Abstract
Bacteriophage (phage) that infect pathogenic bacteria often attach to surface receptors that are coincidentally required for virulence. Receptor loss or modification through mutation renders mutants both attenuated and phage resistant. Such attenuated mutants frequently have no apparent laboratory growth defects, but in the host, they fail to exhibit properties needed to produce disease such as mucosal colonization or survival within professional phagocytic cells. The connection between attenuation and phage resistance has been exploited in experimental demonstrations of phage therapy. In such experiments, phage resistant mutants that arise naturally during therapy are inconsequential because of their attenuated status. A more contemporary approach to exploiting this connection involves identifying small effector molecules, identified in high throughput screens, that inhibit one or more of the steps needed to produce a functioning phage receptor. Since such biosynthetic steps are unique to bacteria, inhibitors can be utilized therapeutically, in lieu of antibiotics. Also, since the inhibitor is specific to a particular bacterium or group of bacteria, no off-target resistance is generated in the host’s commensal bacterial population. This brief review covers examples of how mutations that confer phage resistance produce attenuation, and how this coincidental relationship can be exploited in the search for the next generation of therapeutic agents for bacterial diseases.
Keywords: Bacteriophage, phage, receptor, attenuation, therapy, antibiotic, alternative, antivirulence factor
Review
Ninety years ago, Felix d’Herelle successfully treated fowl typhoid with bacteriophage (phage) delivered orally to chicken flocks in Southern France (d’Herelle 1926). The use of phage to prevent or treat bacterial disease has, in the intervening years, undergone cycles of enthusiasm and skepticism (Domingo-Calap et al. 2016; Eaton and Bayne-Jones 1934; Nobrega et al. 2015). One reason for its checkered reputation is rooted in the mutation rate of bacterial pathogens to phage resistance (Luria and Delbruck 1943). The appearance of a resistant mutant early in therapy is an obvious and unavoidable complication--one well appreciated by d’Herelle himself (d’Herelle 1931). Just over thirty-five years ago a partial solution to that complication was offered.
In a series of elegant studies, H. Williams Smith and colleagues (Smith and Huggins 1980; Smith and Huggins 1982; Smith and Huggins 1983; Smith et al. 1987) demonstrated several successful instances of phage therapy applied to Escherichia coli infections (both enteric and extraintestinal). Whereas Smith employed a number of innovative and novel techniques, the author felt his success (particularly when employing just a single therapeutic phage) was mainly due to the fact that the virulent E. coli strain employed was rendered attenuated as a consequence of acquiring phage resistance. In this particular case, the E. coli K1 capsule was the phage receptor and unencapsulated mutants were attenuated in the extraintestinal infection Smith and his colleagues were modeling. Since a relatively large number of gene products are required to make a K1 capsule, it makes sense that capsular loss would be the most common mechanism (of several: Labrie et al. 2010) to confer phage resistance during an infection.
Smith’s clever and insightful choice of phage for his therapeutic test (the phage performed better than antibiotics: Smith and Huggins 1982) suggested that certain phage would be much better therapeutic candidates than others. However, current phage therapy descriptions typically do not apply Smith’s insights to screen candidate phage for therapeutic use (Loc-Carrillo and Abedon 2011; Nobrega et al. 2015). One impediment may be that it requires additional tests and unless the phage uses a receptor that is known to be required for an obvious step in pathogenesis (e.g., immune evasion, and attachment, reviewed in: Leon and Bastias 2015) the reason for attenuation may not be obvious. Nevertheless, for every pathogenic bacterium, there are many types of phage (Lindberg 1973). Consequently, the likelihood is high that one or more of those phage will utilize a receptor that is also required for virulence.
