Abstract
A bioinformatics analysis of disorder content of proteins from the DisProt database has been performed with respect to position of disordered residues. Each protein chain was divided into three parts: N- and C- terminal parts with each containing 30 amino acid (AA) residues and the middle region containing the remaining AA residues. The results show that in terminal parts, the percentage of disordered AA residues is higher than that of all AA residues (17% of disordered AA residues and 11% of all). We analyzed the percentage of disorder for each of 20 AA residues in the three parts of proteins with respect to their hydropathy and molecular weight. For each AA, the percentage of disorder in the middle part is lower than that in terminal parts which is comparable at the two termini. A new scale of AAs has been introduced according to their disorder content in the middle part of proteins: CIFWMLYHRNVTAGQDSKEP. All big hydrophobic AAs are less frequently disordered, while almost all small hydrophilic AAs are more frequently disordered. The results obtained may be useful for construction and improving predictors for protein disorder.
Keywords: Intrinsically unstructured/disordered proteins, Unstructured/disordered regions, DisProt database
Introduction
In the last decade our paradigm of protein structure has changed. It became evident, based on growing experimental data, that a significant number of proteins do not posses, under physiological conditions, well defined 3D structure [1]. They are known under different names with the most frequently used term being “intrinsically disordered proteins”. In this paper we will use the term “disordered proteins” (DPs). Various aspects of DPs are recently reviewed in detail in [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16].
Since protein disorder seems to be a diverse and complex phenomenon, there is no commonly agreed definition of it [14]. The structure of DPs, as well as their length, is highly varied, ranging (by increasing level of order) from completely unstructured random coils (which resemble the highly unfolded states of globular proteins with no secondary structure) to pre-molten globules (having no well-defined tertiary structure, may contain regions with transient and small amount of secondary structure), or molten globules (compact disordered ensembles that may contain significant secondary structure), as proposed by protein trinity structure [17], or the protein-quartet [18] hypothesis. Any of these states may be the native state – that is, the state relevant to a protein’s biological function (http://www.disprot.org) [2], [18].
At the primary structure level, DPs are characterized by low sequence complexity (i.e. consisting of short repetitive fragments) and are biased toward polar and charged, but against bulky hydrophobic and aromatic AA residues. Using Composition Profiler, Vačić et al. [19] have shown that, in terms of AA composition, DPs are enriched in Ala, Arg, Gly, Gln, Ser, Glu, Lys, and Pro but depleted in order-promoting residues like Trp, Tyr, Phe, Ile, Leu, Val, Cys, and Asn [6], [20], [21]. Using the TOP-IDP scale, based on AA properties such as hydropathy, polarity, volume, etc, Campen et al. [21] provided new ranking tendencies of AA from order to disorder promoting: Trp, Phe, Tyr, Ile, Met, Leu, Val, Asn, Cys, Thr, Ala, Gly, Arg, Asp, His, Gln, Lys, Ser, Glu, and Pro. This new scale is qualitatively consistent with the previous one.
Based on the published experimental data on protein disordered regions in their native state, DisProt database (http://www.disprot.org) [22] currently (February, 2011) contains 643 deposited proteins, originating from various prokaryotic, eukaryotic organisms and their viruses. The length of these proteins varies from 33 to 18,534 AA and length of their disordered regions is 1-3886 AA. For 620 proteins, at least one disordered region is identified while for 26 proteins at least one ordered region is identified. Most proteins contain unmarked regions which are of unknown structure. In total, 96 proteins are completely disordered and have lengths in the range 37–1861 AA (http://www.disprot.org).
Investigation of DPs is of special interest because of growing evidence on its association with various diseases, such as cancer [23], diabetes [24], cardiovascular [25] and neurodegenerative diseases [26]. Experimentally, DPs may be detected by more than 20 various biophysical and biochemical techniques such as: x-ray diffraction crystallography, heteronuclear multidimensional NMR, circular dichroism, optical rotatory dispersion, Fourier transformed infrared spectroscopy, Raman optical activity, etc [3]. DPs are difficult to study experimentally, due to the lack of unique structure in the isolated form [10], [18]. Therefore, a number of prediction tools have been developed [12].
