Significance
The Hedgehog-signaling pathway plays key roles in animal development and physiology. Insufficient Hedgehog signaling causes birth defects, whereas uncontrolled signaling is implicated in cancer. Signaling is triggered by the secreted protein, Sonic Hedgehog, which inhibits the membrane protein Patched1, leading to pathway activation. Despite its fundamental importance, we do not understand how Sonic Hedgehog inhibits Patched1. Here, we uncover a critical interaction between the fatty-acid–modified N-terminal portion of Sonic Hedgehog and Patched1, and we demonstrate that it is necessary and sufficient for Patched1 inhibition during Hedgehog signaling. This interaction explains impairment of a Sonic Hedgehog mutant causing a congenital brain malformation (holoprosencephaly) and oncogenic activity of Patched1 mutants responsible for a human cancer syndrome.
Keywords: Hedgehog, Patched, palmitate, signaling, lipid
Abstract
The Hedgehog cell–cell signaling pathway is crucial for animal development, and its misregulation is implicated in numerous birth defects and cancers. In unstimulated cells, pathway activity is inhibited by the tumor suppressor membrane protein, Patched. Hedgehog signaling is triggered by the secreted Hedgehog ligand, which binds and inhibits Patched, thus setting in motion the downstream events in signal transduction. Despite its critical importance, the mechanism by which Hedgehog antagonizes Patched has remained unknown. Here, we show that vertebrate Patched1 inhibition is caused by direct, palmitate-dependent interaction with the Sonic Hedgehog ligand. We find that a short palmitoylated N-terminal fragment of Sonic Hedgehog binds Patched1 and, strikingly, is sufficient to inhibit it and to activate signaling. The rest of Sonic Hedgehog confers high-affinity Patched1 binding and internalization through a distinct binding site, but, surprisingly, it is not absolutely required for signaling. The palmitate-dependent interaction with Patched1 is specifically impaired in a Sonic Hedgehog mutant causing human holoprosencephaly, the most frequent congenital brain malformation, explaining its drastically reduced potency. The palmitate-dependent interaction is also abolished in constitutively inhibited Patched1 point mutants causing the Gorlin cancer syndrome, suggesting that they might adopt a conformation distinct from the wild type. Our data demonstrate that Sonic Hedgehog signals via the palmitate-dependent arm of a two-pronged contact with Patched1. Furthermore, our results suggest that, during Hedgehog signaling, ligand binding inhibits Patched by trapping it in an inactive conformation, a mechanism that explains the dramatically reduced activity of oncogenic Patched1 mutants.
The Hedgehog (Hh) signaling pathway is essential for embryogenesis in most animals (1, 2), and insufficient Hh activity during development causes many birth defects, including holoprosencephaly (HPE), the most common human congenital brain malformation (3). In adults, Hh signaling is involved in maintenance of tissue stem cells, whereas aberrant activation is implicated in cancers such as basal cell carcinoma and medulloblastoma.
In the absence of Hh stimulation, the multispanning membrane protein, Patched (Ptch) (4), inhibits the seven-spanner protein, Smoothened (Smo) (5, 6), thus keeping the Hh pathway off. Although the mechanism of Smo inhibition by Ptch remains unknown, it is thought to involve a small-molecule intermediate (7), with Ptch either antagonizing a Smo activator or providing Smo with an inhibitor. This model is consistent with Ptch belonging to the RND family of small molecule pumps (7, 8). The prototypical RND protein, AcrB from Escherichia coli, uses the energy of the proton gradient across the plasma membrane to expel small molecules from cells. RND proteins are homotrimeric and pump substrate by a rotating mechanism, in which subunits undergo ordered conformational change, powered by proton flow (9). Whether Ptch undergoes a similar conformational cycle is unknown.
Hh signaling is activated by the secreted Hh ligand, which binds Ptch (10, 11) on the plasma membrane, causing Ptch inactivation and internalization. Although Drosophila Ptch is present throughout the cell surface, vertebrate Patched1 (Ptch1) is concentrated at the primary cilium (12), a cellular structure essential for Hh signal transduction in vertebrates (13). Inhibition of Ptch is followed by Smo activation and translocation to the cell surface [the entire surface in Drosophila (14) or the ciliary membrane in vertebrates (15)], which sets in motion the downstream steps of signal transduction, culminating with a transcriptional program responsible for the cellular effects of the pathway.
A key unanswered question is the mechanism of Ptch inhibition by ligand. Hh ligands are palmitoylated on a conserved N-terminal cysteine by Ski (16), a membrane-bound O-acyl transferase. Blocking ligand palmitoylation—by genetic inactivation of Ski in Drosophila (16) and mouse (17), by mutating the N-terminal acceptor cysteine (16, 18), or by Ski inhibition by small molecules in cells (19)—strongly inhibits signaling. Consistent with these results, unpalmitoylated human Sonic Hedgehog (Shh) is 30 times less potent than palmitoylated Shh (16, 18). Although unpalmitoylated Shh retains some activity, an unpalmitoylated mutant also missing the first nine residues (ShhΔ9) is completely inactive (20), indicating that the palmitoylated N-terminal portion of Shh (comprising the fatty acid moiety and the peptide part) is essential for signaling. Interestingly, palmitoylated Shh, unpalmitoylated Shh, and ShhΔ9 bind Ptch1 with the same high affinity (20), and furthermore, ShhΔ9 acts as a dominant inhibitor toward palmitoylated Shh by competing for Ptch1 binding (20). Together, these results suggest that the palmitoylated N-terminal part of Shh is critical for Ptch1 inhibition at a step distinct from simple binding; however, it is unknown how this occurs.
Here, we investigate the mechanism of Ptch1 inhibition by Shh. We uncover a critical interaction between the palmitoylated N-terminal portion of Shh and an effector site in Ptch1, distinct from the high-affinity site bound by the rest of Shh. We demonstrate that this interaction is necessary and sufficient for Ptch1 inhibition during Hh signaling. Finally, we provide evidence that the interaction is impaired in a Shh mutant causing HPE and in oncogenic Ptch1 mutants responsible for the Gorlin cancer syndrome.
Results
A Short Palmitoylated Shh Peptide Is Sufficient to Activate Hh Signaling.
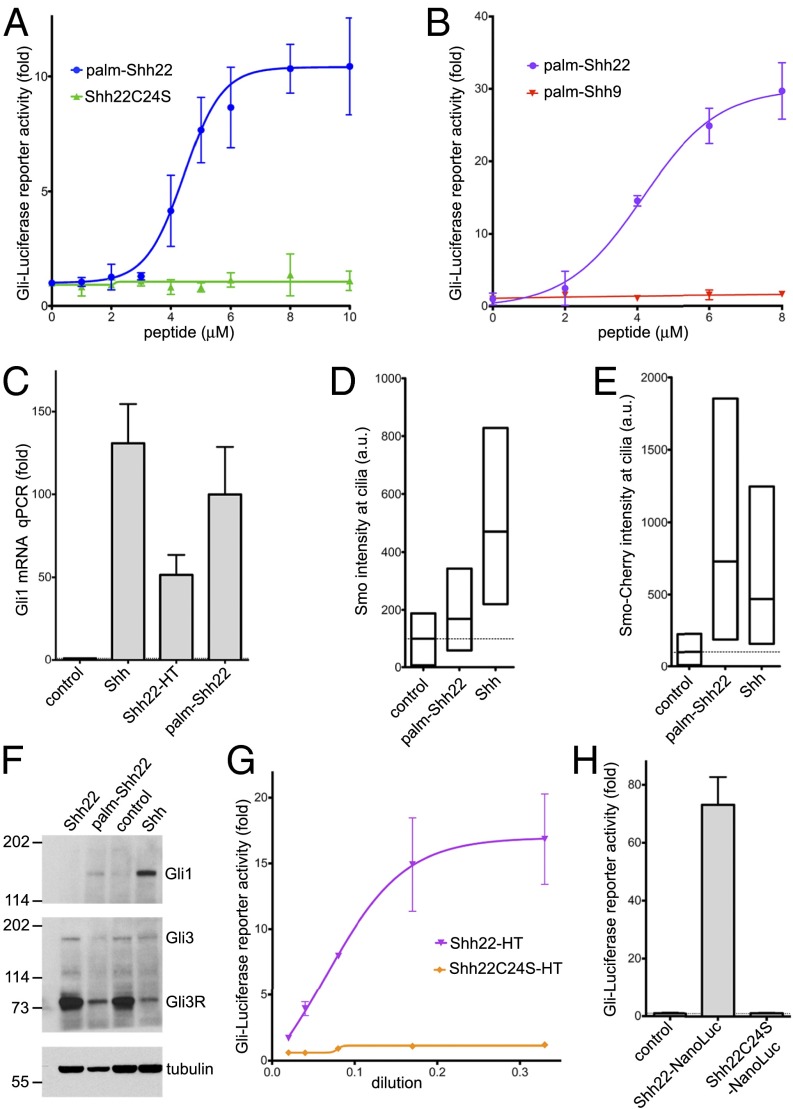
Previous results indicated that the palmitoylated N terminus of Shh is necessary for signaling (20). We asked if an N-terminal portion of Shh might also be sufficient for Hh pathway activation. In the Shh crystal structure (21), the first ∼15 residues adopt an extended conformation and project away from the globular part that binds Ptch1 with high affinity, suggesting the possibility of assaying N-terminal Shh fragments in isolation from the rest of the ligand. We generated palm-Shh22, an N-terminally palmitoylated synthetic peptide comprising the first 22 residues of human Shh (see SI Appendix, Fig. S1, for peptide analysis). Strikingly, palm-Shh22 triggers Hh signaling in mouse NIH 3T3 cells in a dose-dependent manner, by luciferase reporter assay (Fig. 1A). Palm-Shh22 activity is strictly dependent on palmitoylation, as unpalmitoylated Shh22 peptide is completely inactive (Fig. 1A). A shorter palmitoylated peptide, palm-Shh9, consisting of the first nine amino acids of Shh, is inactive (Fig. 1B), indicating that activity requires the N-terminal Shh peptide to be above a certain length.
