Abstract
The DNA replication machinery encounters problems at numerous genomic regions that are inherently difficult to replicate. These genomic regions include telomeres, which contain repetitive DNA and telomere-binding proteins. If not properly regulated, replication of such genomic regions can result in DNA damage, leading to genomic instability. Studies implicated a role of Timeless-related proteins at difficult-to-replicate genomic regions, including telomeres. However, how these proteins maintain telomeres was elusive. In a recent report, we described the role of Swi1, a Timeless-related protein, in telomere maintenance in fission yeast. We demonstrated that Swi1 is required for proper replication of repeat DNA sequences at telomeres. We also showed that Swi1-deficient cells utilize recombination-based ALT (alternative lengthening of telomeres)-like mechanisms to maintain telomeres in the absence of telomerase. Here, we highlight these findings and present additional data to discuss the role of Swi1Timeless in telomere protection and ALT prevention.
Keywords: Swi1, Timeless, FPC, Fork protection complex, Telomeres, Myb/SANT, Tbf1, ALT, Alternative lengthening of telomeres, Replication fork, Genomic integrity, Repeat DNA, Cancer
Role of Swi1Timeless in replication of repeat DNA regions
Numerous chromosomal regions present obstacles for DNA replication. These include fork-blocking sites, DNA secondary structures caused by repeat sequences, highly transcribed regions, and DNA-binding proteins bound to the template DNA. These sites are considered difficult to replicate, because they are susceptible to replication fork arrest or breakage, resulting in replication stress during the normal course of DNA replication. To prevent these occurrences, eukaryotic cells have a dedicated sensor response mechanism, termed the DNA replication checkpoint, responsible for the coordination of DNA repair and cell cycle progression (Leman and Noguchi 2013; Mirkin and Mirkin 2007). Cells also need to protect or stabilize the replication fork when the replisome encounters difficult-to-replication regions. Central to this protection is the replication fork protection complex (FPC) that is comprised of two major components: Swi1 and Swi3 in fission yeast; Tof1 and Csm3 in budding yeast; and Timeless and Tipin in metazoans. The functions of the FPC are conserved among eukaryotes (Leman and Noguchi 2012).
In our recent report, we investigated the role of Swi1, a Timeless-related protein, in telomere DNA replication in fission yeast (Gadaleta et al. 2016). Because telomeres have various features that can hamper replisome progression (Ivessa et al. 2002; Makovets et al. 2004; Millet and Makovets 2016; Verdun and Karlseder 2006), we first sought to narrow down the list of possible telomeric obstacles. This list includes repeat DNA sequences, telomere-binding proteins, heterochromatin, and other secondary structures such as the t-loop. Our genetic studies suggest that telomere-binding proteins and heterochromatin do not present major replication obstacles in the absence of Swi1. Telomerase activity is also intact in swi1Δ cells, indicating that Swi1 does not regulate telomerase. Instead, the repetitive nature of the telomeric DNA sequences was found to be the primary impediment during telomere replication and the main cause of telomere shortening in the absence of Swi1. These observations were further supported by the following findings: (1) An episomal plasmid carrying a 300-bp telomeric repeat tract undergoes extensive recombination when introduced into swi1Δ cells, while no recombination is observed when the same plasmid is introduced into wild-type cells; (2) a single tract of E. coli LacO repeats inserted at chromosome arm regions experience repeat instability in the absence of Swi1; and (3) telomeres, LacO repeats, and rDNA loci, all of which have repeat DNA sequences are enriched with Rad52, a recombinase known to bind ssDNAs at DNA lesions (Gadaleta et al. 2016). Consistently, swi1 deletion also causes contraction of rDNA repeats (Rapp et al. 2010; Sommariva et al. 2005) and fork breakage at these loci (Noguchi et al. 2003). Therefore, Swi1’s role in repeat DNA maintenance is independent of DNA sequence, repeat track length, and genomic location. We therefore propose that Swi1Timeless is a novel regulator of repetitive DNA replication across the genome.
