Abstract
MicroRNAs (miRNAs) selectively localize to subcompartments of the neuron, such as dendrites, axons, and presynaptic terminals, where they regulate the local protein synthesis of their putative target genes. In addition to mature miRNAs, precursor miRNAs (pre-miRNAs) have also been shown to localize to somatodendritic and axonal compartments. miRNA-338 (miR-338) regulates the local expression of several nuclear-encoded mitochondrial mRNAs within axons of sympathetic neurons. Previous work has shown that precursor miR-338 (pre-miR-338) introduced into the axon can locally be processed into mature miR-338, where it can regulate local ATP synthesis. However, the mechanisms underlying the localization of pre-miRNAs to the axonal compartment remain unknown. In this study, we investigated the axonal localization of pre-miR-338. Using proteomic and biochemical approaches, we provide evidence for the localization of pre-miR-338 to distal neuronal compartments and identify several constituents of the pre-miR-338 ribonucleoprotein complex. Furthermore, we found that pre-miR-338 is associated with the mitochondria in axons of superior cervical ganglion (SCG) neurons. The maintenance of mitochondrial function within axons requires the precise spatiotemporal synthesis of nuclear-encoded mRNAs, some of which are regulated by miR-338. Therefore, the association of pre-miR-338 with axonal mitochondria could serve as a reservoir of mature, biologically active miRNAs, which could coordinate the intra-axonal expression of multiple nuclear-encoded mitochondrial mRNAs.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2270-6) contains supplementary material, which is available to authorized users.
Keywords: Subcellular localization, Sympathetic neuron, Superior cervical ganglion, Mitochondrial translation, RNP granule, Proteomics, Local translation, Post-transcriptional regulation
Introduction
MicroRNAs (miRNAs) are highly conserved, short (21–23 bp), single-stranded, non-coding RNAs that function to inhibit translation or induce degradation of their cognate mRNA targets [1, 2]. The biogenesis of miRNAs involves two successive RNase-dependent cleavage steps: (1) processing of pri-miRNA to the precursor form by Drosha in the nucleus, followed by (2) DICER-dependent processing of ~70-bp precursor miRNAs to the mature, biologically active form of the molecule in the cytoplasm [3]. Fully processed mature miRNAs regulate many neurobiological processes, such as neuronal plasticity, development, and axonal guidance [4–7]. Thus, miRNAs play a critical role in maintaining the function of axons (for review, see [8]).
Axons are highly specialized neuronal subcompartments, which must respond to internal and external stimuli with precise spatiotemporal fidelity. To accomplish this, mRNAs are localized into axons in a highly regulated manner [9, 10] and are translated locally [11, 12]. Furthermore, the tightly orchestrated translation of axonal mRNAs is controlled by a heterogeneous population of miRNAs [13, 14]. miRNAs localized to the axons play a critical role in axonal function and viability [8]. In addition to the mature miRNAs, it has also been reported that precursor miRNAs (pre-miRNAs) are localized to axons and dendrites [15–17]. Consistent with pre-miRNAs being present in axons, components of the RISC complex involved in miRNA processing have also been shown to be present in the axon [18, 19]. Together, the presence of pre-miRNA and miRNA processing machinery in axons strongly suggests that a subset of pre-miRNAs are transported and processed in the distal structural/functional domains of neurons. In line with this reasoning, it has been shown that pre-miRNA-134 (pre-miR-134) localizes to dendrites upon binding of DEAH-box helicase DHX36 to its terminal loop sequence [20]. Similarly, in axons, it is likely that the localization of pre-miRNAs is mediated by various RNA-binding proteins (RBPs). However, direct evidence for pre-miRNA trafficking to axons has yet to be elaborated. In addition, the protein complexes involved in regulating pre-miRNA localization to axons are unknown.
In axons of superior cervical ganglion (SCG) neurons, miR-338 regulates the expression of nuclear-encoded mitochondrial mRNAs cytochrome c oxidase IV (COXIV) and ATP synthase 5 gamma 1 (ATP5G1). Modulation of local expression of miR-338 affects mitochondrial oxygen consumption, ATP generation, and the production of reactive oxygen species (ROS) in the axon. The introduction of pre-miR-338 directly into axons results in a marked increase in the levels of mature miRNA-338 within hours after transfection and leads to alterations in the local levels of its target mRNAs [19, 21]. These findings suggest that mechanisms mediating the axonal localization and processing of endogenous pre-miR-338 exist in SCG neurons.
In this study, we aimed to delineate the molecular mechanisms that govern the axonal localization of pre-miR-338. To identify specific pre-miR-338 interacting proteins that might control this process, we performed RNA affinity pulldown assays followed by high-performance liquid chromatography–tandem mass spectrometry (LC/MS). Unexpectedly, proteomic studies revealed that pre-miR-338 interacts with an array of mitochondrial-associated proteins in axons. Quantitative RT-PCR (qRT-PCR) analysis of isolated axonal mitochondria showed that endogenous pre-miR-338 was indeed associated with mitochondria. In addition, DICER protein was also present in mitochondrial fractions obtained from sympathetic neurons. Using compartmentalized culture chambers, we also observed that labeled pre-miR-338 localized to distal axons and was also associated with axonal mitochondria. Taken together, these studies provide direct evidence for the localization of pre-miRNA to distal axons and demonstrate the association of pre-miR-338 with the mitochondria, which may serve to provide a readily accessible source of miRNA to fine-tune the intra-axonal expression of nuclear-encoded mitochondrial mRNAs.
Materials and methods
Neuronal cell cultures
SCG neurons were cultured as described previously [22]. Briefly, SCG dissected from 3-day-old Harlan Sprague–Dawley rat pups was dissociated using the Gentle MACS Dissociator and Neuronal Tissue Dissociation Kit (Miltenyi Biotec) according to the manufacturer’s protocol. Dissociated neurons were plated into the center compartment of a Campenot culture chamber. The neurons are cultured for 2–7 days in serum-free Neurobasal medium (Invitrogen) containing nerve growth factor (NGF; 50 ng/ml; Alamone labs), 20-mM KCl, 20-U/ml penicillin, and 20-mg/ml streptomycin (Hyclone). Culture medium was replaced every 3–4 days. To inhibit the proliferation of non-neuronal cells, 5-fluoro-2′-deoxyuridine (FUDR, 50 μM) was added to the medium after 2 DIV, and remained in the medium for the duration of the experiments. Phase-contrast microscopy was used to determine whether the side compartments, which contained the distal axons used in the experiments, were devoid of neuronal soma and non-neuronal cells. For immunohistochemical studies, dissociated SCG neurons were plated on collagen coated Nunc® Lab-Tek® II-CC2™ glass chamber slides (Sigma-Aldrich) and cultured for two weeks before being processed for immunostaining.
Cytosolic and mitochondrial fraction preparation
Cytosolic extracts were prepared from 4-day-old rat pup brains or 2-week-old compartmentalized SCG cultures using the NE-PER™ Nuclear and Cytoplasmic Extraction Kit (Life Technologies) according to the manufacturer’s instructions. For isolation of mitochondrial fractions from 2-week-old compartmentalized SCG cultures, Mitochondria Isolation Kit for Cultured Cell (Life Technologies) was used according to the manufacturer’s protocol. The cell body and axonal compartments were processed separately.
