Abstract
B cell lymphoma-2 (BCL-2)-related proteins control programmed cell death through a complex network of protein–protein interactions mediated by BCL-2 homology 3 (BH3) domains. Given their roles as dynamic linchpins, the discovery of novel BH3-containing proteins has attracted considerable attention. However, without a clearly defined BH3 signature sequence the BCL-2 family has expanded to include a nebulous group of nonhomologous BH3-only proteins, now justified by an intriguing twist. We present evidence that BH3s from both ordered and disordered proteins represent a new class of short linear motifs (SLiMs) or molecular recognition features (MoRFs) and are diverse in their evolutionary histories. The implied corollaries are that BH3s have a broad phylogenetic distribution and could potentially bind to non-BCL-2-like structural domains with distinct functions.
Keywords: BCL-2 family, BH3, SLiM, MoRF/MoRE, intrinsically disordered proteins, globular domains
Trends
BCL-2 family interactions are mediated by evolutionarily diverse BH3 motifs to regulate apoptosis. Given their key roles, BH3 mimetics are in clinical trials as cancer therapies.
The discovery of novel BH3-only proteins represents a major endeavor in the cell death field. As a result, BH3 motifs are reportedly present in a nebulous conglomerate of different proteins, both structured and intrinsically disordered.
There is no rigorous definition of a BH3 motif. Currently available BH3 signatures are diverse and elusive for predicting new functional BH3-containing proteins.
Redefining the BH3 motif as a new type of short linear motif (SLiM) or molecular recognition feature (MoRF) reconciles many puzzling features of this motif and opens up new avenues for research.
Missing the Forest for the Trees
BCL-2 is the founding member of a group of proteins that regulate apoptotic (programmed) cell death, mitochondrial physiology, and probably other cellular processes 1, 2. This protein group has two main subgroups that share a single sequence motif commonly known as the BH3. The first subgroup comprises a family of homologs related by common ancestry to BCL-2. The second subgroup contains the so-called BH3-only proteins, which are apparently unrelated evolutionarily and structurally to each other and to BCL-2 homologs. Current models of apoptosis regulation exclusively focus on these two subgroups, while ignoring a third, larger collection of even more diverse BH3-containing proteins. The biological impact of this third group and how it integrates into the BCL-2 network is poorly studied, in part because the definition of a BH3 is sketchy. Here we present arguments that BH3s represent a new class of SLiMs. Defining the BH3 in this way relieves constraints on 3D structure requirements and reconciles its many puzzling features. By interpreting the BH3 as a linear sequence motif also provides clues about the emergence of a sophisticated BCL-2-regulated apoptotic pathway in multicellular animals. Rather than seeking to define a subset of true BH3s among the published candidates, this new view broadens the realm of potentially druggable interactions between a BH3 and its target.
The BCL-2 Network: Kite Surfing Over the Mitochondrion
BCL-2-homologous proteins share up to four different BH motifs (BH1–BH4) that are located within a single structural domain, a uniquely folded α-helical bundle. Although many details remain unresolved, BCL-2 homologs are either antiapoptotic (e.g., BCL-2, BCL-xL) or proapoptotic (e.g., BAX, BAK), whereas all eight canonical BH3-only proteins (e.g., BIM, BAD) promote apoptosis after a cell death stimulus. BH3-only proteins have a single region of sequence similarity (see Glossary) with BCL-2, termed the BH3 domain. However, because the BH3 does not fit the criteria for a protein ‘domain’, we refer to the BH3 as a motif rather than a domain. The BH3 motif is present in an α-helical structure but the linear primary sequence signature of the BH3 motif is not conclusively defined 3, 4, 5, 6. The eight canonical BH3-only proteins are further divided into two functional groups, the direct activators and the sensitizers of cell death 7, 8, 9, 10. Despite its critical role in regulating cell death, the BH3 sequence motif currently lacks a unifying definition and is suggested to occur in approximately 40 additional proteins belonging to different protein families, raising the question of whether the BH3 motif is definable 4, 11.