Fig. 1.
A palmitoylated Shh peptide activates Hh signaling. (A) Shh Light II cells were treated with various concentrations of the synthetic peptides palm-Shh22 or Shh22C24S, and Hh pathway activity was measured by luciferase assay. Error bars represent SD (n = 3). Palm-Shh22 activates Hh signaling, whereas nonpalmitoylated Shh22C24S is inactive. (B) As in A, but with treatment with palm-Shh22 or palm-Shh9. Palm-Shh9 does not activate Hh signaling, in contrast to palm-Shh22. (C) NIH 3T3 cells were incubated with control media, Shh ligand, Shh22-HT, or palm-Shh22 (5 μM). Gli1 transcripts were measured by quantitative RT-PCR. Error bars represent SD (n = 3). Shh22-HT, palm-Shh22, and Shh stimulate Gli1 transcription. (D) NIH 3T3 cells were incubated with control media, palm-Shh22 (1 μM), or Shh. Endogenous Smo localization in primary cilia was measured by immunofluorescence and automated image analysis. The graph shows box plots of Smo fluorescence intensity in cilia, indicating the median and the 25th and 75th percentile of the distribution (n > 300 cilia). Palm-Shh22 recruits Smo to cilia, although to a lesser extent than Shh. (E) As in D, but with cells stably expressing mCherry-tagged Smo. (F) As in D, but cells were analyzed by immunoblotting with anti-Gli1 and anti-Gli3 antibodies. Blotting for tubulin served as loading control. Both palm-Shh22 and Shh reduce Gli3R levels and induce Gli1 protein accumulation, while the nonpalmitoylated peptide, Shh22, is inactive. (G) As in A, but cells were treated with various concentrations of Shh22-HT, or the palmitoylation site mutant, Shh22C24S-HT. Shh22-HT activates Hh signaling in a palmitate-dependent manner. (H) As in A, but with incubation with control media, or the secreted fusions Shh22-NanoLuc and Shh22C24S-NanoLuc. Shh22-NanoLuc activates Hh signaling in a palmitate-dependent manner.
We verified that palm-Shh22 triggers bona fide Hh pathway activation by examining three other readouts. First, palm-Shh22 potently induces transcription of the endogenous Hh target gene, Gli1 (Fig. 1C). Second, we assayed the initial steps of Hh signaling, which, in vertebrates, take place in primary cilia. Upon Ptch1 inhibition by Shh, Smo becomes active and accumulates in cilia (15). Palm-Shh22 causes accumulation in cilia of both endogenous and stably overexpressed Smo (Fig. 1 D and E), indicating that it activates Hh signaling upstream of, or at the level of Smo, consistent with Ptch1 inhibition. Third, palm-Shh22 causes a strong decrease in Gli3 repressor (Gli3R) levels and an accumulation of Gli1 protein (Fig. 1F), which are biochemical hallmarks of Hh pathway activation.
Similar results were obtained when Shh22 was expressed as part of a fusion protein. We took advantage of the fact that Ski palmitoylates Shh peptides as short as six residues (22) to generate palmitoylated Shh22 in cells. A fusion consisting of Shh22 attached to the N terminus of the bacterial HaloTag (HT) protein (23) (Shh22-HT) was expressed as secreted protein in 293T cells. Shh22-HT potently activates Hh signaling (Fig. 1 C and G). Activity remains palmitoylation-dependent, as the palmitoylation site mutant, Shh22C24S-HT, is inactive (Fig. 1G). Like synthetic palm-Shh, Shh22-HT recruits Smo to cilia (SI Appendix, Fig. S2A), reduces Gli3R (SI Appendix, Fig. S2B), and increases Gli1 levels (SI Appendix, Fig. S2C).
To rule out the possibility that palmitoylated Shh22 activates Hh signaling by somehow inducing Shh, we used the 5E1 monoclonal antibody, which binds Shh and blocks its interaction with Ptch1 (24), but does not bind Shh22 (21). Addition of 5E1 has no effect on Shh22-HT activity but completely abolishes signaling by Shh; as expected, the small molecule Smo antagonist, SANT1 (25), inhibits both Shh22-HT and Shh (SI Appendix, Fig. S2D). These results are consistent with palmitoylated Shh22 inhibiting Ptch1 directly.
Finally, to exclude the possibility that HT plays a role in Shh22-HT activity, we tested Shh22-NanoLuc, a fusion in which Shh22 is attached to an unrelated protein (shrimp luciferase). As shown in Fig. 1H, Shh22-NanoLuc is strongly active in a palmitate-dependent manner.
Together, these results demonstrate that the palmitoylated N terminus of Shh is sufficient for Ptch1 inhibition and for triggering Hh signaling and that, surprisingly, the rest of Shh is not absolutely required.
Palmitoylated Shh Peptide Binds Ptch1.
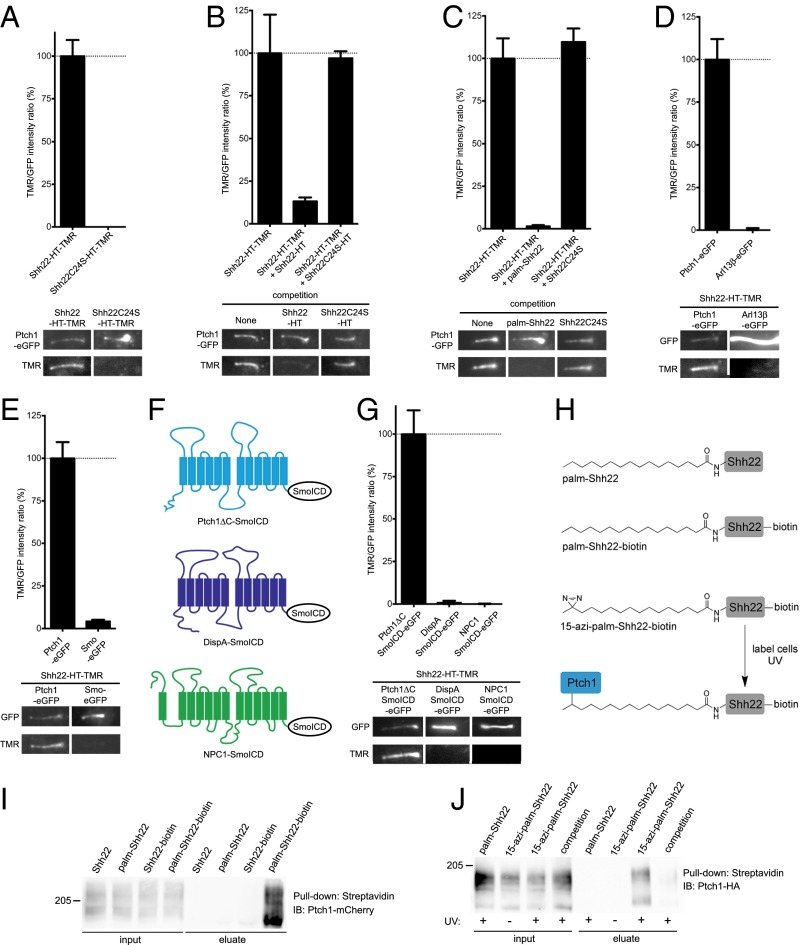
To determine if palm-Shh22 inhibits Ptch1 by direct binding, Shh22-HT was fluorescently labeled by incubation with tetramethylrhodamine (TMR) halo ligand to generate Shh22-HT-TMR, which retains signaling activity (SI Appendix, Fig. S3A). Shh22-HT-TMR was then added to Ptch1-null mouse cells stably expressing low levels of eGFP-tagged Ptch1 (Ptch1-eGFP), and its binding to Ptch1 at primary cilia was assayed by live imaging. Shh22-HT-TMR binds robustly to cilia expressing Ptch1-eGFP (Fig. 2A). Binding is abolished by excess unlabeled Shh22-HT (Fig. 2B) or by synthetic palm-Shh22 (Fig. 2C), demonstrating specificity. The interaction is palmitate-dependent, as shown by two results: (i) Shh22C24S-HT-TMR does not bind Ptch1-eGFP (Fig. 2A) and (ii) Shh22C24S-HT, or unpalmitoylated synthetic Shh22, does not compete binding of Shh22-HT-TMR to Ptch1-eGFP (Fig. 2 B and C).
Fig. 2.
