Significance
Unconventional splicing of unspliced X-box–binding protein 1 (XBP1u) mRNA on endoplasmic reticulum (ER) is an important process by which signals are transferred from the ER to the nucleus to maintain ER homeostasis. Newly synthesized XBP1u protein drags its own mRNA as a ribosome nascent chain complex to the ER; however, its precise location and molecular mechanism of ER recruitment remain unknown. We show that translational pausing of XBP1u is necessary for the recognition of the internal signal sequence of XBP1u by canonical secretory machinery. Interestingly, most XBP1u targeted to the ER was not imported into the ER lumen but was associated with the ER membrane, suggesting a noncanonical mechanism by which mRNA substrates are targeted to the ER for unconventional splicing.
Keywords: XBP1 mRNA, translational pausing, SRP, translocon, unfolded protein response
Abstract
Unconventional mRNA splicing on the endoplasmic reticulum (ER) membrane is the sole conserved mechanism in eukaryotes to transmit information regarding misfolded protein accumulation to the nucleus to activate the stress response. In metazoans, the unspliced form of X-box–binding protein 1 (XBP1u) mRNA is recruited to membranes as a ribosome nascent chain (RNC) complex for efficient splicing. We previously reported that both hydrophobic (HR2) and translational pausing regions of XBP1u are important for the recruitment of its own mRNA to membranes. However, its precise location and the molecular mechanism of translocation are unclear. We show that XBP1u-RNC is specifically recruited to the ER membrane in an HR2- and translational pausing-dependent manner by immunostaining, fluorescent recovery after photobleaching, and biochemical analyses. Notably, translational pausing during XBP1u synthesis is indispensable for the recognition of HR2 by the signal recognition particle (SRP), resulting in efficient ER-specific targeting of the complex, similar to secretory protein targeting to the ER. On the ER, the XBP1u nascent chain is transferred from the SRP to the translocon; however, it cannot pass through the translocon or insert into the membrane. Therefore, our results support a noncanonical mechanism by which mRNA substrates are recruited to the ER for unconventional splicing.
Errors in protein translocation to the endoplasmic reticulum (ER) and subsequent protein folding are associated with a number of diseases. Accordingly, a detailed understanding of the mechanisms that mediate these complex processes is necessary. In eukaryotes, secretory proteins are initially translated by cytosolic ribosomes. When the N-terminal signal peptide reaches outside of the peptide exit channel in the ribosome, the ribosome nascent chain (RNC) complex is recognized by the signal recognition particle (SRP) and peptide elongation is slowed (1–5). This SRP–RNC complex is then recruited to the ER via the affinity between SRP and SRP receptor (SR) that exists in the ER membrane (2, 3). The RNC complex is then delivered to the polypeptide channel in the ER (i.e., the Sec61 translocon) (6). SRP is released from the RNC, which cancels the slowed-down elongation. As a result, the ER-targeted ribosome cotranslationally translocates its synthesizing polypeptide into the luminal space of the ER. The translocated protein is folded into its native 3D structure with the help of molecular chaperones and folding enzymes (7). The folded proteins are sorted to their final destinations to exert their functions.
The burden of new proteins entering the ER varies widely among conditions, such as cell differentiation, environmental conditions, and the physiological state of the cell (8, 9). In addition, the folding capacity in the ER is easily compromised by many stressful conditions, including glucose starvation, virus infection, and perturbations in the Ca2+ concentration. Therefore, it is necessary for cells to manage the imbalance between the load of newly entering proteins and the folding capacity in the ER, and this imbalanced situation is referred to as ER stress. The inositol requiring enzyme 1α (IRE1α)-X-box–binding protein 1 (XBP1) pathway is the most highly conserved regulatory system that mediates ER stress among eukaryotes (8–10). IRE1α is a single-spanning transmembrane protein in the ER with a luminal domain for ER stress sensing and a cytosolic domain harboring kinase-endoribonuclease activity. Upon ER stress, IRE1α forms dimer or oligomer and then transautophosphorylates, resulting in a structural change of its RNase domain to the active form (11–13). Activated IRE1α cleaves two specific sites of the precursor form of XBP1(XBP1u) mRNA (14, 15). The recently identified RtcB joins the 5′ and 3′ fragments, which results in the removal of the 26-base fragment in the middle of the open-reading frame (16–19). This splicing reaction creates a translational frame shift. The spliced XBP1(XBP1s) mRNA encodes the functional transcription factor XBP1s, which induces the transcription of genes to alleviate ER stress, including molecular chaperones and factors for ER-associated degradation (20).
Previously, we reported that the protein encoded by XBP1u cotranslationally recruits its own mRNA to the ER membrane as an RNC complex to guarantee highly efficient splicing upon ER stress (21). Furthermore, we found that XBP1u translation is paused during the synthesis of its C-terminal region, which contributes to ER-targeting efficiency and splicing efficiency (22). In our initial model, we proposed that nascent XBP1u attaches to the ER membrane via the membrane affinity of hydrophobic region 2 (HR2) (22, 23). However, the specific affinity of XBP1u to the ER membrane was not established because an in vitro experiment showed that it also had an affinity to synthetic liposomes (21). Here, we show that the XBP1u–RNC complex is specifically recruited to the ER membrane via the SRP-mediated ER-targeting pathway in an HR2-dependent manner. In addition, translational pausing is indispensable for HR2 recognition by SRP. Interestingly, in contrast to the canonical route of ER targeting, XBP1u is not passed through the Sec61 translocon, but is associated with the cytosolic side of the ER membrane. Taken together, we propose that XBP1u-RNC is targeted to the ER membrane to transmit signals of the proteostatic perturbation of secretory proteins in the ER via a distinct route.
Results
The Cotranslational Targeting of XBP1u Is Specific to the ER Membrane.
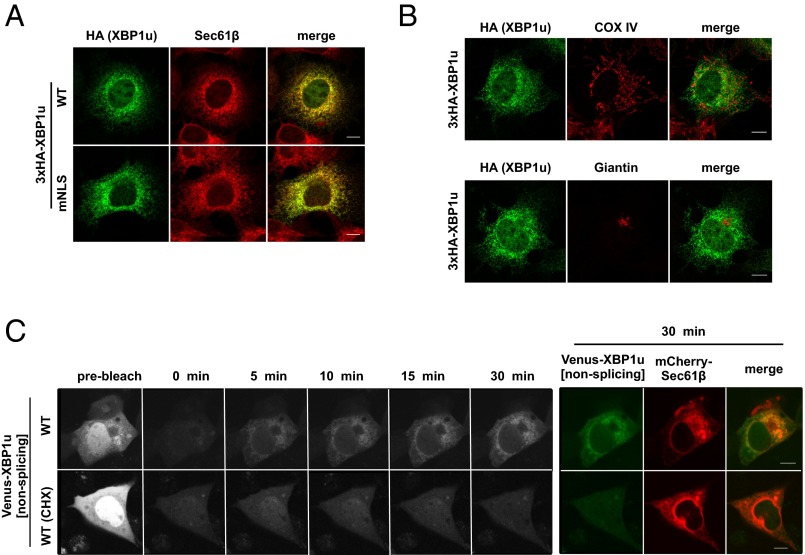
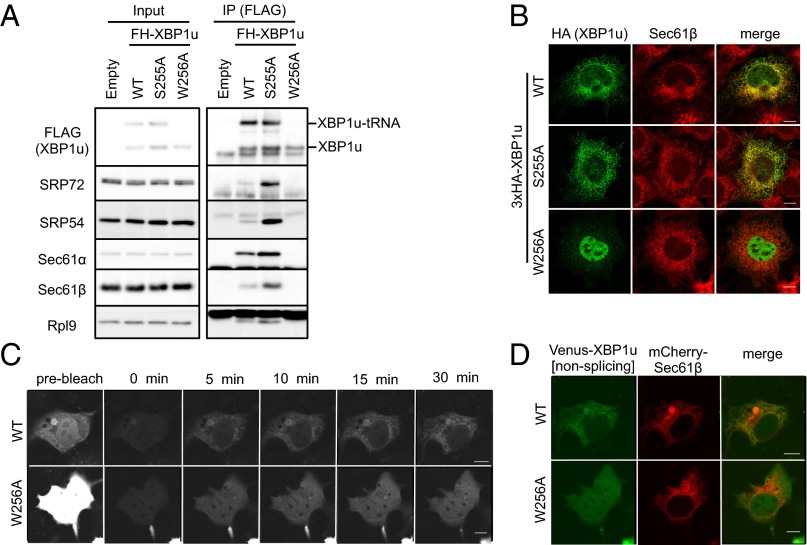
We examined whether the targeting site of the XBP1u–RNC complex is restricted to the ER membrane. Based on immunofluorescence staining, XBP1u is distributed in a typical ER pattern and inside nuclei (Fig. 1A). Importantly, mitochondria and Golgi marker proteins (i.e., COX IV and Giantin, respectively) did not colocalize with XBP1u (Fig. 1B). ER localization became more obvious when the nuclear localization signals of XBP1u were abrogated by mutations (Fig. 1A and Fig. S1). The initial targeting site of XBP1u should be visible in the immunofluorescence images, but it is rather the heterogeneous mixture of the protein as time elapses from synthesis to degradation that is visible. Therefore, we performed fluorescence recovery after photobleaching (FRAP) using Venus-XBP1u to visualize the initial targeting site of XBP1u (Fig. 1C). For this experiment, preexisting fluorescence in COS-7 cells expressing Venus-XBP1u was bleached, and subsequent fluorescence was observed. Given the short lag between protein synthesis and Venus protein fluorescence (24), the emerging fluorescence should represent the distribution of Venus-XBP1u soon after synthesis. We defined the observed distribution as the protein-targeting site. After photobleaching, the signal mainly overlapped with the signal of the fluorescent ER protein mCherry-Sec61β (Fig. 1C). The emerging ER signal reflected the newly synthesized protein; the signal was completely abolished in cells treated with the translational inhibitor cycloheximide (Fig. 1C). These results clearly indicate that the XBP1u protein is specifically recruited to the ER membrane, and not to any other organellar membranes.
