Significance
Glucocorticoid hormones are important mediators of the stress response and implicated in the etiology of stress-related psychiatric disorders. Glucocorticoids act via mineralocorticoid receptors (MRs) and glucocorticoid receptors (GRs) in the hippocampus, resulting in altered transcription of target genes. Currently, however, little information is available about how they interact with the genome after stress in vivo. Here, we show that an acute stressful challenge results in an increased interaction of both MRs and GRs with their genomic recognition sites. Moreover, they may interact with these sites not just as homodimers but also, as heterodimers, the extent of which is highly gene-dependent. These findings provide insight into how MRs and GRs interact with the hippocampal genome after stress in vivo.
Keywords: stress, mineralocorticoid receptor, glucocorticoid receptor, hippocampus, heterodimerization
Abstract
A stressful event results in secretion of glucocorticoid hormones, which bind to mineralocorticoid receptors (MRs) and glucocorticoid receptors (GRs) in the hippocampus to regulate cognitive and affective responses to the challenge. MRs are already highly occupied by low glucocorticoid levels under baseline conditions, whereas GRs only become substantially occupied by stress- or circadian-driven glucocorticoid levels. Currently, however, the binding of MRs and GRs to glucocorticoid-responsive elements (GREs) within hippocampal glucocorticoid target genes under such physiological conditions in vivo is unknown. We found that forced swim (FS) stress evoked increased hippocampal RNA expression levels of the glucocorticoid-responsive genes FK506-binding protein 5 (Fkbp5), Period 1 (Per1), and serum- and glucocorticoid-inducible kinase 1 (Sgk1). Chromatin immunoprecipitation (ChIP) analysis showed that this stressor caused substantial gene-dependent increases in GR binding and surprisingly, also MR binding to GREs within these genes. Different acute challenges, including novelty, restraint, and FS stress, produced distinct glucocorticoid responses but resulted in largely similar MR and GR binding to GREs. Sequential and tandem ChIP analyses showed that, after FS stress, MRs and GRs bind concomitantly to the same GRE sites within Fkbp5 and Per1 but not Sgk1. Thus, after stress, MRs and GRs seem to bind to GREs as homo- and/or heterodimers in a gene-dependent manner. MR binding to GREs at baseline seems to be restricted, whereas after stress, GR binding may facilitate cobinding of MR. This study reveals that the interaction of MRs and GRs with GREs within the genome constitutes an additional level of complexity in hippocampal glucocorticoid action beyond expectancies based on ligand–receptor interactions.
Adrenal glucocorticoid hormones play a pivotal role in orchestrating adaptive responses to stressful challenges to maintain health and wellbeing. Acute surges in glucocorticoid secretion after stress are beneficial for the organism, whereas aberrant secretion, as a result of chronic stress or traumatic experiences, is damaging and increases susceptibility to mental disorders, such as major depression, anxiety, and posttraumatic stress disorder.
Over 40 y ago, McEwen et al. (1) discovered that glucocorticoids act through receptors located in the brain, primarily the hippocampus. In 1985, Reul and de Kloet (2) reported that these steroid hormones bind to two distinct types of receptors, the mineralocorticoid receptor (MR) and the glucocorticoid receptor (GR), in this limbic brain region where these receptors are colocalized in neurons (3). Because of the extraordinary difference in binding affinity of MRs [Kd = 0.1–0.5 nM for binding corticosterone (CORT), the endogenous glucocorticoid of rats and mice] and GRs (Kd = 2–5 nM), there were marked differences in receptor occupancy between these receptors under baseline and stress conditions (2, 4). MRs are already >80% occupied with endogenous glucocorticoids under early morning (AM) baseline conditions, whereas GRs only become substantially occupied by elevated glucocorticoid levels, such as after stress and at the circadian peak of glucocorticoid secretion. These data gave rise to the concept that MRs exert tonic actions on brain, whereas GRs mediate the negative feedback and long-term cognitive changes evoked by glucocorticoids (2, 4, 5).
MR and GR are mainly intracellular receptors that act as ligand-dependent transcription factors. After binding of glucocorticoids, the receptors are translocated to the nucleus with the help of cochaperones and bind to specific glucocorticoid response elements (GREs) within the DNA of glucocorticoid-inducible genes to elicit transcriptional responses (6, 7). The molecular mechanisms underpinning the interaction of MRs and GRs with the genome have been primarily studied in vitro, predominantly using chromatin immunoprecipitation (ChIP), allowing the investigation of transcription factor binding to recognition sites within the genome. ChIP has been used to study the interaction of GR with GREs in glucocorticoid target genes in cell cultures in vitro and pharmacological studies in vivo (8, 9). Until now, however, the binding of MR and GR to GREs within glucocorticoid target genes under physiological conditions in hippocampus tissue in vivo has never been studied. Thus, currently, it is unknown how stressful challenges impact on MR and GR binding to GREs within the genome in vivo.
A long-standing question is whether, in addition to forming MR/MR and GR/GR homodimers, MRs and GRs also form MR/GR heterodimers and interact as such at the genomic level. In fact, the concept of heterodimer formation by these steroid receptors and their ability to bind DNA is based on studies in vitro and has not been shown in vivo. Work using cell cultures and cell-free approaches indicate that MR/GR heterodimers may form under conditions in vitro (10–12). In addition, Trapp et al. (10) found stronger DNA binding and gene transcriptional effects in vitro under conditions that favored MR/GR heterodimerization. These studies did not investigate MR/GR heterodimer binding to GREs within the genome. Moreover, evidence that MR/GR heterodimerization and DNA binding is taking place under physiological conditions in vivo is presently lacking.
We investigated the interaction of MRs and GRs with GREs within the well-known glucocorticoid target genes FK506-binding protein 5 (Fkbp5), Period 1 (Per1), and serum- and glucocorticoid-inducible kinase 1 (Sgk1). These genes are involved in GR ligand binding affinity (13), circadian neuronal activity (14), and neuronal plasticity processes (15), respectively, and were transcriptionally activated after forced swim (FS) stress. Using ChIP, we found that, after stress, MRs and GRs bound transiently to GRE sites within these glucocorticoid target genes, albeit in a gene-dependent manner. For MR, the significant increase in DNA binding after stress was surprising given the high glucocorticoid occupancy of this receptor under baseline AM conditions, challenging the notion that high receptor occupancy would correlate with high DNA binding. Furthermore, as revealed by tandem ChIP, MRs and GRs bind concomitantly to the same GRE sites within Fkbp5 and Per1 but not Sgk1 after stress, indicating that these steroid receptors, in addition to forming homodimers, indeed seem to bind to GREs as heterodimers. Thus, our study shows that, after stress, MRs and GRs may access the genome as homo- and/or heterodimers and in a gene-dependent manner.
Results
Acute Stress Increases Transcription of Glucocorticoid Target Genes Across All Hippocampal Subregions.
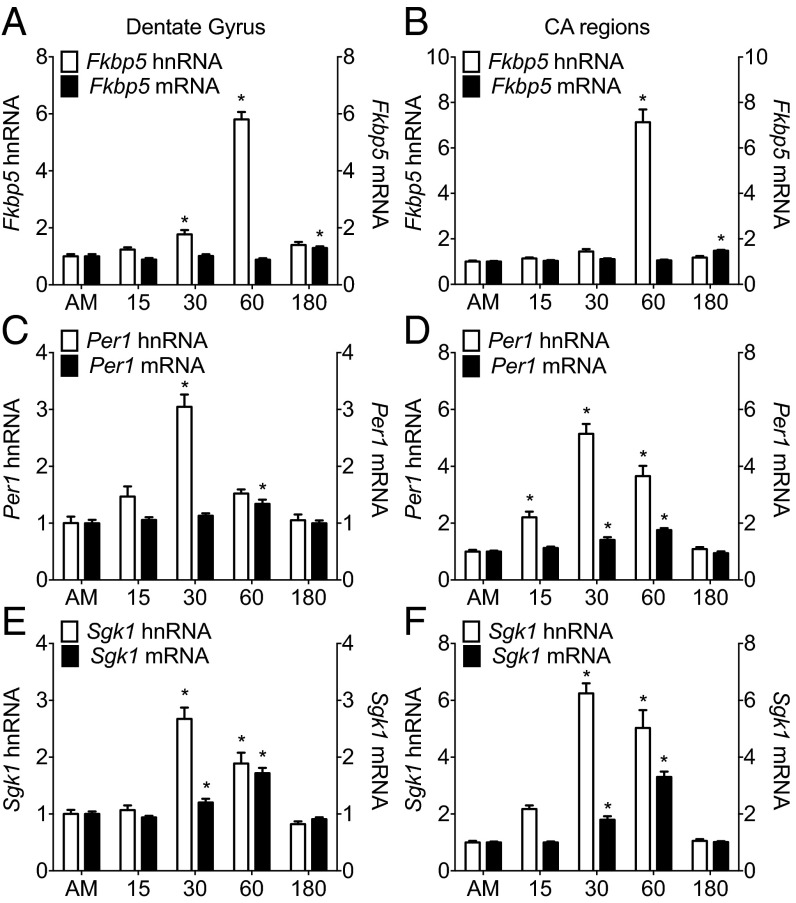
A single 15-min FS challenge resulted in a significant, time-dependent increase in the transcription of the classic glucocorticoid-dependent genes Fkbp5 (Fig. 1A), Per1 (Fig. 1C), and Sgk1 (Fig. 1E) in the dentate gyrus. Very similar patterns of transcriptional activation were found in the Cornu ammonis (CA) regions (Fig. 1 B, D, and F) and ventral hippocampus (Fig. S1). The time course of stress-induced gene transcription and the conversion time for splicing heteronuclear RNA (hnRNA) to mature messenger RNA (mRNA) were gene-specific, with Fkbp5 peaking at 60 min (hnRNA) and significant increases in mRNA expression by 180 min, whereas RNAs of Per1 and Sgk1 peaked earlier (hnRNA, 30 min; mRNA, 60 min). The peaks in Per1 and Sgk1 hnRNA corresponded with the peak in plasma CORT after FS (30 min) (Fig. S2), but the Fkbp5 hnRNA response was clearly delayed. Because the RNA responses were highly similar between the hippocampal subregions, we performed subsequent ChIP analyses on whole-hippocampus tissues.
Fig. 1.
hnRNA and mRNA expression of glucocorticoid-inducible genes in hippocampal subregions under baseline conditions and after stress. Rats were killed direct from home cage (∼7:00 AM; AM baseline) or at 15, 30, 60, or 180 min after the start of FS (15 min, 25 °C water). The graphs show expression of Fkbp5, Per1, and Sgk1 in the (A, C, and E, respectively) dorsal dentate gyrus or (B, D, and F, respectively) CA regions, and they are represented as mean fold change over baseline RNA levels (±SEM; n = 7–9 per group). Expressions of both hnRNA (white bars; left y axis) and mRNA (black bars; right y axis) are shown for individual genes. More information on statistical analyses in Figs. 1, 2, 3, 4, and 5 is in SI Statistics Information to Figs. 1, 2, 3, 4, and 5. *P < 0.05 compared with AM.