The palmitoylated Shh peptide binds Ptch1. (A) Mouse Ptch1-null cells were rescued by stable expression of Ptch1-eGFP and were incubated with fluorescent Shh22-HT-TMR or Shh22C24S-HT-TMR. After washing to remove excess fluorescent protein, cells were imaged by live microscopy. The graph shows the ratio of TMR and GFP fluorescence at primary cilia. Error bars represent SE (n > 40 cilia). Representative images of primary cilia are shown under the graph. Shh22-HT-TMR binds Ptch1 at cilia in a palmitate-dependent manner. (B) As in A, but cells were incubated with Shh22-HT-TMR in the absence or the presence of 40-fold excess Shh22-HT or Shh22C24S-HT. Shh22-HT, but not Shh22C24S-HT, competes binding of Shh22-HT-TMR to Ptch1 at cilia (n > 5 cilia). (C) As in B, but with incubation with the synthetic peptides palm-Shh22 or Shh22C24S (5 μM each). Only palm-Shh22 competes Shh22-HT-TMR binding to Ptch1 (n > 5 cilia). (D) As in A, but with expression of eGFP-tagged Ptch1 or Arl13β. Shh22-HT-TMR does not bind to cilia labeled with Arl13β-eGFP (n = 20 cilia). (E) As in A, but with expression of eGFP-tagged Ptch1 or Smo. Shh22-HT-TMR does not bind to ciliary Smo-eGFP (n > 10 cilia). (F) C-terminally deleted Ptch1 (Ptch1ΔC), DispA, and NPC1 were targeted to cilia by fusion to the intracellular domain of Smo (SmoICD). (G) As in A, but with expression of eGFP-tagged Ptch1ΔC-SmoICD, DispA-SmoICD, or NPC1-SmoICD. Shh22-HT-TMR binds Ptch1ΔC, but not DispA and NPC1 (n > 10 cilia). (H) Photocrosslinking strategy to detect interaction between palmShh22 and Ptch1. The novel palmitate analog, 15-azi-palmitate, was used to synthesize the photoreactive peptide 15-azi-palm-Shh22-biotin. (I) Peptides were added to 293T cells expressing Ptch1-mCherry, and cell lysates were affinity-purified on streptavidin beads under nondenaturing conditions. Ptch1 is pulled down in a palmitate- and biotin-dependent manner. (J) NIH 3T3 cells stably expressing Ptch1-HA were incubated with 15-azi-palm-Shh22-biotin or palm-Shh22-biotin. Palm-Shh22 was used for competition. Cells were UV-irradiated, and photocrosslinking was analyzed by denaturing affinity precipitation with streptavidin. Ptch1 is specifically photocrosslinked to 15-azi-palm-Shh22-biotin.
Shh22-HT-TMR binding to cilia is strictly dependent on Ptch1-eGFP. No binding is observed in cells expressing eGFP-tagged Arl13β or Smo, two other proteins localized to cilia (Fig. 2 D and E). We also tested Shh22-HT-TMR binding to two Ptch1-related proteins, mouse Dispatched-A (DispA) and mouse Niemann-Pick Disease type C Protein 1 (NPC1), which, like Ptch1, contain a sterol-sensing domain (SSD) and belong to the RND family. We generated cilia-localized versions of DispA and NPC1 by C-terminally fusing them to the intracellular domain of Smo (SmoICD) (Fig. 2F). Shh22-HT-TMR does not bind to DispASmoICD or NPC1SmoICD, but binds to Ptch1ΔCSmoICD in cilia (Fig. 2G).
We also examined binding between palm-Shh22 and Ptch1 biochemically, using a palm-Shh22 peptide C-terminally tagged with biotin (palm-Shh22-biotin, Fig. 2H and SI Appendix, Fig. S1). Palm-Shh22-biotin was added to 293T cells expressing mCherry-tagged Ptch1, and the cells were lysed under nondenaturing conditions. The lysates were subjected to affinity purification on streptavidin beads, and precipitated Ptch1 was analyzed by SDS/PAGE and immunoblotting. Ptch1 is pulled down in a palmitate-dependent manner (Fig. 2I), recapitulating the results obtained using our microscopic binding assay.
Although our results strongly imply that palm-Shh22 binds Ptch1 directly, it is conceivable that the interaction is mediated by another protein. We thus developed a photocrosslinking approach (Fig. 2H) to investigate Ptch1 binding. We synthesized 15-azi-palmitic acid (15-azi-palm), a novel analog in which a photoreactive diazirine moiety is attached to the penultimate carbon of palmitic acid, and we used it to generate 15-azi-palm-Shh22-biotin peptide (SI Appendix, Fig. S1). Like palm-Shh22, 15-azi-palm-Shh22-biotin activates Hh signaling (SI Appendix, Fig. S3B), indicating that the diazirine group does not perturb function. To test for direct binding to Ptch1, 15-azi-palm-Shh22-biotin was added to NIH 3T3 cells stably expressing HA-tagged Ptch1, after which cells were UV-irradiated and biotinylated proteins were isolated by denaturing streptavidin affinity to disrupt noncovalent interactions. The precipitated material was then separated by SDS/PAGE and immunoblotted with anti-HA antibodies. 15-azi-palm-Shh22-biotin labels Ptch1 in a UV-dependent manner, indicating photocrosslinking of the two molecules (Fig. 2J and SI Appendix, Fig. S3C). Importantly, this interaction is specific, being abolished by excess palm-Shh22. Together, these results demonstrate that palm-Shh22 binds Ptch1, including a direct contact between the palmitoyl moiety and Ptch1.
Requirements for Palmitate-Dependent Shh-Ptch1 Binding and Activity.
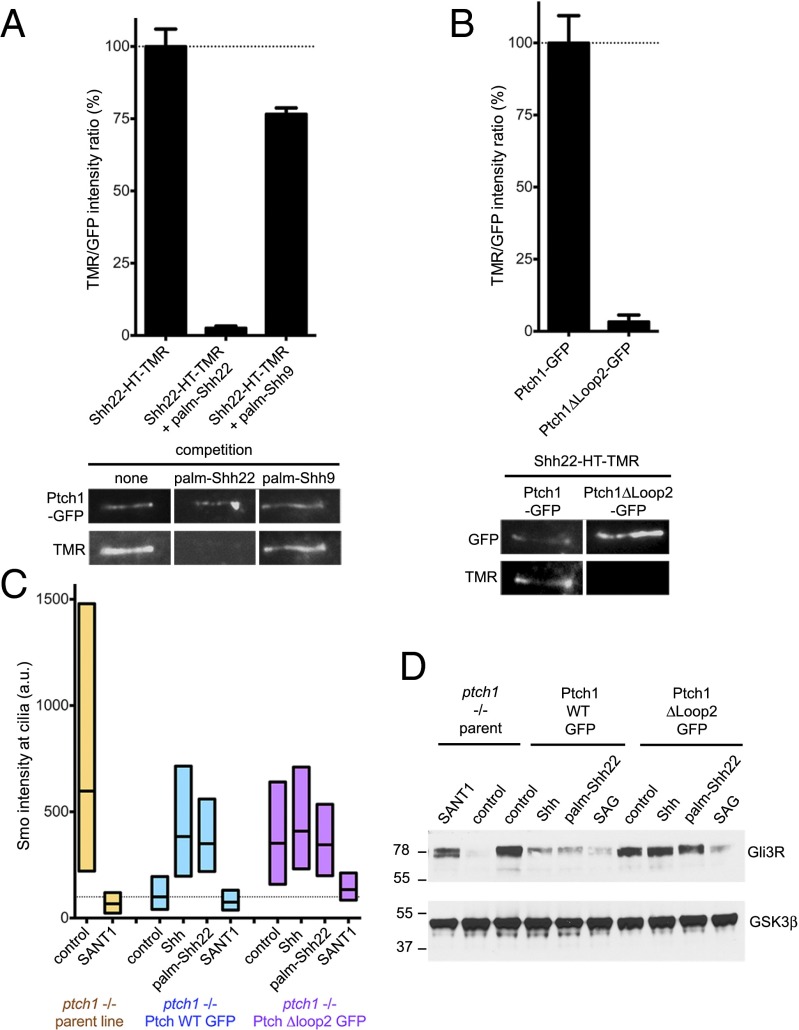
We asked whether palm-Shh9, which is inactive (Fig. 1B), binds Ptch1. As shown in Fig. 3A, palm-Shh9 does not compete binding of Shh22-HT-TMR to Ptch1, indicating that palm-Shh9 does not bind Ptch1. We also asked what portion of Ptch1 is required for interaction with palm-Shh22. Two large extracellular loops (loop1, between transmembrane helices 1 and 2, and loop2, between helices 7 and 8) account for most of the extracellular surface of Ptch1. Ptch1Δloop2, a mutant missing most of loop2, suppresses Hh signaling, but does not bind or respond to Shh (7). When stably expressed in Ptch1-null cells, Ptch1Δloop2 localizes to cilia (Fig. 3B) and reverses constitutive Hh signaling (Fig. 3 C and D and SI Appendix, Fig. S4); as expected, Ptch1Δloop2 does not respond to Shh (Fig. 3 C and D and SI Appendix, Fig. S4). Interestingly, Ptch1Δloop2 does not bind Shh22-HT-TMR (Fig. 3B), indicating that loop2 is required for interaction with palm-Shh22; however, whether loop2 is directly involved in palm-Shh22 binding remains to be determined. Furthermore, cells rescued with Ptch1Δloop2 do not respond to palm-Shh22 (Fig. 3 C and D); importantly, these cells respond robustly to the Smo agonist, SAG (26), indicating that the Hh pathway is functional (Fig. 3D). We could not test the loop1 requirement for palm-Shh22 binding and activity because Ptch1Δloop1 was inactive and did not localize to cilia, likely due to misfolding. Together, these results demonstrate that Hh pathway activation by palmitoylated N-terminal Shh peptides requires binding to Ptch1.
Fig. 3.