Fig. 1.
ER is the initial targeting site of XBP1u. (A and B) HA-XBP1u[nonsplicing] or nuclear localization signal (NLS)-defective mutant (mNLS) HA-XBP1u[mNLS/nonsplicing] transiently expressed in Cos-7 cells was costained with an ER marker (Sec61β), mitochondria marker (COX IV), and Golgi marker (Giantin). (C) Venus-XBP1u[nonsplicing] and mCherry-Sec61β were transiently expressed in Cos-7 cells. The preexisting fluorescence of Venus-XBP1u[nonsplicing] was photobleached, and the emerging fluorescence was observed at the indicated time points. The emerging signal was abrogated by cycloheximide (CHX; 200 μM) treatment for 1 h before the FRAP experiment. The rightmost colored panel shows a merged image of Venus-XBP1u[nonsplicing] and mCherry-Sec61β (ER marker) at 30 min after photobleaching. (Scale bars, 10 μm.)
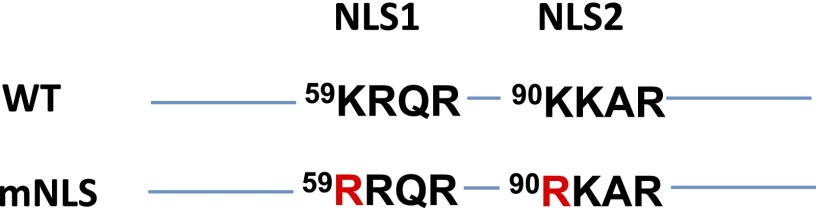
Fig. S1.
Schematic representation of amino acid substitutions in human XBP1u to disrupt two NLS sequences.
ER Targeting of XBP1u Requires Proteinaceous Components.
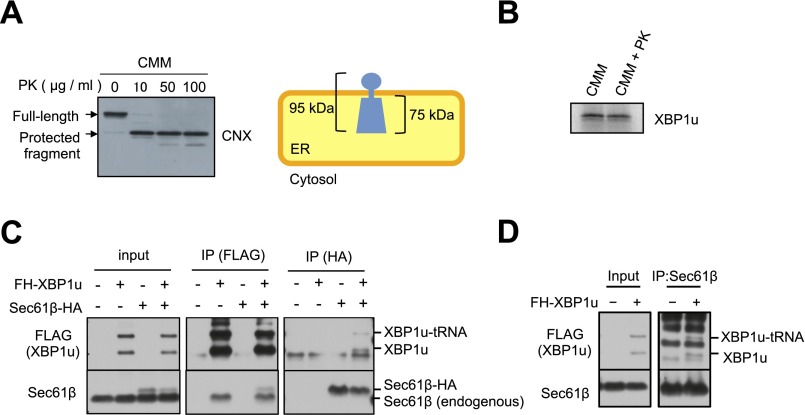
The ER-specific targeting of XBP1u implies the existence of targeting machinery for the ER membrane, but not for the membranes of other organelles. This specificity may be explained by the existence of a receptor for XBP1u on the ER membrane or a specific lipid composition of the ER membrane that confers specificity. To examine these two explanations simultaneously, we prepared a protease-treated microsome, which removes proteins from the cytosolic surface of microsomes without affecting the lipid composition (Fig. S2A). This protease-treated microsome exhibited dramatically decreased affinity to XBP1u (Fig. 2A). This result indicates that a proteinaceous component on the ER membrane is required for the ER targeting of XBP1u.
Fig. S2.
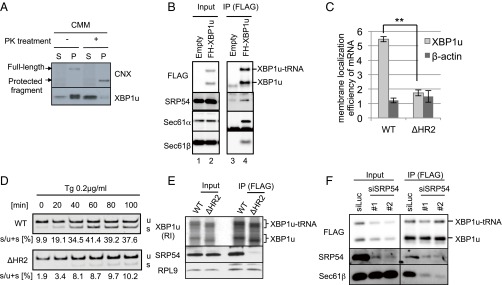
Supporting information for Fig. 2. (A) Preparation of proteinase K (PK)-treated microsomes. CMMs were treated with the indicated concentration of PK. The efficiency of surface protein digestion was evaluated by the removal of a short cytosolic tail of calnexin. (Right) Membrane topology of calnexin is shown. The full-length calnexin and protected fragment of calnexin were detected using the antibody against the ER-luminal portion of calnexin. (B) XBP1u translation was not affected by PK treatment in the presence of a PK inhibitor. XBP1u was synthesized via the in vitro translation system with mock-treated CMMs or PK-treated CMMs with PMSF (2 mM) and benzamidine (2 mM). (C) Endogenous Sec61β and/or HA-tagged Sec61β was coimmunoprecipitated with FH-XBP1u and FH-XBP1u-tRNA using anti-FLAG antibodies. In contrast, FH-XBP1u and/or FH-XBP1u-tRNA was coimmunoprecipitated with Sec61β and/or Sec61β-HA from the cell lysate derived from HEK293T cells transiently expressing FH-XBP1u and/or Sec61β-HA. Anti-FLAG or anti-HA antibody was used for immunoprecipitation (IP). (D) Coimmunoprecipitation of endogenous Sec61β with FH-XBP1u from the cell lysate derived from HEK293T cells transiently expressing FH-XBP1u. Anti-Sec61β antibody was used for immunoprecipitation.
Fig. 2.
ER targeting machinery for secretory proteins interacts with XBP1u. (A) FLAG-XBP1u-HA was synthesized with RRL in the presence or absence of nontreated or proteinase K (PK)-treated CMMs. Then, CMM-associated proteins were separated from free proteins by ultracentrifugation. P, pellet; S, supernatant. Details regarding the PK treatment of microsomes are provided in Fig. S2. FLAG-XBP1u-HA was detected using an anti-FLAG antibody. (B) Coimmunoprecipitation of SRP54 or translocon components with FLAG-His-XBP1u[nonsplicing] (FH-XBP1u) transiently expressed in HEK293T cells. XBP1u and XBP1u-tRNA indicate full-length and translationally paused FH-XBP1u, respectively. IP, immunoprecipitation. (C) Membrane localization efficiencies of both XBP1u and HR2-deleted XBP1u (ΔHR2) mRNA transiently expressed in HEK293T cells. β-Actin was used as a control for cytosolic mRNA. Bars indicate SD. **P < 0.01 (n = 3) using Student’s t test. (D) Splicing of XBP1u or ΔHR2 mRNA transiently expressed in HEK293T cells was determined by RT-PCR after treatment with thapsigargin (Tg; 0.2 μg/mL) for the indicated times. s, spliced form of XBP1 mRNA; u, unspliced form of XBP1 mRNA. (E) FH-XBP1u[WT] or FH-XBP1u[ΔHR2] translated with RRL was coimmunoprecipitated with SRP54 and ribosomal protein L9 (RPL9) using anti-FLAG antibodies. XBP1u was detected by autoradiography, whereas SRP54 and RPL9 were detected by Western blotting. (F) Coimmunoprecipitation of Sec61β with FH-XBP1u[nonsplicing] transiently expressed in HEK293T cells, which were treated with siRNAs against SRP54 or luciferase (control) for 72 h until the immunoprecipitation assay.
The ER-Targeting Machinery for Secretory Proteins Interacts with XBP1u.
