Abstract
Tuberculosis, caused by the bacterium Mycobacterium tuberculosis, remains one of the most serious global health problems. Molecular typing of M. tuberculosis has been used for various epidemiologic purposes as well as for clinical management. Currently, many techniques are available to type M. tuberculosis. Choosing the most appropriate technique in accordance with the existing laboratory conditions and the specific features of the geographic region is important. Insertion sequence IS6110-based restriction fragment length polymorphism (RFLP) analysis is considered the gold standard for the molecular epidemiologic investigations of tuberculosis. However, other polymerase chain reaction-based methods such as spacer oligonucleotide typing (spoligotyping), which detects 43 spacer sequence-interspersing direct repeats (DRs) in the genomic DR region; mycobacterial interspersed repetitive units–variable number tandem repeats, (MIRU-VNTR), which determines the number and size of tandem repetitive DNA sequences; repetitive-sequence-based PCR (rep-PCR), which provides high-throughput genotypic fingerprinting of multiple Mycobacterium species; and the recently developed genome-based whole genome sequencing methods demonstrate similar discriminatory power and greater convenience. This review focuses on techniques frequently used for the molecular typing of M. tuberculosis and discusses their general aspects and applications.
Keywords: Tuberculosis, Mycobacterium tuberculosis, Molecular Strain Typing
INTRODUCTION
Tuberculosis (TB) is one of the most common global health problems and also one of the world’s deadliest communicable diseases. For example, in 2014, an estimated 9.6 million persons developed tuberculosis, and 1.5 million died. More than half (58%) of the estimated affected people were from the South-East Asia and Western Pacific Regions. Another one quarter of cases was from the African region, which also had the highest rates of cases and deaths relative to population. With the emergence of multi-drug-resistant (MDR)-TB, which is defined as Mycobacterium tuberculosis resistant to at least isoniazid and rifampicin, TB has become an even greater threat (1).
As recent molecular typing methods based on the direct analysis of genomic polymorphisms now are widely used and highly valuable, understanding of the evolutionary history, population dynamics, and patterns of dissemination of bacteria has been greatly improved (2). Molecular typing of M. tuberculosis has been used for various epidemiologic purposes such as outbreak investigations in health care settings and communities, to estimate the proportion of epidemiologic links not identified by conventional contact tracing, to identify misdiagnosis resulting from laboratory cross-contamination, to determine the distinction between re-infection and re-activation, to quantify tuberculosis transmission between subpopulations, and to detect the relation between drug resistance and specific genotypes (3,4).
Since the early 1990s, genotyping of M. tuberculosis has been used in molecular epidemiology. Recent worldwide strain distribution of M. tuberculosis lineages demonstrated that the Beijing strain was the most prevalent in East Asia region and found as more than 50% of all strains. Similarly, the Beijing genotypes were the most prevalent in Central Asia, Northern Asia and Southeastern Asia while only small portion appeared in Southern Asia, Western Asia, Central Asia, Austral Africa, Australasia, North America, Northern Europe and South America regions (5). Genotyping not only helps us to understand the epidemiologic aspects of the disease but also is useful for the clinical management of patients by strain classification, which has made association of M. tuberculosis genetic markers with important clinical consequences such as drug-resistance potential, treatment failure, or high transmissibility. Currently, molecular typing is widely used by TB control programs because genotyping results have helped to monitor epidemiologic trends, evaluate program performance, and identify instances of false-positive and cross-contamination of cultures or define the transmission dynamics of drug-resistant organisms (4,6,7).
For the genetic fingerprinting of M. tuberculosis, the gold standard is insertion sequence IS6110-based restriction fragment length polymorphism (RFLP) analysis (8,9,10). However, similar discriminatory power and greater convenience has been demonstrated with other methods such as spacer oligonucleotide typing (spoligotyping), which detects 43 spacer sequences interspersed with direct repeats (DRs) in the genomic region uniquely present in members of M. tuberculosis complex (11); mycobacterial interspersed repetitive units-variable number tandem repeats (MIRU-VNTR) that uses polymerase chain reaction (PCR) amplification and gel electrophoresis to determine the number and size of tandem repetitive DNA sequence in 12 independent loci in the M. tuberculosis genome (12,13); repetitive-sequence-based PCR (rep-PCR), which is a fast and unified method for high-throughput genotypic fingerprinting of multiple Mycobacterium species (14); and whole genome sequencing (WGS), a recent method with high discriminatory power (15,16,17).
Ideally, molecular typing methods should be reproducible and stable, have high discriminatory power and epidemiologic concordance, be adaptable, be easy to perform and interpret, and be cost-effective and time-effective. We have described the currently available typing methods for M. tuberculosis and the current status of strain distribution in Korea (18). In this review, we described the five techniques, including whole genome sequencing (WGS), frequently used for characterizing M. tuberculosis isolates and their applications.
MOLECULAR TYPING
IS6110-RFLP
Mycobacterium tuberculosis insertion sequence IS6110 was first described by Thierry et al. (19), and its value as the standard approach to genotyping of M. tuberculosis isolates based on RFLP analysis has been proved by many studies (8,20,21). IS6110 belongs to the IS3 family of mobile elements and is 1,355 bp long. The IS6110 is found only within the M. tuberculosis complex, and, in most other members of the complex, the sequence is present in multiple copies, although M. bovis normally contains only one copy. In general, the copy number of IS6110 ranges from 0 to 25, depending on the frequency of transposition, which is conditioned largely by the nature of the genomic region at which transposition occurs (22). Differences in the copy number and locations within the genome are responsible for the high degree of IS6110 polymorphism and have predisposed this sequence to be useful as a specific molecular marker for genotyping M. tuberculosis strains (23,24).
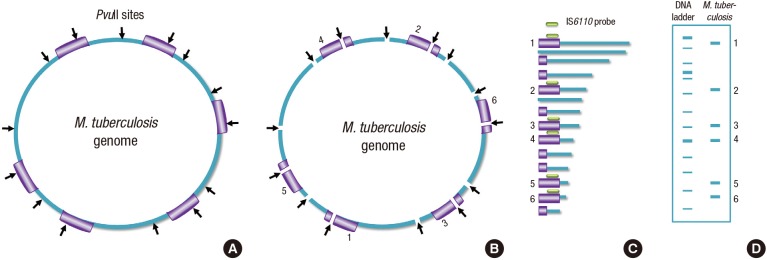
In brief, the methodology includes genomic DNA digestion with the PvuII restriction enzyme that cleaves the IS6110 sequence into fragments, which are separated by gel electrophoresis (Fig. 1C). Then they are transferred to a membrane, and Southern blot hybridization is carried out with a labeled probe complementary to part of the 3′end of the IS6110 sequence. The IS6110 DNA probe is prepared by in vitro amplification of the 245-bp fragment using PCR and labeled by appropriate material. As a result, every visualized fragment represents a single copy of IS6110 surrounded by different lengths of flanking DNA sequences (8). Because the IS6110-RFLP method has been standardized and published, recommendations have been adopted by most research groups; the fingerprints generated in different laboratories can be compared and catalogued (25,26). To analyze the large number of RFLP patterns, a sophisticated pattern-matching computer program has been developed, and RFLP images on film or membrane are scanned and digitized for computer analysis. The sizes of the bands in the image are calculated by comparison with size standards run on the gel. The computer program compares the results from a new isolate with previously analyzed isolates to determine if any matches exist (26,27). A schematic diagram of the RFLP method is shown in Fig. 1.