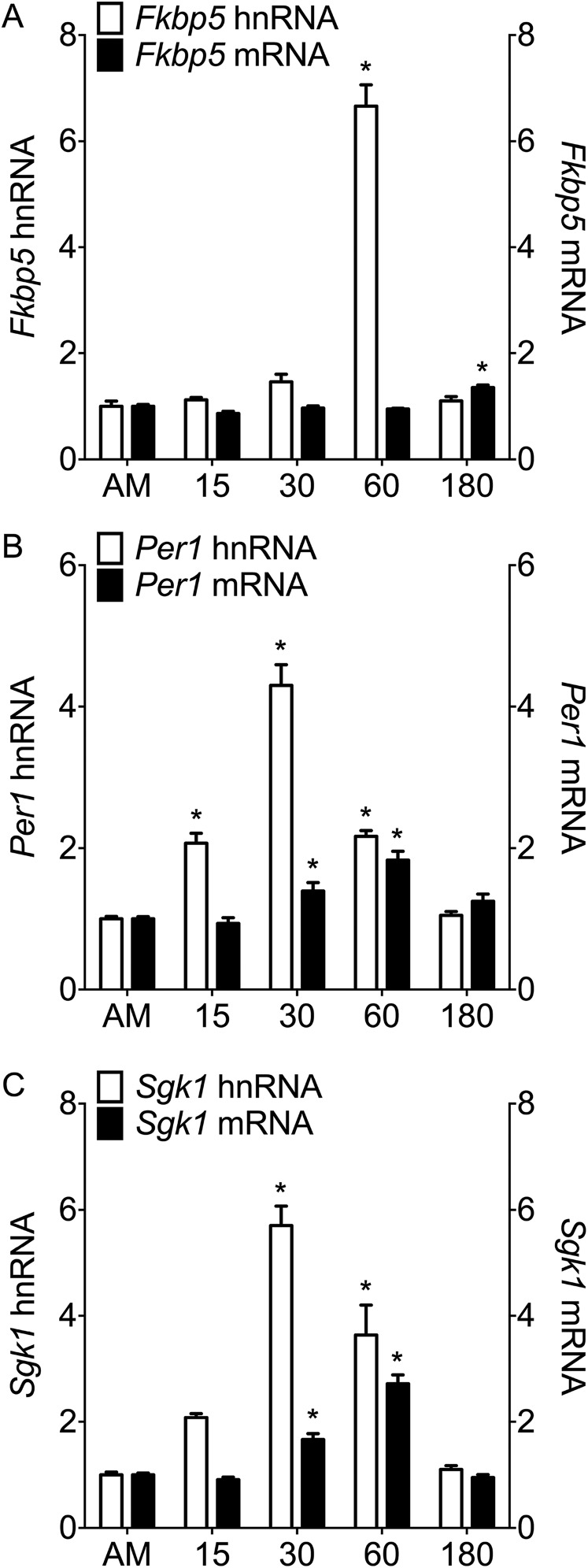
Fig. S1.
hnRNA and mature mRNA expression of glucocorticoid-inducible genes in the ventral hippocampus under baseline conditions and after stress. Rats were killed direct from home cage (∼7:00 AM; AM baseline) or at 15 min (15), 30 min (30), 60 min (60), or 180 min (180) after the start of FS (15 min, 25 °C water). The graphs show expression of (A) Fkbp5, (B) Per1, and (C) Sgk1 in the ventral hippocampus after normalization to the expression of housekeeping genes Hprt1 and Ywhaz, and they are represented as mean fold change over baseline RNA levels (±SEM; n = 8–9 per group). Expressions of both hnRNA (white bars; left y axis) and mature mRNA (black bars; right y axis) are shown for individual genes. Statistical analysis: one-way ANOVA: (A) hnRNA F(4, 35) = 155.30, P < 0.0001; mRNA F(4, 35) = 23.13, P < 0.0001; (B) hnRNA F(4, 35) = 66.36, P < 0.0001; mRNA F(4, 34) = 12.67, P < 0.0001; (C) hnRNA F(4, 36) = 42.22, P < 0.0001; mRNA F(4, 35) = 60.10, P < 0.0001. Bonferroni posthoc test. *P < 0.05 compared with baseline (AM).
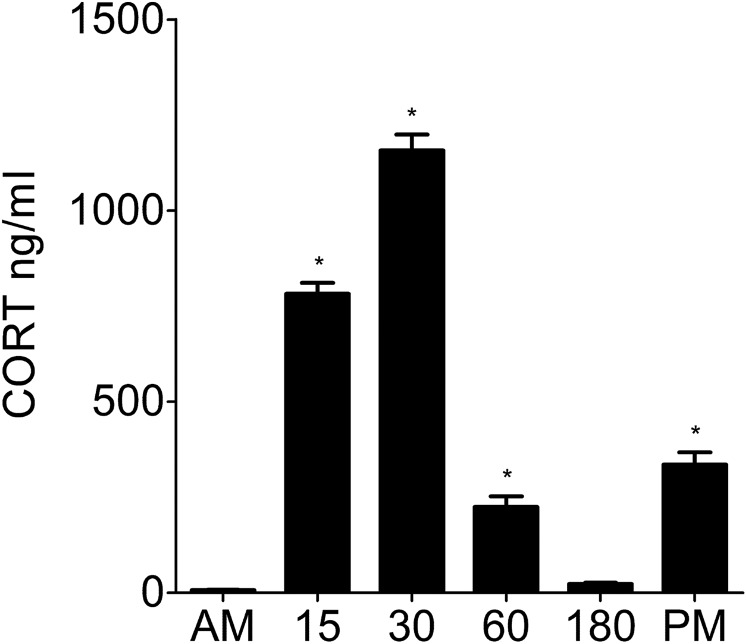
Fig. S2.
Plasma CORT levels under baseline conditions and after stress. Plasma CORT levels (nanograms per milliliter; mean ± SEM) from baseline rats killed direct from home cage in early morning (∼7:00 AM; AM baseline) or late afternoon (∼5:00 PM; PM baseline) or rats killed 15 min (15), 30 min (30), 60 min (60), or 180 min (180) after the start of FS (15 min, 25 °C water) are shown. Statistical analysis: one-way ANOVA: F(5, 51) = 284.6, P < 0.0001. Bonferroni posthoc test. *P < 0.05 compared with early morning baseline (AM).
FS Transiently Increases MR and GR Binding to GREs Within Hippocampal Glucocorticoid Target Genes.
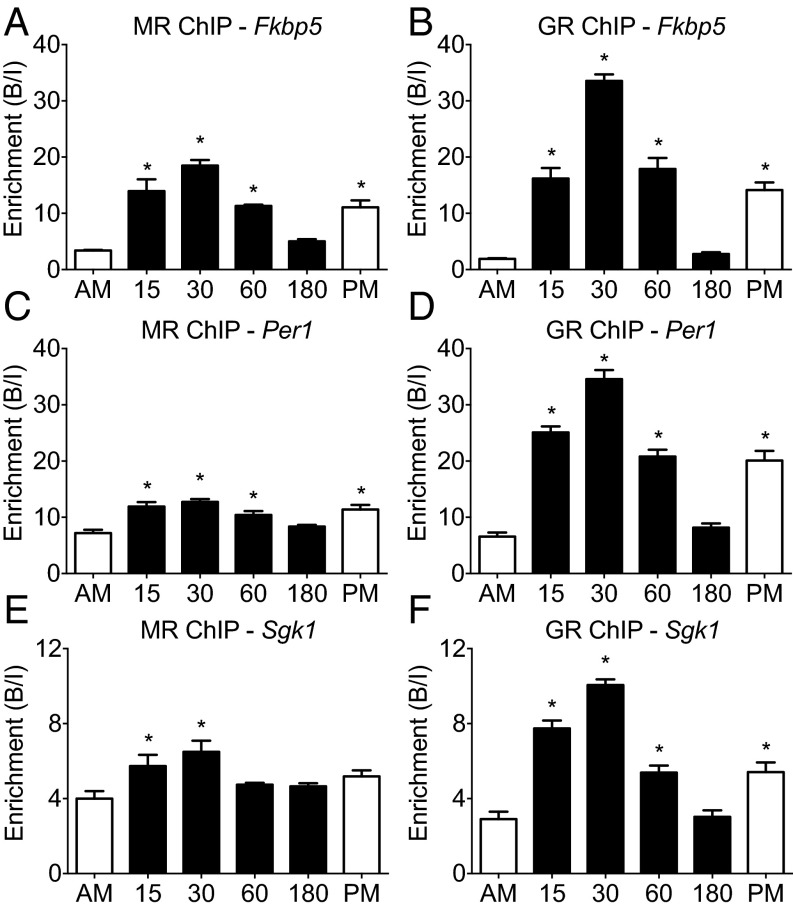
Presently, it is unknown whether stress-induced transcriptional activation of glucocorticoid-dependent genes involves physical interaction of MRs and GRs with GREs within these genes. Using ChIP, we studied MR and GR binding to GREs within Fkbp5 (GRE2), Per1, and Sgk1 (Fig. S3 shows the within-gene location of targeted GREs) in hippocampal chromatin from rats killed under early morning baseline conditions (AM), at various time points after FS stress, or under late afternoon baseline conditions (PM) (Fig. 2). FS stress resulted in a highly significant, transient increase in corticosteroid receptor binding to all glucocorticoid target genes investigated (Fig. 2) that largely paralleled the changes in plasma CORT levels (Fig. S2). The peak in MR and GR binding (at 30 min) coincided with (Per1 and Sgk1) or preceded (Fkbp5) the increases in hnRNA levels after stress (Fig. 1). In contrast to GR (Fig. 2 B, D, and F), MR binding to GREs was already near-maximal at 15 min (Fig. 2 A, C, and E). After stress, MR binding to GREs increased gene dependently between 1.5- (Sgk1) and 6-fold (Fkbp5) (Fig. 2), which was surprising given that MR occupancy by endogenous glucocorticoids is already very high (>80%) under baseline AM conditions (2, 4). Thus, in case of MR, high receptor occupancy does not predict or guarantee high GRE binding. Occupancy of GRs by endogenous glucocorticoids after stress was shown to follow the course of the plasma glucocorticoid concentration (2, 4). Fig. 2 B, D, and F shows that the binding of GRs to the different GREs was highly responsive to stress and also followed the pattern of plasma glucocorticoid levels (Fig. S2) and GR occupancy levels (2, 4). The magnitude of the stress-evoked enhancement in binding of MR and GR to GREs was highly gene-dependent, with highest increments found in Fkbp5 GRE2 and Per1 GRE and smaller increases found in Sgk1 GRE. These observations indicate that, under both baseline and stress conditions, accessibility of GRE sites within glucocorticoid target genes seems to be different within hippocampal cells. Comparison of MR and GR binding between baseline AM and PM presents a clear circadian variation in the interaction of both receptors with the target gene GREs, except for MR binding to the Sgk1 GRE, which failed to reach statistical significance (Fig. 2). These observations show that rises in MR and GR binding can occur in response to circadian-driven increases in circulating glucocorticoids (Fig. S2), independent of stress.