RNA affinity purification
Biotinylated RNA oligonucleotides corresponding to the rat pre-miR-338 sequence was synthesized by IDT technologies. For the RNA affinity pulldown experiments, 5 pmol of biotinylated pre-miR-338 oligomer was incubated with cytosolic brain extracts (5 ug/uL) in a reaction containing 400 ul of binding buffer containing 20-mM HEPES, pH 7.5, 72-mM potassium chloride, 1.5-mM magnesium chloride, 1.6-mM magnesium acetate, 0.5-mM dithiothreitol [DTT], 4-mM glycerol, 1-mM ATP, and 200 units of RNasin (Promega) at room temperature for 30 min. Following incubation, the RNA–protein complexes were incubated with 100 μl of magnetic streptavidin beads (Pierce Corporation) for 2 h at room temperature. The beads were preequilibrated in binding buffer for 1 h at room temperature before use. After incubation, the beads were washed three times with ten volumes of binding buffer, and the bead-bound proteins were eluted by boiling for 10 min in elution buffer (125-mM Tris, pH 6.8, 2 % SDS, 0.02 % bromophenol blue, 0.1-M DTT) for Western analyses or submitted to the NINDS/NIMH clinical proteomics core facility for tandem mass spectrometry.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed using LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) according to manufacturer’s instructions with minor alterations. Briefly, binding reactions contained biotinylated pre-miR-338 oligomer (10 nM) and P4 total brain lysate (10 µg) in binding buffer with RNase inhibitor. For controls, 5 µg of BSA was used. For competition assays, 100-fold unlabeled oligomer was added to the reaction. Binding reactions were then incubated in room temperature for 30 min. The reactions were separated in 4–20 % non-denaturing PAGE gel running in 0.5 × Tris–borate-EDTA buffer for 2 h, then transferred to nitrocellulose membrane at 40 mA for 35 min. Finally, the membrane was UV cross-linked, and the biotinylated oligomer shifts were detected using Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher). The pre-miR-338 oligomer sequence was: 5′-UCCCCAACAAUAUCCUGGUGCUGAGUGGGUGCACAGUGACUCCAGCAUCAGUGAUUUUGUUGAAGA-BIOTIN-3′.
Mass spectrometric analyses
The bead-bound affinity purified proteins were processed using a modified version of a previously described on-bead digestion protocol [23]. The affinity purification and proteomic study was performed in quintuplets as described in [23]. Proteins specific or enriched by greater than 1 unique peptide in pre-miR-338 fraction compared with bead control were considered in each of the 5 biological replicates. Proteins that met this category in at least 3 out of 5 replicates were considered as pre-miR-338 interacting proteins.
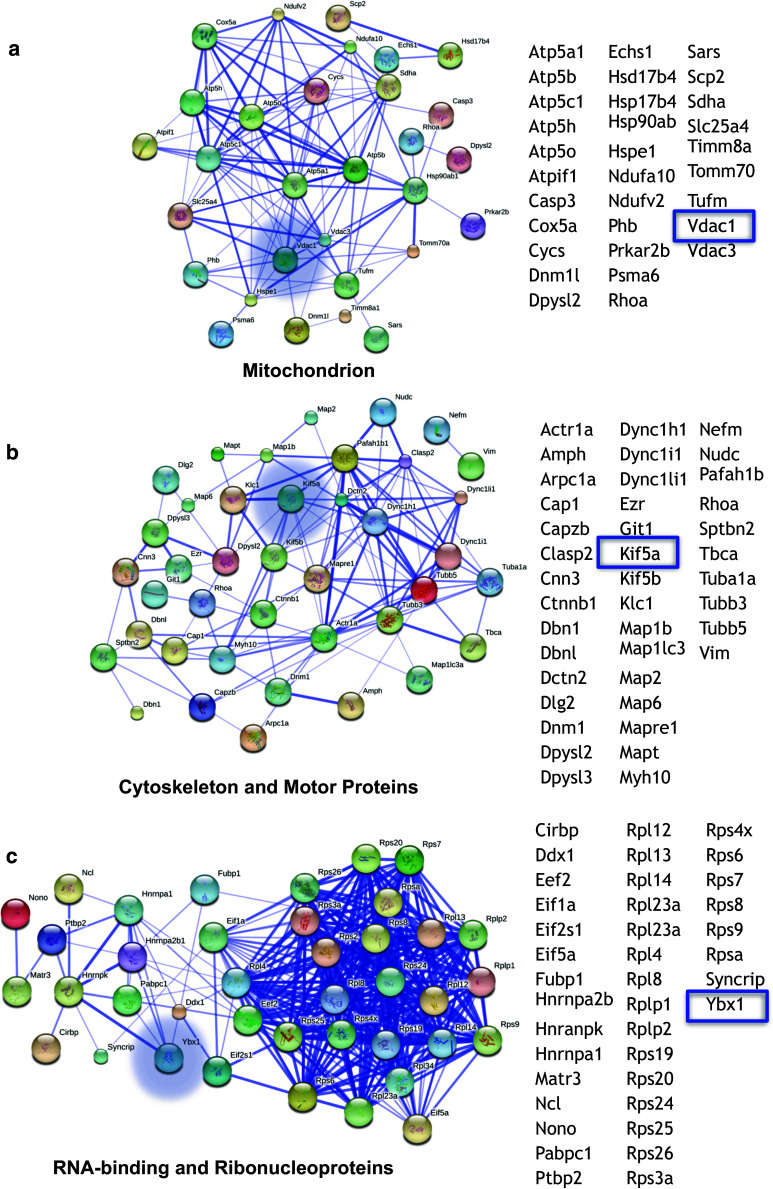
STRING analysis of pre-miR-338 interacting proteins
Pre-miR-338 interacting proteins identified by mass spectrometry were analyzed using DAVID Gene Ontology [24, 25]. Proteins that were under categories: GO:0005753 mitochondrial proton-transporting ATP synthase complex, GO:0005743 mitochondrial inner membrane, GO:0045259 proton-transporting ATP synthase complex, GO:0044455 mitochondrial membrane part, GO:0031966 mitochondrial membrane, GO:0044429 mitochondrial part, GO:0045261 proton-transporting ATP synthase complex, catalytic core F(1), GO:0005740 mitochondrial envelope, GO:0016469 proton-transporting two-sector ATPase complex, GO:0033178 proton-transporting two-sector ATPase complex, catalytic domain, GO:0031980 mitochondrial lumen, GO:0005759 mitochondrial matrix, GO:0000275 mitochondrial proton-transporting ATP synthase complex, catalytic core F(1), and GO:0005739 mitochondrion were collapsed into one list named “Mitochondrion.” It is this collapsed list that served as input for STRING analysis for mitochondrial-related pre-miR-338 interacting proteins (Fig. 5a). Similarly, pre-miR-338 interacting proteins under GO:0005856~cytoskeleton were analyzed (Fig. 5b). Last, proteins under GO:0003723~RNA binding and GO:0030529~ribonucleoprotein complex were collapsed into “RNA-binding and ribonucleoproteins” (Fig. 5c). Disconnected protein nodes were omitted. All GO categories used as inputs for STRING had significant p values ranging from 2.6 × 10−3 to 1.2 × 10−11.