Fig. 1.
Principles of RFLP. (A) M. tuberculosis genome with insertion segment IS6110 showing the PvuII cleavage sites. (B) Digestion of the whole genome with PvuII. (C) DNA segments of different sizes after run in the gel are transferred onto a membrane followed by hybridization. (D) Visualized fragments, which represent a single copy of IS6110 surrounded by flanking DNA of different lengths.
The IS6110-RFLP method is highly discriminatory and reproducible, and its profiles are stable over time. It allows us to distinguish epidemiologically related isolates from unrelated ones. The half-time of the change of the IS6110-RFLP pattern has been estimated in many studies. The rate at which IS6110-RFLP patterns change is essential to know for the correct interpretation of molecular typing in the epidemiology of TB. An estimated rate of change may support the utility of IS6110-RFLP typing for identifying TB cases associated with recent transmission or long term epidemiological study. The half-time of the change of the IS6110-RFLP pattern has been estimated in many studies. de Boer et al. (28) calculated that the half time was 3.2 year. However, Warren et al. (29) calculated the half time as 8.74 years and also revealed different rate of change in RFLP band pattern depending on the stage of the disease. A high rate of change occurred during the early disease phase (half-time of 0.57 years) and a low rate of change in the late disease phase (half-time of 10.69 years). They also reported that changes were observed in 4% of tested strains.
The main advantages of the IS6110-RFLP method are its high discriminatory power, reproducibility, and the stability of its pattern. The main limitation of the method is the low discriminatory power in isolates presenting five or fewer IS6110 copies. In fact, studies have demonstrated that the frequency of clinically confirmed epidemiologic links between cases was decreased in clusters formed by isolates with five or fewer IS6110 copies (30,31,32). Those strains with low copy numbers, which make low discriminatory power of RFLP, have been frequently isolated from Asian, South East Asian and some African countries ranging from 26%–56% (9,33,34) and to improve discrimination on those strains, additional use of other typing method like MIRU-VNTR or spoligotyping is usually needed (35,36).
Furthermore, IS6110-RFLP has significant technical limitations, including the need for 2–3 μg of high-quality (intact) DNA for restriction enzyme digestion, which means it requires time-consuming bacterial culture. Moreover, the method is technically demanding and requires sophisticated and expensive computer software as well as personnel with great technical expertise.
Despite these limitations, the IS6110-RFLP method remains one of the most commonly used approaches for M. tuberculosis typing, indeed still considered the gold standard technique in molecular epidemiologic investigations of TB.
Spoligotyping
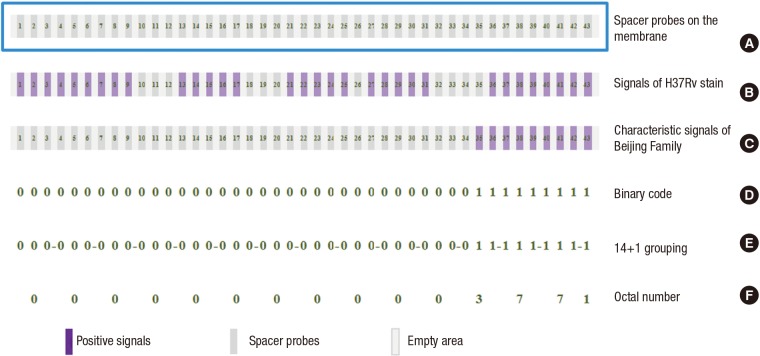
Spoligotyping is a hybridization assay that detects variability in the DR region of the DNA of M. tuberculosis. It is one of the most frequently used PCR-based molecular typing methods. The DR region consists of multiple copies of a conserved 36-bp sequence (the DRs) separated by multiple unique spacer sequences. The entire DR locus is amplified by PCR using primers that are complementary to the sequence of the DRs. The PCR products are hybridized to a membrane with 43 spacer oligonucleotides. Each of the spacers produces either a dark band (indicating the presence of the spacer) or no band (indicating the spacer’s absence). For each M. tuberculosis isolate, the spoligotyping assay produces a series of bands. The pattern is converted to a 43-digit binary code system that has 1 second and 0 seconds (1 means that the band is present and 0 means it is absent). Then the 43-digit binary code is converted to a 15-digit octal numbers. Each octal designation is unique and represents one specific banding pattern. From an octal designation, the binary code of the spoligotyping pattern can be re-created. A simple spreadsheet application can be used to convert binary to octal and octal to binary (4,11,37). Fig. 2 shows the principle of the spoligotyping method.
Fig. 2.
Principle of spoligotyping and the processing of signals. (A) M. tuberculosis genome with well-conserved 36-bp direct repeats (DRs) which are interspersed by 35–43 bp of unique spacer sequences. Genetic diversity depends on the deletion of these spacer regions. The spacer regions are amplified by primers, and the presence of at least one spacer fragment shows a PCR positive reaction. On the membrane, 43 probes targeting each spacer are spotted, and a unique pattern of spoligotyping is visualized after hybridization with PCR product. (B) Signals of reference strain H37Rv. (C) A typical signal pattern of Beijing family M. tuberculosis strain. (D) To analyze signal patterns, the signals are converted to binary code of ‘on (1) and off (0)’. (E) The 43-digit binary code is converted to a 15-digit octal (i.e., base 8, having the digits 0–7) designation by a two-step process. First, the 43-digit binary code is divided into 14 sets of three digits (spacers 1 through 42) plus one additional digit (spacer 43). (F) Each 3-digit binary set is converted to its octal equivalent, with the final additional digit remaining as 1 or 0. The translation of binary numbers to octal numbers is done as follows: 000 = 0; 001 = 1; 010 = 2; 011 = 3; 100 = 4; 101 = 5; 110 = 6; and 111 = 7.