Swi1Timeless as an anti-recombinase at telomeres
Both Rad52 ChIP-seq analysis and telomere-dysfunction induced foci (TIFs) quantification revealed significant enrichment of Rad52 at subtelomeric regions in swi1Δ cells. In addition, swi1Δ cells were also shown to recruit increased levels of Rad52 at LacO and rDNA repeats (Gadaleta et al. 2016). Altogether, these results suggest that Swi1 prevents recombination at multiple loci containing repeat DNA sequences throughout the genome. This function of Swi1 is conserved between fission yeast and mammalian cells. In HeLa cells, telomeres undergo extensive DNA damage and recombination, leading to telomere shortening in Timeless-depleted cells (Leman et al. 2012). Rad51 and Rad52 foci accumulate in mouse NIH3T3 cells and colocalize with PCNA, a marker for the replication fork (Urtishak et al. 2009). Therefore, Swi1Timeless may function as an anti-recombinase at telomeres during DNA replication.
Swi1Timeless may coordinate DNA polymerases at telomeres
How Swi1 loss causes repeat instability remains to be determined. Previous studies showed that the lagging-strand DNA polymerase (pol δ) arrives at telomeres much later than the leading-strand DNA polymerase (pol ε) even in wild-type cells (Moser et al. 2009a). Considering that Swi1 is involved in the coordination of leading- and lagging-strand synthesis (Noguchi et al. 2004; Sommariva et al. 2005), it is reasonable to suggest that swi1Δ cells experience severe uncoupling of the two DNA polymerases. Such uncoupling may result in extensive accumulation of ssDNA and replication fork collapse at telomeres, resulting in hyper-recombination. It is also possible that DNA secondary structures such as hairpins and G quadruplexes may promote replication slippage in the absence of Swi1, resulting in loss of repeats at telomeres and other loci with repeat DNA sequences. Further investigations are warranted to test this interesting possibility and address the role of Swi1 in preventing polymerase slippage at repeat DNA regions including telomeres, rDNA, and LacO repeats.
Role of Swi1–Myb/SANT protein interaction in DNA replication
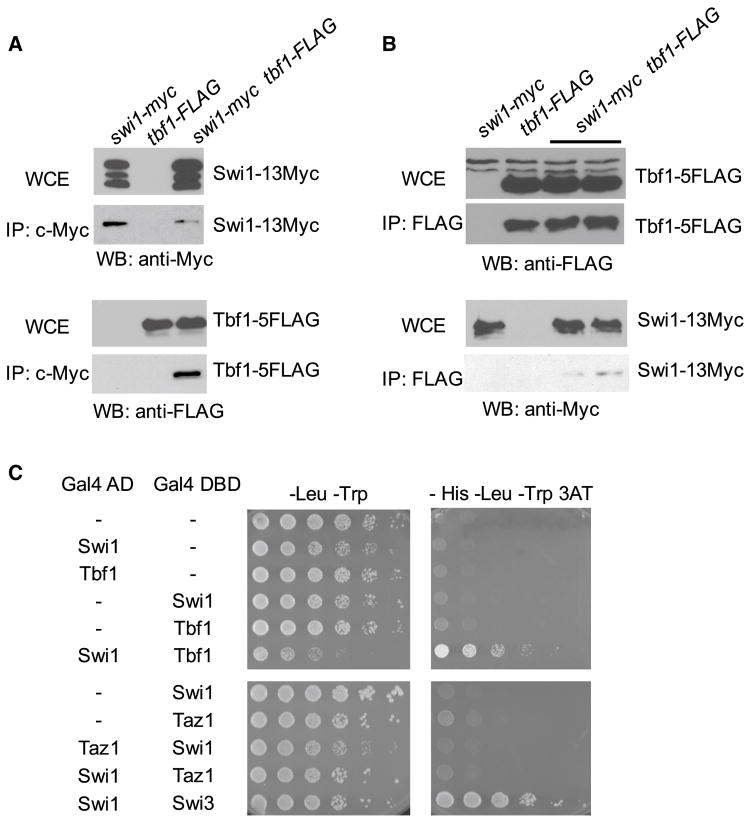
Swi1 and its orthologues are required for replisome stability at natural barriers, including rDNA pausing sites, the fission yeast mating-type locus, highly transcribed loci, and now at telomeres (Cherng et al. 2011; Gadaleta et al. 2016; Leman et al. 2012; Leman and Noguchi 2012, 2013; Liu et al. 2012; Pryce et al. 2009; Razidlo and Lahue 2008; Rozenzhak et al. 2010; Sabouri et al. 2012; Shishkin et al. 2009; Voineagu et al. 2008). Thus, it is straightforward to suggest that Swi1-related proteins are required for the regulation of most difficult-to-replicate