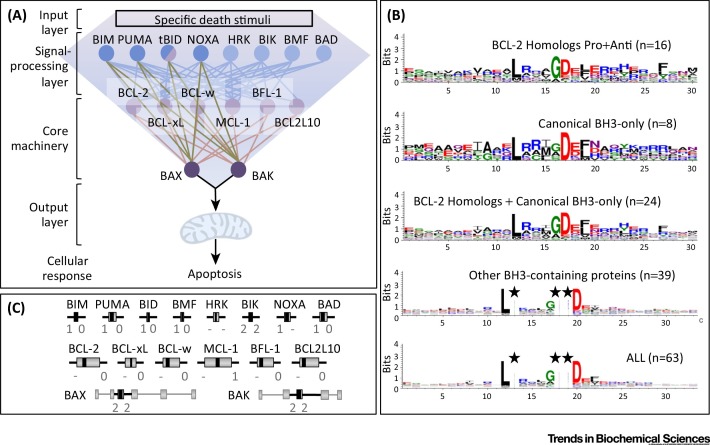
Through a systems-biology lens, the mitochondrial checkpoint of apoptosis comprises a network of highly connected and mutually communicating proteins. In this view, the integration of diverse BH3-only proteins into the BCL-2 network imposes an organized hierarchy with a kite-shaped architecture that is recognizable as a form of bowtie-configured network (Figure 1A). Many robust biological systems share this bowtie (or hourglass) structure in which a central hub (formed here by the BCL-2 homologs) receives and integrates diverse inputs from a signal-processing layer (BH3-only proteins; the kite surfers) to generate one or more outputs (in this case a binary switch: mitochondrial integrity vs mitochondrial permeabilization and death). Consistent with this model, evidence suggests that BH3-only proteins are sentinels that communicate the status of diverse cellular processes to the effector BCL-2-homologous proteins (reviewed in 12, 13). This network structure ensures appropriate responses to multiple signaling pathways and cell stresses to determine cell death.
Figure 1.
B Cell Lymphoma-2 (BCL-2)-Related Proteins, BCL-2 Homology 3 (BH3)-Coding Exons, and BH3 Sequence Motif Logos. (A) Molecular interactions between BH3-only proteins and BCL-2-homologous proteins. BCL-2-related proteins fall into two categories, each with two subcategories: the proapoptotic BH3-only proteins, which are divided into activators (dark blue) and sensitizers (light blue), and the BCL-2-homologous proteins, which can be anti- or proapoptotic (light and dark purple, respectively). BH3-only protein localization and function are regulated at transcriptional, translational, and post-translational levels in response to specific death stimuli (reviewed in [6]). Activator BH3-only proteins can directly activate BAX or BAK by a transient physical interaction leading to insertion into mitochondrial membranes, oligomerization, mitochondrial outer membrane permeabilization (MOMP), and apoptosis. Sensitizer BH3-only proteins induce such phenomena only by docking their BH3 α helix into a hydrophobic groove on the surface of prosurvival BCL-2 homologs, thereby releasing BAX/BAK or activator BH3-only proteins 8, 9, 10. Although it behaves as a direct activator BH3-only protein, the BID protein appears to be a bona fide BCL-2/BAX homolog sharing a similar characteristic 3D fold (hence the two colors: half blue, half purple). All BH3-bearing proteins except BID are natively unfolded proteins (e.g., BIM, BMF) or belong to unrelated protein families having or predicted to have distinct structures. Depicted molecular interactions were extracted from Table S1 in the supplementary material online. (B) Logos resulting from the alignment of BH3 and BH3-like sequences of the various subgroups of BH3-containing proteins. Logo units are bits of information. Each position is displayed as a stack of amino acid letters with the height of the letter representing its proportion of the information content. The conserved Leu and Asp residues are visible in the BH3 core. Sequences are divergent outside this region. Stars indicate alignment gaps. Logos were calculated using Weblogo 3.4 (http://weblogo.threeplusone.com). Aligned sequences are the same as in Figure S1 in the supplementary material online (16 BCL-2-homologous proteins, eight canonical BH3-only proteins, 39 noncanonical BH3-bearing proteins). (C) Phase class of BH3-coding exons. The intron–exon gene structures of BAX and BAK are shown as examples. Exons (grey rectangles) are drawn to scale. The part of the exon encoding the BH3 motif is shown in black. Exons can be classified into nine different groups depending on the phases of their flanking introns: phase 0, reading frame interrupted between codons; phase 1, reading frame interrupted between the first and second nucleotides; phase 2, reading frame interrupted between the second and third nucleotides. See Figure S2 in the supplementary material online for complete intron–exon structures.