The sensitivity of spoligotyping is estimated to be 10 fg of chromosomal DNA, considerably less than what is needed for RFLP (38). The results can easily be interpreted and compared by different laboratories. An international spoligotyping database, which can be accessed online (http://www.pasteur-guadeloupe.fr.8081), describes 1,939 shared types (STs; i.e., spoligotype patterns shared by two or more isolates) and 3,370 orphan types (i.e., spoligotype patterns reported for only singleisolates) from a total of 39,295 M. tuberculosis complex isolates from 122 countries, classified temporarily into 62 clades/lineages. The SITVIT database was incorporated, and it describes a total of 7,105 spoligotype patterns, corresponding to 58,180 clinical isolates, grouped into 2,740 STs representing 53,816 clinical isolates and 4,364 orphan patterns (39,40).
Many studies reported spoligotyping as a simple, cost-effective, and high-throughput method, although it has a lower discriminatory power than IS6110-RFLP typing. However, spoligotyping can differentiate M. tuberculosis strains with low IS6110 copy numbers (≤ 5 bands in RFLP patterns). Spoligotyping could be performed as a first-line screening test, to be followed by another typing method(s) of greater discriminatory power if necessary (30,41).
MIRU-VNTR typing
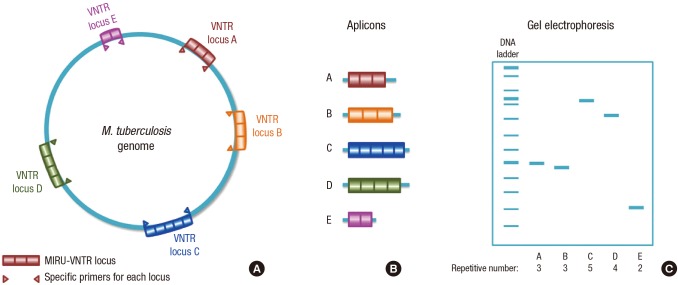
Almost all higher eukaryote genomes possess tandemly repeated sequences that are dispersed by thousands of copies (42). Their repeat numbers are highly variable in many loci and therefore are called “variable number tandem repeat” (VNTR) loci (43,44). Small repetitive DNA sequences with different unique characters were found in M. tuberculosis and other mycobacterial genomes by different scientists (45,46,47,48,49,50). In 1997, Supply et al. (13) identified a novel minisatellite-like structure in the M. tuberculosis genome composed of 40- to 100-bp repetitive sequences and named them “mycobacterial interspersed repetitive units” (MIRU). These are scattered in 41 locations throughout the genome of M. tuberculosis H37Rv. Among those 41 locations, 12 show polymorphisms in copy number of non-related M. tuberculosis isolates (51). These MIRUs are located mainly in intergenic regions and are dispersed throughout the mycobacterial chromosome. They have different characteristics from other repetitive sequences. For example, there are no obvious palindromic sequences; rather, they are direct tandem repeats. Orientation occurs in one direction relative to transcription of the adjacent gene, and they contain small open reading frames (ORFs) (11). In 2001, Supply and colleges proved the usefulness of MIRUs in mycobacterial strain identification for epidemiologic study by developing an automated PCR method with computerized automation of the genotyping. The principle of the typing system is PCR analysis of 12 variable tandem repeat loci with specific primers complementary to the flanking regions followed by gel electrophoresis. The size (bp) of the amplicon reflects the tandem repeat unit and is converted into numerical code to get digital format results in which each digit represents the number of copies at a particular locus (52).
Typing by comparison of these numeric codes makes the method easier to handle a large number of strains. Furthermore, the MIRU-VNTR technique is a reliable genotyping method, as it is 100% reproducible (51,52,53,54), sensitive, and specific for M. tuberculosis complex isolates. At the same time, a website was set up for the analysis of M. tuberculosis MIRU-VNTR genotypes via the Internet (54). Development of this method has been used for the real-time tracing of transmission, comparison of inter-laboratory data, and global database construction (52,55). The way for worldwide epidemiologic surveillance of tuberculosis therefore has been opened.
The MIRU-VNTRs are remarkably stable, and their evolution rate is slightly slower than that of IS6110-RFLP, so that the method is appropriate for long-term epidemiologic analyses (56). The discriminatory power of MIRU-VNTR typing based on 12 loci is slightly less than that of IS6110-RFLP analysis when M. tuberculosis isolates have high copy numbers of IS6110 (36, 54,56) but is more discriminatory than the IS6110-RFLP if isolates have low copy numbers of IS6110 (57). Another drawback of 12 loci MIRU-VNTR is its limited power for discriminating the Beijing family strains of M. tuberculosis (58, 59). In 2006, Supply et al. (60) selected 15 loci (including 6 previously investigated) as the new standard for epidemiologic discrimination of M. tuberculosis as well as 24 loci (including 12 previously investigated) as a high-resolution tool for phylogenetic studies. This newly proposed set of 15 loci of MIRU-VNTR for strain typing has been accepted as highly discriminatory in a population dominated by Beijing family strains (61) and 24 loci as equal in discriminatory power to that of IS6110-RFLP (62, 63). Additional use of hypervariable loci such as VNTRs 3232, 3820, and 4120 has been recommended to be added to standardized loci set for second-line typing if more detailed genotyping is necessary to differentiate Beijing strains (61).
While doing amplification of tandem repeats, PCR failure was found in some loci, which occurred repeatedly even though the tests were performed under different PCR conditions and with different primer sets. This can be assumed to be characteristic of certain strains. It may be attributable to the presence of unexpectedly large numbers of repeat units or deletion of this region (61). In some cases, double alleles are found at a single locus in MIRU-VNTR profiles, and they are considered to be a clonal variant of the same strain. But when double alleles are found at two or more loci of the same isolate, the sample should be considered a mixed infection (64,65).
Identical MIRU-VNTR patterns are considered to be in a cluster. Using the dice coefficient and the un-weighted pair group method with arithmetic averages (UPGMA), dendrograms can be generated. The discriminatory power of strain typing methods is calculated using the method described by Hunter and Gaston (66). Free software named MIRU-VNTRplus is appropriate to analyze the multi-locus variable number tandem repeat analysis (MLVA) and spoligotyping, large sequence polymorphism, and single nucleotide polymorphism data. A weighted combination of these markers has been applicable from the Web since 2010; it is now widely used. By this Web tool, strain similarity search, generating phylogenetic trees and minimum spanning trees, and geographic information mapping all can be done (67). A schematic diagram of the MIRU-VNTR genotyping method is shown in Fig. 3.
Fig. 3.
Principle of MIRU-VNTR genotyping. (A) MIRU-VNTR loci of different repetitive numbers scattered in M. tuberculosis genome are amplify by specific primers for each locus. (B) Different sizes of amplicons after PCR. (C) Amplicons can be seen after gel electrophoresis with different sizes that reflect the repetitive number of each VNTR locus.