regions. However, the underlying mechanism by which Swi1 modulates DNA replication at these genomic regions is not well understood. Key to this mechanism appears to be the Myb/SANT family of DNA-binding proteins. These proteins bind specific sites along the genome, and a subset of them is required for replication fork pausing at natural replication barriers. For instance, Rtf1, a Myb/SANT protein, binds to the RTS1 site at the fission yeast mating-type locus, in order to facilitate fork termination in a Swi1-dependent manner (Eydmann et al. 2008). Reb1, another Myb/SANT protein, is found at Ter1-3 sites in the rDNA repeats and promotes fork pausing, which is also dependent on Swi1 (Dalgaard and Klar 2000, 2001; Krings and Bastia 2004). In addition, fission yeast telomeres also recruit Myb/SANT proteins including TRF1 homologs, Taz1, and Tbf1 (Cooper et al. 1997; Pitt et al. 2008). Therefore, we hypothesized that Swi1 interacts with theses Myb/SANT family proteins at telomeres in order to stabilize replication forks passing along the telomeres. In fission yeast, roles of Taz1 at telomeres are well characterized, whereas the function of Tbf1 is elusive (Cockell et al. 2009; Moser and Nakamura 2009). In budding yeast, Tbf1 binds to telomeric repeats localized at subtelomeres, and it has been shown to play a role in telomere homeostasis by suppressing checkpoint activation at short telomeres (Fukunaga et al. 2012). Tbf1 is an essential protein in S. pombe due to its function for transcription; however, its role in telomere maintenance in fission yeast is unknown (Cockell et al. 2009; Sarda and Hannenhalli 2015; Yan et al. 2015). We tested the idea that Swi1 modulates replication of the telomeric fork-block sites by interacting with the resident Myb/SANT proteins. Interestingly, we found that Swi1 physically interacts with Tbf1 (Fig. 1a–c), but not with Taz1 (Fig. 1c). Therefore, physical interaction between Swi1 and resident Myb/SANT proteins at telomeric repeats such as Tbf1 may ensure efficient replication of telomeric repeat DNA sequences. Such a mechanism also seems to be conserved between fission yeast and humans. We previously showed that Timeless physically interacts with TRF1 and TRF2 (Myb/SANT proteins) in human cells (Leman et al. 2012). Furthermore, TRF1 overexpression can induce replication fork stalling/pausing at telomeres (Ohki and Ishikawa 2004). Consistently, when TRF1 is overexpressed in HeLa cells, replication factors such as Cdc45 and RPA became enriched at telomeres. Strikingly, the enrichment of Cdc45 and RPA was abolished when Timeless was depleted via shRNA (Leman et al. 2012), suggesting that replication fork stalling at human telomeres is also dependent on Timeless-TRF1 interaction.
Fig. 1.