In 1982, Raleigh and Signer suggested the use of phage as selective agents to obtain mutant collections enriched for members defective in host/bacterial interactions (Raleigh and Signer 1982). Using this technique, the authors obtained nodulation defective Rhizobium phaseoli mutants. Subsequently, the group lead by Signer used the phage as a tool to deduce the parts of the phage receptor important for nodulation (Finan et al. 1985). Impressed by their success, I instigated the first of two projects in my laboratory to see if the same strategy could be applied to animal pathogens. Initial work was carried out with Bordetella avium, a gram negative pathogen that causes a pertussis-like upper respiratory tract disease in turkeys called bordetellosis or turkey coryza (Jackwood and Saif 2003). Turkey coryza pathogenesis is rather straightforward: water or aerosol--borne bacteria colonize the trachea and elaborate toxins that produce the characteristic pathogenic features (loss if ciliated tracheal epithelial cells: Arp and Cheville 1984). The pathogen is cleared coincident with the appearance of a humoral immune response (Barnes and Hofstad 1983). We found that 70% of mutants resistant to a phage we had isolated (Ba1: Shelton et al. 2000) were attenuated and all (100%) had assorted lipopolysaccharide (LPS) modifications that prevented direct phage attachment. Analysis of the mutant LPS from attenuated and virulent mutant classes permitted identification of the outer core region of LPS (the point of O-antigen attachment) as crucial to pathogenesis (Shelton et al. 2002; Spears et al. 2000).
Phage were again used in my laboratory, this time to identify and characterize attenuated mutants in the gram positive pathogen Listeria monocytogenes, the causative agent of listeriosis (Gray and Killinger 1966). Listeriosis has a much more complicated pathogenesis than bordetellosis. Listeriae are foodborne pathogens (Farber and Peterkin 1991) that translocate from the intestine to the liver and spleen where they reside intracellularly and spread cell-to-cell by means of a cytosolic motility based on actin polymerization (Portnoy et al. 2002; Tilney and Portnoy 1989). Mutants that fail to spread cell-to-cell effectively are attenuated (Dussurget et al. 2004). Also, many of the immunostimulatory properties of listeriosis are rooted in this pathogenic feature (Spears et al. 2011). A listerial phage, P35h4, was employed as a screening agent and a phage resistant insertion mutant was obtained that failed to adsorb phage and was profoundly attenuated in a mouse oral-infection model (Spears et al. 2008). Subsequently, a collection of P35h4 resistant mutants was systematically assembled. All mutants (each having a lesion in one of four genes required for phage attachment) were attenuated in orally inoculated mice and defective in their ability to spread from cell-to-cell in mouse enterocyte monolayers (Spears et al. 2016). With the help of the phage, we found that each mutant’s wall teichoic acid (WTA; a glycopolymer covalently attached to the cell wall) was modified: each repeat unit was missing a single substituent sugar (galactose). As is true of most WTA “decorating” substituents, the absence produced no change in any of the mutants’ laboratory growth properties (Brown et al. 2013). Nevertheless, in the host cell cytosol, the mutants all exhibited reduced actin polymerization, and decreased cytosolic motility. The enzymatic steps required to add the WTA decoration have been proposed (Spears et al. 2016). Currently, those enzymes provide targets for inhibition by small secondary metabolite derivatives. Such inhibitors are intended for use therapeutically as antiviurlence factors (Clatworthy et al. 2007) that serve in lieu of antibiotics.
Exactly how the absence of a WTA decorating sugar decreases actin-based motility is unclear, but we suspect that the modified WTA composition results in a more elongated polymer (Doyle et al. 1974), and that this may impede the access of cytosolic unpolymerized actin to the listerial cell surface protein (ActA) that nucleates the polymerization process (Spears et al. 2016). A similar elongation of lipopolysaccharide (LPS; a glycopolymer anchored in the outer membrane of gram negative bacteria) is proposed to result from a lack of specific glucosylation of the Shigella flexneri LPS glycopolymeric repeat units (West et al. 2005). In this case, glucosylation is required for the attachment of a set of S. flexneri phage that attach only to glucosylated repeat units (Allison and Verma 2000). The defect in virulence is attributed (West et al. 2005) to the reduce access of the microorganism’s surface-exposed type III secretion system (Ghosh 2004) to host cells. In both L. monocytogenes and S. flexneri, phage receptor modifications reduced virulence. Further, they did so by strikingly similar means: alteration of glycopolymer composition (removal of a substituent sugar), and possibly via a similar alteration in the receptor’s tertiary structure (polymer elongation).