To identify the proteinaceous component for the ER targeting of XBP1u, we performed coimmunoprecipitation and mass spectrometry (Table S1). We identified components of the ER-targeting machinery for secretory proteins, including SRP (SRP72) and Sec61 translocon (Sec61α, Sec61β, and Sec61γ), as factors that interact with XBP1u (Table S1). The interactions of SRP and the Sec61 translocon with XBP1u were further confirmed by coimmunoprecipitation assays using anti-FLAG (FLAG-His-XBP1u) antibody for Sec61α and Sec61β, as well as the other subunits of SRP, SRP54, and SRP72 (Fig. 2B and Fig. S2C; see also Fig. 4A). Importantly, the interactions between XBP1u and the Sec61 translocon components were also confirmed by coimmunoprecipitation assay using anti-HA (Sec61β-HA) or anti-Sec61β antibody (Fig. S2 C and D).
Fig. 4.
Translational pausing enables XBP1u-RNC to be a client for the SRP-mediated ER-targeting pathway. (A) Coimmunoprecipitation of FH-XBP1u[WT], FH-XBP1u[S255A], and FH-XBP1u[W256A] with the indicated proteins in the cell lysate derived from HEK293T cells transiently expressing FH-XBP1u[WT] and its variants. FH-XBP1u[S255A] and FH-XBP1u[W256A] are the prolonged- and pausing-defective mutants, respectively. (B) Wild-type and variants of HA-XBP1u[nonsplicing] transiently expressed in Cos-7 cells were costained with endogenous Sec61β. (C and D) FRAP analysis of Venus-XBP1u[WT/nonsplicing] or Venus-XBP1u[W256A/nonsplicing] transiently expressed in Cos-7 cells is shown. (D) Merged image of Venus-XBP1u[WT/nonsplicing] and mCherry-Sec61β (ER marker) at 30 min after photobleaching. The detailed analysis is the same as described in Fig. 1C. (Scale bars, 10 μm.)
Based on these data, we hypothesized that SRP recruits the XBP1u–RNC complex to the Sec61 translocon in the ER membrane. In the initial step of ER targeting, SRP recognizes a hydrophobic stretch in a nascent polypeptide, such as signal peptides at the N terminus or central signal anchor sequences. Because XBP1u has a hydrophobic region (i.e., HR2), which is indispensable for the membrane targeting of XBP1u mRNA and efficient splicing under ER stress (21) (Fig. 2 C and D), XBP1u is likely to be recognized by SRP via HR2. Consistent with this expectation, in vitro synthesized XBP1u interacted with SRP54 in an HR2-dependent manner (Fig. 2E). Importantly, knocking down SRP54 greatly reduced the interaction between XBP1u and a subunit of the translocon (Sec61β), which strongly suggests that XBP1u-RNC is recruited to the ER by the ER-targeting pathway for secretory proteins (Fig. 2F).
SRP Recruits XBP1u-RNC to the ER.
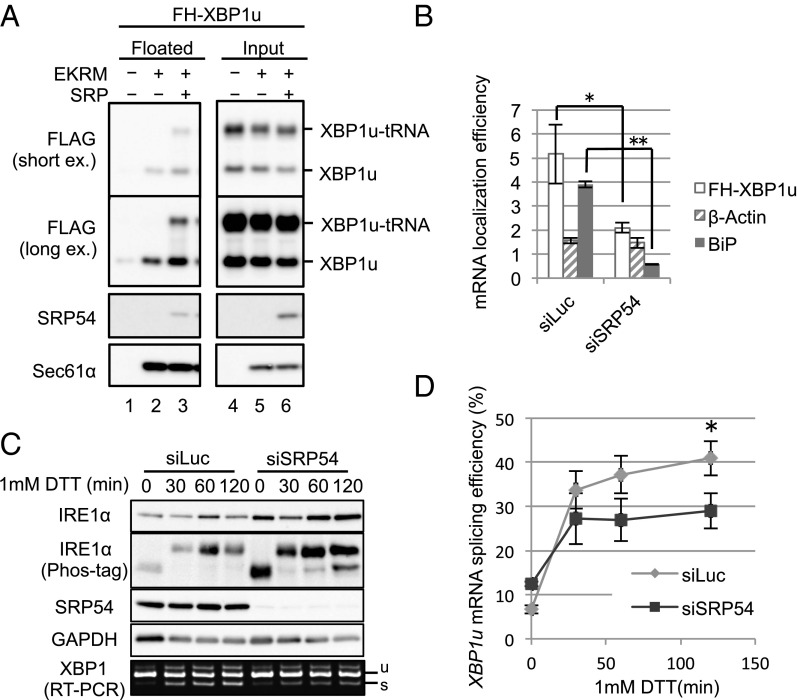
We directly examined the functional role of SRP in the ER targeting of XBP1u-RNC. Specifically, we performed a membrane-flotation assay (25) for XBP1u-RNC using canine pancreatic microsomal membranes (CMMs) and the in vitro translation system composed of wheat germ extract (WGE) in which SRP is depleted (26). As shown in Fig. 3A, in the absence of microsomes, neither the full-length nor the tRNA-attached form (XBP1u-tRNA) of XBP1u was detected in the floating fraction. In the presence of the microsomes, XBP1u-tRNA was not detected in the floating fraction, although a weak signal of the full-length form was observed. In contrast, the addition of SRP with the microsomes resulted in a dramatic increase in XBP1u-tRNA in the floating fraction and a modest increase in the full-length form. This result indicates the absolute requirement of SRP for ER targeting of XBP1u-RNC. Microsome binding of full-length XBP1u, even in the absence of SRP, is presumably due to the direct affinity of this protein to membranes, as evidenced by the affinity of XBP1u to synthetic liposomes (21).
Fig. 3.
SRP recruits XBP1u-RNC to the ER. (A) FH-XBP1u was translated in WGE in the presence or absence of 5 nM purified SRP and CMMs treated with EDTA and high-salt medium (EKRM) to remove preexisting SRP on CMMs. The membrane-bound proteins were separated by a membrane-flotation assay. Proteins in those fractions were detected by immunoblotting. (Top and Middle Top) Same result of immunoblotting exposed for a short time and a long time, respectively, is shown. (B) Membrane localization efficiency (membrane-bound/cytosol) of XBP1u, β-actin, and BiP mRNA in HeLa cells stably expressing FH-XBP1u with SRP54 knockdown for 96 h was quantified as described in Fig. 2C. Bars indicate SD. *P < 0.05, **P < 0.01 (n = 3) in siLuc (Control) vs. siSRP54 using Student’s t test. (C) HeLa cells stably expressing XBP1u-ps with SRP54 knockdown were treated with 1 mM DTT for the indicated times. Western blot analyses of the phosphorylated state of IRE1α and the abundance of indicated proteins are shown. The splicing of XBP1u-ps mRNA was analyzed by RT-PCR. (D) Proportion of the spliced form with respect to the total XBP1u-ps mRNA was calculated from C. Bars indicate SD. *P < 0.05 (n = 3) in siLuc (Control) vs. siSRP54 at 120 min using Student’s t test.
The importance of SRP for ER targeting of XBP1u mRNA in vivo was also obvious. Knocking down SRP54 significantly diminished the ER-targeting efficiency of XBP1u mRNA as well as BiP mRNA, which encodes the ER-luminal protein BiP, whereas the ER-targeting efficiency of cytosolic β-actin mRNA was not affected by the knockdown (Fig. 3B). This result led us to examine the effect of the SRP-mediated ER targeting of XBP1u mRNA on its splicing efficiency under ER stress. SRP54 in HeLa cells was knocked down, and cells were treated with DTT to induce ER stress (Fig. 3C). The activation state of IRE1α was comparable between control and SRP knockdown cells. In contrast, the splicing of XBP1u mRNA was dampened in SRP knockdown cells relative to control cells, although the effect was modest (Fig. 3 C and D). Collectively, these results clearly show that the ER-targeting machinery mediated by SRP is able to recruit XBP1u-RNC to the ER.
Translational Pausing Enables XBP1u-RNC to Be a Client for the SRP-Mediated ER-Targeting Pathway.