Ptch1 binding is required for signaling by palmitoylated Shh peptide. (A) Binding of Shh22-HT-TMR to cilia in Ptch1-null cells rescued with Ptch1-eGFP was measured by live imaging in the absence or presence of palm-Shh22 or palm-Shh9 (5 μM). Graph shows the ratio of ciliary TMR and GFP fluorescence. Error bars represent SE (n > 5 cilia). Representative images are shown below the graph. Palm-Shh9 does not compete binding of Shh22-HT to Ptch1-eGFP, in contrast to palm-Shh22. (B) As in A, but with Ptch1-null cells rescued with Ptch1-eGFP or Ptch1Δloop2-eGFP. Shh22-HT-TMR does not bind Ptch1Δloop2-eGFP. (C) Ptch1-null cells, rescued or not with Ptch1-eGFP or Ptch1Δloop2-eGFP, were incubated with control media, Shh, palm-Shh22 (5 μM), or SANT1 (1 μM), and endogenous Smo localization to cilia was measured by immunofluorescence and automated image analysis (n > 300 cilia). Smo is constitutively at cilia in Ptch1-null cells, which is reversed by Ptch1-eGFP, and partially by Ptch1Δloop2-eGFP. Palm-Shh22 and Shh do not cause Smo accumulation in cilia in cells rescued with Ptch1Δloop2-eGFP, in contrast to Ptch1-eGFP. In all conditions, Smo recruitment to cilia is blocked by SANT1. (D) As in C, but cells were incubated with control media, Shh, palm-Shh22 (5 μM), SAG (1 μM), or SANT1 (1 μM), and endogenous Gli3R was measured by immunoblotting. GSK3β served as loading control. Ptch1-null cells have low Gli3R levels, indicative of constitutive Hh signaling, which is reversed by Ptch1-eGFP, Ptch1Δloop2-eGFP, or SANT1. Cells expressing Ptch1Δloop2-eGFP do not respond to Shh and palm-Shh22, but respond to SAG; in contrast, cells expressing Ptch1-eGFP respond to all three.
Holoprosencephaly-Causing Shh Mutation Abolishes Palmitate-Dependent Interaction with Ptch1.
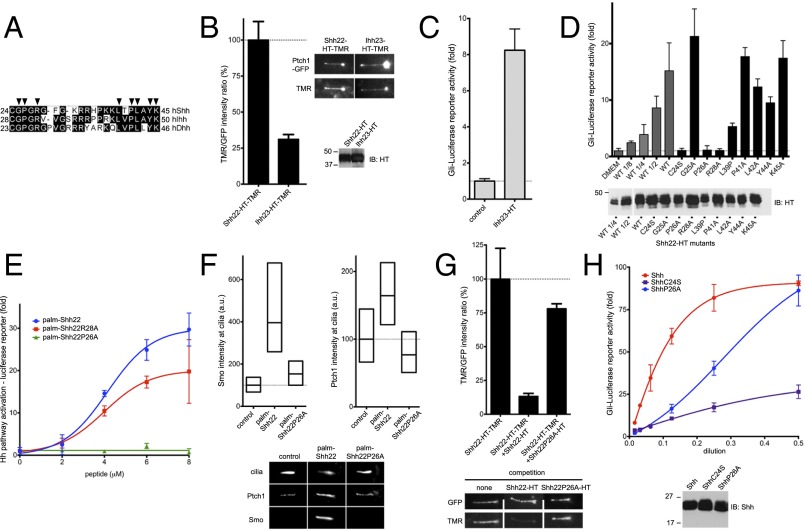
We investigated what amino acids in palm-Shh22 are important for activity. We first asked if the N-terminal cysteine plays a role beyond accepting palmitate during the Ski-catalyzed reaction. The palmitoylated synthetic peptide palm-Shh22C24S has comparable activity to palm-Shh22 (SI Appendix, Fig. S5), indicating that C24 is only critical for enzymatic palmitoylation.
Because all Hh ligands are palmitoylated, we asked if, for other Hh paralogs, the fragment homologous to palm-Shh22, is also active. We generated Ihh23-HT, a fusion carrying the corresponding palmitoylated fragment of human Indian Hedgehog (Ihh) (Fig. 4A). Like Shh22-HT, Ihh23-HT binds Ptch1 (Fig. 4B) and triggers Hh signaling (Fig. 4C), demonstrating that activity of the palmitoylated N-terminal peptide is conserved among Hh ligands.
Fig. 4.
Holoprosencephaly-causing Shh mutation abolishes palmitate-dependent Ptch1 interaction. (A) Alignment of N termini of human Shh, Indian Hh (Ihh), and Desert Hh (Dhh). Arrowheads indicate residues that were tested by mutagenesis. (B) Binding of Shh22-HT-TMR and Ihh23-HT-TMR to cilia in Ptch1-null cells rescued with Ptch1-eGFP was measured by live imaging. Graph shows the ratio of ciliary TMR and GFP fluorescence. Error bars represent SE (n > 5 cilia). Representative images are shown on the right. Equal volumes of HT fusions were analyzed by SDS/PAGE and immunoblotting. Both fusions bind Ptch1. (C) Ihh23-HT activates Hh signaling in Shh Light II cells. Error bars represent SD (n = 3). (D) Wild-type and point mutants of Shh22-HT were expressed as secreted proteins in 293T cells, and their activity was assayed in Shh Light II cells as in C. A portion of 293T-conditioned media was analyzed by SDS/PAGE and immunoblotting to measure protein secretion. All fusions are active, except Shh22P26A-HT and Shh22R28A-HT. (E) Shh Light II cells were treated with palm-Shh22, palm-Shh22P26A, or palm-Shh22R28A, and Hh pathway activity was measured as in C. Palm-Shh22P26A is inactive, in contrast to palm-Shh22R28A. (F) Ptch1-null cells expressing Ptch1-eGFP were incubated with control media, palm-Shh22, or palm-Shh22P26A (5 μM each), and Smo and Ptch1 localization at cilia was measured by immunofluorescence and automated image analysis (n > 300 cilia). Representative cilia micrographs are shown under the graphs. Palm-Shh22P26A is defective in Smo and Ptch1 recruitment to cilia. (G) Ptch1-null cells expressing Ptch1-eGFP were incubated with Shh22-HT-TMR in the presence or the absence of excess Shh22-HT or Shh22P26A-HT. Binding of Shh22-HT-TMR to Ptch1-eGFP at cilia was measured as in B (n > 5 cilia). Shh22P26A-HT is defective in competing Shh22-HT-TMR binding to Ptch1-eGFP. (H) As in E, but cells were treated with Shh, ShhC24S, or ShhP26A. Equal volumes of each ligand were analyzed by SDS/PAGE and immunoblotting. ShhP26A is less active than Shh, but more active than ShhC24S.
We next tested the function of Shh22 residues conserved between human paralogs, Shh, Ihh, and Desert Hedgehog (Dhh) (Fig. 4A) by mutating them to alanine. Point mutants of Shh22-HT were produced in 293T cells, and their activity was measured by reporter assay. All but two mutants, Shh22P26A-HT and Shh22R28A-HT, have significant activity (Fig. 4D), indicating that the mutated residues are not absolutely required. A trivial explanation for inactivity of Shh22P26A-HT and Shh22R28A-HT is that they are defective in palmitoylation by Ski. To exclude this possibility, we synthesized the palmitoylated peptides, palm-Shh22P26A and palm-Shh22R28A (SI Appendix, Fig. S1). Palm-Shh22R28A is active (Fig. 4E), indicating that R28 is not required for signaling and that Shh22R28A-HT is perhaps inactive because it is not palmitoylated. Synthetic palm-Shh22P26A, however, is completely inactive, both by reporter assay (Fig. 4E) and by Smo ciliary recruitment assay (Fig. 4F). Paralleling loss of activity, P26A mutation greatly reduces binding of Shh22P26A-HT to Ptch1 (Fig. 4G). Together, these results indicate that P26 is critical for palm-Shh22 binding to Ptch1 and for activity.
Interestingly, P26 is mutated in some cases of HPE (3), a congenital brain malformation caused most frequently by insufficient Shh signaling during development. To define how the P26A mutation might cause HPE, we measured its effect in the context of the entire Shh ligand. ShhP26A is significantly less active than Shh (Fig. 4H); strikingly, activity is recovered at high doses of ShhP26A. P26A mutation is not expected to affect palmitate-independent interaction of Shh with Ptch1, given that P26 is deleted in ShhΔ9, which binds Ptch1 with high affinity. We interpret activity of ShhP26A at high concentration as being due to palmitate-independent interaction partially rescuing the defective palmitate-dependent interaction.
The palmitoylated N-terminal Shh peptide includes the Cardin–Weintraub motif (27), a short stretch rich in positively charged residues (K32-K38 in human Shh), which contributes to Shh binding to glycosaminoglycans (GAGs) (28). It is conceivable that the P26 mutation might affect Shh interaction with GAGs. However, we find that unpalmitoylated Shh and ShhΔ5 (missing five residues from the N terminus) show identical salt elution profiles by heparin chromatography; the fact that P26 is deleted in ShhΔ5 suggests that P26 does not contribute significantly to interaction with heparin and thus is unlikely to affect Shh binding to GAG. Taken together, our results suggest that P26 mutation causes HPE by a novel mechanism, impairing the palmitate-dependent interaction between the N terminus of Shh and Ptch1.
Separable Parts of Shh Cause Ptch1 Inhibition and Internalization.