The percentage of disordered regions which are longer than 41AA in archaeal, bacterial and eukaryotic proteomes has been analyzed using different predictors [27], [28], [29], [30]. Although direct comparison was not possible due to different DP predictors used, different numbers of genomes and different genomes themselves, all results follow the trend that archaeal proteins have lower disordered structure than bacterial proteins, which in turn have lower percentage of disordered structure than eukaryotic proteins (9–37% [27], 16% [28] and 8–46% [29] for Archaea, 6–33% [27], 20% [28] and 8–53% [29] for Bacteria and 35–51% [27], 43% [28] and 52–61% [30] for Eukaria).
Li et al. [31] and Lobanov et al. [32] investigated the distribution of disorder within different parts of a protein. Li et al. [31] divided protein chains into 3 parts – terminal parts with each 15 AA long and the middle part. They used dataset, consisting of 197 proteins from Protein Data Bank (PDB) (http://www.pdb.org), as training data to construct secondary structure predictor. They tested three different prediction methods on 3 parts of protein mentioned above and found that all of them indicated higher disorder in terminal parts than in the middle part. Lobanov et al. [32] investigated the relationship between AA disorder and the position in protein chains for 28,727 unique protein structures from PDB by dividing proteins into 3 parts similarly except that each terminus containing 30 AA residues. They found that, in terminal parts, the fraction of disordered AA residues is higher than overall fraction of AA residues, while the opposite is true for AA residues in the middle part). These conclusions helped improve the FoldUnfold [33] program for prediction of disordered regions from AA sequences.
The goal of this study was to analyze the DisProt database of experimentally determined disorder with respect to presence of disordered regions in N-terminal, C-terminal and middle parts of protein chains, as well as the AA distribution in these regions. The relationship between disordered AA distribution in these parts and AA physico-chemical characteristics was also investigated.
Results and discussion
Disorder content for proteins from DisProt database was analyzed in respect to the position of AA residues in protein chain. We divided proteins into three parts similarly as indicated by Lobanov et al., including N-terminal parts (containing first 30 AA residues), middle parts and C-terminal parts (containing last 30 AA residues). Fraction of disordered AA residues was calculated in terminal and middle parts of protein, according to protein lengths, AAs as well as their physico-chemical characteristics.
Distribution of AA residues in different parts of protein chains
The fraction of all AA residues was compared to the fraction of disordered AA residues in the three parts of protein chains. As shown in Table 1, the fraction of disordered AA residues in termini is higher than the fraction of all AA residues, which is consistent with the trend shown by Lobanov et al. [32], although the difference was not that distinct.
Table 1.
Distribution of AA residues in different parts of the protein chain
| Protein part | Percentage of all AAs (%) | Percentage of disordered AAs (%) |
|---|---|---|
| N-part (30 AAs near the N-terminus) | 5.62 | 8.45 |
| C-part (30 AAs near the C-terminus) | 5.62 | 8.85 |
| Combined in both terminal parts | 11.23 | 17.30 |
| M-part (all other AA residues) | 88.77 | 82.70 |
Fraction of disordered AA residues in dependence on the distance from terminals of a protein chain
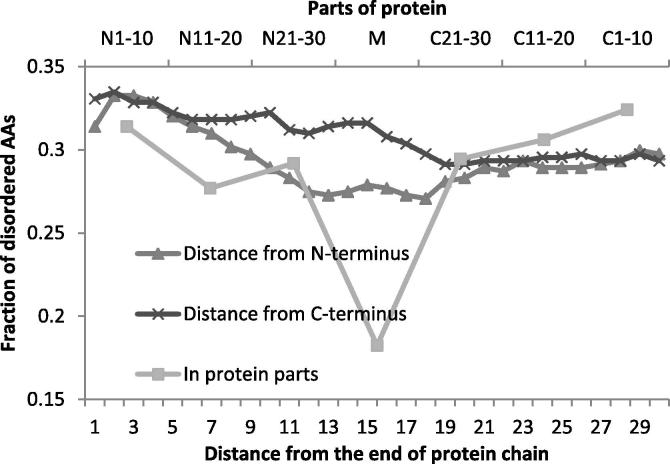
We next analyzed the percentage of disordered AA residues in terms of their distance from N- and C- termini of the protein chain, for distances from 1 to 30. The percentage of disordered AA residues is relatively even within 30 AAs from termini (25–35%) (Figure 1). The terminal parts were further divided into thirds and the percentage of disordered AA residues was calculated in these parts as well as in the remaining middle part. The same result was obtained for terminal parts whereas the middle part contains about 20% disordered AA residues (Figure 1).