According to previous studies, SRP-mediated ER targeting of the RNC complex is predicted to occur not only immediately after the emergence of the signal sequence from the ribosome but also after elongation via the addition of another 60 aa (27). Given that the C-terminal region of XBP1u downstream of HR2 is only 53 aa long, it can be assumed that the normal rate of translation of the C-terminal region makes it difficult for SRP to recognize HR2. We and another group previously reported that the translational elongation of XBP1u is paused near its C terminus (22, 28). Importantly, the paused ribosome exposes HR2 (21). These facts motivated us to examine the contribution of translational pausing to the recognition of HR2 by SRP. When translational pausing was abrogated by the mutation W256A, the interactions between XBP1u and SRP components (SRP54 and SRP72) or translocon subunits (Sec61α and Sec61β) were strongly diminished (Fig. 4A). In contrast, the prolonged-pausing mutation S255A strengthened the interactions. Consistent with this biochemical experiment, an immunofluorescence analysis revealed that the ER localization of XBP1u was accomplished only if the pausing was intact or extended (Fig. 4B). As mentioned above, wild-type XBP1u showed dual localization to the nucleus and the ER (Figs. 1A and 4B). Interestingly, the pausing duration affected the ratio of localization to the ER and the nucleus. In the case of the pausing-defective mutant, W256A, XBP1u tended to accumulate in the nucleus (Fig. 4B). In contrast, the prolonged-pausing mutation S255A caused more ER localization than wild-type XBP1u. A FRAP analysis indicated that the difference in the localization of XBP1u between the wild type and W256A mutant resulted from the efficiency of ER targeting just after synthesis (Fig. 4 C and D). After photobleaching of Venus-XBP1u[WT]-expressing cells, newly emerging fluorescence indicated the typical ER pattern described above (Figs. 1C and 4C). In contrast, the pausing-defective W256A mutant exhibited diffuse distribution throughout the cell without specific ER localization. The FRAP image for the W256A mutant also showed the signal in the nucleus, which is presumably due to the rapid transport of the protein after synthesis (Fig. 4 C and D). Taken together, we concluded that translational pausing enables SRP to recognize HR2 on XBP1u, which allows efficient ER targeting of the XBP1u–RNC complex.
Unusual Mode of the ER Targeting of XBP1u-RNC by SRP.
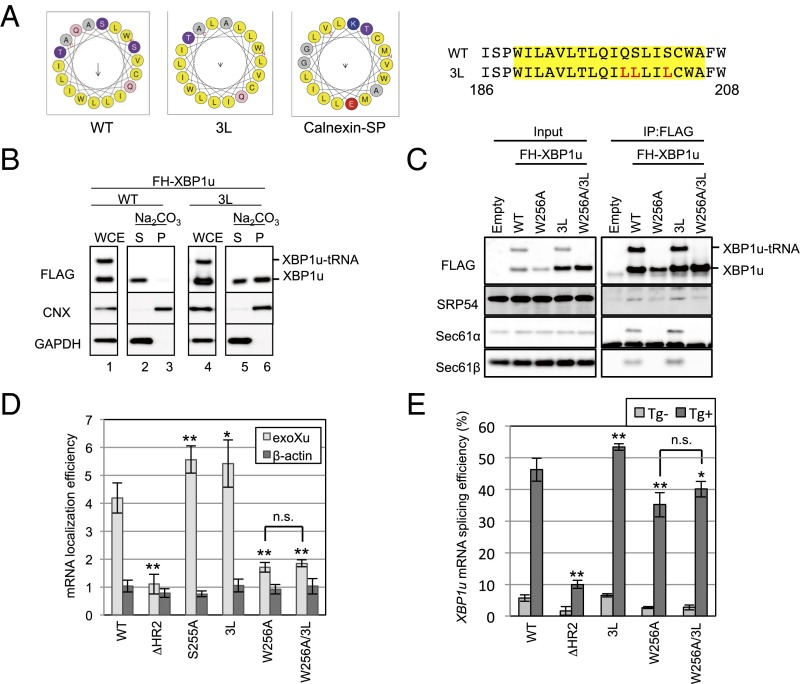
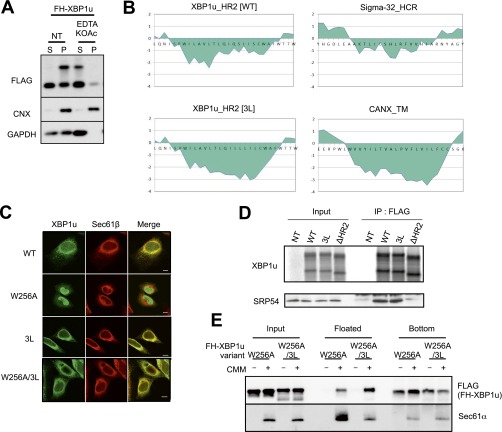
It is generally thought that proteins harboring a signal-anchor sequence are targeted to the translocon in the ER via SRP and inserted into the ER membrane by lateral diffusion from the translocon (29, 30). Based on the location of HR2, XBP1u should be a transmembrane protein if it is targeted to the ER by the canonical SRP-mediated ER-targeting pathway. However, our biochemical analysis indicated that XBP1u exists as a membrane-associated protein rather than a transmembrane protein because membrane attachment of XBP1u was susceptible to alkali or high-salt/EDTA treatment for which transmembrane proteins, such as calnexin, are not extracted (31) (Fig. 5 A and B and Fig. S3A). A possible explanation for these results is that XBP1u is rejected by the translocon after its ER targeting via SRP. Previously, Jungnickel and Rapoport (32) reported a clear example in which a protein harboring the artificial signal sequence is recognized by SRP but is not passed through the channel of the translocon. The artificial signal sequence used in their experiment was less hydrophobic than the canonical sequence. In light of the modest hydrophobicity of HR2, XBP1u may follow the same route as the artificial signal sequence. Accordingly, we examined whether increased hydrophobicity of HR2 affects the fate of XBP1u. As shown in Fig. 5 A and B, additional hydrophobicity of HR2 by the substitution of polar amino acid residues for leucines (designated 3L) converted XBP1u to a transmembrane protein that was resistant to extraction from microsomes by alkaline treatment. Furthermore, immunostaining of XBP1u[3L] showed complete merging of the protein with the ER marker, without the nuclear staining that was observed for XBP1u[WT] (Fig. S3C). Importantly, the affinity to SRP54 was comparable between XBP1u[WT] and XBP1u[3L] (Fig. S3D), which indicates that additional hydrophobicity allows the membrane insertion of the mutant XBP1u via the Sec61 translocon. Interestingly, XBP1u had an affinity to the translocon, even though this protein rejected its insertion into the ER membrane, suggesting that XBP1u localizes to the ER membrane by interacting with the translocon (Fig. 5 B and C and Fig. S3A). Taken together, we concluded that HR2 is an uncanonical signal sequence that is recognized by SRP, but is not capable of opening the pore of the translocon.
Fig. 5.
Unusual mode of ER targeting of XBP1u-RNC via SRP. (A) Helical wheel plots of amino acids in HR2 of XBP1u and the calnexin signal peptide are generated by using HeliQuest (heliquest.ipmc.cnrs.fr) (40). Wild-type and 3L mutant XBP1u are indicated as WT and 3L, respectively. (Right) Amino acid sequences of HR2 of XBP1u are shown. Sequences in the yellow rectangle are used for the helical wheel plot. Red characters in the amino acid sequences are substituted amino acids in the 3L mutant. (B) Microsomes derived from HEK293T cells transiently expressing FH-XBP1u and its variants were treated with sodium carbonate to examine the membrane-binding mode of FH-XBP1u variants. Calnexin (CNX) and GAPDH were used as controls for membrane and cytosolic proteins, respectively. (C) Coimmunoprecipitation of FH-XBP1u and its variants with indicated proteins in the cell lysate derived from HEK293T cells transiently expressing FH-XBP1u and its variants. (D) Membrane localization efficiencies of FH-XBP1u mRNA expressed in HEK293T cells were quantified as described in Fig. 2C. Bars indicate SD. *P < 0.05, **P < 0.01 using ANOVA. (E) Splicing efficiencies (percentage of XBP1s mRNA to total XBP1 mRNA) of XBP1u-ps and its variants transiently expressed in HEK293T cells. ER stress was induced with 0.2 μg/mL Tg for 1 h. Bars indicate SD. *P < 0.05, **P < 0.01 using ANOVA. n.s., nonsignificant difference.
Fig. S3.
Supporting information for Fig. 5. (A) EDTA and high-salt treatment of microsomes in HEK293T cells transiently expressing wild-type or 3L mutant (3L) FH-XBP1u were analyzed by Western blotting. Calnexin (CNX) and GAPDH were used as the control membrane and cytosolic protein, respectively. (B) Kyte and Doolittle hydrophobicity plots for HR2 of WT and 3L mutant XBP1u (window size: 9) are visualized using a ProtScale analysis implemented in ExPASy (web.expasy.org/protscale/). (C) Variants of FH-XBP1u transiently expressed in HeLa cells were costained with Sec61β. (Scale bars, 10 μm.) (D) FH-XBP1u variants translated with RRL were coimmunoprecipitated with SRP54. XBP1u was detected by autoradiography, whereas SRP54 was detected by immunoblotting. (E) Posttranslational ER-targeting assay of FH-XBP1u[W256A] and FH-XBP1u[W256A/3L]. Total protein before fractionation is shown as the input. Floated and bottom fractions are shown. The presence and absence of the ER membrane (EKRM) are indicated by “−” and “+”, respectively. Additional information is provided in Materials and Methods.