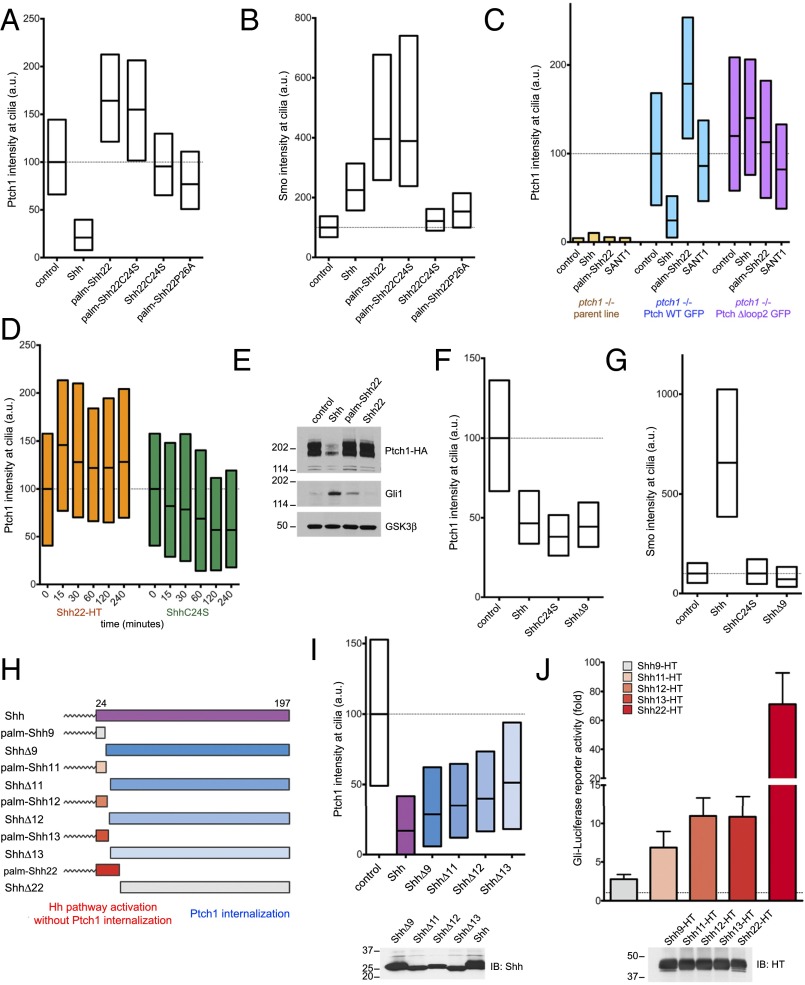
Ptch1 localizes to primary cilia, and, upon Shh binding, is internalized and degraded (12). Surprisingly, palm-Shh22 has the opposite effect on Ptch1, significantly increasing its ciliary levels compared with untreated cells (Fig. 5A). This effect is dependent on palm-Shh22 binding to Ptch1, as shown by two results: (i) Shh22C24S and palm-Shh22P26A have no effect on Ptch1 levels in cilia (Fig. 5A) and (ii) palm-Shh22 has no effect on ciliary levels of Ptch1Δloop2 (Fig. 5C). In contrast to their opposing effects on ciliary Ptch1, both palm-Shh22 and Shh recruit Smo to cilia (Fig. 5B). Ptch1 accumulation in cilia caused by palm-Shh22 is rapid, becoming apparent after 15 min (Fig. 5D). Furthermore, palm-Shh22 activates Hh signaling without causing Ptch1 degradation, in contrast to Shh (Fig. 5E). These results indicate that palm-Shh22 directly inhibits Ptch1 and that Ptch1 internalization or degradation is not absolutely required for Hh pathway activation. However, palm-Shh22 activity is weaker than that of Shh (Fig. 5E), so it remains possible that Ptch1 degradation is important for maximal Hh pathway activation.
Fig. 5.
Separable parts of Shh cause Ptch1 inhibition and internalization. (A) Ptch1-null cells, stably expressing Ptch1-eGFP, were incubated with control media, Shh, or the synthetic peptides palm-Shh22, palm-Shh22C24S, Shh22C24S, and palm-Shh22P26A (5 μM each). Ptch1 localization at cilia was measured by immunofluorescence and automated image analysis (n > 300 cilia). Shh removes Ptch1 from cilia; in contrast, palm-Shh22 and palm-Shh22C24S cause Ptch1 accumulation in cilia. Shh22C24S and palm-Shh22P26A, which do not bind Ptch1, have no effect. (B) As in A, but measuring endogenous Smo at cilia. Shh, palm-Shh22, and palm-Shh22C24S recruit Smo to cilia, in contrast to Shh22C24S and palm-Shh22P26A. (C) As in A, but with cells expressing Ptch1-eGFP or Ptch1Δloop2-eGFP and incubated with control media, Shh, palm-Shh22 (5 μM), or SANT1 (1 μM). Palm-Shh22 does not cause Ptch1Δloop2 accumulation in cilia, in contrast to Ptch1. The graph showing Smo intensity at cilia in this experiment is displayed in Fig. 3B. (D) As in A, but with incubation with Shh22-HT or ShhC24S. Shh22-HT and ShhC24S have opposite effects on Ptch1 levels in cilia, with Shh22-HT causing rapid ciliary accumulation of Ptch1. (E) NIH 3T3 cells stably expressing HA-tagged Ptch1 were incubated with control media, Shh, palm-Shh22 (5 μM), or Shh22 (5 μM). Cell lysates were analyzed by SDS/PAGE and immunoblotting for Ptch1, Gli1, and GSK3β (loading control). Like Shh, palm-Shh22 activates the Hh pathway, but does not cause Ptch1 degradation, in contrast to Shh. Unpalmitoylated Shh22 is inactive. (F) As in A, but with incubation with control media, Shh, ShhC24S, or ShhΔ9. All three proteins reduce Ptch1 levels in cilia. (G) As in F, but measuring endogenous Smo at cilia. Only Shh causes Smo accumulation in cilia. (H) Schematic of constructs used to separate portions of Shh sufficient for Hh pathway activation or Ptch1 internalization. (I) As in A, but with incubation with control media, Shh, or the indicated N-terminal deletion mutants. All mutants induce Ptch1 internalization from cilia. (Lower) Equal volumes of each secreted protein were analyzed by SDS/PAGE and immunoblotting to confirm secretion. (J) N-terminal Shh peptides of various lengths were expressed in 293T cells as secreted HT fusions and were assayed for activity in Shh Light II cells. Error bars represent SD (n = 3). Equal volumes of 293T-conditioned media were analyzed by SDS/PAGE and immunoblotting. Shh11-HT, Shh12-HT, and Shh13-HT retain significant activity.
We next asked what aspects of Shh are important for Ptch1 internalization. Shh, ShhC24S, and ShhΔ9 are equally effective at internalizing Ptch1 (Fig. 5F), indicating that palmitoylation and the first nine residues of Shh are not required. In contrast to Shh, however, ShhC24S and ShhΔ9 do not recruit Smo to cilia (Fig. 5G), and, as expected, signaling activity is greatly reduced (see SI Appendix, Fig. S6 A and B, for ShhC24S) or completely abolished (see SI Appendix, Fig. S6A, for ShhΔ9). Thus, Ptch1 internalization is not sufficient to recruit Smo to cilia and to activate Hh signaling, perhaps because enough Ptch1 remains in cilia to suppress the pathway. Similar results are obtained whether Shh and ShhC24S are cholesterol-modified or not, indicating that the cholesterol moiety is not required for Ptch1 internalization (SI Appendix, Fig. S6C).
Finally, we asked whether the parts of Shh sufficient for Ptch1 inhibition and internalization are separable. Because palm-Shh9 is inactive and ShhΔ17 does not bind Ptch1 (20), we divided Shh into complementary fragments with break points between residues 9 and 17 (Fig. 5H). The N-terminally deleted ligands, ShhΔ11, ShhΔ12, and ShhΔ13, induce Ptch1 internalization (Fig. 5I). The complementary palmitoylated peptides, Shh11-HT, Shh12-HT, and Shh13-HT, activate Hh signaling, although less potently than Shh22-HT (Fig. 5J). These results show that two activities of Shh, Ptch1 inhibition and internalization, are carried out by separable parts of Shh, suggesting a two-pronged contact between Shh and Ptch1.
Palmitate-Dependent Interaction with Shh Is Defective in Oncogenic Ptch1 Mutants.