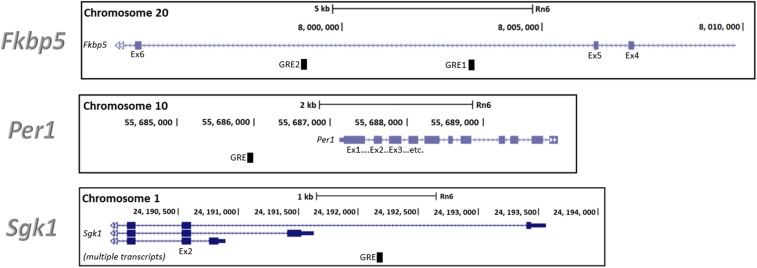
Fig. S3.
Gene maps of glucocorticoid-inducible genes. The locations of GREs within the rat glucocorticoid target genes Fkbp5, Per1, and Sgk1 are shown. Gene traces were adapted from the Rnor_6.0 assembly (genome.ucsc.edu/).
Fig. 2.
MR and GR binding to GREs within glucocorticoid-inducible genes in the hippocampus under baseline conditions and after stress. Rats were killed under AM (∼7:00 AM) or PM (∼5:00 PM) conditions or at 15, 30, 60, or 180 min after the start of FS (15 min, 25 °C water). The graphs show enrichment [bound/input (B/I); mean ± SEM; n = 3–4] at GREs within (A and B) Fkbp5, (C and D) Per1, and (E and F) Sgk1 after MR and GR ChIP on hippocampal chromatin, respectively. *P < 0.05 compared with AM.
Comparison of Different Stressors Regarding MR and GR Binding to Glucocorticoid Target Genes.
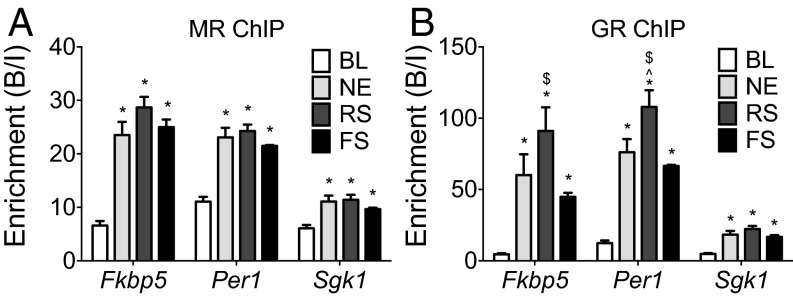
Subsequently, we investigated whether the degree of MR and GR binding to GREs depends on the severity of the stressor. The plasma CORT levels after novelty [novel environment (NE)], restraint (RS), and FS stress were ∼230, 670, and 1,160 ng/mL, respectively (16) (Fig. S2) (baseline AM levels: ∼10 ng/mL). All stressors caused substantial increases in MR and GR binding to all target gene GREs investigated (Fig. 3). Overall, the magnitude of the binding responses was gene-dependent, with Fkbp5 (GRE2) and Per1 showing much higher responses than Sgk1. Remarkably, however, although these stressors are well-known to produce distinct glucocorticoid responses, binding of MRs and GRs to a GRE within a particular gene was very similar. For instance, NE and FS stress evoke very different plasma glucocorticoid levels; nevertheless, binding of MRs and GRs to GREs was not different between these stressors (Fig. 3). RS, generating lower plasma glucocorticoid responses than FS, resulted in significantly higher GR binding to Fkbp5 GRE2 and Per1 GRE (Fig. 3B). Thus, MR and GR binding to GREs after stress is virtually an on/off switch possibly controlled by other factors in addition to glucocorticoids.
Fig. 3.
Effects of different stressors on MR and GR binding to glucocorticoid target genes in the hippocampus. Rats were killed either under baseline AM conditions (BL) or 30 min after stress onset. The graphs show mean enrichment [bound/input (B/I); ±SEM; n = 8 for baseline group; n = 4 for stress groups] at GREs within glucocorticoid target genes after (A) MR or (B) GR ChIP on hippocampal chromatin. *P < 0.05 compared with the respective BL group; ^P < 0.05 compared with the respective NE group; $P < 0.05 compared with the respective FS group.
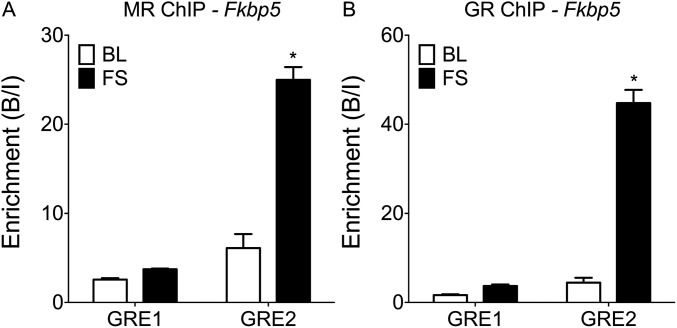
Selective MR and GR Binding to GREs Within Intron 5 of the Fkbp5 Gene.
GR regulation of the Fkbp5 gene occurs predominantly via interaction with intronic GREs (17). Previously, a study in vitro has shown that the intron 5 GRE2 site (Fig. S3) is crucial for glucocorticoid stimulation of Fkbp5 gene transcription, whereas the GRE1 site within this intron was inactive (17). In Figs. 2 and 3, we presented significant increases in MR and GR binding to the Fkbp5 GRE2 site after FS. In Fig. S4, we compared receptor binding to Fkbp5 GRE1 and GRE2 at AM and 30 min after FS. In contrast to GRE2, there was no significant increase in MR or GR binding to GRE1 after FS, which corresponds with reports (17) that this site is not actively involved in transducing glucocorticoid effects on Fkbp5 transcription.
Fig. S4.
Comparison of MR and GR binding to different GRE sites in the Fkbp5 gene in the hippocampus. Rats were killed either direct from home cage under baseline AM conditions (BL) or 30 min after the start of FS stress. The graphs show mean enrichment (B/I; ±SEM; n = 4 per group) at differential GREs within the Fkbp5 gene after (A) MR or (B) GR ChIP on hippocampal chromatin. Statistical analysis: two-way ANOVA: (A) effect of stress: F(1, 12) = 88.07, P < 0.0001; effect of GRE location F(1, 12) = 134.60, P < 0.0001; interaction stress × GRE location F(1, 12) = 69.07, P < 0.0001; (B) effect of stress: F(1, 12) = 180.40, P < 0.0001; effect of GRE location F(1, 12) = 193.10, P < 0.0001; interaction stress × GRE location F(1, 12) = 146.60, P < 0.0001. Bonferroni posthoc test. *P < 0.05 compared with respective BL group.
MR and GR Interaction at GREs Within Glucocorticoid Target Genes.
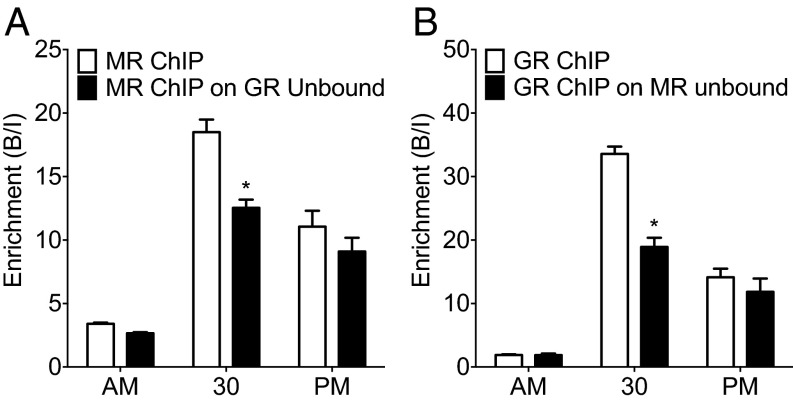
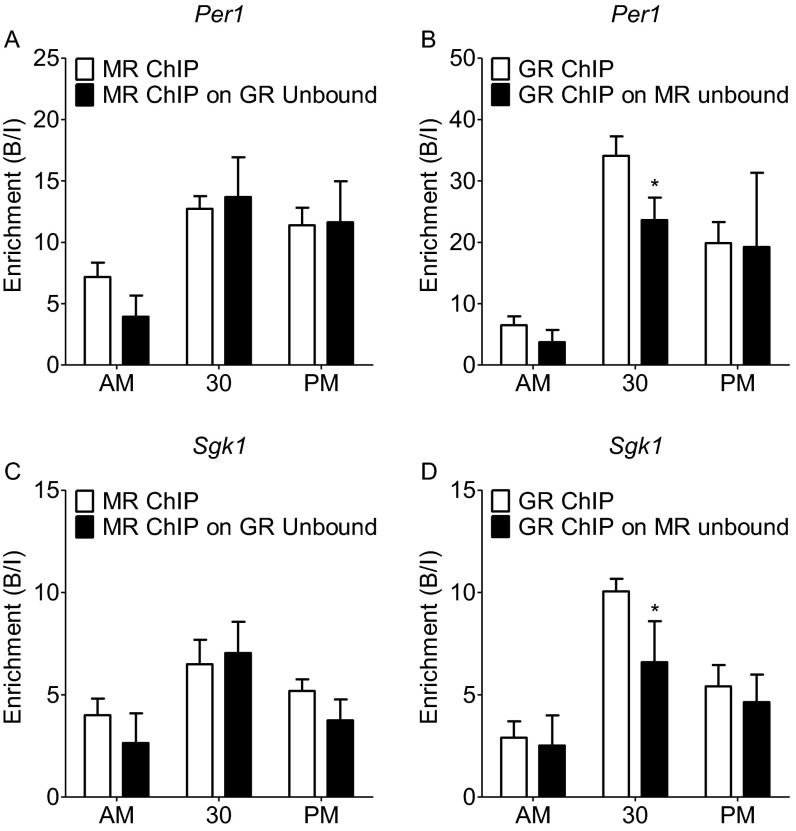
Our results show that both MRs and GRs bind to GREs within target genes after stress and at the circadian peak of glucocorticoid secretion. It is unclear, however, whether the receptors bind to separate GREs, thus strictly as homodimers, or whether they can bind concomitantly at the same GRE site as heterodimers. Although there are indications from cell culture and cell-free studies (10–12) that MRs and GRs may interact with GREs as heterodimers, direct evidence that this may be occurring at the chromatin level under physiological conditions in vivo is lacking. To resolve this question, we adopted a serial ChIP approach. We reasoned that, if MR and GR interact at the same GREs within a given gene, then immunoprecipitation (IP) of one receptor would lead to relative depletion of the other receptor. Fig. 4 shows that, if ChIP was conducted after FS stress for either receptor first followed by a second ChIP for the other receptor on the unbound fraction, then significantly less binding for this receptor at Fkbp5 GRE2 was measured after the second ChIP. This result indicates that ChIP for MR leads to a reduced ChIP outcome for GR and vice versa, providing indirect evidence that MRs and GRs are binding concomitantly to the same GREs. This phenomenon was only observed after FS and not observed in the AM and PM samples. A similar result was found regarding GR binding to the Per1 and Sgk1 GREs conducted after MR ChIPs (Fig. S5 B and D) but not found for MR binding to these GREs after GR ChIP (Fig. S5 A and C), possibly because the stress-induced increases in MR binding to the Per1 and Sgk1 GREs are lower in magnitude than the rise in binding to the Fkbp5 GRE2 (Fig. 2). This experiment provides indirect evidence that MR and GR may interact in part at the same GRE sites within glucocorticoid target genes after stress.