Tbf1, a Myb-like protein, interacts with Swi1 in S. pombe cells. a S. pombe cells were engineered to express Swi1-13Myc and/or Tbf1-5FLAG. Cell extracts were prepared, and Swi1-13Myc was immunoprecipitated with anti-Myc polyclonal antibodies. Whole-cell extracts and precipitated fractions were probed with the anti-Myc (9E10) or anti-FLAG (M2) antibodies, in order to detect Swi1-13Myc and Tbf1-5FLAG by Western blotting (WB). b Tfb1-FLAG was immunoprecipitated from cell extracts expressing Swi1-13Myc and/or Tbf1-5FLAG. Swi1-13Myc and Tbf1-5FLAG in whole-cell extracts and precipitated fractions were detected by Western blotting (WB). c Two-hybrid interactions between Swi1 and Tbf1 or Taz1. Gal4-AD-Swi1 was tested in the Y190 strain for interaction with Gal4-DBD-Tbf1 or Gal4-DBD-Taz1. Growth on the -His-Leu-Trp 3-aminotriazole (3AT) plate indicates protein interaction. The interaction of Gal4-AD-Swi3 and Gal4-DBD-Swi1 was used as a positive control for protein interaction
Role of Swi1 in ALT prevention
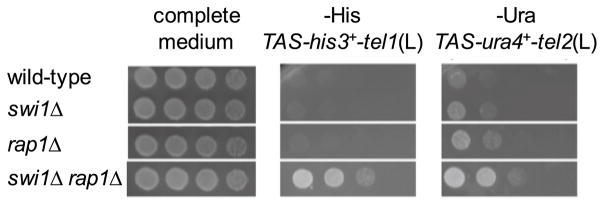
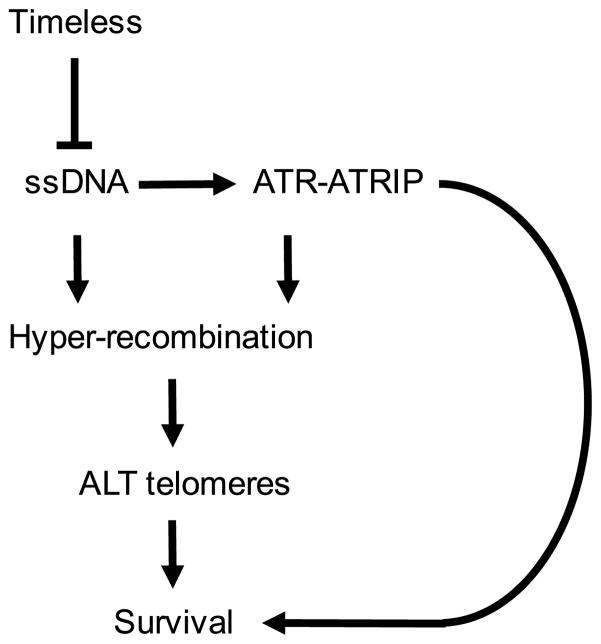
The study of the cellular mechanisms that control telomere length is central topic for the understanding of tumorigenesis and the development of age-related diseases. Approximately 10–15 % of cancer types, especially those of mesenchymal origin, survive without reactivation of telomerase. Instead, they maintain functional telomeres via the activation of ALT pathways; however, the mechanisms underlying ALT activation in these cancer cells are not clear (Dilley and Greenberg 2015). Recently, Zou and colleagues reported that cancer cells with ALT telomeres are hypersensitive to ATR inhibitors. Survival of ALT cells is highly dependent on ATR-ATRIP because ALT telomeres have increased levels of RPA-coated ssDNAs, which activate the ATR-ATRIP checkpoint kinase. Furthermore, ALT telomeres display high levels of telomeric repeat-containing RNA (TERRA) in S phase, which appear to correlate with RPA accumulation at telomeres and ATR-ATRIP activation (Flynn et al. 2015). Importantly, similar telomere defects were also seen in fission yeast swi1Δ cells. We observed that the simultaneous deletion of swi1 and rap1 disrupts the repressive telomeric chromatin state, potentially facilitating transcriptional activity at telomeric repeats (Fig. 2). As mentioned above, telomeres have increased levels of ssDNAs, represented by Rad52 recruitment in swi1Δ cells (Gadaleta et al. 2016). This is consistent with the decreased growth fitness of swi1Δ rad26Δ and swi1Δ chk1Δ cells (Noguchi et al. 2003). Rad26 is the fission yeast homolog of human ATRIP, essential for ATR activation, and Chk1 is a downstream effector of ATR (Hustedt et al. 2013). swi1Δ rad26Δ and swi1Δ chk1Δ cells show increased levels of “cut” phenotype, indicative of mitotic catastrophe and death (Noguchi et al. 2003). These findings indicate that swi1Δ cells hyper-activate ATRRad3-ATRIPRad26 due to increased levels of ssDNAs. Importantly, in telomerase-negative fission yeast cells, Swi1 loss leads to telomere hyper-recombination and an increase in the occurrence of ALT-type survivors (Fig. 3) (Gadaleta et al. 2016). Therefore, it would be interesting to test whether ALT activation in telomerase-negative swi1Δ cells is dependent on ATRRad3-ATRIPRad26. Similar experiments in human cells are also warranted to test the role of Timeless in preventing ALT phenotypes (Fig. 3).
