Recombinant antibodies were expressed transiently in Expi293F cells by cotransfection of equal amounts of heavy- and light-chain plasmid DNA by using ExpiFectamine 293 transfection reagent (Life Technologies). After 7 d of culture, the supernatants were clarified by centrifugation and filtered by using 0.4-µm pore size filter devices. Antibodies were harvested from the supernatants by affinity chromatography on CaptureSelect IgG-CH1 columns (Life Technologies) as previously described (54). Antibodies eluted from affinity columns were concentrated by using Amicon centrifugal filters (Millipore).
P Domain–Fab 5I2 Binding Study Using BLI.
BLI was carried out by using an Octet RED96 instrument (ForteBio). Biotinylation of the P domain for loading onto streptavidin-coated biosensors (ForteBio) was carried out by using EZ-link NHC-LC-LC-biotin (catalog no. 21343; Thermo Scientific) following the instructions of the manufacturer. The P domain was loaded onto streptavidin biosensors at a concentration of 1.25 μg/mL in BLI running buffer (20 mM Hepes, pH 7.8, 150 mM NaCl, 0.05% surfactant P20, and 2 mg/mL BSA) for 600 s, resulting in capture levels of 0.8–1.0 nm within a row of eight tips. P domain–Fab 5I2 association and dissociation curves were obtained through twofold serial dilutions of Fab 5I2 (0.5–0.015 μM) plus buffer blanks by using the Octet acquisition software. The binding data were fitted by using the Octet analysis software.
P Domain–Fab 5I2 Complex Formation and Crystallization.
As crystallographic studies with intact antibodies are technically challenging because of the aggregation they induce as a result of their polyvalent nature, we have used Fabs in our crystallographic studies. Purified P-domain (molecular mass 32 kDa) and Fab 5I2 (molecular mass 50 kDa) proteins were mixed in a 1:1 molar ratio in the P-domain storage buffer and incubated for 2–4 h at 4 °C. The mixture was run through a S75pg 16/60 gel filtration column, and the peak corresponding to the complex (assessed by peak shift compared with the P domain by itself) was collected. The complex eluted at a molecular mass of ∼160 kDa, corresponding to a P-domain dimer bound to two Fab molecules. SDS/PAGE confirmed the presence of both proteins in the complex peak. The peak fractions were then pooled and concentrated to 10 mg/mL for crystallization trials. Crystallization screening using hanging-drop vapor diffusion method at 20 °C was set up by a Mosquito nanoliter handling system (TTP LabTech) with commercially available crystal screens. The P domain–Fab complex crystallized in a buffer containing using 0.2 M sodium formate, 0.1 M Bis-Tris propane, pH 6.5, and 20% wt/vol PEG3350. Initial crystals were small and diffracted to ≥3.5 Å. The initial crystallization conditions were further optimized based on ionic strength, pH, and precipitant concentrations, and microseeding technique was used to obtain larger well diffracting crystals. Crystals measuring 0.1–0.2 mm were obtained in 1–2 wk. The crystals were soaked in the reservoir solution containing 20% (wt/vol) glycerol as cryoprotectant followed by flash freezing in liquid nitrogen.
Diffraction, Data Collection, and Structure Determination.
Diffraction data for the P domain–Fab 5I2 crystals were collected on the 5.0.1 beamline at Advanced Light Source (Berkeley, CA). Diffraction data were processed using IMOSFLM (55). The space group was confirmed using POINTLESS program incorporated in the PHENIX suite (56). An initial electron density map was obtained by molecular replacement using the previously published GI.1 P domain structure (PDB ID 2ZL5) as the starting model using program PHASER (57) in the CCP4i suite (58). The solution from PHASER clearly showed extra electron density for the bound Fab molecule. PHASER was then rerun by using the P-domain structure (PDB ID 2ZL5) and an additional neutralizing Fab structure (PDB ID 4RQQ) (59) as starting models to resolve the Fab density. Using this molecular replacement solution, ab initio automated model building and solvent addition were carried out using AUTOBUILD (60) to reduce model bias. The further model building was carried out by using iterative cycles of refinement and model building based on the FO–FC difference maps. The programs phenix.refine and COOT (61) were used throughout structure determination and refinement. Data collection and refinement statistics are provided in Table 1. The program PyMOL (https://www.pymol.org) was used to generate the final figures. Fab 5I2–P domain interactions were analyzed using COOT and LIGPLOT (62) with the donor to acceptor distances between 2.6 Å and 3.3 Å for hydrogen bonding interactions. The buried surface area of the interaction was calculated by using the PISA server (www.ebi.ac.uk/pdbe/pisa).