Within this bowtie network, the binding interactions of BH3 motifs are the most extensively characterized because of their clinical relevance to diagnostics and therapeutics. The search for small molecules that mimic the biological effects of BH3 motifs enabled the identification of one of the first druggable protein–protein interaction interfaces [14]. The BH3 mimetics ABT-737 and ABT-199 bind in the hydrophobic BH3-binding groove on the surface of antiapoptotic BCL-xL and/or BCL-2 to kill cancer cells [15] (Box 1 ). However, despite having many high-resolution structures of BH3-bound complexes and extensive mutagenesis data to identify critical residues at the interface of both the BH3 and its binding partners 11, 16, 17, 18, the available studies have not exhaustively examined the full range of sequence variability observed in reported BH3 motifs. Without rigorous criteria to accurately predict or exclude candidate BH3-containing proteins, the persistent lumping together of nonhomologous proteins bearing putative BH3-like motifs (e.g., 19, 20, 21) appears to defy logic. However, unforeseen logic may indeed prevail in this madness. Here we argue that BH3 motifs share most of the attributes of SLiMs and, more precisely, of those SLiMs that are associated with binding events (Box 2 ). Thus, BH3 motifs appear to represent a protein–protein interaction module of broader scope that spreads beyond even the extended BCL-2 family and potentially to non-metazoan organisms.
Box 1. BH3 Mimetics Have Therapeutic Value.
The BCL-2-regulated apoptotic pathway is a therapeutic target in degenerative disorders, infectious and immune diseases, and cancer [1]. It was hypothesized over 15 years ago that BH3 mimetics, discovered through rational design or high-throughput screening, would induce cancer cell apoptosis by targeting antiapoptotic members. Consistently, cell-permeable peptides corresponding to BH3 motifs from BID or BAD were shown to induce apoptosis in BCL-2- or BCL-xL-expressing cells 14, 92. Thereafter, synthetic small-molecule inhibitor drugs that bind a hydrophobic pocket on prosurvival BCL-2 proteins (Figure I ) were identified and evaluated for their efficacy as cytotoxic agents used alone or in combination with conventional anticancer drugs. Several such high-affinity antagonists are at various stages of development, the orally bioavailable molecule ABT-199 (which selectively targets BCL-2) being currently in clinical trials with high expectations [93]. Thus, a better appreciation of the complex interplay between the various blocks of BCL-2 proteins successfully identified one of the first α-helix-mediated protein–protein interaction target sites (with p53–hDM2) and demonstrated that affinity-based screening for small molecules mimicking the BH3 motif is a feasible strategy. As SLiMs, BH3 motifs may be found to interact with proteins that do not adopt the BCL-2 structural fold, as recently suggested [91]. Future experiments will have to assess the druggability of such interactions and the potential need to increase the current arsenal of BH3 mimetics.
Box 2. What Are SLiMs?