Fig. 2.
Swi1 is involved in telomere silencing. swi1+ and/or rap1+ genes were deleted from an S. pombe strain bearing reporter genes. Rap1 has been shown to be involved in telomere silencing (Fujita et al. 2012; Moser et al. 2009b). The his3+ and ura4+ genes were inserted at silent subtelomeric regions of chromosome 1 [TAS-tel(L)] and 2 [TAS-tel2(L)], respectively. Fivefold-serial dilutions of the indicated strains were spotted onto YES agar medium or minimal medium lacking histidine or uracil. The plates were then incubated for 2–3 days at 32 °C. Representative images of repeat experiments are shown. Growth on medium lacking nutrients indicates defects in silencing at subtelomeres
Fig. 3.
Model of Timeless and ATR-ATRIP dependent ALT regulation. For details, see text
In summary, our studies have demonstrated a novel and conserved role of the Timeless-related proteins in replication of repetitive DNA and telomere maintenance. Swi1Timeless prevents hyper-recombination and ALT activation in the absence of telomerase activity in fission yeast. Considering the prevalence of human cancers with active ALT pathways as a means to maintain functional telomeres, our study provides a potential mechanism to explain ALT activation during cancer development in humans.
Acknowledgments
This work was supported by the Aging Initiative at Drexel University College of Medicine. We thank Chiaki Noguchi and Grant Grothusen for technical assistance and National BioResource Project Japan for S. pombe strains.
References
- Cherng N, Shishkin AA, Schlager LI, Tuck RH, Sloan L, Matera R, Sarkar PS, Ashizawa T, Freudenreich CH, Mirkin SM. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc Natl Acad Sci USA. 2011;108:2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell MM, Lo Presti L, Cerutti L, Cano Del Rosario E, Hauser PM, Simanis V. Functional differentiation of tbf1 orthologues in fission and budding yeasts. Eukaryot Cell. 2009;8:207–216. doi: 10.1128/EC.00174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 2001;15:2060–2068. doi: 10.1101/gad.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, Greenberg RA. Alternative telomere maintenance and cancer. Trends Cancer. 2015;1:145–156. doi: 10.1016/j.trecan.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eydmann T, Sommariva E, Inagawa T, Mian S, Klar AJ, Dalgaard JZ. Rtf1-mediated eukaryotic site-specific replication termination. Genetics. 2008;180:27–39. doi: 10.1534/genetics.108.089243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, Haber DA, Boussin FD, Zou L. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita I, Tanaka M, Kanoh J. Identification of the functional domains of the telomere protein Rap1 in Schizosaccharomyces pombe. PLoS One. 2012;7:e49151. doi: 10.1371/journal.pone.0049151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K, Hirano Y, Sugimoto K. Subtelomere-binding protein Tbf1 and telomere-binding protein Rap1 collaborate to inhibit localization of the Mre11 complex to DNA ends in budding yeast. Mol Biol Cell. 2012;23:347–359. doi: 10.1091/mbc.E11-06-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta MC, Das MM, Tanizawa H, Chang YT, Noma K, Nakamura TM, Noguchi E. Swi1Timeless prevents repeat instability at fission yeast telomeres. PLoS Genet. 2016;12:e1005943. doi: 10.1371/journal.pgen.1005943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N, Gasser SM, Shimada K. Replication checkpoint: tuning and coordination of replication forks in s phase. Genes (Basel) 2013;4:388–434. doi: 10.3390/genes4030388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosac-charomyces pombe. Proc Natl Acad Sci USA. 2004;101:14085–14090. doi: 10.1073/pnas.0406037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Noguchi E. Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle. 2012;11:3945–3955. doi: 10.4161/cc.