Acknowledgments
We acknowledge the use of the synchrotron beamlines at the Advanced Light Source (5.0.1) (Berkeley, CA) for diffraction data collection and thank their staff for excellent help. This research was funded by Public Health Service Grants NIH P01 AI057788, NIH P30 DK56338, and P30 CA125123; Robert Welch Foundation Grant Q1292; Agriculture and Food Research Initiative Competitive Grant 2011-68003-30395 from the US Department of Agriculture National Institute of Food and Agriculture; and the John S. Dunn Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5KW9).
References
- 1.Ahmed SM, et al. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect Dis. 2014;14(8):725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel MM, et al. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14(8):1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treanor JJ, et al. A novel intramuscular bivalent norovirus virus-like particle vaccine candidate--reactogenicity, safety, and immunogenicity in a phase 1 trial in healthy adults. J Infect Dis. 2014;210(11):1763–1771. doi: 10.1093/infdis/jiu337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atmar RL, et al. Norovirus vaccine against experimental human Norwalk Virus illness. N Engl J Med. 2011;365(23):2178–2187. doi: 10.1056/NEJMoa1101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green KY, et al. Taxonomy of the caliciviruses. J Infect Dis. 2000;181(suppl 2):S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- 6.Ramani S, Atmar RL, Estes MK. Epidemiology of human noroviruses and updates on vaccine development. Curr Opin Gastroenterol. 2014;30(1):25–33. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindesmith LC, Donaldson EF, Baric RS. Norovirus GII.4 strain antigenic variation. J Virol. 2011;85(1):231–242. doi: 10.1128/JVI.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis. 2002;185(9):1335–1337. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- 9.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9(5):548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 10.Nordgren J, Sharma S, Kambhampati A, Lopman B, Svensson L. Innate Resistance and Susceptibility to Norovirus Infection. PLoS Pathog. 2016;12(4):e1005385. doi: 10.1371/journal.ppat.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad BV, et al. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286(5438):287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 12.Singh BK, Leuthold MM, Hansman GS. Human noroviruses’ fondness for histo-blood group antigens. J Virol. 2015;89(4):2024–2040. doi: 10.1128/JVI.02968-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shanker S, et al. Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses. J Virol. 2014;88(11):6168–6180. doi: 10.1128/JVI.00201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P, et al. Norovirus and histo-blood group antigens: Demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol. 2005;79(11):6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanker S, et al. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: Implications for epochal evolution. J Virol. 2011;85(17):8635–8645. doi: 10.1128/JVI.00848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindesmith LC, et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5(2):e31. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi JM, Hutson AM, Estes MK, Prasad BV. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc Natl Acad Sci USA. 2008;105(27):9175–9180. doi: 10.1073/pnas.0803275105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao S, et al. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81(11):5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan M, et al. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology. 2008;379(2):324–334. doi: 10.1016/j.virol.2008.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansman GS, et al. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J Virol. 2011;85(13):6687–6701. doi: 10.1128/JVI.00246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubota T, et al. Structural basis for the recognition of Lewis antigens by genogroup I norovirus. J Virol. 2012;86(20):11138–11150. doi: 10.1128/JVI.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh BK, et al. Structural analysis of a feline norovirus protruding domain. Virology. 2015;474:181–185. doi: 10.1016/j.virol.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Reeck A, et al. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis. 2010;202(8):1212–1218. doi: 10.1086/656364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramani S, et al. Mucosal and cellular immune responses to Norwalk virus. J Infect Dis. 2015;212(3):397–405. doi: 10.1093/infdis/jiv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindesmith LC, et al. Broad blockade antibody responses in human volunteers after immunization with a multivalent norovirus VLP candidate vaccine: Immunological analyses from a phase I clinical trial. PLoS Med. 2015;12(3):e1001807. doi: 10.1371/journal.pmed.1001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z, et al. Development of Norwalk virus-specific monoclonal antibodies with therapeutic potential for the treatment of Norwalk virus gastroenteritis. J Virol. 2013;87(17):9547–9557. doi: 10.1128/JVI.01376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knoll BM, Lindesmith LC, Yount BL, Baric RS, Marty FM. Resolution of diarrhea in an immunocompromised patient with chronic norovirus gastroenteritis correlates with constitution of specific antibody blockade titer. Infection. 2016;44(4):551–554. doi: 10.1007/s15010-016-0875-1. [DOI] [PubMed] [Google Scholar]