A partial list of pathogens whose virulence is influenced by a modification or ablation of a phage receptor has been published recently (Leon and Bastias 2015). A complete list is difficult to assemble in part because phage resistance is often not integral to the screening strategy employed to obtain attenuated isolates (Autret et al. 2001; Bogomolnaya et al. 2008; Eugster et al. 2011; Faith et al. 2009; West et al. 2005). In these cases, the coincident phage resistance, being peripheral to the screen, is not described in detail, and sometimes deduced from bioinformatic or historical data (Autret et al. 2001; Bogomolnaya et al. 2008). How the frequent, if coincidental, connection between phage receptor and virulence can be systematically exploited to obtain new therapeutic agents is outlined below.
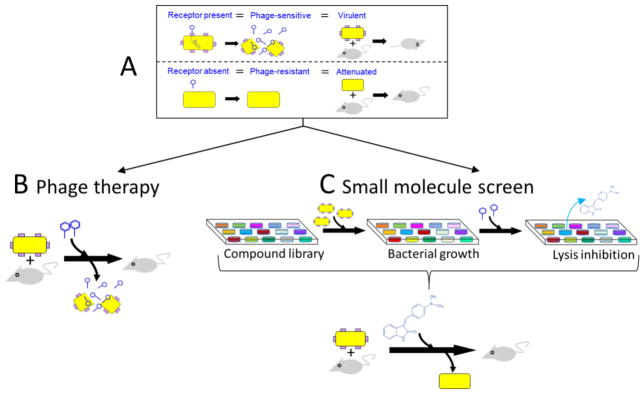
A general procedure to develop new antimicrobials based on phage receptor biosynthesis is outlined in Fig. 1. For a given pathogenic bacterium, the initial step is to employ a collection of phage, select mutants resistant to each, and screen those mutants for attenuation in an appropriate animal model (Fig. 1A). Phage collections are readily available (e.g., Felix d’Herelle Reference Center) or obtainable by traditional isolation procedures (Pelzek et al. 2008). Phage resistant, attenuated mutants are then characterized to identify those that fail to adsorb the phage (i.e., fail to produce a functional phage receptor). In some well-studied pathogens, bioinformatic database queries could be used to reduce (but probably not eliminate) the empirical screening and characterization process.
Fig. 1. Identification and development of therapeutics based upon phage receptors.
The top rectangle (A) depicts the identification of a phage (tailed hexagon) whose receptor (purple squares) is determined, in empirical tests in mice, to be required for virulence. At this point, depending upon the disease under study, phage therapy (B) could be a promising therapeutic approach. Should disease pathogenesis warrant an alternative approach to therapy, a protocol is depicted (C) that utilizes a high-throughput screen of small molecules to identify therapeutic candidates based upon their ability to confer phage resistance. Subsequent experiments, to establish that the compounds identified prevent or alter receptor biosynthesis, are described in the text. Viable candidates (one compound illustrated) are moved forward for tests in mice
Once it is established which members of a phage collection can be use to select attenuated mutants, disease pathogenesis might permit the simple application of phage therapy (Fig. 1B). Such diseases would ideally be relatively superficial (e.g., intestinal, upper respiratory tract) and thus favor phage administration per os or via inhalation (Summers 2001). In those instances where phage therapy is deemed problematical (e.g., systemic infections), a screen of effector molecules that prevent phage growth could yield candidate compounds that inhibit functional receptor biosynthesis (Fig. 1C).