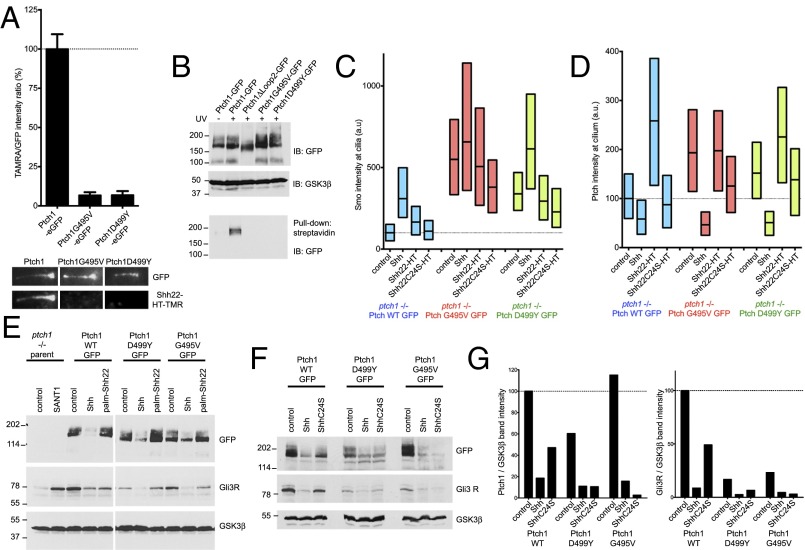
Mutations that impair human Ptch1 cause Gorlin Syndrome (GS), a congenital predisposition to cancers driven by hyperactive Hh signaling, such as basal cell carcinoma and medulloblastoma. Two of the most severe GS mutations in human Ptch1, G509V and D513Y (corresponding to G495V and D499Y in mouse Ptch1), affect conserved residues required for pumping activity in bacterial RND family members (7); it is unclear, however, how these mutations compromise Ptch1. To test for possible defects in mouse Ptch1G495V and Ptch1D499Y, we examined their interaction with palm-Shh22. The two mutants, tagged with eGFP, were stably expressed in Ptch1-null cells. Similar to wild-type Ptch1, both Ptch1G495V and Ptch1D499Y localize to cilia in untreated cells and disappear from cilia upon addition of Shh (SI Appendix, Fig. S7A); this indicates that Shh binding and internalization is normal. Ptch1G495V and Ptch1D499Y reverse constitutive Hh signaling in Ptch1-null cells, reducing ciliary Smo (SI Appendix, Fig. S7B) and Gli1 protein levels (SI Appendix, Fig. S7C); as expected, Ptch1G495V and Ptch1D499Y are less active than Ptch1 in suppressing Hh signaling. Finally, like wild-type Ptch1, the two GS mutants are inhibited by Shh, leading to activation of Hh signaling (SI Appendix, Fig. S7C). Ptch1G495V and Ptch1D499Y, however, have dramatically reduced binding to Shh22-HT-TMR (Fig. 6A) and are not photocrosslinked to 15-azi-palm-Shh22-biotin (Fig. 6B), indicating a profound defect in palmitate-dependent ligand binding. Consistent with this, in Ptch1-null cells rescued with Ptch1G495V or Ptch1D499Y, palm-Shh22 has no effect on Smo (Fig. 6C) or Ptch1 (Fig. 6D) accumulation in cilia and does not activate signaling (Fig. 6E).
Fig. 6.
Defective palmitate-dependent interaction with Shh in Ptch1 mutants causing Gorlin Syndrome. (A) Binding of Shh22-HT-TMR to cilia in Ptch1-null cells rescued with eGFP-tagged Ptch1, Ptch1G495V, or Ptch1D499Y was measured by live imaging. Graph shows ratio of TMR and GFP fluorescence at cilia. Error bars represent SE (n > 14 cilia). Representative images of cilia are shown below the graph. Ptch1G495V and Ptch1D499Y do not bind Shh22-HT-TMR. (B) Ptch1-null cells, stably expressing the indicated Ptch1 constructs, were incubated with 15-azi-palm-Shh22-biotin (3.5 μM). The cells were UV-irradiated, and photocrosslinking was analyzed by denaturing affinity precipitation with streptavidin, followed by SDS/PAGE and immunoblotting. GSK3β was used as loading control. Ptch1G495V and Ptch1D499Y are not photocrosslinked to 15-azi-palm-Shh22-biotin, in contrast to Ptch1. Ptch1Δloop2, which does not bind palm-Shh22, serves as negative control. (C) Ptch1-null cells rescued with eGFP-tagged Ptch1, Ptch1G495V, or Ptch1D499Y were incubated with control media, Shh, Shh22-HT, or Shh22C24S-HT, and endogenous Smo recruitment to cilia was measured by immunofluorescence and automated image analysis (n > 300 cilia). Ptch1G495V and Ptch1D499Y are impaired in their response to Shh22-HT but respond normally to Shh. (D) As in C, but showing Ptch1 localization at cilia. (E) As in C, but cells were treated with control media, Shh, or palm-Shh22 (5 μM), and Gli3R, Ptch1, and GSK3β were analyzed by immunoblotting. In cells expressing Ptch1G495V and Ptch1D499Y, Gli3R levels decrease in response to Shh but not to palm-Shh22. (F) As in E, but cells were treated with control media, Shh, or ShhC24S. Cells expressing Ptch1G495V and Ptch1D499Y respond to both Shh and ShhC24S, whereas Ptch1 responds preferentially to Shh. (G) Quantification of the experiment in F.
A defective palmitate-dependent interaction with Shh predicts that Ptch1 GS mutants should respond equally well to palmitoylated and unpalmitoylated Shh. Indeed, Shh and ShhC24S are similarly effective at activating Hh signaling in Ptch1-null cells expressing Ptch1G495V and Ptch1D499Y (Fig. 6 F and G); in contrast, cells expressing Ptch1 respond very weakly to ShhC24S. Together, these results suggest that the two GS mutants adopt a conformation with greatly decreased affinity for the palmitoylated N-terminal portion of Shh, hinting at a connection between conformational trapping and reduced Ptch1 activity.
Discussion
In contrast to most signaling pathways, in which the ligand activates its receptor, in Hh signaling the ligand inhibits the Ptch receptor. Similar to many other liganded membrane receptors, Hh-bound Ptch is internalized from the cell surface. An important unanswered question has been how Hh inhibits Ptch. The palmitoylated N-terminal part of Shh is critical for activity, but is not required for high-affinity binding to Ptch1, suggesting its involvement in a signaling step distinct from receptor binding; however, the underlying mechanism is unknown. Here, we demonstrate that a key event initiating Hh signaling in mammalian cells is direct inhibition of Ptch1 by the palmitoylated N-terminal part of Shh, mediated via a novel effector site in Ptch1. We find that a short palmitoylated N-terminal Shh peptide is sufficient to bind Ptch1 and activate signaling; in contrast, the rest of Shh binds Ptch1 at a different site and causes its internalization, but is not required for signaling. Hh pathway activation by the Shh peptide occurs without Ptch1 internalization or degradation, consistent with these processes not being absolutely required for signaling. A point mutation that causes HPE (3) maps to a conserved residue in the Shh peptide (P26) and abolishes its binding to Ptch1 and activity; this demonstrates that engagement of the Ptch1 effector site is necessary for proper Hh signaling and explains the signaling defect of the P26-mutated Shh ligand.
Our data point to two contacts between Shh and Ptch1. The first contact is mediated by the globular part of Shh, which confers high-affinity binding to Ptch1 (20). The second contact is mediated by the palmitoylated N-terminal part of Shh and is perhaps of much lower affinity, as suggested by the potency of palmitoylated peptide; this explains why Shh and ShhΔ9 bind Ptch1 with similar affinity (20). This model is also supported by the finding that high-affinity Ptch1-Shh contact can partially rescue a defective interaction with the N-terminal part of Shh, as illustrated by the P26A mutation, which completely eliminates activity of the palm-ShhNP26A peptide yet does not abolish (although it significantly reduces) activity of ShhP26A ligand. The residual activity of unpalmitoylated Shh can be explained similarly. We speculate that unpalmitoylated Shh binds Ptch1 via the high-affinity site, thus greatly increasing the local concentration of the N-terminal peptide; the latter then inhibits Ptch1 via the low-affinity site, although much less efficiently in the absence of palmitoylation. In isolation, however, the unpalmitoylated N-terminal peptide has undetectable Ptch1 binding and activity. Finally, the behavior of ShhΔ9 can be understood in this context. ShhΔ9 binds the high-affinity site but does not inhibit Ptch1 because it cannot engage the low-affinity site; this also explains why ShhΔ9 acts as dominant-negative toward wild-type Shh (20). Interestingly, N-terminal modification with other fatty acids in addition to palmitate also increases Shh potency (29); furthermore, Shh in which the N-terminal cysteine is replaced by two isoleucines (ShhC24II) is significantly more potent than the unmodified ligand (29). We speculate that the palmitate-binding site in Ptch1 can interact with a wide range of hydrophobic moieties, explaining the increased potency of Shh thus modified.
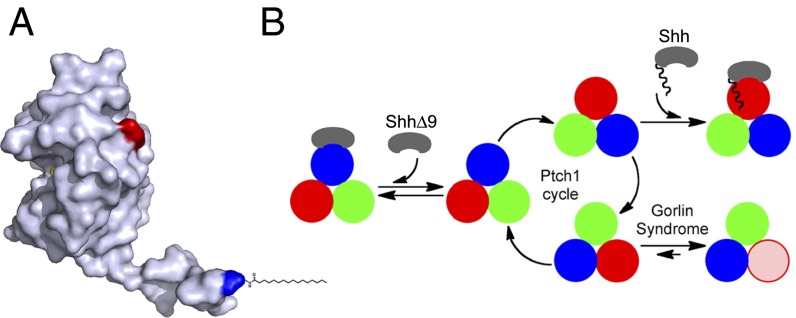
Where is the palmitate-binding site in Ptch1? The crystal structure of Shh (21) shows that the N-terminal peptide adopts an extended conformation (Fig. 7A) such that the first residue projects about 30 Å away from the rest of the protein; this indicates that perhaps the palmitoyl moiety binds Ptch1 at a significant distance from where the globular part of Shh binds. It is possible that the entire Shh-Ptch1 contact occurs over a single, extended site on Ptch1; this is supported by the fact that interactions between Shh and Ptch1 require loop2 of Ptch1. However, structural studies of Shh-Ptch1 interaction will be needed to resolve this issue and to define the palmitate-binding site.
Fig. 7.