Figure 1.
Fraction of disordered AA residues depending on the distance from the end of protein chain and by protein parts
The higher disorder fraction in terminal parts of proteins may be explained by their function. It has been shown that both protein ends may be involved in signaling and molecular recognition functions, that are often connected with protein disorder [6], [34], [35]. Specifically, the N-terminus can determine protein position within the cell, via signal peptide sequences [36], or by posttranslational modification [37], [38]. It is also an important determinant of protein half-life [39] and in general may be involved in molecular recognition and signaling [40]. For example, N-terminal domains of DELLA protein are, under physiological conditions, disordered regions [41]. Within the disordered N-terminal domain of DELLAs, Sun et al. [41] have identified several molecular recognition features (short sequences within DPs responsible for molecular recognition), known to undergo disorder-to-order transitions upon binding to interacting proteins in disordered proteins. Similarly the C-terminus can determine protein position within the cell and can form carboxyl tail domain (often consisting of low complexity repeat sequences) involved in molecular recognition, binding and regulation. For example, Kucera et al. found that a disordered C-terminus allows La, a 3′ RNA binding protein, to assists the biogenesis of diverse non-coding RNA precursors [42].
Fraction of disordered AA residues using two refined protein partitionings
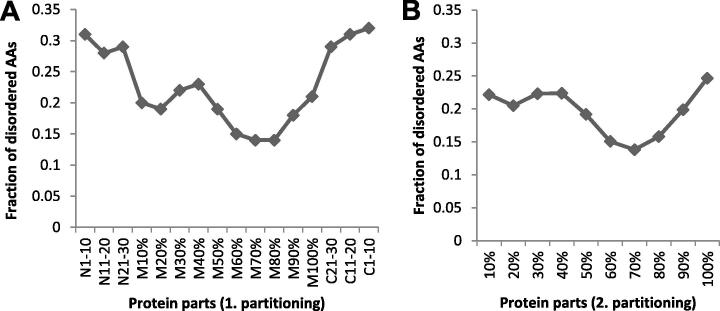
As shown in Figure 2, the difference of disorder fraction between the two neighboring intervals N21-30 and M, as well as C21-30 and M, is as high as 11% in both cases. This discrepancy suggests that the distribution of disordered AA residues in the middle part of protein is not uniform. In order to investigate distribution of disordered AA residues across the entire protein, two new partitionings of protein are introduced. The first partitioning keeps the same fixed terminal parts as the previous partitioning, but divides the middle part into subparts 10% long (N1–10, N11–20, N21–30, M10%, …, M100%, C21–30, C11–20, C1–10). The second partitioning simply divides entire chains into subparts 10% long (10%, …, 100%). The fraction of disordered AA residues in the two partitionings is shown in Figure 2.
Figure 2.
Fraction of disordered AA residues by protein parts using two partitioning Fraction of disordered AA residues by protein parts was analyzed using the first partitioning (A) and second partitioning (B). In the first partitioning, terminal parts contains 30 AAs each and divided into tens, while the middle part is divided into 10% subparts and in the second partitioning, whole proteins are divided into 10% subparts.
The difference between N21-30 and M, and similarly M and C21–30 is 7% and 8%, respectively, whereas the difference between adjoining subparts of middle part is no more than 4% (Figure 2A). A more gradual curve was displayed for the second partitioning and its maximal difference in disorder contents between adjoining subparts (90% to 100% of protein length) is 5% (Figure 2B). Disorder in the N-part did not alter remarkably.