- 28.Lindesmith LC, et al. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 2012;8(5):e1002705. doi: 10.1371/journal.ppat.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lay MK, et al. Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans. Virology. 2010;406(1):1–11. doi: 10.1016/j.virol.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbst-Kralovetz MM, et al. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg Infect Dis. 2013;19(3):431–438. doi: 10.3201/eid1903.121029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong PD, Mascola JR, Nabel GJ. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harb Perspect Med. 2011;1(1):a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329(5995):1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 33.Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: Implications for vaccine development. Cell Host Microbe. 2008;4(3):229–238. doi: 10.1016/j.chom.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sapparapu G, et al. Frequent use of the IgA isotype in human B cells encoding potent norovirus-specific monoclonal antibodies that block HBGA binding. PLoS Pathog. 2016;12(6):e1005719. doi: 10.1371/journal.ppat.1005719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bok K, et al. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc Natl Acad Sci USA. 2011;108(1):325–330. doi: 10.1073/pnas.1014577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindesmith LC, et al. Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002. J Virol. 2012;86(2):873–883. doi: 10.1128/JVI.06200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debbink K, Lindesmith LC, Donaldson EF, Baric RS. Norovirus immunity and the great escape. PLoS Pathog. 2012;8(10):e1002921. doi: 10.1371/journal.ppat.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen DJ, et al. Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding. Virol J. 2009;6:150. doi: 10.1186/1743-422X-6-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parra GI, et al. Multiple antigenic sites are involved in blocking the interaction of GII.4 norovirus capsid with ABH histo-blood group antigens. J Virol. 2012;86(13):7414–7426. doi: 10.1128/JVI.06729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taube S, et al. High-resolution x-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J Virol. 2010;84(11):5695–5705. doi: 10.1128/JVI.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolawole AO, et al. Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization. J Virol. 2014;88(8):4543–4557. doi: 10.1128/JVI.03685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, et al. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog. 2013;9(1):e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuchiya Y, Mizuguchi K. The diversity of H3 loops determines the antigen-binding tendencies of antibody CDR loops. Protein Sci. 2016;25(4):815–825. doi: 10.1002/pro.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirai H, Kidera A, Nakamura H. H3-rules: Identification of CDR-H3 structures in antibodies. FEBS Lett. 1999;455(1-2):188–197. doi: 10.1016/s0014-5793(99)00821-2. [DOI] [PubMed] [Google Scholar]
- 45.North B, Lehmann A, Dunbrack RL., Jr A new clustering of antibody CDR loop conformations. J Mol Biol. 2011;406(2):228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weitzner BD, Dunbrack RL, Jr, Gray JJ. The origin of CDR H3 structural diversity. Structure. 2015;23(2):302–311. doi: 10.1016/j.str.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Czakó R, et al. Experimental human infection with Norwalk virus elicits a surrogate neutralizing antibody response with cross-genogroup activity. Clin Vaccine Immunol. 2015;22(2):221–228. doi: 10.1128/CVI.00516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindesmith LC, et al. Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J Virol. 2013;87(5):2803–2813. doi: 10.1128/JVI.03106-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Payne DC, Parashar UD, Lopman BA. Developments in understanding acquired immunity and innate susceptibility to norovirus and rotavirus gastroenteritis in children. Curr Opin Pediatr. 2015;27(1):105–109. doi: 10.1097/MOP.0000000000000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansman GS, et al. Structural basis for broad detection of genogroup II noroviruses by a monoclonal antibody that binds to a site occluded in the viral particle. J Virol. 2012;86(7):3635–3646. doi: 10.1128/JVI.06868-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bok K, et al. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J Virol. 2009;83(22):11890–11901. doi: 10.1128/JVI.00864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Viral shape-shifting: Norovirus evasion of the human immune system. Nat Rev Microbiol. 2010;8(3):231–241. doi: 10.1038/nrmicro2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLean GR, Nakouzi A, Casadevall A, Green NS. Human and murine immunoglobulin expression vector cassettes. Mol Immunol. 2000;37(14):837–845. doi: 10.1016/s0161-5890(00)00101-2. [DOI] [PubMed] [Google Scholar]
- 54.Aiyegbo MS, et al. Human rotavirus VP6-specific antibodies mediate intracellular neutralization by binding to a quaternary structure in the transcriptional pore. PLoS One. 2013;8(5):e61101. doi: 10.1371/journal.pone.0061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 57.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 59.Sok D, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci USA. 2014;111(49):17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64(pt 1):61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]