Regarding the screening illustrated in Fig. 1C, the compounds identified as conferring phage resistance would necessarily require further characterization to confirm that phage resistance was indeed correlated with a failure to adsorb phage. Also, it would be necessary to determine the target molecule inhibited. This determination could lead directly to the receptor (e.g., a cell surface protein) or could identify an enzyme in a pathway needed for receptor production or proper structure (e.g., a cell surface glycopolymer whose biosynthesis requires several enzymatic steps). Such an identification would allow the inhibitory molecule responsible to be structurally refined to enhance efficacy and therapeutic potential. Modern molecular procedures (Richter et al. 2013) and bioinformatic methods (Fourches et al. 2014) can greatly facilitate target identification procedures. Also, if a multistep pathway is required to produce the phage receptor, the initial target identified could reveal additional potential targets in the receptor biosynthesis pathway using metabolomic data (Nicholson and Wilson 2003). Targets identified in this manner allow the application of in silico docking simulations to be performed that can pre-screen a random small molecule library—reducing and refining the number of candidate inhibitors (Fourches et al. 2013; Spears et al. 2016).
An ideal situation would arise in cases where the pathway for phage receptor production is known before beginning the inhibitor library screen (Brown et al. 2012; Spears et al. 2016; Xia et al. 2010). This would allow the in silico studies to precede a random library screen—reducing the compounds actually tested in the laboratory. A knowledge of the pathway could also be utilized to adjust the pathogen efficacy spectrum by targeting steps in the receptor biosynthesis pathway that are common to several phylogenetically related pathogens, or ones that are more pathogen-specific (Ling et al. 2015; Spears et al. 2016).
Conclusions
This brief review presents examples of how phage can be used to identify surface structures required for pathogenesis, and then employed as tools to characterize compositional and structural features important for host-pathogen interaction. Further, a procedure is outlined for the use of phage to systematically identify bacterial surface structures required for virulence, and two possibilities (phage therapy and receptor biosynthesis inhibition) are described to exploit this therapeutically. Both possibilities serve as replacements for antibiotics. One might think that such a radical departure from antibiotics would be far in the future and encounter a good deal of regulatory delays. However, meaningful tests are likely to begin very soon. Poultry producers and large commercial purchasers of poultry products have expressed their intent that all antibiotic use (even therapeutic use) will be eliminated by 2019. The $48.3 billion/year poultry industry and the USDA have indicated their willingness to aid development of antibiotic alternatives. Initiatives such as these in production animals could emerge as the launching pad for a variety of experimental tests of antibiotic alternatives in humans (Miller 2016).
Acknowledgments
My thanks to Edward A. Havell, Patricia A. Spears, Luke B. Borst and Johanna R. Elfenbein for a critical reading of this manuscript prior to submission. The author declares no conflict of interest. Work in the author’s laboratory is supported by Public Health Service Grants AI103549, AI083838, and AI064333.
Literature Cited
- Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/s0966-842x(99)01646-7. http://dx.doi.org/10.1016/S0966-842X(99)01646-7. [DOI] [PubMed] [Google Scholar]
- Arp LH, Cheville NF. Tracheal lesions in young turkeys infected with Bordetella avium. Am J Vet Res. 1984;45:2196–2200. [PubMed] [Google Scholar]
- Autret N, Dubail I, Trieu-Cuot P, Berche P, Charbit A. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect Immun. 2001;69:2054–2065. doi: 10.1128/IAI.69.4.2054-2065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes HJ, Hofstad MS. Susceptibility of turkey poults from vaccinated and unvaccinated hens to alcaligenes rhinotracheitis (turkey coryza) Avian Dis. 1983;27:378–392. [PubMed] [Google Scholar]