Interestingly, even after the introduction of the 3L mutation, XBP1u[W256A] did not exhibit increased affinity to the ER-targeting machinery (Fig. 5C). This result is consistent with the less efficient ER targeting of XBP1u[W256A/3L] mRNA (Fig. 5D) and its lower splicing efficiency under ER stress (Fig. 5E). Although XBP1u[W256A/3L] was localized to the ER, further analysis showed that the protein targeting was not cotranslational, but posttranslational (Fig. S3 C and E). Collectively, these results indicate that translational pausing is an upstream event in the recognition of HR2 by SRP to recruit XBP1u mRNA to the ER, which is different from the typical order of events, in which translational pausing occurs after the recognition of a signal peptide by SRP. Therefore, these results emphasize the critical role of translational pausing in the recognition of HR2 by SRP.
Discussion
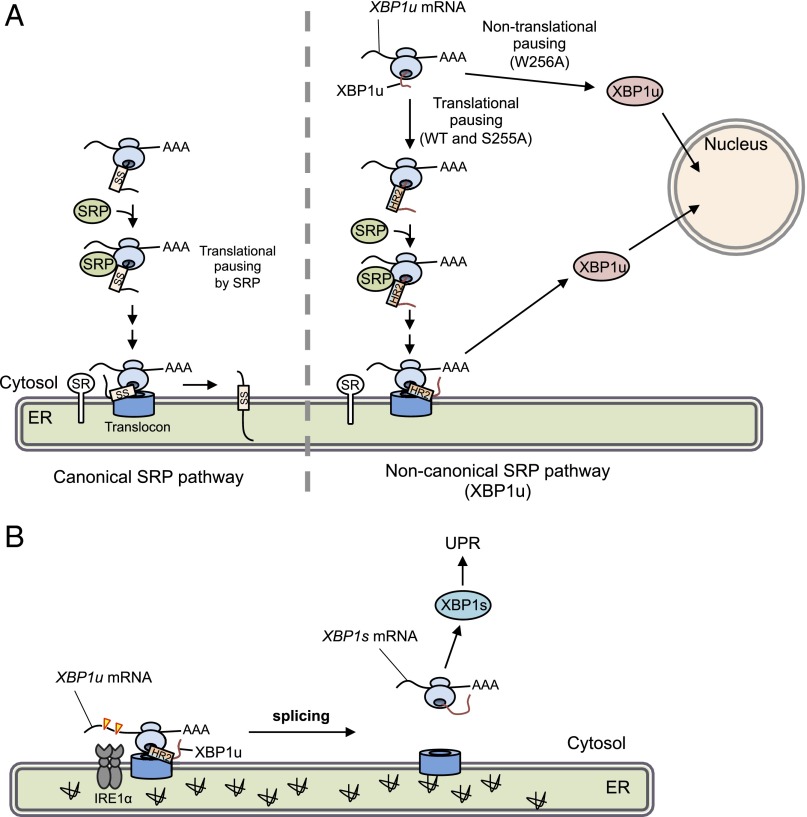
In metazoans, the unconventional splicing of XBP1u mRNA on the cytosolic face of the ER is a key to transmit information regarding misfolded protein accumulation in the ER to the nucleus. This cross-membrane signal transmission is mediated by the membrane-spanning protein IRE1α, which is activated by ER stress and initiates the unconventional splicing of XBP1u mRNA by cleaving two specific sites. Spliced XBP1s mRNA encodes an active transcription factor to activate the transcriptional program to ameliorate ER stress. The low copy number of IRE1α in a cell (e.g., 416 molecules in a HeLa cell) (33) suggests that an active mechanism facilitates the enzyme–substrate interaction to accomplish efficient signal transmission. We previously reported that XBP1u mRNA is actively recruited to membranes via the affinity of its nascent polypeptide to membranes as an RNC complex (21). In a previous study, it was difficult to determine whether the targeting membrane(s) of XBP1u-RNC are nonspecific or limited to the ER membrane because XBP1u was able to associate with synthetic liposomes. In this study, we demonstrated that XBP1u-RNC is specifically recruited to the ER membrane by piggybacking on SRP of the SRP-mediated ER-targeting pathway. The ER-targeting route of XBP1u-RNC is partially different from the general secretory pathway (Fig. 6A). Before engaging the ER-targeting pathway, polypeptide elongation of XBP1u is paused. This pausing fixes XBP1u-RNC in a state that exposes HR2 to the outside of the ribosomal exit tunnel (22). This condition enables SRP to associate with the XBP1u-RNC to recruit it to the SR on the ER. Finally, the XBP1u-RNC is passed to the translocon Sec61 complex. In contrast to the canonical pathway, XBP1u is prevented from entering the luminal space or the ER membrane. Instead, the rejected XBP1u exists as an ER membrane-associated protein. Given that XBP1u has a strong affinity to the translocon (Fig. 2B), it presumably associates with the translocon (Fig. 6). Collectively, we concluded that XBP1u mRNA is specifically recruited to the ER membrane by the SRP-mediated ER-targeting pathway. Our findings regarding SRP involvement in the ER-specific targeting of XBP1u are consistent with previous findings based on different approaches, although the role of translational pausing of XBP1u in the recognition of XBP1u by SRP was not previously evaluated (34).
Fig. 6.
Working model of translational pausing in SRP-mediated localization of XBP1 mRNA. (A) Canonical SRP pathway (Left) and the noncanonical SRP pathway reported here (Right) are indicated. In the latter case, newly synthesized XBP1u is paused at the C terminus region of the nascent XBP1u polypeptide. Under such conditions, HR2 is located just outside of the ribosomal tunnel and is recognized by SRP. The SRP-bound RNC complex associated with its own XBP1u mRNA is recruited to the translocon via SR. After pausing, XBP1u is completely translated, but cannot be inserted into the ER owing to rejection of the translocon. Rejected XBP1u carrying NLS is quickly transported into the nucleus. In contrast, the pausing-defective mutant XBP1u[W256A], whose HR2 cannot be recognized by SRP, is translated and transported into the nucleus. SS, signal sequence, including the signal anchor. (B) Under ER stress, XBP1u mRNA associated with paused RNC on the translocon is efficiently spliced by activated IRE1α, leading to production of the active transcription factor XBP1s, which up-regulates unfolded protein response (UPR) target genes to mitigate ER stress.
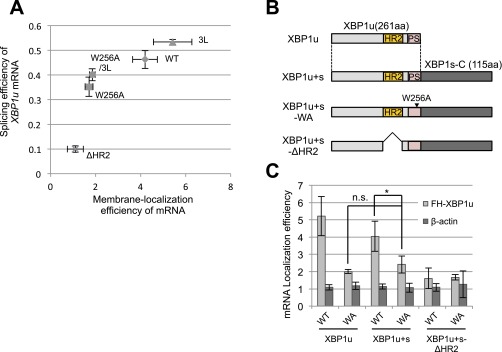
According to the classical viewpoint, cotranslational ER targeting of secretory proteins is accomplished by translational pausing after the recognition of a signal sequence by SRP (2, 3). This process enables the nascent polypeptide to recruit its own mRNA to the ER. In contrast, in the case of XBP1u, translational pausing was a prerequisite for the recognition of XBP1u-RNC by SRP, which enables the ER targeting of XBP1u mRNA. This prerequisite for pausing suggests that SRP is not able to recognize HR2 immediately after exposure from the ribosome. One might suspect that the requirement of translational pausing for HR2 recognition by SRP is related to the relatively modest hydrophobicity of HR2, which extends the time needed for this process relative to the time needed for typical hydrophobic signal sequences (Fig. S4B). However, an increased hydrophobicity of HR2 in the pausing-defective mutant XBP1u[W256A/3L] did not facilitate SRP binding and the ER targeting of its own mRNA, like XBP1u[W256A] (Fig. 5 C and D). Therefore, this impairment is a striking example in which preceding translational pausing is necessary for the recognition of the ER-targeting signal by SRP. Similarly, Frydman and coworkers (35) recently reported that SRP preferentially recognizes a set of proteins harboring a signal sequence with a downstream cluster of suboptimal codons, which slows translational elongation. Further, Weissman and coworkers (36) showed that artificial translational arrest by cycloheximide treatment enables the ER localization of a subset of cytosolic mRNAs encoding the proteins harboring a first transmembrane segment or a signal sequence in the C-terminal region (from 50 to 150 codons before the termination codons). Taken together, these results suggest that translational pausing or the slowdown of elongation extends the time window of the competent state in which SRP is able to recognize the RNC with an exposed signal sequence. However, it is noteworthy that the artificial elongation of the C terminus of XBP1u[W256A] did not rescue the ER-targeting efficiency of its mRNA (Fig. S4 B and C). This result strongly supports the idea that both the specific ribosome configuration caused by translational pausing and the distance from the pausing site to HR2 are critical for SRP to recognize its substrates, at least for HR2 of XBP1u.
Fig. S4.