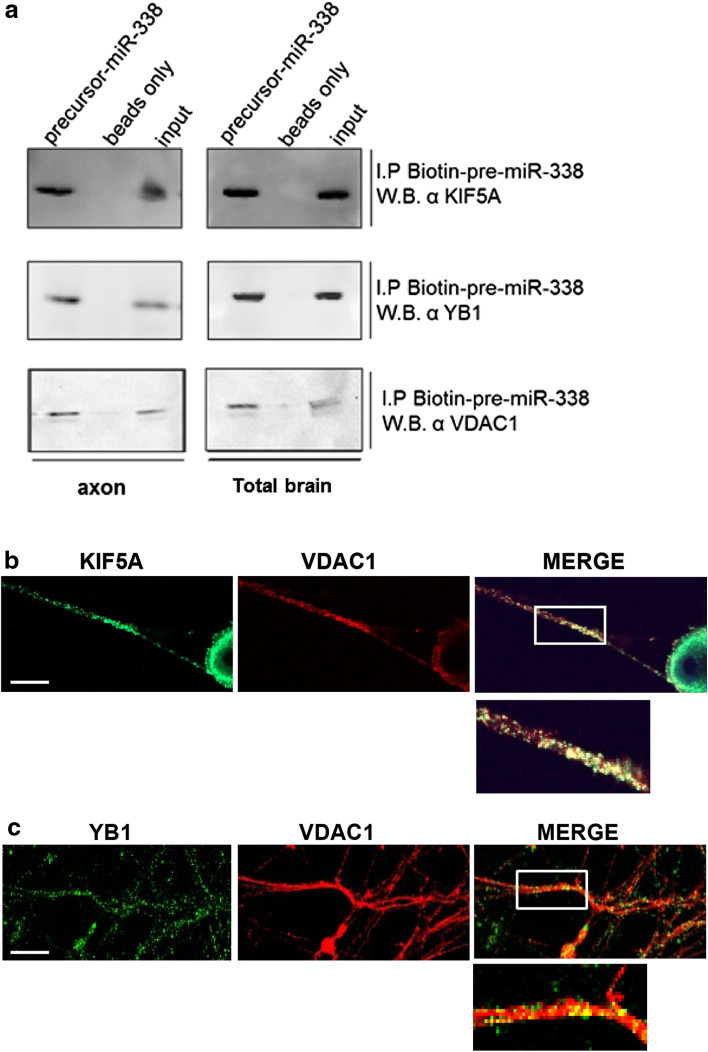
Fig. 5.
Pre-miR-338 interacts with cytoskeletal protein KIF5A, RNA binding protein YB-1, and mitochondrial protein, VDAC1. RNA affinity pulldowns were performed with the biotinylated pre-miR-338 oligomer and P4 brain cytosolic lysates or cytosolic extracts isolated from distal axons present in the side-compartments of Campenot cultures (a). The presence of pre-miR-338 binding proteins KIF5A, YB-1, and VDAC1 (a) was assessed by Western analyses using protein target-specific antibodies. a Western analyses shows the presence of KIF5A, YB-1, and VDAC1 proteins in the pre-miR-338 affinity pulldown fractions derived from brain and axonal cytosolic lysates (b, c) Visualization of pre-miR-338-interacting proteins KIF5A, VDAC1 and YB-1 in SCG axons using fluorescent confocal microscopy. Co-immunostaining studies of dissociated SCG neuron cultures with antibodies specific for KIF5A and VDAC1 (b) and YB-1 and VDAC1 (c). The merge shows the regions of overlap for the indicated protein. Boxes indicate magnified images; arrowheads indicate region of colocalized fluorescent signals. Scale bar 50 µm
Transfection of neurons with pre-miR-338
Fluorescently labeled pre-miR-338 oligomer (Alexa-594) was purchased from IDT. As control RNA, a DY-547 labeled RNA was used (Dharmacon). The oligomers (25 uM) were transfected into cell body compartments of 10-to-14-day-old Campenot cultures using an Accell transfection system according to manufacturer’s instructions (Life Technologies). Cell bodies, located in the central compartments of the chamber, were exposed to media containing the transfection reagents for 24 h, and the media were subsequently changed to media containing culture media containing MitoTracker (see “Materials and methods”, Mitochondrial staining) for mitochodria colocalization studies. For nocodazole treatments, 65 uM of nocodazole was added to side compartments during transfection. Neurons were then washed with PBS and fixed using buffered paraformaldehyde (4 % in PBS) for 15–30 min. After fixation, neurons are washed three times with PBS and, subsequently, imaged using a confocal microscope (Zeiss LSM510). The sequence of the fluorescent pre-miR-338-Alexa-594 oligomer was: 5′-UCCCCAACAAUAUCCUGGUGCUGAGUGGGUGCACAGUGACUCCAGCAUCAGUGAUUUUGUUGAAGA-Alexa-594-3′.
Western analysis
Cytosolic and mitochondrial fractions were prepared as described above. Equal amounts of each lysate were fractionated by 4–12 % gradient Bis–Tris SDS-PAGE gel electrophoresis. For affinity purified proteins, bead-bound proteins were eluted by boiling for 10 min in elution buffer and size-fractionated by electrophoresis using a 4–12 % gradient Bis–Tris SDS-PAGE (Life Technologies). After gel-electrophoresis, the fractionated proteins were electroblotted onto a nitrocellulose-based transfer membrane (Life Technologies). Membranes were blocked with ECL Advance Blocking Agent (GE Healthcare) for 1 h, followed by overnight incubation at 4 °C with antibodies against KIF5A (ThermoFisher), YB-1 (Millipore), VDAC1 and COXII (Abcam), COXIV, β-actin (Cell Signaling Technology), and MAP2 and Tau (Sigma Aldrich) at 1:1000 dilution. Membranes were washed three times with TBS and 0.1 % Tween 20 and incubated with HRP-labeled secondary antibody for 1 h at room temperature. After washing, membranes were developed with SuperSignal West Femto Maximum Sensitivity Substrate Detection Kit (Life Technologies).
RNA isolation and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from SCG axons, parental cell soma, or from cytosolic and mitochondrial fractions using Direct-Zol™ RNA MiniPrep (Zymo Research) according to the manufacturer’s instructions. For reverse transcription, equal amounts of purified RNA were mixed with cDNA SuperMix (Quanta Biosciences™) and incubated in a thermal cycler (MJ Research) for 5 min at 25 °C, followed by 30 min at 42 °C and 5 min at 85 °C. qRT-PCR analysis was performed with gene specific QuantiTect primers (Qiagen) for COXIV or β-actin. Gene specific primers for rat pre-miR-338,-204,-185, and -134 were designed using the OligoPerfect Designer (Life Technologies). The COXII and U6 snRNA primers used in the study were identical to those reported previously [13]. Quantitative PCR was performed using VeriQuest SYBR Green quantitative PCR Master Mix (Affymetrix) and the StepOne Real-Time PCR system (Applied Biosystems). For each experimental RNA sample, the PCR reactions were run in triplicate. The results were analyzed using the StepOne™ Software (Applied Biosystems). Primer sequence list (sense strand, 5′ to 3′):
Pre-miR-204: GGCTACAGCCCTTCTTCATGT
Pre-miR-134: CAGGGTGTGTGACTGGTTGAC
Pre-miR-185: GGGGGTGAGGGATTGGAGAGA
Pre-miR-338: AACAATATCCTGGTGCTGAGTG
Pre-miR-124a: AGGCCTCTCTCTCCGTGTTC
Immunohistochemistry
For double immunostaining experiments, 2-week-old dissociated SCG neurons were stained with KIF5A, YBX1, VDAC1, and DICER-specific antibodies. Dissociated SCG neurons were fixed for 10–15 min in 4 % prewarmed paraformaldehyde solution and permeabilized in 0.2 % Triton X-100 for 10 min. Cells were blocked in the blocking buffer [phosphate buffered saline (PBS) containing 3 % bovine serum albumin (BSA) and 0.1 % Triton X-100] at room temperature for 30 min and incubated with the antibodies (KIF5A 1:500; YB-1 1:200; VDAC 1:500; DICER 1:1000) at 4 °C overnight. After three washes with PBS, cells were subsequently exposed to secondary antibodies at room temperature for 1 h and then fixed with 4 % buffered paraformaldehyde, as described above, for microscopy. Images were obtained using a confocal microscope (Zeiss LSM510).