Rep-PCR
The rep-PCR is one of the PCR amplification-based methods in which spacer fragments lying between repeat motifs of the genome are amplified using two outwardly directed primers at high stringency (68). The fragments are repetitive extragenic palindromic repetitive elements 38 bp long that consist of six degenerate positions and a 5-bp variable loop between each side of the conserved palindromic stem (69). The genetic relatedness is determined by comparing the banding pattern after gel electrophoresis of the amplicons, differences in the number of repetitive elements, and their relative position within the bacterial genome.
For convenience, a semi-automated rep-PCR commercial system (DiversiLab microbial Genotyping System; bio Merieux, Marcy-l’Etoile, France) has been developed using microfluidic chip-based DNA fragment separation that can overcome the reproducibility problem (70). Analysis can be performed with the Web-based DiversiLab software, which uses the Pearson correlation coefficient and an un-weighted pair group method with arithmetic means to perform automatic comparison of the rep-PCR-based DNA fingerprints of unknown isolates. By this system, a dendrogram, a scatter plot for sample comparison electropheogram, gel-like images, and selectable demographic fields can be generated. Like spoligotyping and MIRU-VNTR typing, rep-PCR is a fast time- and labour-saving method that can generate real-time strain typing results. Its discriminatory power is high, and it can be used for samples that are not typable by RFLP because of low copy number of the target insertion elements (14). Above all, this method has proved to discriminate strains of Beijing family members well, whereas discrimination of members of this family is unsatisfactory using the gold standard RFLP method (71). Therefore, by combination with another amplification method such as MIRU-VNTR, the rep-PCR method can be considered to replace the currently accepted gold standard method, especially in countries where Beijing family strains are prevalent. As its significant advantage is its broad applicability, it is especially useful to laboratories that also work on non-tuberculous mycobacteria (NTM).
WGS
In recent years, WGS has been used for genotyping of M. tuberculosis and is especially useful to examine outbreaks by identifying transmission events where strains are genetically indistinguishable by current methods (15,72,73,74). WGS-based genotyping also offers an optimal resolution of M. tuberculosis complex isolates in molecular epidemiologic studies and can provide additional information (e.g., on drug resistance) (73,75). Bryant et al. (76) performed a randomized controlled trial of tuberculosis treatment using WGS and MIRU typing as molecular tools and reported that WGS enables the differentiation of relapse and re-infection with better resolution than MIRU-VNTR. A recent population-based study using a long-term large-scale WGS approach in a high-prevalence area provided strong evidence for differences in transmission patterns and virulence in various M. tuberculosis lineages (77).
One major obstacle to WGS-based genotyping is the difficulty of data standardization and integration into a readily accessible and expandable database. A number of studies were carried out to improve the standardization of WGS genotyping and the creation of Web-assessable databases for global TB surveillance (78). Bio-informative programs such as CLC Genomics Workbench (www.clcbio.com/products/clc-main-workbench/), and UniproUGENE (http://ugene.unipro.ru/) can be used for WGS data analysis (75). With the decreasing price of WGS, which has a higher discriminatory power and a single rapid analysis for identification, drug resistance prediction, and epidemiologic typing, the method is expected to become a new standard for routine typing of M. tuberculosis in the near future.
Other molecular typing methods
Apart from WGS, the methods described above belong to methods based on repetitive sequences. There are many other methods based on repetitive sequences that are less frequently used for M. tuberculosis typing. One of them is the polymorphic GC-rich repetitive PGRS-RFLP method which has the same principle as IS6110-RFLP with the exception of the use of a different restriction enzyme and a recombinant plasmid pTBN12 for the probe. The pTBN12 fingerprinting method has limitations similar to those of IS6110-RFLP typing, and in addition, the hybridization patterns produced by PGRS typing are more complex and difficult to interpret (79,80). Before development of WGS, the following are the methods based on nonrepetitive sequences also used for TB genotyping. During the 1990s, there were reports that randomly amplified polymorphic DNA (RAPD) analysis, which uses the PCR with arbitrarily designed primers, can provide highly pleomorphic and easily interpretable result profiles. However, its poor reproducibility limits its use for molecular typing (81,82). Pulse-field gel electrophoresis (PFGE) is of limited use because of its requirement for a large amount of DNA and lack of a standard protocol for analysis of M. tuberculosis patterns (83). Amplified fragment length polymorphism (AFLP) can be used as a complement to RFLP because AFLP can differentiate between clusters identified by RFLP (84,85). The conventional radioactive AFLP method has a lower discriminatory potential than IS6110-RFLP typing, but the improved fluorescent AFLP (fAFLP) provides greater discriminatory ability, and these methods are widely used for species differentiation of NTM isolates (86,87). In 1998, Maiden et al. (88), proposed a typing scheme for pathogenic microorganisms called multilocus sequence typing (MLST) which enables electronic portability of nucleotide sequence data for the characterization of microorganisms. MLST method has proved to be valuable in investigations of MTB when seven high discrimination gene loci (rec X, rps L, rmlC, rpmG1, mprA, gcvH, ideR) were chosen (89) and also useful in detecting NTM in laboratory outbreak settings (90). Although single nucleotide polymorphisms (SNPs) represent the most reliable markers for lineage classification of M. tuberculosis complex, their use is hampered by the need to test a large set of genes to achieve satisfactory resolution (2,91).
DISCUSSION
Molecular strain-typing methods have been rapidly advancing toward computer-assisted design and increasing automation, resolution, throughput, and reproducibility. Tools of bioinformatics, such as computer-based programs for genotyping markers, genotyping design and comparison of genotyping data, development of software analysis, and phylogenetic analysis lead to convenience in retrospective studies, inter-laboratory comparison, and long-term epidemiologic surveillance. In this review, we have provided an overview of five molecular typing techniques that are used mostly for the differentiation of M. tuberculosis isolates. We compare the advantages and disadvantages of each method in Table 1. Although we have said that IS6110-RFLP typing is universally accepted as the gold standard for tuberculosis genotyping, one of its drawbacks is insufficient discrimination for those strains whose copy number of IS6110 is 5 or less. For those strains, additional typing method(s) such as spoligotyping or MIRU-VNTR usually is needed to complement the genotype pattern. Moreover, some NTM have multiple copies of sequences homologous to IS6110 and may hybridize with the IS6110 probe (92). Thus, PCR-based typing is more reliable than other genotyping methods.