Fig. 4.
Comparison of MR and GR ChIP on original vs. GR/MR unbound hippocampal chromatin at the Fkbp5 GRE2 under baseline conditions and after stress. Rats were killed under AM or PM conditions or at 30 min after the start of FS (15 min, 25 °C water). The graphs show mean enrichment [bound/input (B/I); ±SEM; n = 3–4 per group] at Fkbp5 GRE2 after (A) MR ChIP on original chromatin (white bars) and on the unbound fraction of chromatin after GR ChIP (black bars) or (B) GR ChIP on original chromatin (white bars) and on the unbound fraction of chromatin after MR ChIP (black bars). *P < 0.05 compared with the respective ChIP at the same time point.
Fig. S5.
Comparison of MR and GR ChIP on original vs. GR/MR unbound hippocampal chromatin at GREs within glucocorticoid-inducible genes under baseline conditions and after stress. Rats were killed direct from home cage in early morning (∼7:00 AM; AM baseline) or late afternoon (∼5:00 PM; PM baseline) or at 30 min after the start of FS (15 min, 25 °C water). The graphs show mean enrichment (B/I; ±SEM; n = 3–4 per group) of GREs within the (A and B) Per1 or (C and D) Sgk1 genes after MR ChIP on original chromatin (white bars) and the unbound fraction of chromatin after GR ChIP (black bars) or GR ChIP on original chromatin (white bars) and the unbound fraction of chromatin after MR ChIP (black bars), respectively. Two-way ANOVA: (A) effect of stress: F(2, 16) = 25.92, P < 0.0001; effect of ChIP F(1, 16) = 0.53, P = 0.4774; interaction stress × ChIP: F(2, 16) = 2.04, P = 0.1624; (B) effect of stress: F(2, 16) = 36.45, P < 0.0001; effect of ChIP: F(1, 16) = 4.07, P = 0.0587; interaction stress × ChIP: F(2, 16) = 1.702, P = 0.2104; (C) effect of stress: F(2, 17) = 17.88, P < 0.0001; effect of ChIP: F(1, 17) = 2.31, P = 0.1471; interaction stress × ChIP: F(2, 17) = 1.79, P = 0.1978; (D) effect of stress: F(2, 17) = 37.66, P < 0.0001; effect of ChIP: F(1, 17) = 8.48, P = 0.0093; interaction stress × ChIP: F(2, 17) = 3.325, P = 0.0590. Bonferroni posthoc test. *P < 0.05 compared with respective ChIP at same time point.
Evidence Supporting MR/GR Heterodimerization at GREs Within Glucocorticoid Target Genes.
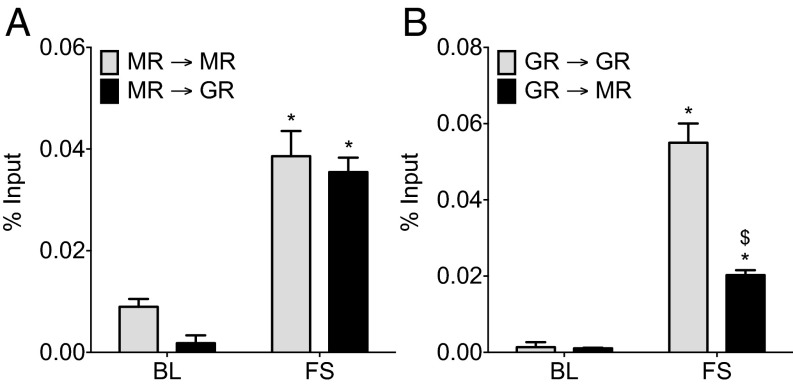
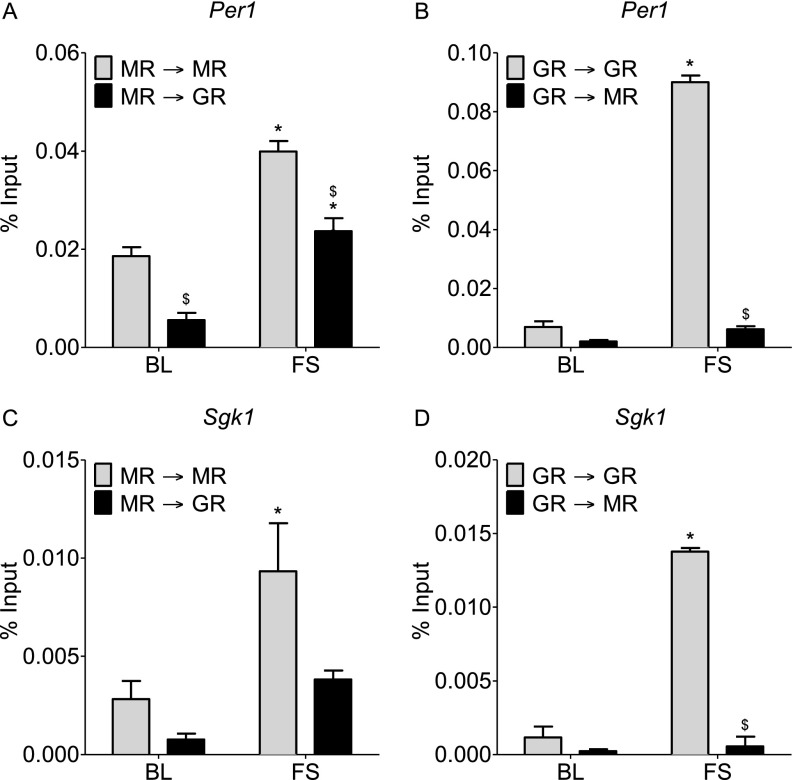
To provide direct evidence for concomitant binding of MR and GR to the same GRE sites within glucocorticoid target genes, we applied a tandem ChIP protocol. We conducted an MR ChIP or a GR ChIP on hippocampal chromatin of AM or FS (30 min) rats, and subsequently, we re-chromatin immunoprecipitated the eluted immunoprecipitated chromatin with an antibody against the same (ChIP and re-ChIP samples: MR→MR and GR→GR) or the other receptor (MR→GR and GR→MR). This procedure was followed by quantitative PCR (qPCR) analysis of GREs within Fkbp5 (GRE2) (Fig. 5), Per1, and Sgk1 (Fig. S6). Based on the different tandem ChIP combinations, the MR→GR and GR→MR tandem ChIP selectively revealed concomitantly bound MR and GR (indicating MR/GR heterodimer formation) at the qPCR-targeted GRE. The MR→MR and the GR→GR combinations determine the binding of the respective homodimers (MR/MR and GR/GR, respectively) plus the binding of MR/GR heterodimers. Accordingly, the difference between the MR→MR or the GR→GR combination and the parallel MR→GR or GR→MR combinations would provide an estimate for the contribution of the respective MR/MR and GR/GR homodimers to the ChIP result. These tandem ChIPs provide tangible but not absolute evidence for MR/GR heterodimerization, because MR and GR cooccupancy of GREs could possibly be occurring in conjunction with other proteins.
Fig. 5.
Tandem ChIP for MR and GR binding to Fkbp5 GRE2 in the hippocampus under baseline conditions and after stress. Rats were killed either under AM conditions [baseline (BL)] or 30 min after the start of FS stress. Graphs show percentage input (mean ± SEM; n = 3 per group) at Fkbp5 GRE2 after (A) MR ChIP immediately followed by MR, GR, or IgG (negative control) binding to the MR bound chromatin or (B) GR ChIP followed by GR, MR, or IgG binding to the GR bound chromatin. The IgG levels (percentage input) were deducted from the MR and GR bound data. *P < 0.05 compared with the respective BL ChIP; $P < 0.05 compared with GR→GR FS ChIP.
Fig. S6.
Tandem ChIP for MR and GR binding to GREs within Per1 and Sgk1 in the hippocampus under baseline conditions and after stress. Rats were killed either direct from home cage under baseline AM conditions (BL) or 30 min after the start of FS stress. Graphs show binding (percentage input ± SEM; n = 3 per group) at specific GREs within the (A and B) Per1 and (C and D) Sgk1 genes after (A and C) MR ChIP immediately followed by MR, GR, or IgG (negative control) binding to the MR bound chromatin or (B and D) GR ChIP followed by GR, MR, or IgG (negative control) binding to the GR bound chromatin. The IgG levels (percentage input) were deducted from the MR and GR bound data. Two-way ANOVA: (A) MR ChIP − Per1: effect of stress: F(1, 8) = 91.56, P < 0.0001; effect of ChIP F(1, 8) = 50.37, P = 0.0001; interaction stress × ChIP: F(1, 8) = 0.60, P = 0.4622; (B) GR ChIP − Per1: effect of stress: F(1, 8) = 997.34, P < 0.0001; effect of ChIP F(1, 8) = 1,032.59, P < 0.0001; interaction stress × ChIP: F(1, 8) = 815.8, P < 0.0001; (C) MR ChIP − Sgk1: effect of stress: F(1, 8) = 12.88, P = 0.0071; effect of ChIP: F(1, 8) = 8.09, P = 0.0217; interaction stress × ChIP: F(1, 8) = 1.68, P = 0.2316; (D) GR ChIP − Sgk1: effect of stress: F(1, 8) = 239.38, P < 0.0001; effect of ChIP: F(1, 8) = 287.04, P < 0.0001; interaction stress × ChIP: F(1, 8) = 216.98, P < 0.0001. Bonferroni posthoc test. *P < 0.05 compared with respective BL ChIP; $P < 0.05 compared with GR→GR FS ChIP.
Our results show that the extent of putative homodimer and heterodimer formation at GREs under AM and stress conditions was highly gene-dependent (Fig. 5 and Fig. S6). Regarding Fkbp5 GRE2, under AM conditions, it appeared that there was higher MR/MR homodimer binding than MR/GR or GR/GR binding, but differences were not statistically significant (Fig. 5). FS resulted in a significant increase in the binding of putative MR/GR heterodimers to Fkbp5 GRE2 as revealed by both MR→GR and GR→MR tandem ChIPs (Fig. 5). Because there was no significant difference between the MR→MR and the MR→GR results, it is likely that MRs participate in binding to this GRE only together with GRs as heterodimers and not as MR/MR homodimers (Fig. 5A). After stress, the substantial difference between the GR→GR and the GR→MR results (Fig. 5B) indicates that, in addition to forming heterodimers with MRs, GRs also bound significantly as GR/GR homodimers.
Under baseline conditions and after FS stress, the MR→GR ChIP result was significantly lower than the MR→MR result at Per1 GRE, indicating that, under these conditions, corticosteroid receptors may be binding as both MR/MR homodimers and MR/GR heterodimers (Fig. S6A). In addition, the rise in GR→GR binding after stress at Per1 GRE was highly significant, but the increase in GR→MR binding failed to reach statistical significance (Fig. S6B). In conjunction, these tandem ChIP data suggest that, after stress, MRs and GRs bind to the Per1 GRE largely as GR/GR homodimers and to a lesser extent, as MR/MR homodimers and MR/GR heterodimers (Fig. S6 A and B). At the Sgk1 GRE, FS increased both MR→MR and GR→GR binding (Fig. S6 C and D). Given that FS-induced binding in MR→GR and GR→MR failed to reach statistical significance, it is likely that this gene is regulated predominantly by homodimers (Fig. S6 C and D).