Mitochondrial staining
Mitochondria were identified by staining with MitoTracker® Deep Red CMXRos (Life Technologies), an organelle-specific fluorescent dye. In these experiments, MitoTracker was added to the culture media at a final concentration of 25 nM for 15 min. The cells were incubated under normal culture conditions for 30 min, then washed with PBS, and visualized by fluorescence confocal microscopy (Zeiss LSM510).
Statistical analyses
All statistical analyses were performed in Excel. Results are expressed as mean ± SEM. p values were calculated using the Student’s two-tailed t test, and p values ≤0.05 were considered as significant.
Results
Pre-miR-338 and other pre-miRs are present in SCG axons
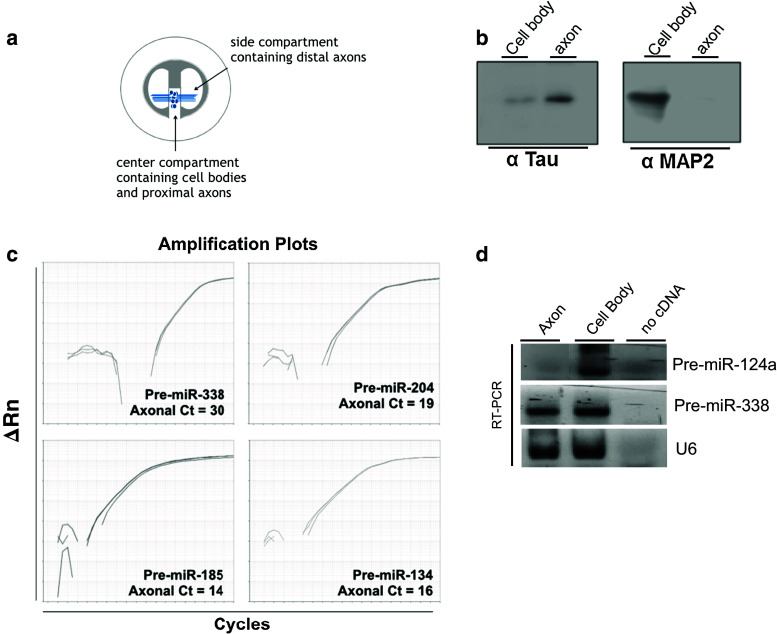
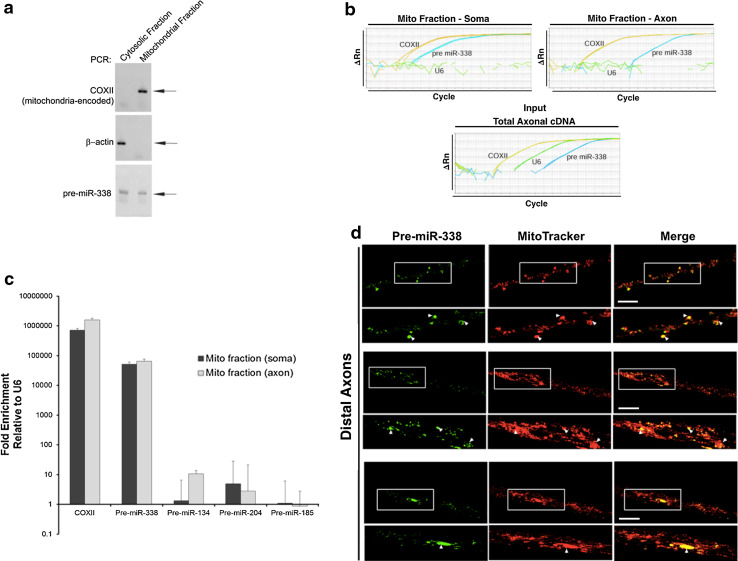
To study the localization of pre-miR-338 in SCG axons, SCG neurons were cultured in compartmentalized Campenot chambers (Fig. 1a). Campenot chambers allow the isolation of axonal samples from the side compartments devoid of neuronal cell bodies and other non-neuronal cells. To verify that the side compartments contained only axons, Western blot analyses were performed to detect the presence of soma-restricted protein MAP2 and the axonally enriched protein TAU. MAP2 and low levels of TAU can be detected using samples from the center compartment containing cell bodies and proximal axons (Fig. 1b), while only TAU protein is detectable in distal axon lysates obtained from the side compartment (Fig. 1b). To test for the presence of pre-miR-338 in SCG axons, we extracted axonal RNA from side compartments and performed qRT-PCR using pre-miR-338-specific primers. The results show the presence of pre-miR-338 in SCG axons (Fig. 1c). In addition, the presence of three other pre-miRNAs (Fig. 1c), whose mature forms have previously been identified in SCG axons, was detected [13]. In contrast to pre-miR-338 and other axonally localized pre-miRs, we did not detect pre-miR-124a in axons via RT-PCR after 50 cycles (Fig. 2d). Indeed, our group and others have previously reported that mature miR-124a appears to be restricted to the cell body [ 13, 26, 27, 28]. The presence of pre-miR-338 and the lack of pre-miR-124a in sympathetic axons suggest that pre-miRs are selectively localized in the axon.
Fig. 1.
Pre-miR-338 is present in axons of superior cervical ganglion neurons. a Schematic showing the compartmentalized Campenot cultures used in this study. b Western analyses of protein lysates extracted from central and side-compartments of Campenot cultures, containing the parental cell soma and distal axons, respectively. Levels of MAP2 and TAU were used to assess the purity of the protein lysates obtained from distal axons located in the side compartments of the compartmentalized cultures. c Quantitative RT-PCR analysis was conducted using total RNA obtained from distal axons to assess the levels of several precursor miRNAs. Cycle threshold (Ct) value, the number of amplification cycles required to exceed background signal, is shown for each pre-miRNA indicating their presence in SCG axons. d Gel electrophoresis of RT-PCR amplicons (50 cycle amplification) of pre-miR-124a, pre-miR-338, and U6 in the cell body or axon of SCG neurons
Fig. 2.