Table 1. Comparison of frequently used molecular typing methods for M. tuberculosis .
| Method | Advantages | Disadvantages |
|---|---|---|
| RFLP | High discriminatory power | Lower discriminatory power in isolates with ≤ 5 copies |
| Reproducible | Need large amount of high-quality DNA (2–3 µg) | |
| Stable | Laborious and time consuming | |
| Requires sophisticated and expensive computer software and experienced personnel with considerable technical expertise | ||
| Spoligotyping | Simple | Lower discriminatory power than RFLP |
| Need only small amount of chromosomal DNA | Single pattern in Beijing family strains | |
| Cost effective and high throughput | ||
| Results are easy to interpret | ||
| Useful for the differentiation of low IS6110 copy-number strains | ||
| Can be compared between laboratories using online database | ||
| MIRU-VNTR | Simple to perform, fast and labor saving | 12 loci and 15 loci MIRU VNTR has lower discriminatory power than IS6110 RFLP |
| Stable, and evolution rate is slightly slower than that of RFLP | ||
| Need only small amount of DNA | ||
| Easier to handle large number of strains by comparison of numeric codes | ||
| Cost effective and 100% reproducible | ||
| Real-time tracing of spreading strains | ||
| A free web tool is available to compare the strains internationally | ||
| Rep-RCR | Simple to perform, fast and labor saving | Relatively less reproducible |
| Generate real-time strain-typing results | ||
| Analysis can be performed with Web-based software | ||
| Highly discriminatory for Beijing strains | ||
| Broadly applicable to different strains of mycobacteria | ||
| WGS | High discriminatory power | Expensive |
| Precise genetic information can be obtained | Difficulty of data standardization | |
| Do not have readily accessible and expandable database |
RFLP = restriction fragment length polymorphism, MIRU-VNTR = mycobacterial interspersed repetitive unit-variable number of tandem repeat, Rep-PCR = repetitive element palindromic PCR, WGS = whole genome sequencing.
Spoligotyping, for which the discriminatory power is less than that of IS6110-RFLP and MIRU-VNTR (93,94), nevertheless has advantages over IS6110-based genotyping, as it requires only small amounts of DNA so that it can be performed on clinical samples or liquid culture shortly after inoculation. The method is simple, the results can be obtained from a M. tuberculosis culture within one day, and they are expressed in digital format. Because of its rapidity, this method is useful in the clinical setting to detect causative bacteria and to provide epidemiologic information on strain identities (95). In recent years, some researchers have proved that VNTR typing has a discriminatory power equal to that of IS6110-RFLP (96), and the combination of MIRU-VNTR typing and spoligotyping is accepted as an appropriate way to obtain a rapid and reliable genotyping result (97). Automated MIRU typing is now available, which is based on multiplex PCR in which one of each primer set is labeled with a different fluorescent dye. To estimate the PCR size, an automated sequencer is used to electrophorese the fluorescently labeled amplicons (54). This method is highly reproducible and fast, as the results are obtained through a computerized analysis of the signals, although it has the disadvantage of a need for a particular sequencer and a specialized software package (60).
Spread of MDR- and extensively drug resistant (XDR)-TB is endangering the world; real-time tracing of tuberculosis strains therefore is of major importance for control of these super spreaders. This aspect makes PCR-based genotyping a suitable method for tuberculosis surveillance systems and tracing current spreading routes. Moreover, WGS, a recently developed high-cost technique, is expected to become a new standard method for routine typing of M. tuberculosis in the near future because it provides a single rapid analysis that contains identification, drug resistance profiles, selection of locus-specific typing markers, and rational design of genotyping strategies.
In a short term outbreak of tuberculosis, the strains might be derived from one main family like Beijing or Haarlam, and discrimination and tracing of source and the transmission route may be difficult to work out. Studies working on the differentiation of clustered strains from one method with other methods revealed different results and give us a light to choose the typing methods when a short term outbreak occur (75,98). In the study of Kremer et al. (98), 51 isolates (74% of the study samples) were found to be Beijing genotype by spoligotyping with HGDI of 0.253. However, the HGDI of MIRU-VNTR and IS6110 RFLP for those isolates were 0.982 and 0.997 respectively, and discriminating powers were much better than that of spoligotyping. In another study, a large cluster of Haarlem lineage was identified by two classical strain typing methods, IS6110 DNA fingerprinting and 24-loci MIRU-VNTR typing, and clustered strains from those typing results had undergone WGS. The analysis of WGS data showed SNP and small deletions which sub divided 53% of that 86 cluster isolates (75). A study on a 3-year period outbreak in Canada also revealed that WGS could differentiate two genetically distinct lineages of M. tuberculosis with identical MIRU-VNTR genotypes, and suggesting that there were two concomitant outbreaks (15). Those researches not only revealed that WGS has higher discriminatory power than classical genotyping, but also showed better correlation with the spatial distribution of the cases, contact tracing data, and involvement of multiple source cases (15,75).
CONCLUSION
Each technique has its own advantages and disadvantages, and we have to consider these facts when we select, implement, and combine those techniques according to our specific laboratory conditions and the particular features of the geographic region. Currently, molecular typing is widely used by TB control programs because it offers a powerful tool to monitor epidemiologic trends and transmission dynamics of drug-resistant strains. It has been witnessed that simple conventional technologies moved to more advanced ones, such as molecular diagnostics, and now we are facing a brand new technology of ‘WGS’ even though still there are many things to be solved; normalization of the data, mining meaningful data sets from enormous amounts of data, high costs, etc. WGS will soon replace most of the typing methods that are currently being used. However, it will take quite a long time to replace the whole typing settings in the world. Meanwhile, we need to, and will, use current molecular typing methods in combination with WGS. It will be important not only for the transition phase of moving to the new technology, but also important that through such a combination, we will get deeper knowledge that relies on the genetic characters and differences of each M. tuberculosis strain. We support the fact that TB molecular typing may play as an essential role in monitoring of the progress toward eliminating TB transmission.
Footnotes
Funding: This study was supported financially by the Korea International Co-operation Agency (KOICA) (2015 APP Track 1-1).
DISCLOSURE: The authors have no potential conflicts of interest to disclose.
AUTHOR CONTRIBUTION: Prepared first manuscript and drawing figures: Ei PW. Contributed some parts of manuscript writing and revised the first manuscript: Aung WW. Revised the final manuscript: Lee JS. Finalized and revised the figures: Choi GE. Supervised the review outline and proof read the final version of the manuscript: Chang CL. Approval of final manuscript: all authors.