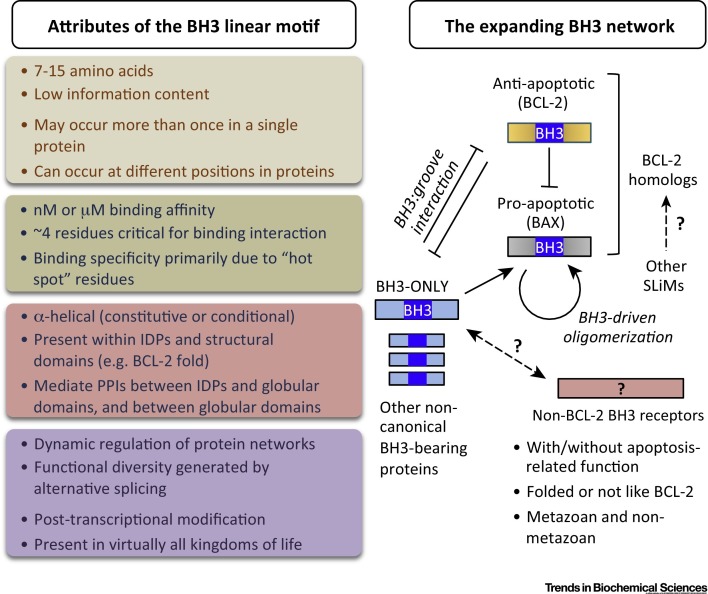
SLiMs/eukaryotic motifs (ELMs) are short, modular functional units of proteins, typically comprising a stretch of 3–11 contiguous amino acid residues with only three to five core positions conferring most of their binding specificity 41, 81. SLiMs serve as post-translational modification sites or ligands for other molecules (protein domains or nucleic acids) 35, 39, 40, 41, 43, 81. Due to their short length, they constitute only a small binding surface area and engage in low-affinity, transient, and modulatable interactions, typically in the low-micromolar range. The best known examples are the binding motifs of typical modular protein–protein interaction domains such as the SH3, SH2, and PDZ domains and they are usually described by a short consensus sequence (also termed a ‘regular expression’; e.g., [P-X-X-P] for the SH3 domain-binding motif). Because the number of residues involved in the activity of motifs is very small, they exhibit high evolutionary plasticity, can be generated by a single point mutation, often arise de novo, and evolve convergently. Therefore, motifs are evolutionarily highly variable and are much less conserved than globular domains in the same sequences. SLiMs tend to occur in intrinsically disordered regions of proteins 42, 43 and, owing to this fact, their distinction from MoRFs is not clear cut. MoRFs are on average longer than SLiMs and are defined on a structural basis (by their ability to fold on interacting with partners) rather than on a sequence basis 44, 45. Due to their potential high density and evolutionary agility, SLiMs are thought to be actively involved in wiring and rewiring the connectivity of the interactome. Established motif instances are deposited in the ELM resource [55], where their major functional classes are ligand binding (LIG), docking (DOC), degradation (DEG), targeting (TAR), cleavage (CLV), and post-translational modification (MOD). BCL-2 homologs have never been described before as proteins able to bind linear motifs, although the attributes of SLiMs clearly suggest that BH3 should be considered as such a motif. Although some amount of sequence variation is observed in the BH1 and BH2 motifs of BCL-2-homologous proteins, these regions are not SLiMs as they constitute the borders of the structural hairpin lying at the center of the BCL-2 helical bundle and form the hydrophobic receptor cleft essential for BH3 binding (rather than binding by themselves to a hydrophobic pocket).
Canonical versus Noncanonical BH3-Containing Proteins
The BH3 motif was originally defined in 1995 as a region of ∼7–15 amino acids with sequence similarity shared by the pro- and antideath BCL-2 homologs BAK, BAX, BCL-2, and BCL-xL plus a shorter, nonhomologous protein, BIK (reviewed in [5]). In the following 5 years, BH3 motifs were identified in additional proteins, eventually settling on the full repertoire of 14 BCL-2 homologs and eight canonical BH3-only proteins in humans (Table S1 in the supplementary material online) comprising four direct activators (BIM, PUMA, truncated tBID, and NOXA) and four sensitizers (BAD, BIK, BMF, and HRK) (Figure 1A). The eight BH3 motifs in these eight proteins were identified only on the basis of visual inspection of their sequences and were confirmed by experimental evidence of proapoptotic activity on binding to BCL-2 homologs. These proteins (plus worm EGL-1) constitute the canonical BH3-only subgroup.