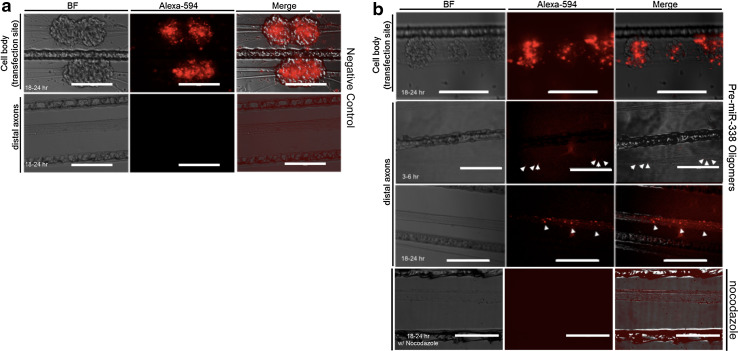
Pre-miR-338 is transported to the distal axons of superior cervical ganglion neurons. Alexa-594-labeled pre-miR-338 oligomers were transfected into cell bodies and proximal axons present in the central compartment of 2-week-old SCG neurons cultured in compartmentalized Campenot chambers. 3–6 and 18–24 h after transfection, the localization of the fluorescent oligomers was determined by fluorescence microscopy. As a control for passive diffusion, fluorescently labeled bacterial RNA was used. a Representative images of central and side compartments after transfection show efficient transfection of control RNA in the parental soma. However, no signal was detected in distal axons even after 18–24 h. b Representative images showing transfection of pre-miR-338 in the cell bodies, which is comparable with negative control RNA (see Fig. 2a). Specific punctate labeling is observed in distal axon bundles of SCG neurons after 3–6 h and more robust labeling after 18–24 h indicating the transport of pre-miR-338 oligomers (arrowheads). Pre-miR-338 signal is absent in distal axons when axons are treated with nocodazole. Arrowheads indicate pre-miR-338 puncta in distal axon bundles. Scale bars 200 µm. BF bright field
Moreover, to visualize the presence of pre-miR-338 in distal axons, fluorescently labeled pre-miR-338 oligomers were transfected into SCG cell bodies located in the center compartment of Campenot chambers. Since the cell bodies and distal axons are in separate chambers, any fluorescent probe that is visualized in the distal axons is likely due to the relocalization of the transfected probe. Indeed, 6 h after transfection of pre-miR-338 in the center compartment, we detected low levels of the probe in distal axons; 18–24 h after transfection, fluorescent pre-miR-338 signal was readily detected in distal axons (Fig. 2b). This fluorescent signal appeared as puncta along the entire length of the axon, suggesting the presence of RNA granules containing pre-miR-338 (Fig. 2b). To control for passive diffusion of fluorescent oligomers, labeled-RNA derived from bacterial genome was used. Unlike pre-miR-338, control bacterial RNA fluorescent signal was not detected in the distal axons after being transfected into the parental soma, suggesting that only specific RNA molecules, such as pre-miR-338, are localized to axons (Fig. 2a). Furthermore, to determine whether pre-miR-338 may undergo microtubule-dependent transport to the axoplasm, we treated the side compartment with nocodazole (65 uM), a compound that destabilizes microtubules. In the presence of nocodazole, pre-miR-338 oligomers were not observable in the distal axons, suggesting that pre-miR-338 may be localized to the axon in a microtubule-dependent manner (Fig. 2b). These results suggest that transfected pre-miR-338 oligomers are localized into the distal axon, and that this event is unlikely due to passive diffusion of RNA molecules. Together, these results illustrate the presence of some pre-miRNAs in axons of SCG neurons, including pre-miR-338.
Identification of pre-miR-338 interacting proteins
The localization of RNA into axons often requires the formation of RNP complexes to stabilize the RNA molecule [9, 29, 30]. The presence of endogenous pre-miR-338 (Fig. 1c, d) and the localization of exogenous pre-miR-338 to the axon (Fig. 2b) suggest that pre-miR-338 may form RNP complexes that facilitate its localization to distal axons.
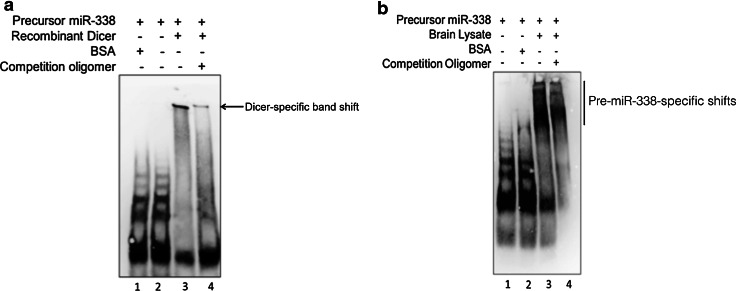
To investigate the interaction of pre-miR-338 with proteins that may control its axonal localization, a 3′-biotinylated-pre-miR-338 oligomer was synthesized for a series of proteomic studies. To evaluate the ability of the 3′-biotinylated-pre-miR-338 oligomer to bind miRNA-associating proteins, we tested its capacity to bind DICER, a pre-miRNA processing enzyme, by performing an EMSA. When the RNA probe was incubated with recombinant DICER, it produced a band shift distinct from the RNA probe alone. (Fig. 3a, compare lane 2 and 3). This shift was significantly diminished by competition with an unlabeled pre-miR-338 probe (Fig. 3a, lanes 3 and 4). In contrast, there is no shift observed when bovine serum albumin (BSA) was used (Fig. 3a, lane 1). Clearly, the pre-miR-338 oligomer interacts with DICER protein. Next, the ability of pre-miR-338 to form protein complexes was evaluated using cytosolic protein lysates derived from P4 rat brain. We observed a gel shift that produced high molecular weight bands in the presence of protein lysates as compared with probe alone (Fig. 3b, lanes 1 and 3). Furthermore, incubation of pre-miR-338 with BSA did not result in a similar gel shift (Fig. 3b, lane 2) illustrating the specificity of the pre-miR-338 gel shift. The addition of unlabeled probe to the reaction (Fig. 3b, lane 4) diminished the intensity of the shifted bands. Together, these findings demonstrate that pre-miR-338 forms RNP complexes.
Fig. 3.
Pre-miR-338 forms ribonucleoprotein complexes. 3′-biotin labeled pre-miR-338 probes were incubated with recombinant DICER protein (a) or cytosolic proteins derived from total brains of 4-day-old rat pups (b). After incubation, the RNA–protein complexes were resolved on 4–20 % polyacrylamide gels and visualized using LightShift Chemiluminescent RNA EMSA kit. In competitive binding assays, non-biotinylated pre-miR-338 oligomer was added in 100-fold molar excess. a Incubation of pre-miR-338 with BSA (lane 1), free pre-miR-338 probe (lane 2), or recombinant DICER protein (lane 3). A gel-shift band was obtained in the presence of recombinant DICER protein, lane 3 (arrow). No specific gel-shift complexes were observed in the presence of BSA, lane 1. Addition of non-labeled pre-miR-338 into the binding reaction significantly diminished the gel-shift observed with DICER, (lane 4). b Incubation of pre-miR-338 with brain cytosolic fractions derived from 4-day-old rat pups. Lane 1 unbound pre-miR-338 probe; lane 2 no band shift observed in the presence of BSA; lane 3 pre-miR-338 specific gel-shift bands observed after incubation with rat brain cytosolic extract. Lane 4 reduced gel-shift bands observed with rat brain lysate in presence of 100-fold excess of unlabeled pre-miR-338 oligomer
To identify the components of the pre-miR-338 RNP complex, we employed RNA affinity pulldown experiments, using rat total brain protein extracts and 3′-biotinylated-pre-miR-338 coupled with streptavidin beads. This allowed for the isolation of proteins that interact with pre-miR-338. High-performance liquid chromatography–tandem mass spectroscopy (LC/MS) was then performed on the affinity-purified fraction to identify the pre-miR-338 interacting proteins. The pulldown experiments were repeated five times and identified 212 proteins that were either specific or enriched in the pre-miR-338 pulldown fractions relative to the bead-only control in at least 3 of 5 runs (Fig. S1). Gene Ontology (GO) enrichment analysis was conducted to interrogate the functional clustering of the pre-miR-338 interacting proteins [24, 25]. Proteins that interacted with pre-miR-338 clustered under categories, such as “RNA-binding proteins,” “ribonucleoprotein complex,” “cytoskeleton,” and “motor proteins.” The proteins identified included RBPs, such as: Y box binding protein 1 (YBX1 or YB1), various heterogeneous nuclear ribonucleoproteins (hnRNP), CIRBP, and PTBP2; translational machinery constituents, such as EIF2S1, EEF1A, EIF5A, and components of the 60s and 40s ribosomal subunits; and motor proteins, such as: KIF5A, KIF5B, DYNC1H1, and DYNC1I1.