References
- 1.World Health Organization. Global tuberculosis report 2015 [Internet] [accessed on 9 September 2016]. Available at http://www.who.int/tb/publications/global_report/en/
- 2.Gutacker MM, Mathema B, Soini H, Shashkina E, Kreiswirth BN, Graviss EA, Musser JM. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis. 2006;193:121–128. doi: 10.1086/498574. [DOI] [PubMed] [Google Scholar]
- 3.van Soolingen D, Borgdorff MW, de Haas PE, Sebek MM, Veen J, Dessens M, Kremer K, van Embden JD. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J Infect Dis. 1999;180:726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- 4.National Tuberculosis Controllers Association (US); Centers for Disease Control and Prevention Advisory Group on Tuberculosis Genotyping (US) Guide to the Application of Genotyping to Tuberculosis Prevention and Control. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention; 2004. [Google Scholar]
- 5.Jagielski T, Minias A, van Ingen J, Rastogi N, Brzostek A, Żaczek A, Dziadek J. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and other mycobacteria. Clin Microbiol Rev. 2016;29:239–290. doi: 10.1128/CMR.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foxman B, Riley L. Molecular epidemiology: focus on infection. Am J Epidemiol. 2001;153:1135–1141. doi: 10.1093/aje/153.12.1135. [DOI] [PubMed] [Google Scholar]
- 7.Cattamanchi A, Hopewell PC, Gonzalez LC, Osmond DH, Masae Kawamura L, Daley CL, Jasmer RM. A 13-year molecular epidemiological analysis of tuberculosis in San Francisco. Int J Tuberc Lung Dis. 2006;10:297–304. [PubMed] [Google Scholar]
- 8.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park YK, Bai GH, Kim SJ. Restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolated from countries in the Western Pacific region. J Clin Microbiol. 2000;38:191–197. doi: 10.1128/jcm.38.1.191-197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi GE, Jang MH, Song EJ, Jeong SH, Kim JS, Lee WG, Uh Y, Roh KH, Lee HS, Shin JH, et al. IS6110-restriction fragment length polymorphism and spoligotyping analysis of Mycobacterium tuberculosis clinical isolates for investigating epidemiologic distribution in Korea. J Korean Med Sci. 2010;25:1716–1721. doi: 10.3346/jkms.2010.25.12.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yun KW, Song EJ, Choi GE, Hwang IK, Lee EY, Chang CL. Strain typing of Mycobacterium tuberculosis isolates from Korea by mycobacterial interspersed repetitive units-variable number of tandem repeats. Korean J Lab Med. 2009;29:314–319. doi: 10.3343/kjlm.2009.29.4.314. [DOI] [PubMed] [Google Scholar]
- 13.Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 14.Cangelosi GA, Freeman RJ, Lewis KN, Livingston-Rosanoff D, Shah KS, Milan SJ, Goldberg SV. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J Clin Microbiol. 2004;42:2685–2693. doi: 10.1128/JCM.42.6.2685-2693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 16.Schürch AC, van Soolingen D. DNA fingerprinting of Mycobacterium tuberculosis: from phage typing to whole-genome sequencing. Infect Genet Evol. 2012;12:602–609. doi: 10.1016/j.meegid.2011.08.032. [DOI] [PubMed] [Google Scholar]
- 17.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang MH, Choi GE, Chang CL, Kim YD. Characteristics of molecular strain typing of Mycobacterium tuberculosis isolated from Korea. Korean J Clin Microbiol. 2011;14:41–47. [Google Scholar]
- 19.Thierry D, Brisson-Noël A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon JL, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol. 1990;28:2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asgharzadeh M, Shahbabian K, Majidi J, Aghazadeh AM, Amini C, Jahantabi AR, Rafi A. IS6110 restriction fragment length polymorphism typing of Mycobacterium tuberculosis isolates from East Azerbaijan Province of Iran. Mem Inst Oswaldo Cruz. 2006;101:517–521. doi: 10.1590/s0074-02762006000500006. [DOI] [PubMed] [Google Scholar]
- 22.Wall S, Ghanekar K, McFadden J, Dale JW. Context-sensitive transposition of IS6110 in mycobacteria. Microbiology. 1999;145:3169–3176. doi: 10.1099/00221287-145-11-3169. [DOI] [PubMed] [Google Scholar]
- 23.Cave MD, Eisenach KD, McDermott PF, Bates JH, Crawford JT. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 24.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Embden JD, Van Soolingen D, Heersma HF, De Neeling AJ, Jones ME, Steiert M, Grek V, Mooi FR, Verhoef J. Establishment of a European network for the surveillance of Mycobacterium tuberculosis, MRSA and penicillin-resistant pneumococci. J Antimicrob Chemother. 1996;38:905–907. doi: 10.1093/jac/38.5.905. [DOI] [PubMed] [Google Scholar]
- 26.Heersma HF, Kremer K, van Embden JD. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis . Methods Mol Biol. 1998;101:395–422. doi: 10.1385/0-89603-471-2:395. [DOI] [PubMed] [Google Scholar]
- 27.Skuce RA, McCorry TP, McCarroll JF, Roring SM, Scott AN, Brittain D, Hughes SL, Hewinson RG, Neill SD. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology. 2002;148:519–528. doi: 10.1099/00221287-148-2-519. [DOI] [PubMed] [Google Scholar]
- 28.de Boer AS, Borgdorff MW, de Haas PE, Nagelkerke NJ, van Embden JD, van Soolingen D. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J Infect Dis. 1999;180:1238–1244. doi: 10.1086/314979. [DOI] [PubMed] [Google Scholar]
- 29.Warren RM, van der Spuy GD, Richardson M, Beyers N, Borgdorff MW, Behr MA, van Helden PD. Calculation of the stability of the IS6110 banding pattern in patients with persistent Mycobacterium tuberculosis disease. J Clin Microbiol. 2002;40:1705–1708. doi: 10.1128/JCM.40.5.1705-1708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barlow RE, Gascoyne-Binzi DM, Gillespie SH, Dickens A, Qamer S, Hawkey PM. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J Clin Microbiol. 2001;39:2453–2457. doi: 10.1128/JCM.39.7.2453-2457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauer J, Andersen AB, Kremer K, Miörner H. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microbiol. 1999;37:2602–2606. doi: 10.1128/jcm.37.8.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang ZH, Ijaz K, Bates JH, Eisenach KD, Cave MD. Spoligotyping and polymorphic GC-rich repetitive sequence fingerprinting of Mycobacterium tuberculosis strains having few copies of IS6110. J Clin Microbiol. 2000;38:3572–3576. doi: 10.1128/jcm.38.10.3572-3576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S, Paramasivan CN, Lowrie DB, Prabhakar R, Narayanan PR. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, South India. Tuber Lung Dis. 1995;76:550–554. doi: 10.1016/0962-8479(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 34.Yang ZH, Mtoni I, Chonde M, Mwasekaga M, Fuursted K, Askgård DS, Bennedsen J, de Haas PE, van Soolingen D, van Embden JD, et al. DNA fingerprinting and phenotyping of Mycobacterium tuberculosis isolates from human immunodeficiency virus (HIV)-seropositive and HIV-seronegative patients in Tanzania. J Clin Microbiol. 