The opportunity to discover new BH3-only proteins is attractive given their roles as linchpins connecting the cellular interactome to the apoptosis checkpoint. However, computational approaches have not validated the BH3 sequence signature 4, 22, 23, 24. Most of the additional cellular and viral proteins claimed to contain functional BH3-like motifs belong to unrelated (non-BCL-2) protein families and are referred to here as ‘noncanonical’ BH3-containing proteins due to limited experimental data relative to the eight canonical BH3-only proteins included in the extended BCL-2 family (Figure S1 and Table S1 in the supplementary material online). The overwhelming majority of these noncanonical BH3-bearing proteins are sensitizer BH3s rather than direct activator BH3s because they trigger cell death by engaging prosurvival BCL-2 family members. Although most noncanonical BH3 motifs are untested for direct BAX/BAK activator function, exceptions have been reported 25, 26, 27.
The Elusive BH3 Motif
The BH3 motif is traditionally defined as an amphipathic α helix that interacts with the hydrophobic cleft of antiapoptotic BCL-2 family proteins. Whereas this structural perspective has been highly useful for understanding interaction specificity 11, 16, 17, 18, the available studies have tested relatively limited numbers of BH3 peptides/proteins compared with the large number of possible sequences at each amino acid position that occurs in natural BH3 motifs. Due to this sequence diversity, a validated BH3 signature sequence is elusive [4]. However, the BH3 is widely denoted as the hexameric sequence L-X(3)-G-D. Despite considerable progress in defining experimentally the key positions within the BH3 that specify its binding partners, computational methods applied to only the obvious BCL-2 homologs have produced a mixture of BH3 motif signatures [4]. Graphical BH3 alignments derived from each of the traditional subgroups of the extended BCL-2 family, the antiapoptotic BCL-2 homologs (eight proteins: six from humans plus worm CED9 and fly Buffy), the proapoptotic homologs (eight human proteins plus fly Debcl), and the canonical BH3-only proteins (eight human proteins plus worm EGL-1), indicate some composition bias in that some amino acids occur more frequently at some positions, but only in the central hexameric region (Figure 1B). If all three of these protein groups are combined, it is possible to derive a significant phylogenetic signal (Figure 1B). However, the signal degenerates to L-x(4,6)-D, which has no predictive value, when the 39 reported noncanonical BH3 motifs are aligned, all of which have at least some functional validation (Figures 1B and S1). Alignment of the grand total of 63 reported BH3s illustrates only what is already generally agreed on: that BH3s share two crucial residues (Leu and Asp) separated by four residues L-x(3)-[GA]-D, suggesting that BH3s lack sequence conservation (Figure S1). Attempts thus far to further define BH3 motifs have yielded signatures that are too strict (i.e., they exclude many BCL-2 homologs) or too inclusive (i.e., they fail to discriminate scrambled proteomes) [4]. Furthermore, the borders of BH3 motifs also have not been defined, and some reports include 20-amino-acid stretches 11, 28.
Difficulties with computational definitions of BH3 motifs pose limitations on large-scale in silico screens, invariably resulting in long lists of likely false positives [4]. Thus, BH3 motifs pose two challenging computational problems of de novo identification: motif finding and functional-site prediction (the so-called Futility Theorem [29]).
What Are the Evolutionary Origins of BH3 Motifs?