Splicing efficiency and membrane localization efficiency of XBP1u mRNA. (A) Relationship between splicing efficiency and membrane localization efficiency of XBP1u mRNA according to Figs. 2C and 5 D and E is shown. The bar indicates SD. (B) Primary structure of variants of XBP1u and C-terminal extended XBP1u, XBP1u+s. PS, pausing sequence; WA, W256A. (C) Membrane localization efficiencies of XBP1u variant mRNA transiently expressed in HEK293T cells were quantified as described in Fig. 2C. Bars indicate SD. *P < 0.05 (n = 4) using ANOVA. n.s., nonsignificant difference.
The strong affinity of XBP1u-RNC to the translocon implies the splicing of XBP1u mRNA on the translocon. The results of our previous study support this notion (21). Specifically, we previously demonstrated that the splicing of XBP1u mRNA lacking HR2 (Int [+A]) was abrogated under ER stress. However, the splicing efficiency was restored by introducing the calreticulin signal sequence at the N terminus, and the levels exceeded the levels observed for the wild-type XBP1u. Because an N-terminal signal sequence causes cotranslational translocation of the protein into the luminal space of the ER, the mRNA should remain on the translocon, which indicates that the mRNA is able to be spliced on the translocon with high efficiency. This notion is consistent with a recently published paper showing that IRE1α directly associates with a translocon that could splice XBP1u mRNA (34).
According to a recent report, a significant proportion of XBP1u is integrated into the ER membrane as a type II transmembrane protein. It is then cleaved by signal peptide peptidase (SPP), which triggers the degradation of XBP1u via the ER-associated protein degradation pathway (37). Although our results indicate that XBP1u is not a transmembrane protein but a membrane-associated protein, it is possible that a small portion of XBP1u is misintegrated in the ER membrane as a type II membrane protein, and this type II membrane protein might be degraded by an SPP-mediated pathway. With respect to the quality control of misintegrated membrane-associated proteins, the SPP-mediated degradation pathway for XBP1u is consistent with our model.
We showed that the ER-targeting efficiency of XBP1u mRNA depends on the presence of SRP (Fig. 3B). The mRNA targeting efficiency in the SRP54 knockdown cells was reduced by ∼59% relative to the control cells (Fig. 3B). In this condition, the splicing efficiency of XBP1u mRNA exhibited a 29% reduction relative to the control, at most (Fig. 3 C and D). In contrast, XBP1u with an HR2 deletion exhibited a 74% reduction in the ER-targeting efficiency of the mRNA and an 80% reduction in splicing efficiency under ER stress (Figs. 2C and 5D). The correlation between the ER-targeting efficiency of mRNA and splicing efficiency was not linear (Fig. S4A). Rather, there appeared to be a threshold. This threshold for the splicing efficiency may explain why SRP54 knockdown had a modest effect on the splicing of XBP1u mRNA under ER stress, even though it affected the ER-targeting efficiency of XBP1u mRNA (Fig. 3 B and D). Alternatively, the subcellular fractionation of mRNA used in this study might not capture brief, transient ER associations of mRNA from the pausing mutant, which might explain the discrepancy between the ER-targeting efficiency of mRNA and its splicing efficiency.
In this study, we characterized a unique instance of the ER targeting of a membrane-associated protein mediated by SRP. Compared with transmembrane proteins or soluble secretory proteins, the ER-targeting route of ER-associated proteins is poorly understood. The uncanonical route toward the ER membrane for XBP1u may explain the targeting specificity of ER-associated proteins. Previously, Sigma-32, a bacterial transcription factor activated only under heat shock stress, was reported to be recruited to the cell membrane of Escherichia coli by SRP as a membrane-associated protein (38). Importantly, the responsive region of Sigma-32 for SRP recognition is modestly hydrophobic, which is similar to HR2 in XBP1u (Fig. S3B). These examples imply the existence of a novel type of SRP client that harbors a modestly hydrophobic region and is delivered as an ER-associated protein (39).
Materials and Methods
Plasmids and Antibodies.
The pcDNA3.1 plasmids encoding XBP1u, XBP1s, XBP1u-ps, XBP1u-ps[ΔHR2], XBP1u-ps[S255A], and XBP1u-ps[W256A] were previously described (21). The G519C mutation of XBP1u was XBP1u[nonsplicing] as previously described (21). N-terminal 3× tandem HA epitope tagging (described as HA-) or 3× tandem FLAG epitope tagging followed by eight histidine tagging constructs (described as FH-) was generated by standard procedures. For the nuclear localization signal (NLS) mutant of XBP1u, lysines 59 and 90 were substituted with arginines by standard procedures (Fig. S1). For the 3L mutation of XBP1u, glutamine 199, serine 200, and serine 203 were substituted with leucines (Fig. 5A). For XBP1u+s, XBP1s cDNA was obtained with the mutations T490A, C491G, and G519C to prevent splicing by IRE1α, and a cytosine was inserted at the stop codon of the XBP1u ORF (TAA to TAcA, lowercase c indicates the inserted cytosine) to obtain a +1 frame-shift, resulting in the fusion of XBP1u and the C-terminal region of XBP1s. As XBP1u+s variants, XBP1u+s[W256A] and XBP1u-ps[ΔHR2] were made as described above. pMXs-puro-XBP1u-ps and pMXs-puro-FH-XBP1u-ps were made by the insertion of PCR products of XBP1u-ps and FH-XBP1u-ps into pMXs-puro (kindly provided by Dr. Toshio Kitamura, University of Tokyo, Tokyo, Japan) at PacI and BamHI sites to establish HeLa cells stably expressing XBP1u-ps or FH-XBP1u-ps. The C-terminal HA epitope-tagged Sec61β (described as Sec61β-HA) was inserted into pcDNA3.1(+) at KpnI and EcoRI sites. For in vitro translation experiments, FH-XBP1u and its mutants, ΔHR2, W256A, and W256A-3L, were amplified by PCR. Venus-fused XBP1u at the N-terminal region of XBP1u (referred to as Venus-XBP1u) was inserted into pcDNA3.1(+) at KpnI and BamHI sites. To make mCherry-Sec61β and EGFP-Sec61β, Sec61β, with the exception of the first ATG codon, was amplified by PCR for insertion into pmCherry-N1 (Clontech) or pEGFP-N1 (Clontech).
Commercial antibodies were as follows: mouse anti–FLAG-M2 (Sigma–Aldrich), mouse anti-HA (Roche) for immunoprecipitation and Western blotting (Fig. S2C), mouse anti-HA (Covance) for immunofluorescence (Fig. 1 A and B), rabbit anti-Giantin (Abcam,), rabbit anti-COX IV (Thermo Scientific), mouse anti-SRP54 (BD Biosciences), rabbit anti-SRP72 (Atlas Antibodies), rabbit anti-Sec61α (Millipore), rabbit anti-Sec61β (Millipore), mouse anti-Rpl9 (Abnova), rabbit anti-GAPDH (CST), rabbit anti-IRE1α (CST), rabbit anti-PERK (CST), rabbit anti-Calnexin N terminus (Enzo Life Sciences), rabbit anti-Calnexin C terminus (Enzo Life Sciences), anti-rabbit IgG-HRP (MBL), anti-mouse IgG-HRP (Jackson ImmunoResearch), anti-mouse IgG-Alexa 488 (Molecular Probes), and anti-rabbit IgG-Alexa 647 (Molecular Probes).
Cell Culture.
HeLa (RIKEN BRC), HEK293T (RIKEN BRC), and Cos-7 cells were maintained in DMEM containing 4.5 g/L glucose, l-glutamine, nonessential amino acids, sodium pyruvate (Nacalai), and 10% (vol/vol) FBS (12E183-A; Sigma) at 37 °C and 5% CO2. Transfection was performed using Lipofectamine 2000 (Invitrogen) or Lipofectamine LTX (Invitrogen) for plasmids or using Lipofectamine RNAiMAX (Invitrogen) for siRNAs according to the manufacturer’s procedures. Furthermore, polyethylenimine (PEI) Max (Polysciences) was prepared for plasmid transfection according to previously described procedures (39). For knockdown experiments, cells were transfected with siRNAs at a final concentration 10 nM by reverse transfection using Lipofectamine RNAiMAX. After incubation for 2 d, cells were replated once by reverse transfection. After 4 d from the first transfection, cells were collected or used for further experiments. The following control siRNA and stealth siRNAs (Invitrogen) were used: siLuc (catalog no. S20C-0200; Cosmo Bio), siSRP54 no. 1 sense (5′-GCUUCUGAAGGAGUAGAGAAAUUUA-3′) and antisense (5′-UAAAUUUCUCUACUCCUUCAGAAGC-3′), siSRP54 no. 2 sense (5′-UGCGAGACAUGUAUGAGCAAUUUCA-3′) and antisense (5′-UGAAAUUGCUCAUACAUGUCUCGCA-3′), and siSRP54 no. 3 sense (5′-CGCUUUGUUGGAAGCAGAUGUUAAU-3′) and antisense (5′-AUUAACAUCUGCUUCCAACAAAGCG-3′).