1995;33:1064–1069. doi: 10.1128/jcm.33.5.1064-1069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cronin WA, Golub JE, Magder LS, Baruch NG, Lathan MJ, Mukasa LN, Hooper N, Razeq JH, Mulcahy D, Benjamin WH, et al. Epidemiologic usefulness of spoligotyping for secondary typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J Clin Microbiol. 2001;39:3709–3711. doi: 10.1128/JCM.39.10.3709-3711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J Clin Microbiol. 2002;40:1592–1602. doi: 10.1128/JCM.40.5.1592-1602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jagielski T, van Ingen J, Rastogi N, Dziadek J, Mazur PK, Bielecki J. Current methods in the molecular typing of Mycobacterium tuberculosis and other mycobacteria. Biomed Res Int. 2014;2014:645802. doi: 10.1155/2014/645802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dale JW, Brittain D, Cataldi AA, Cousins D, Crawford JT, Driscoll J, Heersma H, Lillebaek T, Quitugua T, Rastogi N, et al. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int J Tuberc Lung Dis. 2001;5:216–219. [PubMed] [Google Scholar]
- 39.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demay C, Liens B, Burguière T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N. SITVITWEB--a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Goguet de la Salmonière YO, Li HM, Torrea G, Bunschoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis . J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox R, Mirkin SM. Characteristic enrichment of DNA repeats in different genomes. Proc Natl Acad Sci USA. 1997;94:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura Y, Leppert M, O’Connell P, Wolff R, Holm T, Culver M, Martin C, Fujimoto E, Hoff M, Kumlin E, et al. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987;235:1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura Y, Carlson M, Krapcho K, Kanamori M, White R. New approach for isolation of VNTR markers. Am J Hum Genet. 1988;43:854–859. [PMC free article] [PubMed] [Google Scholar]
- 45.Woods SA, Cole ST. A family of dispersed repeats in Mycobacterium leprae . Mol Microbiol. 1990;4:1745–1751. doi: 10.1111/j.1365-2958.1990.tb00552.x. [DOI] [PubMed] [Google Scholar]
- 46.Hermans PW, van Soolingen D, Bik EM, de Haas PE, Dale JW, van Embden JD. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermans PW, van Soolingen D, van Embden JD. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae . J Bacteriol. 1992;174:4157–4165. doi: 10.1128/jb.174.12.4157-4165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Soolingen D, de Haas PE, Hermans PW, Groenen PM, van Embden JD. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis . J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bigi F, Romano MI, Alito A, Cataldi A. Cloning of a novel polymorphic GC-rich repetitive DNA from Mycobacterium bovis . Res Microbiol. 1995;146:341–348. doi: 10.1016/0923-2508(96)81057-6. [DOI] [PubMed] [Google Scholar]
- 50.Philipp WJ, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom BR, Jacobs WR, Jr, Cole ST. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae . Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 52.Han H, Wang F, Xiao Y, Ren Y, Chao Y, Guo A, Ye L. Utility of mycobacterial interspersed repetitive unit typing for differentiating Mycobacterium tuberculosis isolates in Wuhan, China. J Med Microbiol. 2007;56:1219–1223. doi: 10.1099/jmm.0.47005-0. [DOI] [PubMed] [Google Scholar]
- 53.Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans PW, Martín C, Palittapongarnpim P, Plikaytis BB, Riley LW, Yakrus MA, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001;39:3563–3571. doi: 10.1128/JCM.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frothingham R, Meeker-O’Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 56.Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, Tibayrenc M, Locht C, Supply P. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci USA. 2001;98:1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee AS, Tang LL, Lim IH, Bellamy R, Wong SY. Discrimination of single-copy IS6110 DNA fingerprints of Mycobacterium tuberculosis isolates by high-resolution minisatellite-based typing. J Clin Microbiol. 2002;40:657–659. doi: 10.1128/JCM.40.2.657-659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kam KM, Yip CW, Tse LW, Leung KL, Wong KL, Ko WM, Wong WS. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol Lett. 2006;256:258–265. doi: 10.1111/j.1574-6968.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- 59.Surikova OV, Voitech DS, Kuzmicheva G, Tatkov SI, Mokrousov IV, Narvskaya OV, Rot MA, van Soolingen D, Filipenko ML. Efficient differentiation of Mycobacterium tuberculosis strains of the W-Beijing family from Russia using highly polymorphic VNTR loci. Eur J Epidemiol. 2005;20:963–974. doi: 10.1007/s10654-005-3636-5. [DOI] [PubMed] [Google Scholar]
- 60.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis . J Clin Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwamoto T, Yoshida S, Suzuki K, Tomita M, Fujiyama R, Tanaka N, Kawakami Y, Ito M. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol Lett. 2007;270:67–74. doi: 10.1111/j.1574-6968.2007.00658.x. [DOI] [PubMed] [Google Scholar]
- 62.Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, Niemann S, Supply P. Assessment of an optimized mycobacterial interspersed repetitive- unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol. 2007;45:691–697. doi: 10.1128/JCM.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alonso-Rodríguez N, Martínez-Lirola M, Herránz M, Sanchez-Benitez M, Barroso P, INDAL-TB group Bouza E, García de Viedma D. Evaluation of the new advanced 15-loci MIRU-VNTR genotyping tool in Mycobacterium tuberculosis molecular epidemiology studies. BMC Microbiol. 2008;8:34. doi: 10.1186/1471-2180-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shamputa IC, Rigouts L, Eyongeta LA, El Aila NA, van Deun A, Salim AH, Willery E, Locht C, Supply P, Portaels F. Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J Clin Microbiol. 2004;42:5528–5536. doi: 10.1128/JCM.42.12.5528-5536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shamputa IC, Jugheli L, Sadradze N, Willery E, Portaels F, Supply P, Rigouts L. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir Res. 2006;7:99. doi: 10.1186/1465-9921-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326-31. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farber JM. An introduction to the hows and whys of molecular typing. J Food Prot. 1996;59:1091–1101. doi: 10.4315/0362-028X-59.10.1091. [DOI] [PubMed] [Google Scholar]