The proteins homologous to BCL-2 have a traceable origin and therefore their BH3 most probably evolved from the ancestral proto-BCL-2 protein through divergent evolution. This is supported by our observation that BCL-2-like proteins from species at the base of the metazoan tree such as sponges and cnidarians clearly exhibit a recognizable BH3 motif (four proteins labeled ‘BCL2-like’ in Figure S1), indicating that this motif was already present in BCL-2 proteins from stem metazoans. A similar situation exists for each of the eight canonical BH3-only genes, although these are mainly limited to vertebrates (except Caenorhabditis elegans EGL-1). By contrast, the conservation of BH3 motifs in noncanonical proteins is variable. Although the eukaryotic proteins BECN1/ATG6, ATG12, and BLM-s arose before chordates, their BH3 motif appeared after the emergence of chordates and has been subsequently conserved (Table S1). The controversial yeast BH3 protein is conserved in humans (GRINA/TMBIM3) but the BH3 motif is conserved only between closely related yeast species, and the BH3 motif of mammalian MOAP-1 is present in primates only [27]. Noncanonical BH3-containing genes were also described in several unrelated microbial species: DNA viruses (HHV8, HBV, HCV), RNA viruses (SARS coronavirus and NDV), and a bacterial endosymbiont (Photorhabdus luminescens) (Table S1). Based on amino acid sequence only, it could appear that BH3 motifs arose independently in several evolutionary lines, either randomly or by convergence.
To gain further insight into the evolutionary origins of BH3 motifs, we analyzed the exons that encode BH3 motifs. Information about exon size and the reading frames (phase class) at exon/intron boundaries can be useful in revealing evolutionary relationships between fast-evolving proteins or remote homologs, as gene structures evolve much slower than sequences [30]. Strikingly, all eight canonical BH3-only proteins encode their BH3 on a short exon (between 99 and 276 bp) and five of the eight (BIM, PUMA, BID, BMF, and BAD) share the same exon phase structure. In this case, the BH3-encoding exon starts with the last two nucleotides of the first codon and ends with a complete codon, designated phase [1,0] by convention (Figure 1C and Figure S2 and Table S2 in the supplementary material online). This phase preservation raises the possibility that there is an evolutionary link between these proteins. In addition, EGL1 (canonical BH3-only) and CED13 (noncanonical), which are restricted to nematodes, also have a short BH3-coding exon of the same [1,0] phase. This suggests that, in addition to functional conservation, evolutionary homology between these two molecules may exist, further supported by their 60% sequence identity shared over 21 amino acids in the BH3 region. Furthermore, the genes for BCL-2 homologs belonging to the ‘BID clade’ (i.e., BCL2L12, BCL2L14, BCL2L15, and BID [4]) and several additional noncanonical BH3-containing genes (ATG12, CHMP5, CLU, CUL7, HUWE1, RAD9A) all have a BH3-coding region of the same phase class [1,0] (Figure S2 and Table S2). Intriguingly, ten of these 16 with the same BH3 exon phase class (BIM, PUMA, BID, BAD, BMF, BCL2L15, ATG12, CHMP5, CUL7, and RAD9) also have both 5′ and 3′ flanking exons with identical phases, forming the phase triad [0,1]-[1,0]BH3-[0,0] for three consecutive exons. This topology might reflect the conserved exon/intron structure of a common ancestor or, more likely, the shuffling of a symmetric two- or three-exon ancestral unit.
One other exon-phase type is shared between a different group of BH3-containing proteins. The canonical BH3-only member BIK and the homologs BAX and BAK have a BH3 exon with the phase 2, 2, a very rare exon class [31]. Thus, the BH3 of BAX, BAK, and BIK may not share any evolutionary relationship with the larger group of BH3s with phase [1,0]. Furthermore, the [2,2]BH3 exon of BIK could have been recruited from a BAX/BAK-like progenitor gene, as BIK arose more recently than BAX/BAK [3]. In support of this hypothesis is that the BH3 exon and the following two exons display identical phases in BIK and BAK ([2,2]BH3-[2,0]-[0,−]). Furthermore, there is unusually high resemblance (for a BH3-only protein) between BIK and BAK at the protein-sequence level in a 60-amino-acid region encompassing the BH3 motif (e.g., ∼60% similarity between chicken BIK and murine BAK). Collectively, these observations indicate that BH3 motifs may have evolved from distinct progenitors, potentially involving processes of divergent evolution, random/convergent evolution, and transfer events (e.g., exon shuffling).