Fraction of disordered AA residues by protein lengths in two partitionings
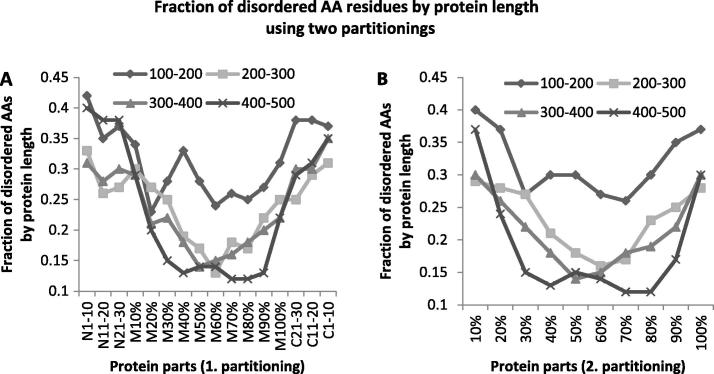
Being aware that protein length in DisProt varies substantially (92 AA – 18,543 AA), we analyzed the fraction of disordered AA residues in proteins by length intervals. The results on both scales are displayed in Figure 3.
Figure 3.
Fraction of disordered AA residues by protein length using two partitioning Fraction of disordered AA residues was analyzed according to protein length using two partitionings. A. on the first partitioning, for proteins shorter than 500 AA. B. Fraction of disordered AA residues by protein length on the second partitioning for proteins shorter than 500 AA.
For proteins of length >100 AA and ⩽500 AA, a more gradual decrease of fraction of disordered AAs was noticed from the N-parts towards the middle part and an increase from the middle part to the C-parts (Figure 3A) than for proteins of all lengths (Figure 2A). Furthermore, in some parts there are high differences in adjoining subparts of the middle part (for example, for proteins 100–200 AA, 11% difference between M10% and M20%, and a steep slope in 400–500 AA between N21–30, M10%, and M20%, and similarly M90%, M100% and C21–30). Figure 3B shows more distinctive difference between terminal and middle subparts when compared with Figure 2B. Disorder distribution for proteins longer than 500 AA is different (see more details in Suplementary materials).
Fraction of disordered AA residues for each of 20 AAs
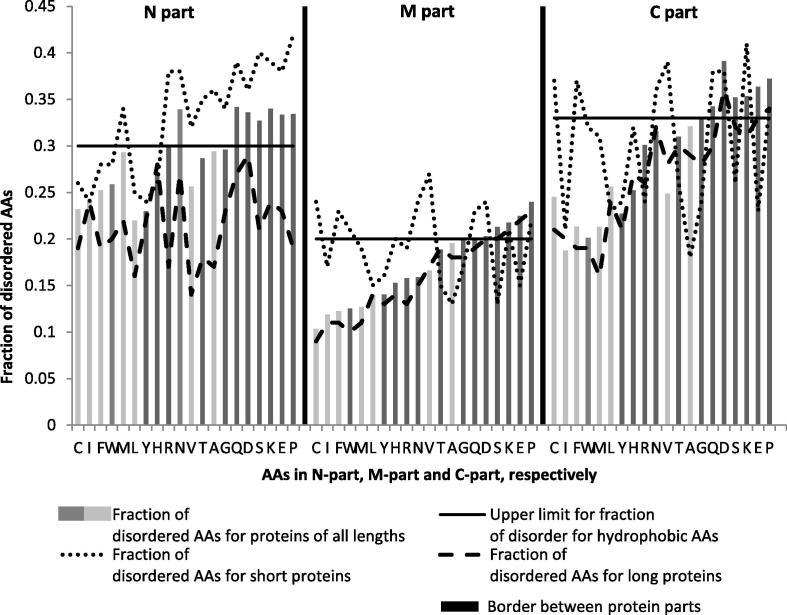
Fractions of disordered AA residues for each of 20 AAs in the N-part (the first 30 AAs at N terminus), middle part and the C-part (the last 30 AAs at C terminus), respectively, are displayed in Figure 4. All AAs show higher disorder fraction in terminal parts than in the middle part.
Figure 4.
Fraction of disordered AA residues for each of 20 AA in N, M and C part for proteins of all length Dark vertical lines separate parts. In all parts, AAs are ordered by the disorder fraction in the middle part (the new scale). Fractionall differences for hydrophobic AAs are in light grey.