- 69.Stern MJ, Ames GF, Smith NH, Robinson EC, Higgins CF. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 70.Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, Schrock R, Manry J, Renwick A, Nieto R, et al. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005;43:199–207. doi: 10.1128/JCM.43.1.199-207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jang MH, Choi GE, Shin BM, Lee SH, Kim SR, Chang CL, Kim JM. Comparison of an automated repetitive sequence-based PCR microbial typing system with IS6110-restriction fragment length polymorphism for epidemiologic investigation of clinical Mycobacterium tuberculosis isolates in Korea. Korean J Lab Med. 2011;31:282–284. doi: 10.3343/kjlm.2011.31.4.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niemann S, Köser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA, et al. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One. 2009;4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009;93:105–111. doi: 10.1016/j.ygeno.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 74.Kato-Maeda M, Ho C, Passarelli B, Banaei N, Grinsdale J, Flores L, Anderson J, Murray M, Rose G, Kawamura LM, et al. Use of whole genome sequencing to determine the microevolution of Mycobacterium tuberculosis during an outbreak. PLoS One. 2013;8:e58235. doi: 10.1371/journal.pone.0058235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bryant JM, Schürch AC, van Deutekom H, Harris SR, de Beer JL, de Jager V, Kremer K, van Hijum SA, Siezen RJ, Borgdorff M, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis. 2013;13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guerra-Assunção JA, Houben RM, Crampin AC, Mzembe T, Mallard K, Coll F, Khan P, Banda L, Chiwaya A, Pereira RP, et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis. 2015;211:1154–1163. doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kohl TA, Diel R, Harmsen D, Rothgänger J, Walter KM, Merker M, Weniger T, Niemann S. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol. 2014;52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ross BC, Raios K, Jackson K, Dwyer B. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J Clin Microbiol. 1992;30:942–946. doi: 10.1128/jcm.30.4.942-946.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chaves F, Yang Z, el Hajj H, Alonso M, Burman WJ, Eisenach KD, Dronda F, Bates JH, Cave MD. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis . J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abed Y, Davin-Regli A, Bollet C, De Micco P. Efficient discrimination of Mycobacterium tuberculosis strains by 16S-23S spacer region-based random amplified polymorphic DNA analysis. J Clin Microbiol. 1995;33:1418–1420. doi: 10.1128/jcm.33.5.1418-1420.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glennon M, Smith T. Can random amplified polymorphic DNA analysis of the 16S-23S spacer region of Mycobacterium tuberculosis differentiate between isolates? J Clin Microbiol. 1995;33:3359–3360. doi: 10.1128/jcm.33.12.3359-3360.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh SP, Salamon H, Lahti CJ, Farid-Moyer M, Small PM. Use of pulsed-field gel electrophoresis for molecular epidemiologic and population genetic studies of Mycobacterium tuberculosis . J Clin Microbiol. 1999;37:1927–1931. doi: 10.1128/jcm.37.6.1927-1931.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruiz M, Rodríguez JC, Rodríguez-Valera F, Royo G. Amplified-fragment length polymorphism as a complement to IS6110-based restriction fragment length polymorphism analysis for molecular typing of Mycobacterium tuberculosis . J Clin Microbiol. 2003;41:4820–4822. doi: 10.1128/JCM.41.10.4820-4822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krishnan MY, Radhakrishnan I, Joseph BV, Madhavi Latha GK, Ajay Kumar R, Mundayoor S. Combined use of amplified fragment length polymorphism and IS6110-RFLP in fingerprinting clinical isolates of Mycobacterium tuberculosis from Kerala, South India. BMC Infect Dis. 2007;7:86. doi: 10.1186/1471-2334-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goulding JN, Stanley J, Saunders N, Arnold C. Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis . J Clin Microbiol. 2000;38:1121–1126. doi: 10.1128/jcm.38.3.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sims EJ, Goyal M, Arnold C. Experimental versus in silico fluorescent amplified fragment length polymorphism analysis of Mycobacterium tuberculosis: improved typing with an extended fragment range. J Clin Microbiol. 2002;40:4072–4076. doi: 10.1128/JCM.40.11.4072-4076.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu B, Dong HY, Zhao XQ, Liu ZG, Liu HC, Zhang YY, Jiang Y, Wan KL. A new multilocus sequence analysis scheme for Mycobacterium tuberculosis . Biomed Environ Sci. 2012;25:620–629. doi: 10.3967/0895-3988.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 90.Cooksey RC, Jhung MA, Yakrus MA, Butler WR, Adékambi T, Morlock GP, Williams M, Shams AM, Jensen BJ, Morey RE, et al. Multiphasic approach reveals genetic diversity of environmental and patient isolates of Mycobacterium mucogenicum and Mycobacterium phocaicum associated with an outbreak of bacteremias at a Texas hospital. Appl Environ Microbiol. 2008;74:2480–2487. doi: 10.1128/AEM.02476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baker L, Brown T, Maiden MC, Drobniewski F. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis . Emerg Infect Dis. 2004;10:1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McHugh TD, Newport LE, Gillespie SH. IS6110 homologs are present in multiple copies in mycobacteria other than tuberculosis-causing mycobacteria. J Clin Microbiol. 1997;35:1769–1771. doi: 10.1128/jcm.35.7.1769-1771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349:1149–1156. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 94.Mulenga C, Shamputa IC, Mwakazanga D, Kapata N, Portaels F, Rigouts L. Diversity of Mycobacterium tuberculosis genotypes circulating in Ndola, Zambia. BMC Infect Dis. 2010;10:177. doi: 10.1186/1471-2334-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gori A, Bandera A, Marchetti G, Degli Esposti A, Catozzi L, Nardi GP, Gazzola L, Ferrario G, van Embden JD, van Soolingen D, et al. Spoligotyping and Mycobacterium tuberculosis . Emerg Infect Dis. 2005;11:1242–1248. doi: 10.3201/1108.040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Beer JL, van Ingen J, de Vries G, Erkens C, Sebek M, Mulder A, Sloot R, van den Brandt AM, Enaimi M, Kremer K, et al. Comparative study of IS6110 restriction fragment length polymorphism and variable-number tandem-repeat typing of Mycobacterium tuberculosis isolates in the Netherlands, based on a 5-year nationwide survey. J Clin Microbiol. 2013;51:1193–1198. doi: 10.1128/JCM.03061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jonsson J, Hoffner S, Berggren I, Bruchfeld J, Ghebremichael S, Pennhag A, Groenheit R. Comparison between RFLP and MIRU-VNTR genotyping of Mycobacterium tuberculosis strains isolated in Stockholm 2009 to 2011. PLoS One. 2014;9:e95159. doi: 10.1371/journal.pone.0095159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kremer K, Au BK, Yip PC, Skuce R, Supply P, Kam KM, van Soolingen D. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J Clin Microbiol. 2005;43:314–320. doi: 10.1128/JCM.43.1.314-320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]