Model for Ptch1 inhibition by Shh and by oncogenic mutations. (A) Crystal structure of human Shh (21), showing the N-terminal peptide protruding from the globular part of the protein. The N and C termini are colored blue and red, respectively. The palmitoyl residue was added manually. Shh can be divided into two parts: a short, palmitoylated N-terminal peptide that inhibits Ptch1 via a low-affinity binding site and a globular part that causes Ptch1 internalization via a high-affinity binding site. (B) Speculative model for Ptch1 function and inhibition. Although its oligomeric structure is unknown, Ptch1 is shown as a homotrimer undergoing conformational cycling (different conformations are shown in red, blue, and green) by analogy to bacterial RND pumps. This cycle is proposed to be required for Hh pathway-suppressing activity of Ptch1, perhaps via a small-molecule Smo modulator. Shh binds Ptch1 and, via its palmitoylated N-terminal portion, interrupts cycling by conformational trapping. ShhΔ9 binds Ptch1 with high affinity but cannot interrupt cycling and is thus inactive. Oncogenic Ptch1 mutants that cause GS adopt a conformation (pink) defective in interaction with the palmitoylated N-terminal portion of Shh. This conformational trapping could explain reduced activity of these mutants.
The Shh-Ptch1 interaction is reminiscent of the interaction between Wnt ligands and Frizzled (Fz) receptors. Like Shh, Wnt proteins are palmitoylated, which is critical for their activity (30). Structural analysis of Xenopus Wnt8 (xWnt8) bound to the extracellular, cysteine-rich domain (CRD) of mouse Frizzled 8 (mFz8) (31) shows that the high-affinity interaction results from a two-pronged contact: the palmitoyl moiety of xWnt8, located at the tip of a “thumb,” occupies a groove on mFz8CRD (site 1), whereas the C-terminal domain of xWnt8 forms an “index finger” that binds to the opposite surface of mFz8CRD (site 2). A possible difference between Wnt-Fz and Shh-Ptch1 interactions might be the role of palmitoyl moieties. Palmitate is important for xWnt8 binding to mFz8CRD, but mFz8CRD conformation does not change upon ligand binding (31); this suggests that palmitate is perhaps not directly involved in Fz activation. In contrast, the palmitoylated N terminus of Shh is critical for inhibition of Ptch1, and we speculate that it induces a conformational change in Ptch1 (see below).
How does Shh inhibit Ptch1? We envision the following possible scenario. Secreted Shh reaches the responding cell, where perhaps it first binds to the coreceptors Cdon, Boc, or Gas1 (32, 33) on the cell surface; interestingly, the coreceptors do not localize to cilia (34), where Ptch1 is concentrated. Shh is then somehow delivered to Ptch1 at the primary cilium (12), binding Ptch1 via high-affinity palmitate-independent interaction. Shh subsequently uses its palmitoylated N-terminal portion to contact the effector site of Ptch1. The high-affinity interaction ensures that Shh is active at low concentration, but it is interaction via the effector site that inhibits Ptch1; indeed, high concentration of palm-Shh22 peptide is sufficient to activate Hh signaling, bypassing palmitate-independent interaction.
Based on subcellular localization and activity, different forms of Ptch1 can be distinguished: (i) unliganded Ptch1, which is active and localizes to cilia; (ii) Ptch1 bound to ShhΔ9, which is active but is internalized; (iii) Ptch1 bound to palm-Shh22, which is inactive and accumulates in cilia above levels of unliganded Ptch1; and (iv) Ptch1 bound to Shh, which is inactive and internalized. We speculate that these forms of Ptch1 correspond to distinct conformations and that, reminiscent of bacterial RND pumps, Ptch1 might exert its Smo-suppressing activity by conformational cycling. In this model (Fig. 7B), Shh binds Ptch1 and engages the effector site, which traps Ptch1 in one conformation, thus inhibiting it by interrupting its hypothetical conformational cycle.
The behavior of Ptch1 mutants that cause GS can also be understood in light of this model. We find that two of the most oncogenic mutants, mouse Ptch1G495V and Ptch1D499Y, which retain just over 10% of wild-type activity (7), are severely compromised in binding palm-Shh22, although they bind Shh. We speculate that this indicates the mutants are trapped in a conformation distinct from wild-type Ptch1 (Fig. 7B); it must be emphasized that trapping is not complete, as the mutants retain significant activity and they still respond to ligand. However, we propose that stabilizing any Ptch1 conformation relative to others will reduce activity. Many aspects of this mechanism remain to be elucidated, especially the structural basis of different Ptch1 conformations.
An unresolved issue is the role of Ptch1 internalization in Hh signaling. In principle, Shh could inhibit Ptch1 by a dual mechanism: direct inhibition and removal from cilia by internalization. However, internalization is not necessary for Ptch1 inhibition (35). Furthermore, internalization is insufficient for Ptch1 inhibition, as demonstrated by ShhΔ9, which induces robust Ptch1 internalization but is inactive; perhaps low levels of Ptch1 remaining at cilia are sufficient to repress Smo. We speculate that ligand-induced Ptch1 internalization might modulate Hh signaling, particularly at nonsaturating levels of ligand or in cells expressing lower levels of Ptch1. Quantitative studies of signaling in different cell types will be important for determining if Ptch1 internalization contributes to Hh pathway output.
Materials and Methods
Reagents.
The following small molecules were obtained commercially: SAG from Axxora (≥98%) and SANT1 from Calbiochem (≥95%).
Chemical Synthesis.
Synthesis and characterization of the photoreactive palmitic acid analog, 15-azi-hexadecanoic acid, is described in SI Appendix, SI Materials and Methods.
Synthetic Peptides.
Peptides were custom-synthesized by solid-phase synthesis and were purified by HPLC to greater than 90% purity (Biomatik and Massachusetts General Hospital Peptide/Protein Core Facility). To generate photoreactive peptides, the peptides were first synthesized on solid support with a biotinyl moiety attached to the ε-amino group of a C-terminal lysine residue. The deprotected N-terminal α-amino group of the peptide was then acylated with fivefold excess of 15-azi-hexadecanoic acid under standard conditions. The peptide was deprotected and released from solid phase, after which it was HPLC-purified and converted to chloride salt (final purity ≥98%). HPLC traces and mass spectrometric analysis of synthetic peptides is shown in SI Appendix, Fig. S1.
Cell Culture and Stable Cell Lines.
3T3Flp-In (Life Technologies) and Shh Light II reporter cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% bovine calf serum, penicillin, and streptomycin. Human 293T cells and mouse embryonic fibroblasts (MEFs) were grown in DMEM with 10% FBS, penicillin, and streptomycin. Lentiviruses were packaged in 293T using standard methods and were used to generate stable 3T3 and MEF lines by infection followed by selection with blasticidin, as described (36). Alternatively, 3T3Flp-In cells stably expressing various constructs were generated using the Flp-In System (Life Technologies), according to manufacturer’s instructions. Expression of tagged constructs was confirmed by immunofluorescence, immunoblotting, and by Hh activity assays.
Antibodies.
Anti-mouse Smo and anti-mCherry antibodies have been described before (37). Other antibodies used in this study were the following: mouse anti-acetylated tubulin (Sigma), rat anti-HA (Roche), goat anti-GFP (Rockland), goat anti-human Gli1 (R&D Systems), goat anti-human Gli3 (R&D Systems), mouse anti-GSK3β (BD Biosciences).
DNA Constructs.
Expression constructs were generated by overlapping PCR and were subcloned into lentiviral production vector or into pEF5-FRT (Life Technologies). Membrane proteins were C-terminally tagged with mCherry, eGFP, or one copy of the influenza HA epitope, as indicated. The constructs encoding membrane proteins used in this study were the following: full-length mouse Patched1 (Ptch1) and point mutants thereof; Ptch1Δloop2 (mouse Ptch1 with amino acids 787–998 deleted); DispASmoICD (amino acids 1–1128 of mouse DispA fused to amino acids 543–793 of mouse Smo); NPC1SmoICD (amino acids 1–1,253 of mouse NPC1 fused to amino acids 543–793 of mouse Smo); and full-length mouse Smo. Secreted N-terminal peptides of human Shh were C-terminally fused to HT and one HA tag or to NanoLuc luciferase and an HA tag. Unless otherwise noted, all Shh proteins used in this study were not cholesterol-modified, being expressed from constructs with the C-terminal auto-processing domain deleted. The signal sequence of human calreticulin was used for secretion of N-terminally truncated Shh proteins.
Production of Secreted Fusions and Shh Ligands.
Secreted proteins were produced in 293T cells by transient transfection and were collected for 48 h in DMEM, without serum. The conditioned media was concentrated, and was used in reporter assays diluted in fresh DMEM. For the experiment in SI Appendix, Fig. S6C, cholesterol-modified Shh proteins were released in serum-free media by Xenopus tropicalis Scube2 protein, as described (36). For comparing different secreted proteins, conditioned media were normalized based on Western blotting with anti-HA or anti-Shh antibodies. To generate fluorescent HT fusions, the concentrated conditioned media was incubated with 100 μM TMR Halo ligand (Promega) for 30 min at room temperature. Labeled HT fusion was separated from excess Halo ligand on a NAP-5 desalting column (GE Healthcare).
Hh Reporter Assays.
Hh pathway activity was measured using Shh Light II cells, which express firefly luciferase from an Hh-responsive promoter and Renilla luciferase from a constitutive promoter. Confluent cell cultures were treated for 30 h with the desired agents in DMEM. Firefly and Renilla luciferase were measured using Dual-Glo system (Promega) and a VICTOR2 plate reader (Wallac). Activity is expressed as a ratio of firefly to Renilla counts, normalized to 1 for untreated cells. All reporter assay experiments were performed at least twice. For all experiments, three biological replicates were performed for each treatment. Data points represent the mean, with error bars showing the SD. Dose–response curves were plotted in Prism (GraphPad), using nonlinear regression to a four-parameter curve.