As shown in Figure 4, there is a correlation between fractional differences of each AA and its hydropathy. Using the Kyte-Doolitle scale of hydropathy [43], we identified upper limit of fraction of disorder for hydrophobic AAs. In the middle part, all hydrophobic AAs have a fraction lower than 0.2, whereas some hydrophilic AAs (P, Q, D, K, E and S) have a disorder fraction higher than 0.2 and others (W, Y, R, N, H, T and G) do not. In the N-part, all hydrophobic AAs have a fraction lower than 0.3, some hydrophilic AAs (P, Q, D, K, E, S, R and N) have a fraction higher than 0.3 and others (W, Y, H, T and G) do not. In the C-part, all hydrophobic AAs have a fraction lower than 0.33, some hydrophilic AAs (P, Q, D, K, E, S) have a fraction higher than 0.33 and others (W, Y, H, R, N, T and G) have a fraction lower than 0.33, which precisely corresponds to hydropathy distribution in the middle part.
Using values from Figure 4, we constructed an AA scale based on disorder fraction of AA residues in the middle part. The obtained AA scale is similar to the one presented by Lobanov et al. based on the same calculation [32]. Table 2 presents the new scale (second row), the positions of AAs in that scale (first row) and positions of AAs in Lobanov’s scale (third row). We can see that the difference between positions of the corresponding AAs in both scales (first and third row) for almost all AAs is ⩽3 (except C, V, G and P).
Table 2.
New scale based on disorder fraction in the this study in comparison with those published previously
| Positions in new scale | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAs | C | I | F | W | M | L | Y | H | R | N | V | T | A | G | Q | D | S | K | E | P |
| Positions in Lobanov’s scale | 5 | 3 | 2 | 1 | 8 | 6 | 4 | 9 | 12 | 13 | 7 | 11 | 10 | 19 | 15 | 16 | 20 | 17 | 18 | 14 |
| Positions in Campen’s scale | 9 | 4 | 2 | 1 | 5 | 6 | 3 | 15 | 13 | 8 | 7 | 10 | 11 | 12 | 16 | 14 | 18 | 17 | 19 | 20 |
Fraction of disordered AA residues for each of 20 AAs
Figure 4 also displays the fraction of disordered AA residues for each AA in short and long proteins (shorter and longer than 500 AA, respectively). The border value 500 AA was chosen since proteins containing less than 500 AAs showed relatively higher disorder in terminal parts comparing to those containing more than 500 AAs as shown in Figures 3 and S1.
For proteins of all lengths, the trend of disorder for AAs in the C-part is more similar to that in the M-part than to that in the N-part. For short proteins, disorder fractions for the N-part and the C-part are approximate, whereas for long proteins the disorder fraction for the N-part is closer to that for the M-part.
Fractional differences between disordered and undefined sets of regions for each of 20 AAs
DisProt database contains information on disordered regions of proteins. For most of the proteins, some AAs are marked as disordered and the remaining AAs are marked neither as ordered nor as disordered. In this analysis, such unmarked AA residues in proteins will be referred to as structure-undefined, undefined for short (see Materials and methods for more details).
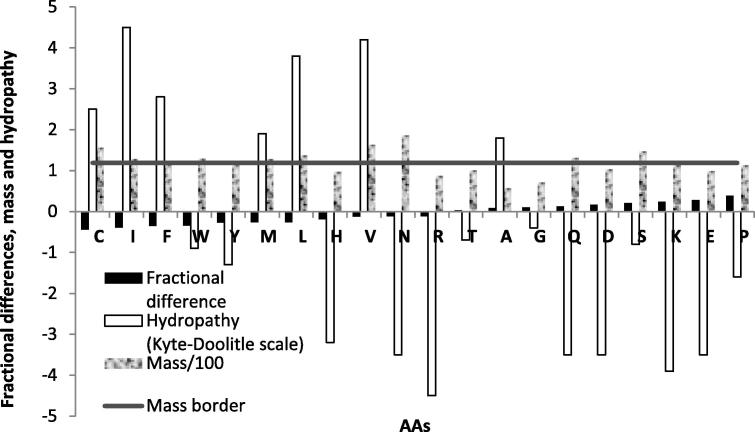
The fractional difference between disordered and undefined sets of regions for each of 20 AAs is shown in Figure 5. The AAs were ordered from left to right according to the new scale presented in Table 2 and Figure 4. It is notable that the fractional difference of AAs increased along with their positions on the new scale (except Y). According to the Kyte-Doolitle scale of hydropathy [43], almost all hydrophobic AAs have negative fractional difference (except A), and most hydrophilic AAs have positive fractional difference (except W, Y, H, R and N).