Discussion
This study shows that, in the hippocampus, an acute stressful challenge transiently increases MR and GR binding to GREs within the glucocorticoid target genes Fkbp5, Per1, and Sgk1 and enhances transcription of these genes. Surprisingly, despite the high occupancy level of MRs under baseline conditions (2, 4), a relatively low binding of this receptor to GREs was observed under these conditions. Different stressors, although evoking different glucocorticoid peak levels, resulted in largely similar increases in MR and GR binding to GREs within these genes. Overall, we observed that the interaction of MRs and GRs with GREs under the various conditions investigated was highly gene-dependent. Sequential and tandem ChIP analyses showed that, after stress, MRs and GRs may bind as homodimers as well as heterodimers to Fkbp5 and Per1 GREs, whereas Sgk1 GRE appeared only to be bound by the respective homodimers. These data show that the interaction of GRs with the genome seems to be a reflection of circulating glucocorticoid levels and expected receptor occupancy levels, whereas MRs’ genomic interaction may be restricted under baseline AM conditions and/or depend on cobinding with GRs. Together, these results reveal gene-dependent and receptor-specific modes of interactions with the genome, which cannot be predicted solely on the basis of hormone concentrations and receptor occupancy levels.
FS caused a significant increase in RNA expression of Fkbp5, Per1, and Sgk1, which is consistent with their well-known responsiveness to glucocorticoids (17–20). The hnRNA levels for Per1 and Sgk1 peaked at 30 min, whereas Fkbp5 hnRNA levels reached their maximal levels later at 60 min. The mRNA expression levels after stress followed the hnRNA responses with a delay of at least 30 min in all genes, indicative of time required for the splicing process. Whereas GREs in the Per1 and Sgk1 genes are located in their proximal promoter region, in the Fkbp5 gene, the transcriptionally active GREs are located within intronic sequences (6, 17–19, 21, 22). In the rat, GRE2 within intron 5 has been identified as being particularly important for glucocorticoid-induced Fkbp5 transcription (17). Receptor-bound GREs within introns of the Fkbp5 gene are thought to stimulate gene transcription through chromatin remodeling including loop formation that allows the direct interaction of the intronic region with the transcriptional start site (23, 24). Such a process is anticipated to require more time than the direct transactivational stimulation originating from promoter-located GREs, like in Per1 and Sgk1, which may explain the differences in the time course of the stress-induced hnRNA (and mRNA) responses, despite similar time courses of MR and GR binding to these GREs.
Although corticosteroid receptor interaction with glucocorticoid target genes has been studied under pharmacological conditions [e.g., glucocorticoid injections in ADX rats (9)], the interaction of MRs and GRs with such genes in the hippocampus under glucocorticoid-relevant physiological conditions has not been studied to date. Under baseline conditions, MR and GR binding levels at GREs were relatively low in the early morning but rose significantly during the day, reaching significantly elevated levels in late afternoon, except for MR binding at the Sgk1 GRE. An acute FS challenge evoked a substantial rise in receptor binding to GREs, with MRs reaching near-maximal level at 15 min and GR binding peaking at 30 min. These peak binding levels superseded the respective levels observed at baseline PM. The receptor binding profiles at baseline and after stress largely followed the circulating CORT levels. Regarding GR binding to GREs, this result may have been expected given that studies had shown that GR occupancy by endogenous glucocorticoids critically depends on circulating hormone concentrations (2, 4). The peak in GR binding at 30 min after stress concurs with the peak in stress-induced plasma glucocorticoid levels, but in view of recent findings on stress-induced free CORT levels (16), this finding was unexpected. Recent microdialysis studies in vivo have shown that, after FS stress, the peak in free CORT in the hippocampus is delayed 20–30 min compared with the plasma hormone response (16, 25). Thus, because the free CORT concentration is the critical parameter for hormone–receptor interaction, maximal GR binding to GREs after stress would be expected to occur at 60 min rather than at 30 min. At 60 min poststress, however, GR (and MR) binding levels were substantially lower than at 30 min. Thus, GR’s interaction with GREs (and MR’s interaction as well) is only partly determined by glucocorticoid levels, indicating the on and off status regarding GREs is actively regulated by additional molecular factors (26).
The binding profile of MRs to GREs under baseline and stress conditions is remarkable, because Reul and de Kloet (2) reported 30 y ago that hippocampal MRs are at least 80% occupied with endogenous glucocorticoids under all physiological conditions studied (4) and that occupancy levels only dropped after adrenalectomy (4). Therefore, we had expected MR binding to GREs to be relatively high under baseline AM conditions, with only small increases after stress. Our results, however, show a different picture: relatively low binding at baseline AM and substantial increases after stress and at baseline PM (except Sgk1 GRE). Thus, high occupancy of MR does not translate into high binding to GREs. One reason may be that, under baseline AM conditions, MR binding to GREs is restricted because of an action of a steroid receptor corepressor like death-associated protein (DAXX), which after stress, is expunged and/or exchanged for a steroid receptor coactivator like Fas-associated factor 1 (FAF-1). DAXX and FAF-1 are hippocampal proteins that have been shown to modulate MR transcriptional activity in hippocampal cells in vitro (27). Alternatively, MR binding to GREs may be weak as supported by early transfection studies in vitro (28). Trapp et al. (10) showed that DNA binding of MR was low in monkey kidney COS-1 cells solely transfected with MR (compared with GR) but could be increased dramatically when both receptors were transfected together. Furthermore, transcriptional activity of cotransfected receptors was higher than that of separately transfected receptors (10). Thus, MR binding in the absence of activated GRs, such as is the case under baseline AM conditions, is weak, which changes considerably after GRs become activated because of stress-induced glucocorticoid production. In other words, MRs seem to require GRs for substantial binding to GREs to occur. This notion is consistent with MRs and GRs heterodimerizing after stress.
In absolute terms, MR and GR binding to GREs within Fkbp5 and Per1 after stress was overall substantially higher than receptor binding observed to Sgk1 GRE. Because results are from the same ChIP DNA samples, these binding profiles are directly comparable. The observed differences in receptor binding were consistent across the different stressors. Because each hippocampal cell contains two copies of each gene (if located on autosomal chromosomes), theoretically, MR and GR binding to GREs could be similar, but this situation is clearly not the case. Possibly, there is less availability of the Sgk1 GRE for binding compared with the Fkbp5 GRE2 and Per1 GRE, because in a significant number of hippocampal cells, the gene may be located within inactive, condensed chromatin. In situ hybridization analysis suggests that Sgk1 mRNA is indeed primarily expressed in hippocampal pyramidal neurons, whereas Fkbp5 and Per1 mRNA is ubiquitously expressed in hippocampal neurons (29). Alternatively, there may be differences at the GRE level in terms of its nucleotide sequence as well as involvement of local modulators (e.g., steroid receptor coregulators) affecting the affinity and stability of receptor–GRE interactions. Our work points to a role of other molecular mechanisms in vivo in addition to GRE nucleotide sequence, because although both GRE1 and GRE2 within the Fkbp5 present ideal nucleotide consensus sequences, MR and GR binding to these GREs is dramatically different. Whereas GRE2 shows significant increases in MR and GR binding after FS, GRE1 shows no significant change in binding. Our findings correspond with transcriptional analyses in vitro showing that GRE2 is a transactivationally active site, whereas GRE1 is not (17) and may be explained by distinct accessibility of GRE1 vs. GRE2 as a result of epigenetic and other molecular (e.g., steroid receptor coregulators) mechanisms (23, 24, 26). These observations are consistent with our notion that MR and GR binding to GREs within the genome is highly controlled at the cellular level as well as the single-gene level.
To investigate stressor specificity and the role of different levels of stress-induced glucocorticoid levels, we compared the effects of FS with those of NE and RS. FS and RS are strong stressors, resulting in high glucocorticoid responses, whereas NE is regarded as a mild psychological stressor, leading to moderate increases in plasma hormone levels (16). MR binding to GREs was very similar after the different stressors, albeit with consistent intergene differences. Apparently, the mechanisms triggering MR binding to GREs are independent of the extent of stress-induced glucocorticoid responses and other stressor-specific mediators. The independence of glucocorticoid responses is not surprising, because MRs are already highly occupied at baseline AM glucocorticoid levels (2, 4). Our findings regarding GR binding to GREs were, however, surprising, because despite the substantial difference in glucocorticoid responses between stressors, the interactions of GRs with GREs were largely similar. These observations underscore that the glucocorticoid response is not an all-determining factor in the genomic action of GRs (and MRs). It seems that additional stressor-specific factors are involved in determining GR binding to GREs in hippocampal cells, including signaling pathways, epigenetic factors, and local modulators. An additional factor may be the duration of the stressful experience: the RS (and NE) experience lasted the full 30 min until death, whereas FS lasted 15 min, after which the rats were returned to their home cages for the remaining 15 min. Therefore, the shorter-lasting FS challenge may have allowed rats to shut down the stress response, resulting in lower GR binding levels. Elucidation of the factors determining GR (and MR) interaction with the genome should be an intriguing challenge for future research.
To study if MR and GR are acting separately (as homodimers) to stimulate transcription of glucocorticoid target genes or possibly, together in a complex, we initially performed a serial ChIP first with one antibody (anti-MR or anti-GR) and then, rechromatin immunoprecipitated the unbound fraction using the opposite receptor antibody (either MR or GR). The rationale was that, if GR and MR were interacting at the same GRE, IP of one DNA-bound receptor with the first antibody would also result in IP of the other receptor into the bound fraction and deplete it from the unbound fraction. A subsequent ChIP for the “other” receptor on the unbound fraction from the first ChIP would recover less target DNA compared with the amount recovered in the original ChIP. If, however, MR and GR were not bound to the same DNA strand but instead, bound to the same GRE location but on different strands, recovered DNA (covering this GRE site) would be comparable between ChIPs performed on both the original chromatin and the unbound fraction of opposite receptor ChIP. We found that MR binding to GREs after stress was substantially reduced if GRs had been removed from the sample by IP previously and vice versa. These effects were most clear for the Fkbp5 GRE2, most likely because this GRE presented the largest stress-induced MR binding response. Cross-receptor depletion was only observed in hippocampal chromatin samples from stressed rats but was not observed in chromatin from baseline PM animals, ruling out that depletion is the result of an assay artifact and indicating that MRs and GRs interact concomitantly with GREs within glucocorticoid target genes specifically after stress. We used a more direct approach to investigate MR and GR interaction at GREs in vivo using MR→GR and GR→MR tandem ChIPs, which only immunoprecipitate GRE DNA bound to both MR and GR at the same time. The results show that, after stress, MRs and GRs bind concomitantly to the same GRE sites within Fkbp5 and Per1 genes, possibly through the formation of heterodimers. Previous co-immunoprecipitation (co-IP) experiments in a cell-free system have shown that MRs and GRs can heterodimerize in solution (11); our work provides evidence to support that this can occur at the DNA template in vivo. Moreover, after stress, MRs and GRs may bind to GREs as heterodimers as well as homodimers but with striking gene-dependent differences. After stress, there appeared to be a strong recruitment of MR/GR heterodimers at the Fkbp5 GRE2, less at the Per1 GRE, and very low recruitment at the Sgk1 GRE. Thus, local chromatin status possibly defined by epigenetic factors in conjunction with coregulatory factors may determine the level of recruitment as well as preference for homo- vs. heterodimer binding. MR and GR cotransfection studies indicated that formation of the MR/GR heterodimer results in stronger GRE binding and greater reporter gene responses than shown by the respective homodimers (10). Presently, the gene transcriptional significance of MR/GR heterodimer formation is unclear. Our findings allow for the study of the significance of heterodimer formation for gene transcriptional responses using pharmacological approaches as well as high-throughput sequencing methods.