Much to our surprise, a significant clustering of the pre-miR-338 interacting proteins with functions related to mitochondrial processes, such as: “ATP synthesis coupled proton transport,” “oxidative phosphorylation,” and “mitochondrion,” was also identified. The mitochondrion proteins that interacted with pre-miR-338 include VDAC1, a voltage-dependent anion channel found in the mitochondrial outer membrane; COX5A, a subunit of the cytochrome c oxidase complex; ATP5B, a subunit of mitochondrial ATP synthase; TIMM8A1, which is a mitochondrial inner membrane chaperone; and TOMM7A, a mitochondrial translocase. Next, a subset of the pre-miR-338 interacting proteins was subjected to STRING analysis to produce a gene association network based on known and predicted protein–protein interactions [31]. The pre-miR-338 interacting proteins chosen for the STRING analysis were from GO categories related to mitochondrion, cytoskeleton and motor proteins, RNA-binding proteins, and ribonucleoproteins (see “Materials and methods”: “STRING analysis of pre-miR-338 interacting proteins” for exact list of GO category inputs). The results showed highly interrelated protein–protein association networks, indicating complex interactions between pre-miR-338-binding proteins (Fig. 4a–c). Together, these results reveal that pre-miR-338 interacts with proteins associated with RNA stability, trafficking, and the translational machinery. Importantly, the data raise the possibility that the pre-miR-338 RNP particle is associated with mitochondria.
Fig. 4.
Pre-miR-338 interacting proteins form functional networks. Pre-miR338-associated proteins identified by mass spectrometry were analyzed using STRING to assess predicted protein–protein interactions. Protein enriched in Gene Ontology (GO) clusters mitochondrion (a), cytoskeletal and motor proteins (b), and RNA binding and ribonucleoprotein complex (c) were used as input. In the networks shown, the nodes are the proteins and the lines represent the predicted functional associations. The width of lines indicates the confidence of the predicted functional interactions between proteins. Pre-miR-338-binding proteins enriched in the selected GO categories form extensive protein–protein interaction networks. The names of proteins in each cluster are provided in the columns. Blue boxes and blue halo around specific nodes indicate the candidate proteins selected for validation studies (see Fig. 5)
To validate the findings of the LC/MS studies, we performed Western blot analyses on pre-miR-338 pulldown fractions using protein lysates from rat total brain, SCG cell bodies, and distal axons. The RNA pulldown experiments were performed using the same parameters as those performed for LC/MS analysis: three candidate pre-miR-338-interacting proteins were selected for further study: KIF5A, YB1, and VDAC1, a mitochondrial protein. KIF5A is a microtubule-dependent anterograde motor protein required for axonal transport of mitochondria [32]; YB1 has been shown to regulate the translation and stability of nuclear-encoded mitochondrial mRNAs [33]. The results reveal the presence of all three proteins in pre-miR-338 pulldown fractions, but not in bead-only control fractions using total brain and axonal lysates (Fig. 5a; Fig. S2). Thus, the results from pre-miR-338 RNA pulldown followed by Western analyses of candidate pre-miR-338 interacting proteins confirm the proteomic findings and establish that the pre-miR-338 RNP is present in SCG axons.
To visualize the candidate pre-miR-338 interacting proteins in SCG axons, we performed immunohistochemical experiments using dissociated SCG neurons. In these studies, the colocalization of VDAC1 with KIF5A and YB1 in SCG axons was observed (Fig. 5b, c), suggesting that the candidate proteins may interact in SCG axons as a part of the pre-miR-338 RNP complex. This postulate is in agreement with the results of the pre-miR-338 RNA pulldown followed by either LC/MS or Western analyses demonstrating that VDAC1, KIF5A, and YB1 are all part of the pre-miR-338 RNP complex. Taken together with the observation that pre-miR-338 is localized to distal axons (Fig. 2b), and the formation of the pre-miR-338 complex using axon-derived protein lysates (Fig. 5a), the data raise the possibility that the pre-miR-338 RNP complex is also present within SCG axons.
Endogenous pre-miR-338 is present in mitochondrial fractions and localizes with mitochondria in axons
The mass spectrometry and biochemical studies raised the possibility that endogenous pre-miR-338 localizes to the mitochondria. To evaluate the postulate that endogenous pre-miR-338 does indeed localize to the organelle, RT-PCR analysis was performed using RNA obtained from mitochondrial fractions. To demonstrate the purity of the mitochondrial fractions, we assessed the presence of COXII mRNA, a mitochondrial-encoded gene, and β-actin mRNA, a nuclear-encoded cytoplasmic message, in the samples. COXII, but not β-actin, was present in mitochondrial fractions. Conversely, only β-actin mRNA was detected in cytosolic fractions (Fig. 6a). These results indicate the enrichment of the mitochondria and the absence of cytosolic contamination in the mitochondrial fraction. Importantly, we detected pre-miR-338 in the mitochondrial fraction, as well as the cytosolic fraction indicating that a subset of pre-miR-338 localizes to the mitochondria (Fig. 6a). To investigate whether the interaction of pre-miR-338 with the mitochondria holds true in SCG axons, qRT-PCR was performed on RNA purified from mitochondrial fractions obtained from SCG cell body or distal axons grown in compartmentalized cultures. In this experiment, U6 snRNA was used as a negative control. This RNA is a ubiquitous, non-coding RNA molecule, comparable in length to pre-miR-338. The results show that COXII and pre-miR-338, but not U6 can be detected in SCG neuron-derived mitochondrial fractions (Fig. 6b). To investigate whether the interaction between pre-miR-338 and the mitochondria is specific, we tested for the presence of three other axonally localized pre-miRNAs in mitochondrial fractions from SCG neurons. The results show that only pre-miR-338 is enriched in mitochondrial fractions relative to U6 snRNA (Fig. 6c), suggesting that the localization of pre-miR-338 to the organelle is specific relative to other axonally abundant pre-miRs. Moreover, an overlap between axonally localized pre-miR-338, and the mitochondria in SCG distal axons was observed by confocal microscopy (Fig. 6d). These data suggest that pre-miR-338 is localized to mitochondria in SCG neurons.
Fig. 6.