To establish HeLa cells stably expressing XBP1u-ps or FH-XBP1u-ps, HeLa cells were infected with retrovirus. The retrovirus produced in Platinum GP cells (Cell Biolabs) was transfected to produce pMXs-puro-XBP1u-ps, or FH-XBP1u-G519C-ps and pCMV-VSV-G using PEI Max. After 4 h of transfection, the transfection medium was changed to fresh medium and cells were incubated for 24 h. The medium containing retrovirus was collected, filtered through a 0.20-μm Minisart syringe filter (Sartorius), and mixed with 4 μg/mL Polybrene. HeLa cells were infected with retrovirus mixtures. After 24 h of infection, cells were selected with fresh medium containing 1 μg/mL puromycin for 48 h.
SDS/PAGE and Western Blotting.
XBP1u was separated by neutral SDS/PAGE with a Bis-Tris polyacrylamide gel [0.36 M Bis-Tris (pH 6.5 with HCl)] and MES running buffer (50 mM MES, 50 mM Tris, and 0.1% SDS). Other proteins were separated by Laemmli SDS/PAGE, which was prepared as a mixture of acrylamide and Bis-acrylamide at a ratio of 30:0.8.
Coimmunoprecipitation.
HEK293T cells were transfected with pcDNA3.1-FH-XBP1u using PEI Max. After 24 h, the cells were lysed with lysis buffer [20 mM Hepes⋅KOH (pH 7.5), 150 mM KOAc, 2.5 mM Mg(OAc)2, 1% Nonidet P-40, 10 μg/mL leupeptin, 1 mM benzamidine, 10 μg/mL pepstatin A, and 1 mM phenylmethylsulfonyl fluoride] for 30 min on ice. Then, insoluble materials were removed by centrifugation at 17,000 × g for 20 min at 4 °C. The supernatant was incubated with anti-Flag antibody, anti-HA antibody, or anti-Sec61β antibody for 30 min followed by incubation with 20 μL of a 50% slurry of protein A Sepharose beads (GE Healthcare) for 1 h at 4 °C. The beads were washed four times with lysis buffer without protease inhibitors. The coimmunoprecipitated proteins were eluted by incubation in 2× sample buffer [125 mM Tris⋅HCl (pH 6.8), 4% SDS, and 15% sucrose] containing 50 mM DTT and analyzed by Western blotting. For immunoprecipitation of in vitro-translated product, proteins were synthesized by in vitro translation with rabbit reticulocyte lysate (RRL) in the presence of 35S-labeled methionine and cysteine using EXPRE35S35S Protein Labeling Mix (PerkinElmer) (Fig. 2E and Fig. S3D). FH-XBP1u and its variants were translated in 10 μL of RRL prepared according to the manufacturer’s instructions at 30 °C for 10 min and resolved in 500 μL of lysis buffer. The subsequent immunoprecipitation procedure was the same as described above. FH-XBP1u was separated with a neutral Bis-Tris polyacrylamide gel and detected by autoradiography. The other proteins were detected by Western blotting using normal Laemmli SDS/PAGE.
Identification of Proteins That Interact with XBP1u.
HEK293T cells cultured in 10-cm-diameter dishes were transfected with pcDNA3.1-FH-XBP1u using the calcium phosphate coprecipitation method. After 24 h, the cells were lysed following the procedure described above in the section on coimmunoprecipitation. After centrifugation, the supernatant was incubated with 80 μL of anti-Flag agarose (50% slurry; Sigma) for 1 h at 4 °C. The agarose beads were washed five times with lysis buffer. The coimmunoprecipitated proteins were eluted with 250 μg/mL 3× FLAG peptide in lysis buffer for 30 min at 4 °C. The resultant elution (500 μL) was further incubated with 40 μL of 50% slurry; nickel-nitrilotriacetic agarose beads (Qiagen) were incubated for 1 h at 4 °C. The beads were washed three times with lysis buffer containing 20 mM imidazole followed by elution with lysis buffer. Finally, the purified proteins were concentrated using ultrafiltration (14,000 × g, 4 °C for 2.5 h) with a Microcon YM-3 (Millipore). The concentrated sample was denatured in 2× SDS sample buffer containing 50 mM DTT at 37 °C for 30 min. Purified FH-XBP1u and its interacting proteins were separated with a Nu-PAGE Bis-Tris gradient acrylamide 4–12% gel (Novex) in MES running buffer. Whole lanes of each sample were divided into four pieces, and gels were digested with trypsin. The digested proteins were fractionated using liquid chromatography with Paradigm MS4 (Michrom) and analyzed using tandem mass spectrometry with an LTQ-Orbitrap XL (Thermo Scientific). Peptide mass fingerprinting was performed with Mascot (Matrix Science) using NCBInr 20121013 as a peptide database.
XBP1 mRNA Splicing Assay.
RNA was purified with RNAiso Plus (TaKaRa) according to the manufacturer’s instructions, followed by reverse transcription to generate cDNA with M-MLV RNase H-point Mutation (Promega). DNA fragments derived from unspliced or spliced XBP1 mRNA were amplified by PCR as previously described (21). The DNA fragments were separated with a 1× Tris-Borate-EDTA (TBE) polyacrylamide gel, stained with ethidium bromide, and detected using Gel-Doc XR (Bio-Rad). The ratio of XBP1s/XBP1u mRNA was calculated from the intensities of the respective bands quantified using ImageJ (NIH).
Immunofluorescence.
HeLa and Cos-7 cells were transfected with plasmids using Lipofectamine LTX. After 4 h of transfection, cells were replated on a coverslip (Matsunami Glass). After 24 h of transfection, cells were fixed with 4% paraformaldehyde for 15 min at 4 °C, permeabilized with 0.1% Triton-X100 for 30 s at room temperature, and incubated in 5% BSA in PBS overnight or for 1 h. Immunoreactions of primary and secondary antibodies diluted in blocking buffer were performed at room temperature for 1 h. After immunoreaction, cells were embedded with Prolong Gold (Invitrogen). The FV1000 confocal microscopy system (Olympus) equipped with a UPLSAPO 60XO 1.35 (Olympus) or the LSM700 confocal microscopy system (Zeiss) equipped with a Plan-Apochromat 63× oil 1.40 M27 objective was used. Image processing was performed using ImageJ.
FRAP Analysis.
Cos-7 cells plated on a 35-mm glass-bottomed dish (Matsunami) were transiently transfected to express Venus or Venus-XBP1u[nonsplicing] and mCherry-Sec61β. During FRAP experiments, cells were incubated in Leibovitz’s L-15 Medium (Gibco) at 37 °C. To detect the newly synthesized Venus, preexisting fluorescence in a whole cell was bleached by light with the excitation wavelength, and time-lapse images of Venus and mCherry were obtained at 2-min intervals after photobleaching. Observations were performed using the FV1000 confocal microscopy system described above.
In Vitro Transcription.
To make templates for in vitro transcription, pcDNA3.1(+)-FH-XBP1u was amplified by PCR using the forward primer 5′-ATTTAGGTGACACTATAGAAGAGacccaagtggctagc-3′, where uppercase and lowercase letters indicate the SP6 promoter and annealing part to pcDNA3.1(+), respectively, and the reverse primer hybridized to the poly-A sequence [5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTcacctactcagacaatgcgatgc-3′, where the uppercase and lowercase letters indicate the poly-A sequence and the part that anneals to pcDNA3.1(+), respectively]. Capped mRNA was transcribed from purified template DNA using SP6 RNA Polymerase (Promega) in reaction buffer according to the manufacturer’s instructions at 37 °C for 2 h. The template DNA was degraded by DNase I (TaKaRa) at 37 °C for 20 min. The synthesized RNA was purified with ISOGEN-LS (Nippon Gene) following the manufacturer’s instructions.
Cotranslational ER Targeting Assay.