Based on receptor occupancy studies, 30 y ago, de Kloet and Reul (2,5) proposed a concept on the role of MRs and GRs in the effects of glucocorticoids on the brain. In view of its constant high occupancy, it was thought that MRs exert a tonic influence on hippocampus function, including neuronal excitability, hypothalamic–pituitary–adrenal (HPA) axis activity, sympathetic outflow, and cognitive behavior (2, 5, 30). GRs only became significantly occupied by elevated glucocorticoid levels and were thought to exert negative feedback action on HPA axis activity and facilitate memory formation of stressful events (2, 5, 31). In view of our data, this concept may require adjustments. The interaction of MRs and GRs with GREs under baseline and stress conditions and its gene transcriptional consequences seem much more complex than originally thought. The terminology tonic and feedback fall short in view of the multitude of mechanisms controlling the interaction of these steroid receptors with GRE sites, including the highly diverse molecular processes governing accessibility of such sites within different genes and different cells and moreover, their distinct, gene-specific way to interact with GREs, probably as homodimers and heterodimers. This complexity may further grow after MR and GR binding has been conducted across the entire genome by ChIP sequencing. This work has provided the basis to continue elucidating the critical question of how glucocorticoids affect brain function. The answer to this question may hold the key to resolving stress-related disorders.
Materials and Methods
Animals and Stress Procedures.
Male Wistar rats (150–175 g) were purchased from Harlan and group-housed. Rats were forced to swim for 15 min in 25 °C water or subjected to RS or NE or left undisturbed (32). Rats were killed under baseline conditions or at the indicated times after stress (shown in the figures). All animal procedures were approved by the University of Bristol Ethical Committee and the Home Office of the United Kingdom (Animal Scientific Procedures Act, 1986, UK).
Tissue Preparation.
After decapitation, the entire hippocampus was dissected, or the dentate gyrus and CA regions were microdissected from the dorsal hippocampus. Tissues were snap-frozen in liquid N2 and stored at −80 °C.
ChIP, RNA Analysis, and qPCR.
Hippocampal chromatin preparation (Fig. S7), ChIP, and RNA extraction were performed using published methods (33). Analysis by qPCR was conducted using primer and probe sets listed in Dataset S1 (34). Control experiments validated our ChIP method (Fig. S8).
Fig. S7.
Confirmation of the sonication efficiency in reversed and nonreversed hippocampal chromatin. This figure shows a representative blot of sheared hippocampal chromatin from all FS time course groups before (odd numbers) or after (even numbers) reversal of cross-links. Well allocations: 1 and 2, baseline AM; 3 and 4, FS15; 5 and 6, FS30; 7 and 8, FS60; 9 and 10, FS180; 11 and 12, baseline PM.
Fig. S8.
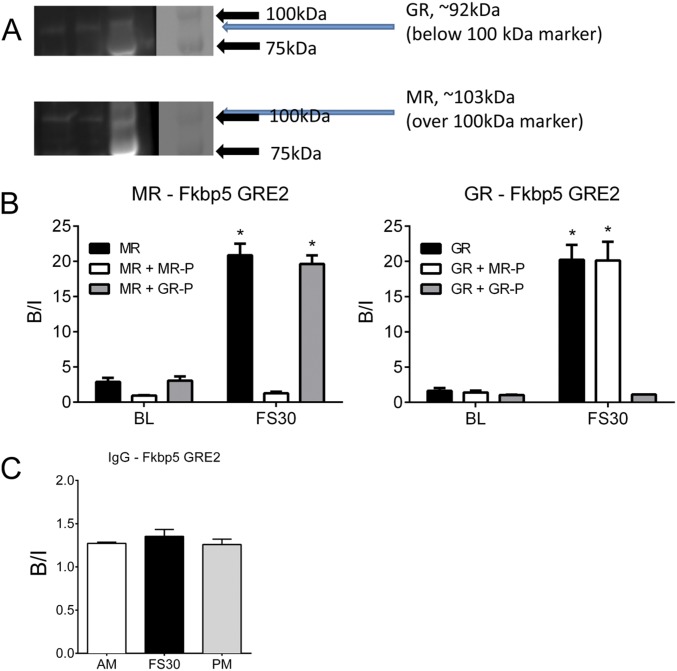
Antibody specificity control experiments. (A) Western blot analysis of the (Upper) GR H-300 and (Lower) MR H-300 antibodies. From left, lanes 1 and 2, nuclear fractions of FS30 hippocampus tissue; lane 3, protein ladder under UV illumination; lane 4, ladder under normal light. The ladder contains markers at 75 and 100 kDa. The lanes containing the nuclear fractions show bands generated by the GR antibody and the MR antibody at ∼92 and 103 kDa, respectively, which correspond with the predicted protein molecular masses. There was no cross-over in the staining pattern, because the GR antibody did not visualize a protein band at 103 kDa and the MR antibody did not recognize a band at 92 kDa. (B) Preabsorption analysis and ChIP on hippocampal chromatin using MR N-17 and GR M-20 antibodies. Chromatin was prepared from rats killed under baseline AM conditions (BL) or at 30 min after FS (FS30). Aliquots of MR N-17 and GR M-20 antibody were preincubated with peptide preparations against which the antibodies had been raised for 3 h at 4 °C as indicated in B. Next, the antibody–peptide mixture was added to chromatin samples, and the ChIP procedure continued as described. qPCR analysis of bound DNA was conducted using primers for Fkbp5 GRE2. Data are expressed as B/I (mean ± SEM; n = 3). Statistical analysis: two-way ANOVA: MR (B, Left): effect of stress: F(1, 12) = 244.1, P < 0.0001; effect of antibody treatment: F(2, 12) = 88.63, P < 0.0001; interaction: F(2, 12) = 57.82, P < 0.0001; GR (B, Right): effect of stress: F(1, 12) = 119.8, P < 0.0001; effect of antibody treatment: F(2, 12) = 32.7, P < 0.0001; interaction: F(2, 12) = 29.64, P < 0.0001. Bonferroni posthoc test. *P < 0.05 compared with the respective BL control. (C) IgG binding to Fkbp5 GRE2 under baseline conditions and after stress. ChIP with normal IgG antibody was conducted with hippocampal chromatin prepared from rats killed under baseline AM or PM conditions or at 30 min after FS. The graph shows lack of IgG enrichment (B/I; mean ± SEM; n = 3) at Fkbp5 GRE2 under all conditions. Statistical analysis: one-way ANOVA: F(2, 8) = 0.7582, P = 0.5087.
Statistical Analysis.
Data were analyzed by ANOVA and appropriate posthoc tests. More information on materials and methods is in SI Materials and Methods.
SI Materials and Methods
Animal Experiments.
Male Wistar rats were purchased from Harlan and weighed 150–175 g on arrival. Animals were housed two to three per cage under standard light (80–100 lx; lights on from 5:00 AM to 7:00 PM) with environmentally controlled conditions (temperature 22 °C ± 2 °C; relative humidity: 50 ± 10%) and food and water available ad libitum. After arrival, the animals were given 5 d to habituate to the housing conditions and then, handled (2–3 min per rat per day) for a week before the experiment to reduce nonspecific stress. All animal procedures were approved by the University of Bristol Ethical Committee and the Home Office of the United Kingdom (United Kingdom Animal Scientific Procedures Act, 1986). Experiments were carried out between 7:00 AM and 1:00 PM as previously described (16, 32). Rats were forced to swim in a glass beaker filled with water at 25 °C for 15 min. Rats were then dried and either killed immediately (time point: 15 min) or returned to their home cage and killed at 30, 60, or 180 min after the start of FS. Exposure to an NE comprised placing the rats individually in a clean, empty, novel cage with increased light intensity (∼500 lx) for 30 min. RS involved placing the rat in a Plexiglas restrainer with ventilation holes for 30 min. Immediately after NE and RS, rats were killed. Rats were killed quickly by isoflurane (<15 s) exposure followed by decapitation and removal of the brain. For mRNA studies, brains were microdissected on ice-cold steel boxes as previously described (33), and dorsal dentate gyrus (DG) and CA regions as well as the whole-ventral hippocampus samples were collected. For whole-hippocampal dissections (ChIP studies), brains were also dissected on ice-cold steel boxes. All samples were snap-frozen in liquid N2 and stored at −80 °C until analysis. Unless otherwise described, all reagents used in sample preparation were from Sigma.
RNA Analysis.
RNA was extracted using TRI Reagent (Sigma) following the manufacturer’s guidelines. The RNA pellet was air-dried, resuspended in nuclease-free water (Life Technologies), and quantified using a NanoPhotometer P300 (Implen). RNA integrity was assessed in random samples with an Agilent 2100 Bioanalyser to confirm that high-quality RNA was extracted [RNA integrity numbers: 8.5 ± 0.1 (mean ± SEM); n = 12]. Total RNA was reverse-transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen) as per the manufacturer’s instructions (15 min, 42 °C; 5 min, 95 °C) using a BioRad T1000 Thermal Cycler. cDNA was diluted fourfold, and 2 µL diluted cDNA was used per reaction in the qPCR analysis detailed below. Expression of mRNA in samples was calculated based on the Pfaffl method of relative quantification (33) using primer/probes listed in Dataset S1 and standardized to the expression of housekeeping genes hypoxanthine phosphoribosyltransferase 1 (Hprt1) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta (Ywhaz). The data were expressed as fold change over baseline.
qPCR Analysis.
Mastermix for qPCR was prepared containing 900 nM forward and reverse primers, 200 nM probe, and 1× TaqMan Fast Mastermix (Life Technologies) and made up to volume with nuclease-free water. Primers and dual-labeled probe with 6-Carboxyfluorescein (6-FAM) as the fluorescent dye and tetramethylrhodamine (TAMRA) as the quencher were designed using Primer Express software (version 3.0.1; Life Technologies) (Dataset S1). Standard curves were performed for each primer pair, and the qPCR efficiency was calculated using the equation E = ((10 − 1/slope) − 1) × 100 (where E is qPCR efficiency, and the slope is the gradient of the standard curve). Only primer pairs with efficiencies greater that 90% were used. qPCR was performed using a StepOne Plus Machine (Life Technologies). Taq enzymes were activated at 95 °C for 20 s, and then, 40 cycles of 95 °C (1 s) to 60 °C (20 s) were performed to amplify samples.
Chromatin Preparation and ChIP Analysis.