Endogenous pre-miR-338 specifically localizes with mitochondria. Mitochondrial fractionation was performed on P4 rat total brain lysates or from cell body and axons of SCG neurons, and the presence of RNA transcripts was assessed by RT-PCR (a) and qRT-PCR (b, c). a Gel electrophoreses of RT-PCR amplicons show the presence or absence of COXII, β-actin mRNA, and pre-miR-338 in either mitochondrial or cytosolic fractions obtained from P4 total rat brain as indicated by arrows. b qRT-PCR analysis of mitochondrial fractions isolated from SCG cell bodies or distal axons. Results show the presence of pre-miR-338 in mitochondrial fractions from SCG neurons. U6 snRNA was not detected in mitochondrial preparations. c Quantification of qRT-PCR analyses for pre-miR-338, 134, 204, and 185 in SCG cell bodies or axons. Values represent fold enrichment of pre-miRNA relative to U6. COXII was also quantified as a control for mitochondrial enrichment. d Representative image of pre-miR-338 oligomer (green) and mitochondria (mitoTracker, red) in distal axons of SCG neurons in Campenot chambers after 18–24 h after transfection of pre-miR-338 oligomers in the center compartment. Merge image shows robust colocalization of mitochondria and pre-miR-338 oligomers. Arrowheads indicate colocalization between pre-miR-338 and mitochondria. Boxes indicate magnified regions to illustrate colocalization. Scale bars 50 µm
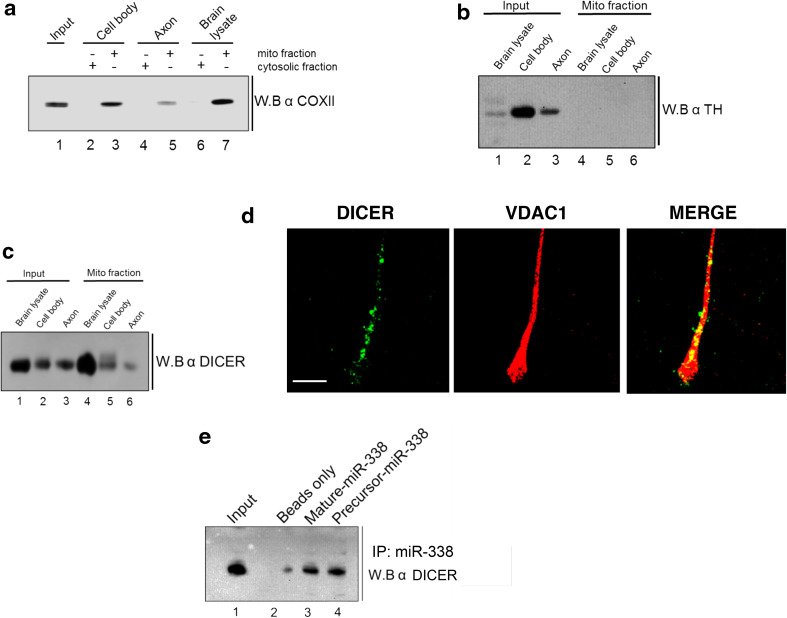
DICER colocalizes with mitochondria in axons of sympathetic neurons
DICER-mediated cleavage is required for the processing of pre-miR-338 to mature miR-338. Since pre-miR-338 is enriched in mitochondrial fractions, the association of DICER with the mitochondria was assessed. To investigate the interaction between DICER and the mitochondria, a Western blot analyses was performed on mitochondrial fractions obtained from rat total brain and SCG neurons. As shown in Fig. 7a and b, COXII protein is present in mitochondrial fractions but not in cytosolic fractions (Fig. 7a). In contrast, tyrosine hydroxylase (TH) protein, which is highly expressed in SCG neurons, is absent in mitochondrial fractions but is enriched in cytosolic fractions (Fig. 7b). Importantly, DICER was found to be present in mitochondrial fractions from whole brain and SCG cell bodies, as well as in axons (Fig. 7c). Furthermore, fluorescence immunohistochemical colocalization studies revealed that DICER colocalizes with VDAC1, a protein used as a mitochondrial marker, in SCG axons (Fig. 7d). Gel shift studies had shown that pre-miR-338 binds DICER protein (Fig. 3a); surprisingly, however, the mass spectrometry results did not identify DICER in the pre-miR-338 pulldown fraction. However, Western blot analysis, under some circumstances, may be more informative than LC/MS. Therefore, a Western blot analysis was performed following pre-miR-338 affinity pulldown assay on total brain lysates to test for the presence of DICER. The pulldown using mature miR-338 was performed as positive control (Fig. 7e). Taken together, these results suggest that along with the pre-miR-338 RNP complex, DICER protein also colocalizes with the mitochondria in SCG axons.
Fig. 7.
DICER colocalizes with mitochondria in SCG axons. Western blot analyses of mitochondrial fractions from P4 total brain, SCG cell bodies, and distal axons using antibodies against a COXII, b tyrosine hydroxylase, and c DICER protein. The results show the presence of DICER in mitochondrial fractions. d Co-immunostaining studies using antibodies against DICER and VDAC1 protein in SCG axons. The representative images show the co-localization of DICER with VDAC. e Western blot analysis of RNA affinity pulldown using mature and pre-miR-338 as probes. Results show the presence of DICER protein in pulldowns of both mature and pre-miR-338. Scale bars 50 µm
Discussion
It has been hypothesized that miRNAs are trafficked in their mature or precursor forms, contained within RNP particles composed of RBPs and motor proteins [29]. Recently, a subset of pre-miRNAs has been identified in axons of sympathetic neurons that respond to axonal injury, suggesting that there are active mechanisms that control the trafficking of pre-miRNAs [17]. However, direct evidence for the trafficking of pre-miRNAs into axons has yet to be reported. In this study, we show that fluorescently labeled-pre-miR-338 transfected into cell bodies of SCG neurons readily localizes to distal axons, a finding that suggests that pre-miRNAs can be transported to the axon from their parental cell bodies. Furthermore, the proteomic studies suggest that a subset of proteins that interact with pre-miR-338 may be involved in the mechanism(s) that direct the axonal trafficking of pre-miRNAs. Further experiments are needed to determine the role played by these pre-miR-338 interacting proteins in axonal transport.
In addition to demonstrating the axonal localization of pre-miR-338, multiple lines of evidence provided in this study show that pre-miR-338 associates with the mitochondria in axons of SCG neurons. Consistent with these results, certain pre-miRNAs have previously been shown to localize to the mitochondria [34]. Previous studies have shown that miR-338 regulates the axonal translation of COXIV and ATP5G1 mRNAs [19–21], and that these nuclear-encoded mRNAs, similar to pre-miR-338, can localize to the mitochondria in SCG axons. Furthermore, the intra-axonal translation of COXIV and ATP5G1 plays a vital role in axonal bioenergetics [19, 35]. Recent studies demonstrate the subcellular translation of nuclear-encoded mitochondrial mRNAs occurs while tethered to the mitochondrial outer membrane protein TOM20, in a PINK1/PARKIN-dependent manner [36]. Moreover, electron microscopy experiments reveal that ribosomes and RNA granules, such as P-bodies, come in contact with the mitochondria [23, 37]. Indeed, qRT-PCR analysis of RNA bound to polysomes associated with the organelle revealed various nuclear-encoded mitochondrial mRNAs [33]. Taken together, these studies raise the distinct possibility that nuclear-encoded mitochondrial mRNAs are translated on the surface of the mitochondria.
In axons, the upkeep of the mitochondria requires the highly coordinated expression of nuclear- and mitochondrial-encoded genes. miRNAs that regulate mitochondrial genes, such as miR-338, function to coordinate the subcellular translation of proteins essential to mitochondrial maintenance in axons [19, 21]. In addition to regulating nuclear-encoded mitochondrial mRNAs, miRNAs have also been shown to control the expression of mRNAs derived from the mitochondrial genome, such as COXI [38, 39]. Interestingly, miR-1 has been shown to function within the mitochondrial matrix during myogenesis, where it enhances, rather than attenuates, the expression of mitochondrial-encoded transcripts [26]. These surprising results provide insights into the complexity of posttranscriptional regulation of both nuclear- and mitochondrial-encoded mRNAs via the action of miRNAs on the surface or within the organelle itself.