Figure 5.
Fractional differences between disordered and undefined sets of regions for each of 20 AAs in respect to their mass and hydropathy For details about calculating fractional differences, see Materials and methods, Processing steps, Step 4.
Campen et al. [21] presented a scale of AAs according to hydropathy. As shown in Table 2, for most AAs the difference between positions of an AA in the two scales is ⩽3 (except C, Y, H, R and V).
Fractional differences between disordered and undefined sets of regions for each AA in respect to their mass and hydropathy
It was expected that big hydrophobic AAs would be more ordered and small hydrophilic ones more disordered [6], [15], [21], which implies that big hydrophobic/small hydrophilic AAs would have negative/positive fractional differences between disordered and ordered sets of regions, respectively. In this analysis, we investigated whether the same pattern occurs for fractional differences between disordered and undefined sets of regions in DisProt database. An AA can be classified as big or small according to parameters such as volume, surface, mass etc. For each parameter, the border between big and small AAs can be determined as an average value or as a median. Of the three suggested parameters, the most complete classification of AAs into big hydrophobic and small hydrophilic categories was achieved based on mass (Table S1). Here, an AA with mass less than 114.6 Daltons, which is a median mass for all AAs, is considered to be small. The mass border is presented as a horizontal solid line in Figure 5. As shown in Figure 5, out of 7 hydrophobic AAs, 6 are big and their fractional difference is negative as expected, except A, which is not big and has a positive fractional difference. Of all hydrophilic AAs, only 9 of them are small, and 6 of them have a positive fractional difference, as expected. Y, N and R are small and hydrophilic, but have a negative fractional difference.
Conclusion
The database of disordered proteins, DisProt, has been analyzed for the first time with respect to distribution of disordered regions according to their position in protein chains. The dataset of proteins analyzed is quite different from those analyzed previously [32]. Our dataset is smaller, the average size of proteins as well as the number of disordered regions per protein is smaller. More importantly, the disordered regions in this database were determined by different experimental methods. Results obtained show the same trend as those previously published with certain differences in specific values and positions possibly due to differences in datasets examined. The new scale of amino acids proposed according to their disorder content in the middle part of proteins correlates well with previous scales [21], [32], especially with respect to physico-chemical characteristics such as mass and hydropathy. This study refines the existing scales due to specific credibility of the dataset used. Such analyses provide useful hints for the better understanding of protein disorder and may be also useful to improve the performance of protein disorder predictors.
Materials and methods
Dataset
The dataset includes 484 proteins. These are almost all the proteins in the DisProt database, except proteins with 100% undefined or disordered residues and those containing less than 90 residues. The DisProt database contains proteins with overlapping disordered regions, as a result of different experimental methods for structure determination. In such cases, the union of the regions was taken into account. Most proteins (99%) from the dataset contain unmarked residues, neither disordered nor ordered. In this paper, such residues will be referred to as structure-undefined, undefined for short. Around 5% of proteins contain regions marked as ordered. Since ordered content of the dataset is quite poor, it was included in the undefined class rather than considered as a separate class. Distribution of proteins by protein length in the DisProt database is shown in Table S2 The number of proteins and their average length by super kingdoms is shown in Table S3.
Processing steps
-
(1)
Release 5.7 of DisProt was used in this research. A Perl program has been developed for preparing the data on protein sequences for further analysis.
-
(2)
A database has been designed and populated with proteins and disorder regions data.
-
(3)
In order to examine the relationship between disorder and its location in a protein chain, protein is divided into three parts: the N-part, which contains first 30 residues of a protein, the C-part, which contains last 30 residues, and the middle part (M-part), which contains remaining residues [32]. Further subdivision of the N- and C- parts into three equal-length parts has also been considered: N1–10, N11–20, N21–30 and similarly C1–10, C11–20, C21–30 [32]. Two new scales were introduced: one involving division of the middle part into tens and another involving division of entire protein into tens.