Standard ChIP was performed as previously described (33). We added 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) or 0.1 mM PMSF, 5 mM Na+-Butyrate (NaBut), and PhosSTOP Phosphatase Inhibitor Mixture Tablets (one per 10 mL; Roche) to all solutions unless otherwise stated. Briefly, hippocampal tissues from two rats were cross-linked for 10 min in 1% formaldehyde in PBS. Cross-linking was terminated by adding glycine (5 min; final concentration: 200 µM) and centrifuging (5 min at 6,000 × g at 4 °C). Pellets were washed three times with ice-cold PBS. Next, the pellets were resuspended in ice-cold lysis buffer [50 mM Tris⋅HCl, pH 8, 150 mM NaCl, 5 mM EDTA, pH 8.0, 0.5% (vol/vol) Igepal, 0.5% Na-deoxycholate, 1% SDS, 5 mM NaBut, 2 mM AEBSF, 1 mM Na3VO4, Complete Ultra EDTA-Free Protease Inhibitor Tablets, PhosSTOP Phosphatase Inhibitor Mixture Tablets (both one per 10 mL; Roche)] and rotated for 15 min at 4 °C. Samples were aliquoted, sonicated (high power; 3 × 10 cycles; 30 s on and 60 s off) using a water-cooled (4 °C) Bioruptor (UCD-300; Diagenode), and centrifuged (10 min at 20,000 × g at 4 °C). Supernatants (containing the sheared chromatin) were recombined and realiquoted into fresh tubes for subsequent ChIP analysis and assessment of input DNA (i.e., the starting material). Chromatin was sonicated to ∼3-nucleosome lengths (∼450 bp) as confirmed by agarose gel electrophoresis (Fig. S7).
For ChIP analysis, aliquots of chromatin were diluted 10 times in ice-cold dilution buffer [50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, pH 8.0, 1% (vol/vol) Triton, 0.1% Na-deoxycholate, 5 mM NaBut, 1 mM AEBSF, Complete Ultra EDTA-Free Protease Inhibitor Tablets, PhosSTOP Phosphatase Inhibitor Mixture Tablets (both one per 10 mL; Roche)]. Ten microliters antibody, MR (MR H-300 antibody; sc11412X; Santa Cruz), GR (GR H-300 antibody, sc8992X; Santa Cruz), or IgG control (normal rabbit IgG; sc2027X; Santa Cruz), was added to each sample, and tubes were rotated overnight at 4 °C. The anti-MR and anti-GR antibodies were checked for specificity by Western blotting (Fig. S8A) and have been used in published studies (8, 9). In addition, we conducted ChIP assays with alternative MR and GR antibodies (MR N-17; sc-6860X; Santa Cruz; and GR M-20; sc-1004X; Santa Cruz) on baseline AM and FS30 hippocampal chromatin, producing similar results to those obtained previously with the MR H-300 and GR H-300 antibodies (Fig. S8B). Preabsorption tests using preparations of peptide [MR-peptide (sc-6860 P) and GR-peptide (sc-1004 P)] against which the antibodies MR N-17 and GR M-20 had been raised were performed as well (Fig. S8B). These control experiments showed that the MR and GR antibodies showed selective specificity for the respective receptor against which they had been produced. The IgG antibody (normal rabbit control IgG; sc-2027X; Santa Cruz) was used to check for nonspecific binding. As shown in Fig. S8C, the IgG ChIP resulted in an enrichment virtually equaling one [bound/input (B/I) ∼1], indicating no significant nonspecific binding. Protein A-coated Dynabeads (Life Technologies; for MR N-17 antibody, Protein G-Coated Dynabeads were used) were washed once in ice-cold 0.5% BSA/PBS before blocking overnight at 4 °C. Preblocked beads were washed once in ice-cold dilution buffer, resuspended in the antibody–chromatin mix, and allowed to incubate for 3 h at 4 °C to allow binding of beads to antibody–chromatin complexes. After 3 h, the samples were placed in a magnetic stand to allow the beads (with the bound fraction bound) to separate from the liquid unbound fraction. In some ChIP assays, this unbound fraction was collected in separate tubes for additional ChIP assays. In these assays, anti-MR antibody was added to chromatin that had been previously incubated with anti-GR antibody (a GR unbound fraction); vice versa, anti-GR antibody was added to an MR unbound fraction. After addition of antibody, ChIP on unbound fraction proceeded as usual.
Beads carrying the bound chromatin were washed three times with ice-cold RIPA buffer (10 mM Tris⋅HCl, pH 7.5, 1 mM EDTA, pH 7.5, 0.1% SDS, 0.5 mM EGTA, 1% Triton, 0.1% Na-deoxycholate, 140 mM NaCl, inhibitors) and washed twice with ice-cold Tris⋅EDTA buffer. Bound DNA was eluted in two steps at room temperature: first with 200 µL elution buffer 1 (10 mM Tris⋅HCl, pH 7.4, 50 mM NaCl, 1.5% SDS) and second with 100 µL elution buffer 2 (10 mM Tris⋅HCl, pH 7.4, 50 mM NaCl, 0.5% SDS). Cross-links were reversed by addition of NaCl (final concentration of 200 mM) and overnight incubation at 65 °C. The next day, samples were incubated first with RNase A (60 µg/mL, 37 °C, 1 h) followed by incubation with proteinase K (250 µg/mL, 37 °C, 3.5 h). DNA was purified using a QIAquick PCR Purification Kit (Qiagen) as per the manufacturer’s instructions. Input samples were incubated overnight at 65 °C with 200 mM NaCl to reverse cross-links and incubated with RNase A and proteinase K (overnight), and DNA was purified using a Qiagen PCR Purification Kit. Total dsDNA content was determined with a High-Sensitivity Qubit DNA Assay Kit (Life Technologies) as per the manufacturer’s instructions and quantified using a Qubit 2.0 Fluorometer. All samples (bounds and inputs) were diluted to a standardized concentration with nuclease-free water and analyzed by qPCR as described above using primers/probes listed in Dataset S1. A standard curve, created from serial dilutions of rat brain genomic DNA (Biochain), was included in each qPCR run. Data are expressed as quantity of bound DNA divided by the respective quantity of input DNA (i.e., B/I), which is a measure of the enrichment of steroid receptor bound to specific genomic sequences.
For tandem ChIP, the protocol of ChIP explained above was used, albeit with modifications. To attain n = 3 independent samples, three pools of chromatin were prepared of hippocampus tissue from four rats (baseline or killed 30 min after FS) for each pool. From each pool of chromatin, aliquots were incubated with anti-GR H-300 antibody overnight at 4 °C, after which the mixture was incubated for 3 h with Protein A-coated Dynabeads at 4 °C. Subsequently, the beads were washed as described above and next, incubated with 1% SDS buffer (50 mM Tris, 10 mM EDTA, 1% SDS) at 68 °C for 15 min. After incubation in this hot SDS buffer, samples were allowed to cool and then, diluted 10 times in ice-cold dilution buffer. The samples were placed in ice, and 10 µL anti-GR H-300 (GR→GR), anti-MR H-300 (GR→MR), or anti-IgG (background control; GR→IgG) antibody was added to start the second ChIP. After overnight incubation at 4 °C, the mixture was incubated with Protein A-coated Dynabeads, which were subsequently washed and eluted as described for the normal ChIP protocol above.
In a separate tandem ChIP assay, for the reverse protocol, aliquots of chromatin (n = 3) were incubated first with anti-MR antibody (first ChIP) and subsequently, with anti-MR (MR→MR), anti-GR (MR→GR), or anti-IgG (MR→IgG) antibody (second ChIP).
The amount of recovered DNA (after reversal of cross-links, RNase and proteinase K treatment, and purification using Qiagen PCR Purification Columns) was too low to quantify using the Qubit dsDNA Assay Kit used above. Therefore, equal volumes of bound and input samples were subjected to qPCR using the GRE-spanning primers for the three genes (Fkbp5, Per1, and Sgk1) listed in Dataset S1. Binding was then calculated as percentage of input. The percentages of input values of the GR→IgG and MR→IgG ChIP samples were deducted from the values of the respective GR and MR tandem ChIP outcomes.
Western Blotting.
Hippocampal nuclear and cytoplasmic fractions were prepared using the Universal Magnetic Co-IP Kit (Active Motif) according to the manufacturer’s instructions. Protein was quantified using the Qubit Protein Assay Kit (Life Technologies) as per the manufacturer’s instructions using a Qubit 2.0. Nuclear protein samples (20 µg per well) and Precision Plus Protein Dual Color Standard Protein Ladder (BioRad) were loaded onto 4–15% (wt/vol) acrylamide Mini-PROTEAN TGX Precast Gels (BioRad) and separated by electrophoresis for ∼45 min at 200 V. Proteins were transferred to a PVDF membrane (BioRad) and blocked overnight [GR: 5% (wt/vol) BSA/PBS-T; MR: 5% (wt/vol) milk/phosphate buffered saline/Tween-20 (PBS-T)] at 4 °C. Membranes were incubated with either anti-GR (1:500; H-300; sc-8992X; Santa Cruz) or anti-MR (1:500; H-300; sc-11412X; Santa Cruz) in 1% blocking solution for 2 h at room temperature. Membranes were washed three times (PBS-T), incubated with goat αRb HRP (1:5,000 in 0.5% blocking solution; Jackson ImmunoResearch) for 1 h, and then, finally washed another five times. Antibody binding was visualized by ECL using reagents (GE Healthcare) and a processor (G-Box) following the manufacturers’ guidelines.
Measurement of Corticosterone by RIA.
Plasma CORT concentrations were measured using a commercial CORT RIA Kit (MP Biomedicals) as described previously (16).
Statistical Analysis.
Data were statistically analyzed using SPSS (IBM). Results are presented as group means ± SEM; sample sizes are indicated in the figures. Multiple statistical comparisons were conducted with one- or two-way ANOVA, and if significant, a Bonferroni posthoc test was performed. Statistical results are provided in the figures. P < 0.05 was considered statistically significant.
SI Statistics Information to Figs. 1, 2, 3, 4, and 5
hnRNA and Mature RNA Expression of Glucocorticoid-Inducible Genes in Hippocampal Subregions Under Baseline Conditions and After Stress (Fig. 1).
Statistical analysis was as follows. One-way ANOVA: (Fig. 1A) hnRNA F(4, 38) = 167.50, P < 0.0001; mRNA F(4, 38) = 7.97, P < 0.0001; (Fig. 1B) hnRNA F(4, 39) = 102.10, P < 0.0001; mRNA F(4, 39) = 35.02, P < 0.0001; (Fig. 1C) hnRNA F(4, 37) = 32.87, P < 0.0001; mRNA F(4, 38) = 6.45, P < 0.0001; (Fig. 1D) hnRNA F(4, 39) = 50.23, P < 0.0001; mRNA F(4, 36) = 25.07, P < 0.0001; (Fig. 1E) hnRNA F(4, 38) = 32.12, P < 0.0001; mRNA F(4, 36) = 37.50, P < 0.0001; (Fig. 1F) hnRNA F(4, 39) = 51.28, P < 0.0001; mRNA F(4, 39) = 91.56, P < 0.0001. Bonferroni posthoc test: *P < 0.05 compared with early morning baseline (AM).