To prepare EDTA and high-salt–treated rough microsomes (EKRM), CMMs (Promega) were treated with 50 mM EDTA and 500 mM KOAc in 250 mM sucrose solution containing 50 mM Hepes⋅KOH and 5 mM Mg(OAc)2 for 15 min on ice. For purification, the EKRM solution was layered on 500 mM sucrose in HKM buffer [120 mM KOAc, 50 mM Hepes⋅KOH, and 5 mM Mg(OAc)2] followed by centrifugation at 4 °C, 140,000 × g for 30 min. The pelleted membrane was washed by centrifugation at 4 °C, 140,000 × g for 30 min after suspension in 250 mM sucrose in HKM buffer. Finally, pelleted EKRM was suspended in the original volume of 250 mM sucrose in HKM buffer. RRL (Promega) or WGE (Promega) was used as an in vitro translation system. Capped FH-XBP1u mRNA was translated for 10 min in the in vitro translation mix with or without a 1/10 volume of EKRM and/or purified SRP (final = 5 nM; tRNA Probes). For translation with RRL, 0.02 μg of mRNA was translated at 30 °C in 10 μL for 10 min. For translation with WGA, 0.1 μg of mRNA was translated at 25 °C in 10 μL for 10 min. Then, 1 μL of translation mix was used as the input sample. To fractionate membrane-bound components, 10 μL of translation mixture was mixed with 40 μL of 250 mM sucrose solution in HKM buffer containing 1 mM cycloheximide. Moreover, the mixture was mixed with 500 μL of 2.5 M sucrose in HKM buffer, and put on the bottom of a polycarbonate ultracentrifuge tube (Beckman). It was then layered on 1 mL of 1.9 M sucrose, and 250 mM sucrose was layered on the 2 M sucrose mix. The membranes were floated to the interphase between 1.9 M sucrose and 250 mM sucrose by centrifugation at 4 °C, 55,000 rpm for 4 h with a SW55 Ti rotor (Beckman). After centrifugation, 600 μL of the interphase between 1.9 M and 250 mM sucrose was collected as the floating fraction, and 600 μL was collected from the bottom of the tube and referred to as the bottom fraction. Each fraction and input sample was desalted and concentrated by TCA precipitation and analyzed by Western blotting.
Posttranslational ER-Targeting Assay.
FH-XBP1u[W256A] and FH-XBP1u[W256A/3L] mutants were translated in RRL at 30 °C for 10 min. After translation, 1 mM puromycin was added to the translation mixture and incubated at 30 °C for 10 min. Then, EKRM was added to the translation mixture and incubated at 30 °C for 20 min to test the posttranslational translocation activity. To fractionate membrane-bound components, the floating method described for the cotranslational translocation assay was used.
Subcellular Fractionation and Sodium Carbonate Treatment.
After 24 h of transfection, HEK293T cells in a single 10-cm-diameter dish were collected and disrupted by passage through a 27-gauge needle in hypotonic buffer [10 mM Hepes⋅KOH, 2.5 mM Mg(OAc)2, 10 μg/mL leupeptin, 1 mM benzamidine, 10 μg/mL pepstatin A, and 1 mM phenylmethylsulfonyl fluoride]. The cell lysate was centrifuged at 8,000 × g at 4 °C for 10 min to isolate the postmitochondrial fraction collected as the supernatant. The postmitochondrial fraction was centrifuged at 140,000 × g and 4 °C for 1 h with a TLA100.3 rotor to obtain the cytosolic fraction pellet and microsome fraction. For sodium carbonate treatment, the postmitochondrial fraction was treated with 200 mM sodium carbonate for 30 min on ice, and transmembrane proteins were separated following the same procedure described above. The supernatant and pellet were concentrated and desalted by TCA precipitation. Each sample was analyzed by Western blotting.
Analysis of mRNA Membrane Localization Efficiency.
To separate membrane-bound mRNA from cytosolic RNA, HEK293T cells in one well of a 12-well plate at 90% confluence were incubated with 200 μL of buffer A [50 mM Hepes⋅KOH, 140 mM KOAc, 2.5 mM Mg(OAc)2, 0.025 μg/mL digitonin (D5628; Sigma), and protease inhibitors] for 5 min on ice and centrifuged at 3,000 × g at 4 °C. The supernatant was collected as the cytosolic fraction. The pellet was washed with 200 μL of buffer A without digitonin. The pellet was incubated with 200 μL of buffer B [50 mM Hepes⋅KOH, 500 mM KOAc, 2.5 mM Mg(OAc)2, 1% Triton-X100, and protease inhibitors] for 10 min on ice followed by centrifugation at 8,000 × g at 4 °C. The supernatant was collected as the membrane fraction. HeLa cells were collected from one well of a six-well plate at 90% confluence, and the digitonin concentration in buffer A was changed to 0.1 μg/mL. Cytosolic and membrane-bound mRNAs were extracted from each fraction with ISOGEN-LS (Nippon Gene) according to the manufacturer’s procedure. The mRNAs were treated with DNase I (TaKaRa) and purified again with ISOGEN-LS. Then, the concentration of RNAs in the cytosolic fraction of each sample was determined by estimating absorbance at OD260. For reverse transcription, 250 ng of cytosolic mRNA and the same volume of membrane-bound mRNA from each sample were used. cDNAs were synthesized with M-MLV RNase H-point Mutant (Promega) and random hexamers (Promega) according to the manufacturer’s instructions. To quantify cDNAs, quantitative PCR (qPCR) was performed with Thunderbird SYBR qPCR mix (Toyobo) and LightCycler 480 (Roche). The ratio of membrane-bound mRNA to cytosolic mRNA was calculated using the following formula (where Cp is the crossing point):
The primer sets for real-time PCR were as follows: BiP, forward primer (5′-CATCAAGTTCTTGCCGTTCA-3′), reverse primer (5′-TTCAGGAGCAAATGTCTTTGTTT-3′); β-actin, forward primer (5′-CCAACCGCGAGAAGATGA-3′), reverse primer (5′-CCAGAGGCGTACAGGGATAG-3′); and exogenous XBP1u, forward primer (5′-GTTCCTTACCAGCCTCCCTT-3′), reverse primer (5′-ATCCCGTGAACAGCTCCTCG-3′).
Supplementary Material
Acknowledgments
We thank Yoichiro Fukao, Rie Kurata, and Masayuki Fujiwara for assistance with the mass spectrometry analysis and Michinori Toriyama for helping with the FRAP analysis. We also thank members of the K.K. laboratory for helpful discussions and Azumi Wada, Junko Iida, Yumiko Kawakami, and Masami Yoshida for technical assistance. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 24228002 and 26116006 (to K.K.), the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) KAKENHI Grant 19058010 (to K.K.), and the Takeda Science Foundation (K.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604435113/-/DCSupplemental.
References
- 1.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91(2 Pt 1):557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: An essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 4.Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133(3):440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427(6977):808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 6.Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 7.Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 8.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 9.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 10.Mori K. Signalling pathways in the unfolded protein response: Development from yeast to mammals. J Biochem. 2009;146(6):743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 11.Kimata Y, et al. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179(1):75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA. 2010;107(37):16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohno K. Stress-sensing mechanisms in the unfolded protein response: Similarities and differences between yeast and mammals. J Biochem. 2010;147(1):27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 15.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Liang FX, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell. 2014;55(5):758–770. doi: 10.1016/j.molcel.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurkin J, et al. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 2014;33(24):2922–2936. doi: 10.15252/embj.201490332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosmaczewski SG, et al. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 2014;15(12):1278–1285. doi: 10.15252/embr.201439531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinya S, et al. Reconstitution and characterization of the unconventional splicing of XBP1u mRNA in vitro. Nucleic Acids Res. 2011;39(12):5245–5254. doi: 10.1093/nar/gkr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida H, et al. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4(2):265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 21.Yanagitani K, et al. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34(2):191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 22.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331(6017):586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 23.Yanagitani K, Kohno K. In: Regulatory Nascent Polypeptides. Ito K, editor. Springer; Tokyo: 2014. pp. 291–310. [Google Scholar]
- 24.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 25.Potter MD, Nicchitta CV. Endoplasmic reticulum-bound ribosomes reside in stable association with the translocon following termination of protein synthesis. J Biol Chem. 2002;277(26):23314–23320. doi: 10.1074/jbc.M202559200. [DOI] [PubMed] [Google Scholar]
- 26.Walter P, Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci USA. 1980;77(12):7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goder V, Crottet P, Spiess M. In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. EMBO J. 2000;19(24):6704–6712. doi: 10.1093/emboj/19.24.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.High S, Flint N, Dobberstein B. Requirements for the membrane insertion of signal-anchor type proteins. J Cell Biol. 1991;113(1):25–34. doi: 10.1083/jcb.113.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormick PJ, Miao Y, Shao Y, Lin J, Johnson AE. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol Cell. 2003;12(2):329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- 31.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93(1):97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82(2):261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 33.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11(3):319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 34.Plumb R, Zhang ZR, Appathurai S, Mariappan M. A functional link between the co-translational protein translocation pathway and the UPR. eLife. 2015;4:e07426. doi: 10.7554/eLife.07426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pechmann S, Chartron JW, Frydman J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol. 2014;21(12):1100–1105. doi: 10.1038/nsmb.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346(6210):1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CY, et al. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014;33(21):2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim B, et al. Heat shock transcription factor σ32 co-opts the signal recognition particle to regulate protein homeostasis in E. coli. PLoS Biol. 2013;11(12):e1001735. doi: 10.1371/journal.pbio.1001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods. 2006;138(1-2):85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 40.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24(18):2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.