-
(4)
SQL queries were developed for analysis of protein disorder: Analysis of disordered regions: Distribution of disordered regions of different length (1–30 AA) in entire proteins as well as in the N-, C-, M- parts of protein chains have been calculated. Analysis of disordered AAs: Distribution of AAs and disordered AAs in the N-, C-, M- parts of protein chains, as well as in their subparts described in “Division of protein chains” paragraph, has been determined; the distribution of disordered AAs by length of proteins and by specific AAs has been calculated. Mole fractions and fractional differences: Mole fractions for AAs have been calculated for entire proteins as well as fractional differences between disordered and undefined sets of regions. The mole fraction for the j-th AA (j = 1,20) in the i-th sequence (e.g., i-th protein) is determined as Pj = sum(ni·Pji)/sum(ni), where ni is the length of the i-th sequence and Pji – frequency of the j-th AA in the i-th sequence. The fractional difference is calculated by the formula (Pj(a) − Pj(b))/Pj(b), where Pj(a) is the mole fraction of the j-th AA in the set of disordered regions in proteins (set a), and Pj(b) is the corresponding mole fraction in the set of undefined regions in proteins (set b). Fractional differences were analyzed in respect to hydropathy and mass of AAs.
Competing interests
The authors have no competing interests to declare.
Acknowledgements
The work presented has been financially supported by the Ministry of Education and Science, Republic of Serbia (Project No. 174021). I would like to thank my supervisors Drs Gordana M. Pavlović-Lažetić and Miloš V. Beljanski for their precious advice during this research.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gpb.2012.01.002.
Supplementary material
Supplementary material.
Fig. S1. Fraction of disordered AA residues by protein length using two partitioning.
Fig. S2. Distribution of disordered and undefined regions by region length.
Fig. S3. Length distribution of disordered regions.
Fig. S4. Fraction of disordered AA residues for each of the 20 AAs on two partitionings.
References
- 1.Tompa P., Fersht A. CRC Press/Taylor & Francis Group; Florida: 2010. Structure and function of intrinsically disordered proteins. [Google Scholar]
- 2.Sigalov A.B. Protein intrinsic disorder and oligomericity in cell signaling. Mol BioSyst. 2010;6:451–461. doi: 10.1039/b916030m. [DOI] [PubMed] [Google Scholar]
- 3.Uversky V.N., Longhi S. John Wiley & Sons, Inc.; New Jersey: 2010. Instrumental analysis of intrinsically disordered proteins: assessing structure and conformation. [Google Scholar]
- 4.Turoverov K.K., Kuznetsova I.M., Uversky V.N. The protein kingdom extended: ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog Biophys Mol Biol. 2010;102:73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uversky V.N. The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/568068. 568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uversky V.N., Dunker A.K. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishikawa K. Natively unfolded proteins: an overview. Biophysics. 2009;95:53–58. doi: 10.2142/biophysics.5.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruscher S., Pomes R. Molecular simulations of protein disorder. Biochem Cell Biol. 2010;88:269–290. doi: 10.1139/o09-169. [DOI] [PubMed] [Google Scholar]
- 10.Tompa P., Kovacs D. Intrinsically disordered chaperones in plants and animals. Biochem Cell Biol. 2010;88:167–174. doi: 10.1139/o09-163. [DOI] [PubMed] [Google Scholar]
- 11.Dosztanyi Z., Mezsaros B., Simon I. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief Bioinform. 2010;11:225–243. doi: 10.1093/bib/bbp061. [DOI] [PubMed] [Google Scholar]
- 12.He B., Wang K., Liu Y., Xue B., Uversky V.N., Dunker A.K. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 13.Dyson HJ, Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys 44:467–518. [DOI] [PMC free article] [PubMed]
- 14.Orosz F, Ovádi, J. Proteins without 3D structure: definition, detection and beyond. Bioinformatics 2011;27:1449–54. [DOI] [PubMed]
- 15.Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 2011;43:1090–103. [DOI] [PubMed]