Analysis of MR and GR Binding to GREs Within Glucocorticoid-Inducible Genes in the Hippocampus Under Baseline Conditions and After Stress (Fig. 2).
Statistical analysis was as follows. One-way ANOVA: (Fig. 2A) F(5, 17) = 27.88, P < 0.0001; (Fig. 2B) F(5, 17) = 68.19, P < 0.0001; (Fig. 2C) F(5, 17) = 11.94, P < 0.0001; (Fig. 2D) F(5, 17) = 66.62, P < 0.0001; (Fig. 2E) F(5, 17) = 4.49, P = 0.0086; (Fig. 2F) F(5, 17) = 45.44, P < 0.0001. Bonferroni posthoc test: *P < 0.05 compared with early morning baseline (AM).
Effects of Different Stressors on MR and GR Receptor Binding to Glucocorticoid-Inducible Genes in the Hippocampus (Fig. 3).
Statistical analysis was as follows. One-way ANOVA: (Fig. 3A) Fkbp5, F(3, 16) = 52.81, P < 0.0001; Per1, F(3, 16) = 36.74, P < 0.0001; Sgk1, F(3, 16) = 12.35, P < 0.0001; (Fig. 3B) Fkbp5, F(3, 16) = 19.79, P < 0.0001; Per1, F(3, 16) = 52.37, P < 0.0001; Sgk1, F(3, 16) = 32.35, P < 0.0001. Bonferroni posthoc test: *P < 0.05 compared with the respective baseline group; ^P < 0.05 compared with the respective NE group; $P < 0.05 compared with the respective FS group.
Comparison of MR and GR ChIP on Original Vs. GR/MR Unbound Hippocampal Chromatin at an Intronic GRE Within the Fkbp5 Gene Under Baseline Conditions and After Stress (Fig. 4).
Statistical analysis was as follows. Two-way ANOVA: (Fig. 4A) effect of stress: F(2, 18) = 114.40, P < 0.0001; effect of ChIP F(1, 18) = 18.24, P < 0.0005; interaction stress × ChIP F(2, 18) = 5.440, P = 0.0142; (Fig. 4B) effect of stress: F(2, 18) = 183.60, P < 0.0001; effect of ChIP F(1, 18) = 29.57, P < 0.0001; interaction stress × ChIP F(2, 18) = 19.11, P < 0.0001. Bonferroni posthoc test: *P < 0.05 compared with respective ChIP at same time point.
Tandem ChIP for MR and GR Binding to GRE2 Within Fkbp5 in the Hippocampus Under Baseline Conditions and Stress (Fig. 5).
Statistical analysis was as follows. Two-way ANOVA: (Fig. 5A) effect of stress: F(1, 8) = 106.9, P < 0.0001; effect of ChIP F(1, 8) = 2.81, P = 0.132; interaction stress × ChIP: F(1, 8) = 0.44, P = 0.527; (Fig. 5B) effect of stress: F(1, 8) = 191.52, P < 0.0001; effect of ChIP F(1, 8) = 44.26, P = 0.0002; interaction stress × ChIP: F(1, 8) = 42.79, P = 0.0002. Bonferroni posthoc test: *P < 0.05 compared with the respective baseline ChIP; $P < 0.05 compared with GR→GR FS ChIP.
Supplementary Material
Acknowledgments
We thank Dr. S. D. Carter for her help and reading the manuscript and Biotechnology and Biological Sciences Research Council (BBSRC) for Grant BB/K007408/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605246113/-/DCSupplemental.
References
- 1.McEwen BS, Weiss JM, Schwartz LS. Selective retention of corticosterone by limbic structures in rat brain. Nature. 1968;220(5170):911–912. doi: 10.1038/220911a0. [DOI] [PubMed] [Google Scholar]
- 2.Reul JMHM, de Kloet ER. Two receptor systems for corticosterone in rat brain: Microdistribution and differential occupation. Endocrinology. 1985;117(6):2505–2511. doi: 10.1210/endo-117-6-2505. [DOI] [PubMed] [Google Scholar]
- 3.Reul JMHM, de Kloet ER. Anatomical resolution of two types of corticosterone receptor sites in rat brain with in vitro autoradiography and computerized image analysis. J Steroid Biochem. 1986;24(1):269–272. doi: 10.1016/0022-4731(86)90063-4. [DOI] [PubMed] [Google Scholar]
- 4.Reul JMHM, van den Bosch FR, de Kloet ER. Relative occupation of type-I and type-II corticosteroid receptors in rat brain following stress and dexamethasone treatment: Functional implications. J Endocrinol. 1987;115(3):459–467. doi: 10.1677/joe.0.1150459. [DOI] [PubMed] [Google Scholar]
- 5.De Kloet ER, Reul JMHM. Feedback action and tonic influence of corticosteroids on brain function: A concept arising from the heterogeneity of brain receptor systems. Psychoneuroendocrinology. 1987;12(2):83–105. doi: 10.1016/0306-4530(87)90040-0. [DOI] [PubMed] [Google Scholar]
- 6.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240(4854):889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beato M. Gene regulation by steroid hormones. Cell. 1989;56(3):335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 8.Datson NA, et al. Specific regulatory motifs predict glucocorticoid responsiveness of hippocampal gene expression. Endocrinology. 2011;152(10):3749–3757. doi: 10.1210/en.2011-0287. [DOI] [PubMed] [Google Scholar]
- 9.Polman JA, de Kloet ER, Datson NA. Two populations of glucocorticoid receptor-binding sites in the male rat hippocampal genome. Endocrinology. 2013;154(5):1832–1844. doi: 10.1210/en.2012-2187. [DOI] [PubMed] [Google Scholar]
- 10.Trapp T, Rupprecht R, Castrén M, Reul JMHM, Holsboer F. Heterodimerization between mineralocorticoid and glucocorticoid receptor: A new principle of glucocorticoid action in the CNS. Neuron. 1994;13(6):1457–1462. doi: 10.1016/0896-6273(94)90431-6. [DOI] [PubMed] [Google Scholar]
- 11.Savory JG, et al. Glucocorticoid receptor homodimers and glucocorticoid-mineralocorticoid receptor heterodimers form in the cytoplasm through alternative dimerization interfaces. Mol Cell Biol. 2001;21(3):781–793. doi: 10.1128/MCB.21.3.781-793.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Wang J, Sauter NK, Pearce D. Steroid receptor heterodimerization demonstrated in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92(26):12480–12484. doi: 10.1073/pnas.92.26.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jääskeläinen T, Makkonen H, Palvimo JJ. Steroid up-regulation of FKBP51 and its role in hormone signaling. Curr Opin Pharmacol. 2011;11(4):326–331. doi: 10.1016/j.coph.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Rawashdeh O, et al. PERIOD1 coordinates hippocampal rhythms and memory processing with daytime. Hippocampus. 2014;24(6):712–723. doi: 10.1002/hipo.22262. [DOI] [PubMed] [Google Scholar]
- 15.Tsai KJ, Chen SK, Ma YL, Hsu WL, Lee EH. sgk, a primary glucocorticoid-induced gene, facilitates memory consolidation of spatial learning in rats. Proc Natl Acad Sci USA. 2002;99(6):3990–3995. doi: 10.1073/pnas.062405399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian X, et al. A rapid release of corticosteroid-binding globulin from the liver restrains the glucocorticoid hormone response to acute stress. Endocrinology. 2011;152(10):3738–3748. doi: 10.1210/en.2011-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubler TR, Scammell JG. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones. 2004;9(3):243–252. doi: 10.1379/CSC-32R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webster MK, Goya L, Ge Y, Maiyar AC, Firestone GL. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol Cell Biol. 1993;13(4):2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itani OA, Liu KZ, Cornish KL, Campbell JR, Thomas CP. Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5′-flanking region. Am J Physiol Endocrinol Metab. 2002;283(5):E971–E979. doi: 10.1152/ajpendo.00021.2002. [DOI] [PubMed] [Google Scholar]
- 20.Cheon S, Park N, Cho S, Kim K. Glucocorticoid-mediated Period2 induction delays the phase of circadian rhythm. Nucleic Acids Res. 2013;41(12):6161–6174. doi: 10.1093/nar/gkt307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Bilang-Bleuel A, et al. Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: Involvement in a glucocorticoid receptor-dependent behavioural response. Eur J Neurosci. 2005;22(7):1691–1700. doi: 10.1111/j.1460-9568.2005.04358.x. [DOI] [PubMed] [Google Scholar]
- 23.Klengel T, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zannas AS, Binder EB. Gene-environment interactions at the FKBP5 locus: Sensitive periods, mechanisms and pleiotropism. Genes Brain Behav. 2014;13(1):25–37. doi: 10.1111/gbb.12104. [DOI] [PubMed] [Google Scholar]
- 25.Droste SK, et al. Corticosterone levels in the brain show a distinct ultradian rhythm but a delayed response to forced swim stress. Endocrinology. 2008;149(7):3244–3253. doi: 10.1210/en.2008-0103. [DOI] [PubMed] [Google Scholar]
- 26.Miranda TB, Morris SA, Hager GL. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol Cell Endocrinol. 2013;380(1-2):16–24. doi: 10.1016/j.mce.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obradović D, et al. DAXX, FLASH, and FAF-1 modulate mineralocorticoid and glucocorticoid receptor-mediated transcription in hippocampal cells--toward a basis for the opposite actions elicited by two nuclear receptors? Mol Pharmacol. 2004;65(3):761–769. doi: 10.1124/mol.65.3.761. [DOI] [PubMed] [Google Scholar]
- 28.Arriza JL, Simerly RB, Swanson LW, Evans RM. The neuronal mineralocorticoid receptor as a mediator of glucocorticoid response. Neuron. 1988;1(9):887–900. doi: 10.1016/0896-6273(88)90136-5. [DOI] [PubMed] [Google Scholar]
- 29.Allen Institute for Brain Science 2015 Allen Brain Atlas. Available at www.brain-map.org. Accessed March 14, 2016.
- 30.De Kloet ER, Vreugdenhil E, Oitzl MS, Joëls M. Brain corticosteroid receptor balance in health and disease. Endocr Rev. 1998;19(3):269–301. doi: 10.1210/edrv.19.3.0331. [DOI] [PubMed] [Google Scholar]
- 31.Reul JMHM. Making memories of stressful events: A journey along epigenetic, gene transcription, and signaling pathways. Front Psychiatry. 2014;5:5. doi: 10.3389/fpsyt.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutièrrez-Mecinas M, et al. Long-lasting behavioral responses to stress involve a direct interaction of glucocorticoid receptors with ERK1/2-MSK1-Elk-1 signaling. Proc Natl Acad Sci USA. 2011;108(33):13806–13811. doi: 10.1073/pnas.1104383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter SD, Mifsud KR, Reul JM. Distinct epigenetic and gene expression changes in rat hippocampal neurons after Morris water maze training. Front Behav Neurosci. 2015;9:156. doi: 10.3389/fnbeh.2015.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guidotti G, et al. Glucocorticoid receptor and FKBP5 expression is altered following exposure to chronic stress: modulation by antidepressant treatment. Neuropsychopharmacology. 2013;38(4):616–627. doi: 10.1038/npp.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.