In this study, we provide evidence for the localization of pre-miR-338 in SCG axons and begin to delineate, for the first time, the composition of a pre-miRNA RNP complex, which we found to contain various RBPs and motor proteins. We also provide convergent evidence, using biochemical and proteomic approaches, which together confirm the subcellular localization of pre-miR-338 to the mitochondria. Together with the well-established function of miR-338 in coordinating the expression of nuclear-encoded mitochondrial mRNAs in axons [19, 21], the present findings raise the intriguing possibility that mitochondrially localized pre-miR-338 may serve as a reservoir of miRNAs to regulate subcellular translation of nuclear-encoded mitochondrial mRNAs, thereby coordinating mitochondrial function in distal neuronal compartments. These observations offer new understanding on possible mechanisms of subcellular gene regulation and long-term maintenance of the mitochondrial function within axons.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1. A schematic of the RNA affinity purification protocol to identify pre-miR-338 interacting proteins. Biotinylated pre-miR-338 was incubated with P4 rat brain cytosolic extracts. The formation of RNP granules/complexes was monitored by EMSA gel shift assay. After incubation, the RNA-protein complexes were isolated using streptavidin beads. The identity of the pre-miR-338 binding proteins was determined by mass spectrometry and validated by Western analyses (pptx 178 kb)
Fig. S2. Full blot images of Western analyses performed to confirm pre-miR-338 RNP identified via LC/MS. RNA affinity pulldowns were performed with the biotinylated pre-miR-338 oligomer and P4 brain cytosolic lysates or cytosolic extracts isolated from distal axons present in the side-compartments of Campenot cultures (see Fig. 6). Arrows indicate bands corresponding to protein of interest. Asterisks indicate non-specific protein band (pdf 150 kb)
Table S1. Pre-miR-338 interacting protein list. Proteins identified by LC/MS to be enriched in pre-miR-338 pulldowns (XLSX 47 kb)
Acknowledgments
The work was supported by the Division of Intramural Research Programs of the National Institute of Mental Health (MH002768). The authors would like to thank Mrs. Ching-Yu Sun, Ms. Miranda Tompkins, Dr. Noreen Gervasi, Mr. Shane Scott, and Ms. Anna-Leigh Brown for their scientific insights and engaging conversations, which contributed to the fruition of this work.
Abbreviations
- miR
MicroRNA
- Pre-miRNA
Precursor microRNA
- RBPs
RNA-binding proteins
- SCG
Superior cervical ganglion
- COXIV
Cytochrome C oxidase IV
- ATP5G1
ATP synthase 5 gamma 1
- LC/MS
Liquid chromatography–tandem mass spectrometry
- ROS
Reactive oxygen species
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interests.
References
- 1.Lee Y, Jeon K, Lee J-T, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 3.Leung AKL, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harb Symp Quant Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- 4.Ashraf SI, Kunes S. A trace of silence: memory and microRNA at the synapse. Curr Opin Neurobiol. 2006;16:535–539. doi: 10.1016/j.conb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Chiu H, Alqadah A, Chang C. The role of microRNAs in regulating neuronal connectivity. Front Cell Neurosci. 2014 doi: 10.3389/fncel.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer AN, Bellon A, Baudet M-L. MicroRNAs in axon guidance. Front Cell Neurosci. 2014;8:78. doi: 10.3389/fncel.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olde Loohuis NFM, Kos A, Martens GJM, et al. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan BB, Kar AN, Gioio AE, Aschrafi A. MicroRNAs in the axon and presynaptic nerve terminal. Front Cell Neurosci. 2013;7:126. doi: 10.3389/fncel.2013.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo S, van Niekerk EA, Merianda TT, Twiss JL. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp Neurol. 2010;223:19–27. doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gumy LF, Katrukha EA, Kapitein LC, Hoogenraad CC. New insights into mRNA trafficking in axons. Dev Neurobiol. 2014 doi: 10.1002/dneu.22121. [DOI] [PubMed] [Google Scholar]
- 11.Scott S, Gervasi N, Kaplan BB. Subcellular compartmentalization of neuronal RNAs: an overview. Trends in Cell Mol Bio. 2015;10:1–36. [Google Scholar]
- 12.Kim E, Jung H. Local protein synthesis in neuronal axons: why and how we study. BMB Rep. 2015;48:139–146. doi: 10.5483/BMBRep.2015.48.3.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010 doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki Y, Gross C, Xing L, et al. Identification of axon-enriched microRNAs localized to growth cones of cortical neurons. Dev Neurobiol. 2014;74:397–406. doi: 10.1002/dneu.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lugli G, Larson J, Demars MP, Smalheiser NR. Primary microRNA precursor transcripts are localized at post-synaptic densities in adult mouse forebrain. J Neurochem. 2012;123:459–466. doi: 10.1111/j.1471-4159.2012.07921.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim HH, Kim P, Phay M, Yoo S. Identification of precursor microRNAs within distal axons of sensory neuron. J Neurochem. 2015 doi: 10.1111/jnc.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aschrafi A, Schwechter AD, Mameza MG, et al. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008 doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bicker S, Khudayberdiev S, Weiß K, et al. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 2013 doi: 10.1101/gad.211243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aschrafi A, Kar AN, Natera-Naranjo O, et al. MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007 doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berberich MJ, Kowalak JA, Makusky AJ, Martin D, Vullhorst A, Buonanno A, Markey SP. Development of an on-bead digestion procedure for immunoprecipitated proteins. In: Ivanov AR, Lazarev AV, editors. Sample preparation in biological mass spectrometry. Netherlands: Springer; 2011. pp. 109–124. [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Zuo X, Yang B, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kye M-J, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 30.Tolino M, Köhrmann M, Kiebler MA. RNA-binding proteins involved in RNA localization and their implications in neuronal diseases. Eur J Neurosci. 2012;35:1818–1836. doi: 10.1111/j.1460-9568.2012.08160.x. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucl Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell PD, Shen K, Sapio MR, et al. Unique function of Kinesin Kif5A in localization of mitochondria in axons. J Neurosci. 2014;34:14717–14732. doi: 10.1523/JNEUROSCI.2770-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto K, Wolffe AP. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 1998;8:318–323. doi: 10.1016/S0962-8924(98)01300-2. [DOI] [PubMed] [Google Scholar]
- 34.Barrey E, Saint-Auret G, Bonnamy B, et al. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011 doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kar AN, Sun CY, Reichard K, et al. Dysregulation of the axonal trafficking of nuclear-encoded mitochondrial mRNA alters neuronal mitochondrial activity and mouse behavior. Dev Neurobiol. 2014 doi: 10.1002/dneu.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gehrke S, Wu Z, Klinkenberg M, et al. PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab. 2015;21:95–108. doi: 10.1016/j.cmet.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L, Mollet S, Souquere S, et al. Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J Biol Chem. 2011;286:24219–24230. doi: 10.1074/jbc.M111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das S, Ferlito M, Kent OA, et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandiera S, Matégot R, Girard M, et al. MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic Biol Med. 2013 doi: 10.1016/j.freeradbiomed.2013.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A schematic of the RNA affinity purification protocol to identify pre-miR-338 interacting proteins. Biotinylated pre-miR-338 was incubated with P4 rat brain cytosolic extracts. The formation of RNP granules/complexes was monitored by EMSA gel shift assay. After incubation, the RNA-protein complexes were isolated using streptavidin beads. The identity of the pre-miR-338 binding proteins was determined by mass spectrometry and validated by Western analyses (pptx 178 kb)
Fig. S2. Full blot images of Western analyses performed to confirm pre-miR-338 RNP identified via LC/MS. RNA affinity pulldowns were performed with the biotinylated pre-miR-338 oligomer and P4 brain cytosolic lysates or cytosolic extracts isolated from distal axons present in the side-compartments of Campenot cultures (see Fig. 6). Arrows indicate bands corresponding to protein of interest. Asterisks indicate non-specific protein band (pdf 150 kb)
Table S1. Pre-miR-338 interacting protein list. Proteins identified by LC/MS to be enriched in pre-miR-338 pulldowns (XLSX 47 kb)