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Noguchi E. The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes (Basel) 2013;4:1–32. doi: 10.3390/genes4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Dheekollu J, Deng Z, Lee SW, Das MM, Lieberman PM, Noguchi E. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle. 2012;11:2337–2347. doi: 10.4161/cc.20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X, Gao Y, Lewis T, Barthelemy J, Leffak M. Altered replication in human cells promotes DMPK (CTG) (n). (CAG)(n) repeat instability. Mol Cell Biol. 2012;32:1618–1632. doi: 10.1128/MCB.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet C, Makovets S. Aneuploidy as a mechanism of adaptation to telomerase insufficiency. Curr Genet. 2016 doi: 10.1007/s00294-015-0559-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Nakamura TM. Protection and replication of telomeres in fission yeast. Biochem Cell Biol. 2009;87:747–758. doi: 10.1139/O09-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009a;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. Fission yeast Tel1(ATM) and Rad3(ATR) promote telomere protection and telomerase recruitment. PLoS Genet. 2009b;5:e1000622. doi: 10.1371/journal.pgen.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, McDonald WH, Yates JR, 3rd, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Ishikawa F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt CW, Valente LP, Rhodes D, Simonsson T. Identification and characterization of an essential telomeric repeat binding factor in fission yeast. J Biol Chem. 2008;283:2693–2701. doi: 10.1074/jbc.M708784200. [DOI] [PubMed] [Google Scholar]
- Pryce DW, Ramayah S, Jaendling A, McFarlane RJ. Recombination at DNA replication fork barriers is not universal and is differentially regulated by Swi1. Proc Natl Acad Sci USA. 2009;106:4770–4775. doi: 10.1073/pnas.0807739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp JB, Noguchi C, Das MM, Wong LK, Ansbach AB, Holmes AM, Arcangioli B, Noguchi E. Checkpoint-dependent and -independent roles of Swi3 in replication fork recovery and sister chromatid cohesion in fission yeast. PLoS One. 2010;5:e13379. doi: 10.1371/journal.pone.0013379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razidlo DF, Lahue RS. Mrc1, Tof1 and Csm3 inhibit CAG. CTG repeat instability by at least two mechanisms. DNA Repair (Amst) 2008;7:633–640. doi: 10.1016/j.dnarep.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenzhak S, Mejia-Ramirez E, Williams JS, Schaffer L, Hammond JA, Head SR, Russell P. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 2010;6:e1001032. doi: 10.1371/journal.pgen.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev. 2012;26:581–593. doi: 10.1101/gad.184697.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda S, Hannenhalli S. High-throughput identification of cis-regulatory rewiring events in yeast. Mol Biol Evol. 2015;32:3047–3063. doi: 10.1093/molbev/msv203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishkin AA, Voineagu I, Matera R, Cherng N, Chernet BT, Krasilnikova MM, Narayanan V, Lobachev KS, Mirkin SM. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol. 2005;25:2770–2784. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtishak KA, Smith KD, Chanoux RA, Greenberg RA, Johnson FB, Brown EJ. Timeless maintains genomic stability and suppresses sister chromatid exchange during unperturbed DNA replication. J Biol Chem. 2009;284:8768–8776. doi: 10.1074/jbc.M806103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci USA. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Zhang D, Raygoza Garay JA, Mwangi MM, Bai L. Decoupling of divergent gene regulation by sequence-specific DNA binding factors. Nucleic Acids Res. 2015;43:7292–7305. doi: 10.1093/nar/gkv618. [DOI] [PMC free article] [PubMed] [Google Scholar]