- Bogomolnaya LM, Santiviago CA, Yang HJ, Baumler AJ, Andrews-Polymenis HL. ‘Form variation’ of the O12 antigen is critical for persistence of Salmonella Typhimurium in the murine intestine. Mol Microbiol. 2008;70:1105–1119. doi: 10.1111/j.1365-2958.2008.06461.x. MMI6461 [pii] [DOI] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci U S A. 2012;109:18909–18914. doi: 10.1073/pnas.1209126109. 1209126109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- d’Herelle F. The Behavior of the Bacteriophage in Epidemics. Chapter IV. The Williams & Wilkins Company; Baltimore, Md: 1926. pp. 490–497. [DOI] [Google Scholar]
- d’Herelle F. Bacterial Mutations Yale. J Biol Med. 1931;4:55–61. [PMC free article] [PubMed] [Google Scholar]
- Domingo-Calap P, Georgel P, Bahram S. Back to the future: bacteriophages as promising therapeutic tools. HLA. 2016;87:133–140. doi: 10.1111/tan.12742. [DOI] [PubMed] [Google Scholar]
- Doyle RJ, McDannel ML, Streips UN, Birdsell DC, Young FE. Polyelectrolyte nature of bacterial teichoic acids. J Bacteriol. 1974;118:606–615. doi: 10.1128/jb.118.2.606-615.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget O, Pizarro-Cerda J, Cossart P. Molecular determinants of Listeria monocytogenes virulence. Annu Rev Microbiol. 2004;58:587–610. doi: 10.1146/annurev.micro.57.030502.090934. [DOI] [PubMed] [Google Scholar]
- Eaton MD, Bayne-Jones S. Bacteriophage therapy: Review of the principles and results of the use of bacteriophage in the treatment of infections. JAMA. 1934;103:1847–1853. [Google Scholar]
- Eugster MR, Haug MC, Huwiler SG, Loessner MJ. The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol Microbiol. 2011;81:1419–1432. doi: 10.1111/j.1365-2958.2011.07774.x. [DOI] [PubMed] [Google Scholar]
- Faith N, et al. The role of L. monocytogenes serotype 4b gtcA in gastrointestinal listeriosis in A/J mice. Foodborne Pathog Dis. 2009;6:39–48. doi: 10.1089/fpd.2008.0154. [DOI] [PubMed] [Google Scholar]
- Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, et al. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985;40:869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- Fourches D, Muratov E, Ding F, Dokholyan NV, Tropsha A. Predicting binding affinity of CSAR ligands using both structure-based and ligand-based approaches. J Chem Inf Model. 2013;53:1915–1922. doi: 10.1021/ci400216q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D, Sassano MF, Roth BL, Tropsha A. HTS navigator: freely accessible cheminformatics software for analyzing high-throughput screening data. Bioinformatics. 2014;30:588–589. doi: 10.1093/bioinformatics/btt718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. Process of Protein Transport by the Type III Secretion System Microbiol. Mol Biol Rev. 2004;68:771–795. doi: 10.1128/mmbr.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ML, Killinger AH. Listeria monocytogenes and listeric infections. Bacteriol Rev. 1966;30:309–382. doi: 10.1128/br.30.2.309-382.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood MW, Saif YM. Bordetellosis. In: Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougal LR, Swayne DE, editors. Diseases of Poultry. 11. Iowa State University Press; Ames: 2003. [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Micro. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- Leon M, Bastias R. Virulence reduction in Bacteriophage resistant bacteria. Frontiers in Microbiology. 2015;6:1–7. doi: 10.3389/fmicb.2015.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg AA. Bacteriophage receptors. Annu Rev Microbiol. 1973;27:205–241. doi: 10.1146/annurev.mi.27.100173.001225. [DOI] [PubMed] [Google Scholar]
- Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. Antibacterial Development: a Changing Landscape. Microbe Magazine. 2016;11:111–118. doi: 10.1128/microbe.11.111.1. [DOI] [Google Scholar]