- 16.Dunker A.K., Gough J. Sequences and topology: intrinsic disorder in the evolving universe of protein structure. Curr Opin Struct Biol. 2011;21:379–381. doi: 10.1016/j.sbi.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Dunker A.K., Obradović Z. The protein trinity—linking function and disorder. Nat Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 18.Uversky V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vačić V., Uversky V.N., Dunker A.K., Lonardi S. Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radivojac P., Iakoucheva L.M., Oldfield C.J., Obradovic Z., Uversky V.N., Dunker A.K. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campen A., Williams R.M., Brown C.J., Meng J., Uversky V.N., Dunker A.K. TOP-IDP-Scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept Lett. 2008;15:956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sickmeier M., Hamilton J.A., LeGall T., Vacic V., Cortese M.S., Tantos A. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iakoucheva L.M., Brown C.J., Lawson J.D., Obradović Z., Dunker A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 24.Uversky V.N., Oldfield C.J., Dunker A.K. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys Biomol Struct. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Y., LeGall T., Oldfield C.J., Dunker A.K., Uversky V.N. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45:10448–10460. doi: 10.1021/bi060981d. [DOI] [PubMed] [Google Scholar]
- 26.Uversky V.N. Intrinsic disorder in proteins associated with neurodegenerative diseases. In: Ovádi J., Orosz F., editors. Protein folding and misfolding: neurodegenerative diseases. Springer; New York, USA: 2008. pp. 21–75. [Google Scholar]
- 27.Dunker A.K., Obradović Z., Romero P., Garner E.C., Brown C.J. Intrinsic protein disorder in complete genomes. In: Miyano S., Takagi T., editors. Vol 11. Japan; Tokyo: 2000. pp. 161–171. (Proc Genome Informatics). [PubMed] [Google Scholar]
- 28.Bogatyreva N.S., Finkelstein A.V., Galzitskaya O.V. Trend of amino acid composition of proteins of different taxa. J Bioinform Comput Biol. 2006;4:597–608. doi: 10.1142/s0219720006002016. [DOI] [PubMed] [Google Scholar]
- 29.Pavlović-Lažetić G., Mitić N., Kovačević J., Obradovićc Z., Malkov S., Beljanski M. Bioinformatics analysis of disordered proteins in prokaryotes. BMC Bioinformatics. 2011;12:66. doi: 10.1186/1471-2105-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vučetić S., Brown C.J., Dunker A.K., Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 31.Li X., Romero P., Rani M., Dunker A.K., Obradovic Z. Predicting protein disorder for N-, C- and internal regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 32.Lobanov M.Y., Garbuzynskiy S.O., Galzitskaya O.V. Statistical analysis of unstructured amino acid residues in protein structures. Biochemistry (Mosc) 2010;75:192–200. doi: 10.1134/s0006297910020094. [DOI] [PubMed] [Google Scholar]
- 33.Galzitskaya O.V., Garbuzynskiy S.O., Lobanov M.Y. FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006;22:2948–2949. doi: 10.1093/bioinformatics/btl504. [DOI] [PubMed] [Google Scholar]
- 34.Dunker A.K., Brown C.J., Lawson J.D., Iakoucheva L.M., Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 35.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 36.Martoglio B., Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 37.Podell S., Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004;5:37. doi: 10.1186/1471-2164-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basu J. Protein palmitoylation and dynamic modulation of protein function. Curr Sci. 2004;87:212–217. [Google Scholar]
- 39.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1992;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 40.Sun X, Xue B, Jones WT, Rikkerink E, Dunker AK, Uversky VN. A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol Biol 2011;77:205–23. [DOI] [PubMed]
- 41.Sun X., Jones W.T., Harvey D., Edwards P.J., Pascal S.M., Kirk C. N-terminal Domains of DELLA proteins are intrinsically unstructured in the absence of interaction with GID1/gibberellic acid receptors. J Biol Chem. 2010;285:11557–11571. doi: 10.1074/jbc.M109.027011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kucera N. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. PNAS. 2011;108:1308–1313. doi: 10.1073/pnas.1017085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyte J., Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Fig. S1. Fraction of disordered AA residues by protein length using two partitioning.
Fig. S2. Distribution of disordered and undefined regions by region length.
Fig. S3. Length distribution of disordered regions.
Fig. S4. Fraction of disordered AA residues for each of the 20 AAs on two partitionings.