Quantitative RT-PCR.
Confluent cell cultures were starved overnight and were then treated with the desired agents in DMEM for 24 h. Total RNA was extracted using RNA-Bee (TelTest), treated with RQ1 DNase (Promega), and purified by another round of RNA-Bee extraction. cDNA was synthesized using Transcriptor reverse transcriptase and random hexamers (Roche). Real-time PCR was performed using FastStart SYBR Green Master reagent (Roche) on a Rotor-Gene 6000 (Corbett Robotics). Gli1 transcript was used as a measure of Hh pathway activity, normalized to Rpl27 transcript. The primers used for amplification of mouse Gli1 transcript were 5′-GGCCAATCACAAGTCAAGGT-3′ and 5′-TTCAGGAGGAGGGTACAACG-3′; for amplification of mouse L27 transcript, they primers were 5′-GTCGAGATGGGCAAGTTCAT-3′ and 5′-GCTTGGCGATCTTCTTCTTG-3′. For all quantitative PCR experiments, three biological replicates were performed for each treatment. Data points show the mean with error bars indicating SD.
Immunofluorescence.
Cells plated on glass coverslips were incubated overnight with the desired factors in DMEM. For acute treatments (<6 h), cells were starved overnight before treatment. Cells were fixed in 3.7% (wt/vol) formaldehyde in PBS, permeabilized with TBST (TBS + 0.2% Triton X-100), and blocked with 25 mg/mL BSA in TBST (TBST-BSA). Antibody incubations were performed in TBST-BSA with intervening TBST washes. The primary antibodies used were the following: mouse anti-acetylated tubulin (Sigma, 1:3,000 dilution), rabbit anti-mCherry (2 μg/mL), goat anti-GFP (2 μg/mL), and goat anti-mouse Smo (2 μg/mL). Alexa-conjugated secondary antibodies (Life Sciences) were used at 1 μg/mL.
Ciliary Localization Measurements.
Immunofluorescence images were acquired on an automated TE2000E microscope (Nikon) equipped with an OrcaER camera (Hamamatsu), using 40× Plan Apo 0.95 N.A. or 100× Plan Apo 1.4 N.A. objectives (Nikon). For quantifying ciliary localization, cilia were segmented based on acetylated tubulin images, and fluorescence intensities of Ptch1-eGFP or endogenous mouse Smo at cilia were calculated using custom image analysis software implemented in MATLAB, as previously described (37). Ptch1-eGFP and Smo intensities at cilia are presented as box plots that indicate the median and the 25th and 75th percentiles of a population of n > 300 cilia for each condition. For select experiments, cilia were manually scored and represented as the mean of three separate counts of n = 30 cilia for each condition. Error bars represent SD.
Cell-Binding Assays.
Binding of fluorescent HT fusions to primary cilia was assayed by live imaging, using a 100× PlanApo 1.4 N.A. objective (Nikon). Cells stably expressing C-terminally eGFP-tagged ciliary protein were incubated with TMR-labeled HT fusion (750 ng/mL final concentration in phenol red-free DMEM) for 30 min before imaging. For competition, synthetic peptides were added to 5 μM, and unlabeled HT fusions were added in 40-fold excess over TMR-labeled HT fusion. GFP and TMR fluorescence were measured in a region manually drawn around each cilium. For background subtraction, fluorescence was measured in an identical region elsewhere in the image. Relative TMR intensity, defined as the average TMR intensity divided by GFP intensity, was calculated for each cilium. Relative TMR intensity for each condition was normalized to binding of Shh22-HT-TMR to Ptch1-eGFP, which was designated as 100%. Each data point represents the mean relative TMR intensity of five or more cilia with error bars representing SE.
Immunoblotting.
Harvested cells were resuspended in TBS supplemented with protease inhibitors (Roche), 5 mM magnesium chloride, and benzonase. Cells were lysed in 1% SDS for 15 min at room temperature. The lysate was clarified by centrifugation at 20,000 × g for 10 min. The supernatant was mixed with SDS/PAGE sample buffer and 50 mM DTT, separated on a 5–15% (wt/vol) polyacrylamide gradient gel, and transferred to nitrocellulose membrane. The primary antibodies for immunoblotting were used at a concentration of 1 μg/mL in TBST with 5% (wt/vol) nonfat dry milk.
Photocrosslinking.
Confluent cultures of NIH 3T3 or Ptch1-null cells, stably expressing various Ptch1 constructs, were starved overnight and incubated in DMEM with 3.5 μM photoreactive peptide (15-azi-palm-Shh22-biotin) or negative control peptide (palm-Shh22-biotin) in the absence or the presence of competitor peptide (palm-Shh22, 10 μM), for 1 h at 37 °C. Cells were washed with DMEM to remove excess peptide, UV-irradiated on ice for 15 min, and then lysed in TBS with 1% SDS, as described for immunoblotting. The clarified lysate was diluted with TBS with Triton X-100 to final concentrations of 2% (vol/vol) Triton X-100 and 0.2% SDS, and equal amounts of protein were incubated with streptavidin beads (Pierce). After washing with TBS with 2% (vol/vol) Triton X-100, material bound to beads was eluted by boiling in sample buffer supplemented with 50 mM DTT. The eluate was analyzed by SDS/PAGE, followed by immunoblotting with anti-GFP (Rockland) or anti-HA (Roche) antibodies. A lysate portion was analyzed for input.
Nondenaturing Pull Down.
293T cells expressing Ptch1-mCherry were incubated with peptides (3.5 μM), as described for photocrosslinking. The cells were lysed in TBS with 1% Triton X-100, and the clarified lysate was bound to streptavidin beads. After washing with lysis buffer, precipitated material was eluted and analyzed as described for photocrosslinking, using anti-mCherry antibodies.
Supplementary Material
Acknowledgments
We thank members of the A. Salic laboratory for helpful discussions and Ashok Khatri (Massachusetts General Hospital) for photoreactive peptide synthesis. This work was supported by NIH grants R01 GM092924 and GM110041.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606719113/-/DCSupplemental.
References
- 1.Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science. 2004;304(5678):1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 3.Roessler E, et al. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009;30(10):E921–E935. doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakano Y, et al. A protein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature. 1989;341(6242):508–513. doi: 10.1038/341508a0. [DOI] [PubMed] [Google Scholar]
- 5.van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382(6591):547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 6.Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86(2):221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 7.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418(6900):892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 8.Tseng TT, et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1(1):107–125. [PubMed] [Google Scholar]
- 9.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 10.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384(6605):176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 11.Stone DM, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384(6605):129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 12.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317(5836):372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 13.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102(32):11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu AJ, Zheng L, Suyama K, Scott MP. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 2003;17(10):1240–1252. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437(7061):1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 16.Chamoun Z, et al. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science. 2001;293(5537):2080–2084. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- 17.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18(6):641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepinsky RB, et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273(22):14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- 19.Petrova E, Rios-Esteves J, Ouerfelli O, Glickman JF, Resh MD. Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat Chem Biol. 2013;9(4):247–249. doi: 10.1038/nchembio.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams KP, et al. Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J Cell Sci. 1999;112(Pt 23):4405–4414. doi: 10.1242/jcs.112.23.4405. [DOI] [PubMed] [Google Scholar]
- 21.Pepinsky RB, et al. Mapping sonic hedgehog-receptor interactions by steric interference. J Biol Chem. 2000;275(15):10995–11001. doi: 10.1074/jbc.275.15.10995. [DOI] [PubMed] [Google Scholar]
- 22.Hardy RY, Resh MD. Identification of N-terminal residues of Sonic Hedgehog important for palmitoylation by Hedgehog acyltransferase. J Biol Chem. 2012;287(51):42881–42889. doi: 10.1074/jbc.M112.426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Los GV, Wood K. The HaloTag: A novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 24.Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87(4):661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- 25.Frank-Kamenetsky M, et al. Small-molecule modulators of Hedgehog signaling: Identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1(2):10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99(22):14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9(1):21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 28.Whalen DM, Malinauskas T, Gilbert RJ, Siebold C. Structural insights into proteoglycan-shaped Hedgehog signaling. Proc Natl Acad Sci USA. 2013;110(41):16420–16425. doi: 10.1073/pnas.1310097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor FR, et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry. 2001;40(14):4359–4371. doi: 10.1021/bi002487u. [DOI] [PubMed] [Google Scholar]
- 30.Nusse R. Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development. 2003;130(22):5297–5305. doi: 10.1242/dev.00821. [DOI] [PubMed] [Google Scholar]
- 31.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenzen T, et al. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10(5):647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Lum L, et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299(5615):2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 34.Song JY, Holtz AM, Pinskey JM, Allen BL. Distinct structural requirements for CDON and BOC in the promotion of Hedgehog signaling. Dev Biol. 2015;402(2):239–252. doi: 10.1016/j.ydbio.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torroja C, Gorfinkiel N, Guerrero I. Patched controls the Hedgehog gradient by endocytosis in a dynamin-dependent manner, but this internalization does not play a major role in signal transduction. Development. 2004;131(10):2395–2408. doi: 10.1242/dev.01102. [DOI] [PubMed] [Google Scholar]
- 36.Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Reports. 2012;2(2):308–320. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedelcu D, Liu J, Xu Y, Jao C, Salic A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat Chem Biol. 2013;9(9):557–564. doi: 10.1038/nchembio.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.