- Nicholson JK, Wilson ID. Understanding ‘Global’ Systems Biology: Metabonomics and the Continuum of Metabolism. Nat Rev Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- Nobrega FL, Costa AR, Kluskens LD, Azeredo J. Revisiting phage therapy: new applications for old resources. Trends Microbiol. 2015;23:185–191. doi: 10.1016/j.tim.2015.01.006. http://dx.doi.org/10.1016/j.tim.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Pelzek AJ, Schuch R, Schmitz JE, Fischetti VA. Current Protocols Essential Laboratory Techniques. John Wiley & Sons, Inc; 2008. Isolation, Culture, and Characterization of Bacteriophages. [DOI] [Google Scholar]
- Portnoy DA, Auerbuch V, Glomski IJ. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol. 2002;158:409–414. doi: 10.1083/jcb.200205009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh EA, Signer ER. Positive selection of nodulation-deficient Rhizobium phaseoli. J Bacteriol. 1982;151:83–88. doi: 10.1128/jb.151.1.83-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter SG, Elli D, Kim HK, Hendrickx AP, Sorg JA, Schneewind O, Missiakas D. Small molecule inhibitor of lipoteichoic acid synthesis is an antibiotic for gram-positive bacteria. Proc Natl Acad Sci U S A. 2013;110:3531–3536. doi: 10.1073/pnas.1217337110. 1217337110 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton CB, Crosslin DR, Casey JL, Ng S, Temple LM, Orndorff PE. Discovery, purification, and characterization of a temperate transducing bacteriophage for Bordetella avium. J Bacteriol. 2000;182:6130–6136. doi: 10.1128/jb.182.21.6130-6136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton CB, Temple LM, Orndorff PE. Use of bacteriophage Ba1 to identify properties associated with Bordetella avium virulence. Infect Immun. 2002;70:1219–1224. doi: 10.1128/IAI.70.3.1219-1224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HW, Huggins MB. The association of the O18, K1 and H7 antigens and the Co1V plasmid of a strain of Escherichia coli with its virulence and immunogenicity. J Gen Microbiol. 1980;121:387–400. doi: 10.1099/00221287-121-2-387. [DOI] [PubMed] [Google Scholar]
- Smith HW, Huggins MB. Successful treatment of experimental Escherichia coli infections in mice using phage: its general superiority over antibiotics. J Gen Microbiol. 1982;128:307–318. doi: 10.1099/00221287-128-2-307. [DOI] [PubMed] [Google Scholar]
- Smith HW, Huggins MB. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J Gen Microbiol. 1983;129:2659–2675. doi: 10.1099/00221287-129-8-2659. [DOI] [PubMed] [Google Scholar]
- Smith HW, Huggins MB, Shaw KM. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J Gen Microbiol. 1987;133:1111–1126. doi: 10.1099/00221287-133-5-1111. [DOI] [PubMed] [Google Scholar]
- Spears PA, et al. Listeria monocytogenes wall teichoic acid decoration in virulence and cell-to-cell spread. Mol Microbiol. 2016 doi: 10.1111/mmi.13353. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- Spears PA, Suyemoto MM, Hamrick TS, Wolf RL, Havell EA, Orndorff PE. In vitro properties of a Listeria monocytogenes bacteriophage-resistant mutant predict Its efficacy as a live oral vaccine strain. Infect Immun. 2011;79:5001–5009. doi: 10.1128/IAI.05700-11. IAI.05700-11 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spears PA, Suyemoto MM, Palermo AM, Horton JR, Hamrick TS, Havell EA, Orndorff PE. A Listeria monocytogenes mutant defective in bacteriophage attachment is attenuated in orally inoculated mice and impaired in enterocyte intracellular growth. Infect Immun. 2008;76:4046–4054. doi: 10.1128/IAI.00283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spears PA, Temple LM, Orndorff PE. A role for lipopolysaccharide in turkey tracheal colonization by Bordetella avium as demonstrated in vivo and in vitro. Mol Microbiol. 2000;36:1425–1435. doi: 10.1046/j.1365-2958.2000.01963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers WC. Bacteriophage therapy. Annu Rev Microbiol. 2001;55:437–451. doi: 10.1146/annurev.micro.55.1.43755/1/437. [pii] [DOI] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West NP, et al. Optimization of virulence functions through glucosylation of Shigella LPS. Science. 2005;307:1313–1317. doi: 10.1126/science.1108472. [DOI] [PubMed] [Google Scholar]
- Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M, Holst O, Peschel A. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J Biol Chem. 2010;285:13405–13415. doi: 10.1074/jbc.M109.096172. M109.096172 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]