The BH3 Motif Goes on a SLiM Diet
Difficulties in defining biologically important protein motifs are not unique to the BH3 motif. The P-X-X-P motif that binds to SH3 domains [32], the endoplasmic reticulum (ER)-retention sequence K-D-E-L (defined by [KRHQSA]-[DENQ]-E-L) [33], and the L-X-X-L-L motif that binds transcription regulators [34] are difficult to accurately predict due to their low information content. These example motifs are short linear sequence signatures that, like BH3s, bind to specific globular proteins. Such motifs have been termed eukaryotic linear motifs (ELMs) [35], miniMotifs [36], SLiMs [37], or simply ‘linear motifs’ [38]. Many proteins contain short linear motifs, referred to here collectively as SLiMs (Box 2). SLiMs serve as either modification sites (e.g., phosphorylation) or protein-binding motifs analogous to BH3s, which bind into pockets on BCL-2 family proteins. Despite the difficulties in defining their sequence signatures, SLiMs appear to be the most abundant type of functional element in the proteome 35, 39, 40, 41. SLiMs overlap with MoRFs 42, 43, a type of recognition element that originated from research in the intrinsic disorder field (Box 2) 44, 45. Although not previously proposed, BH3 motifs of both homologous and nonhomologous proteins exhibit multiple attributes of SLiMs/MoRFs, potentially expanding the concept of a BH3 (Figure 2 , Key Figure).
Figure 2.
Key Figure: Attributes and Interactome of the B Cell Lymphoma-2 (BCL-2) Homology 3 (BH3) Linear Motif
Some or all BH3 motifs have the attributes of short linear motifs (SLiMs) associated with protein–protein binding events (left). However, BH3 is an unusual SLiM type as it is found in ordered proteins as well as intrinsically disordered proteins (IDPs) and achieves high-affinity binding to globular BCL-2-homologous proteins. BH3-mediated protein–protein interactions (PPIs) form an intricate network that controls the process of apoptotic cell death in metazoan organisms (right). Both pro- and antiapoptotic BCL-2 homologs have a globular domain structure, share common ancestry, and constitute binding sites (receptors) for BH3 motifs, which act as ligands to either activate the executioner proteins BAX/BAK or inhibit the antiapoptotic BCL-2 proteins. Both pro- and antiapoptotic BCL-2 homologs can receive a BH3 from BH3-only proteins and can also receive the BH3 from BAX/BAK (e.g., BAX–BAX or BAX–BCL-2 interactions). Redefining the BH3 cell death domain as a SLiM/molecular recognition feature (MoRF) leads to several testable predictions, including the possible existence of: (i) BH3-mediated protein–protein interactions in non-metazoan organisms (including prokaryotes); and (ii) non-BCL-2-like receptors for BH3 motifs. Because globular domains often bind to multiple linear motif classes, the question of whether BCL-2-homologous proteins can interact with other types of SLiMs, in addition to the BH3, is also open. The color scheme used to group the various attributes of the BH3 SLiM is as follows: grey, sequence features; green, binding features; pink, structural features; violet, other features.
Motif Size and Positioning
SLiMs generally are less than 10–12 residues with three to five hot-spot residues critical for binding their targets, consistent with the number of critical interacting residues observed in high-resolution structures of BH3s bound to their globular partners 9, 28. Also like BH3s, well-established SLiMs are degenerate, with variable sequences at most of their amino acid positions, and thus can occur by random chance at high frequencies, sometimes several times in a single protein. Although not a general rule, the mouse BH3-only protein Noxa has two BH3 motifs and BID is reported to have a second BH3-like motif termed BH3-B [46]. Like SLiMs, BH3 motifs are found in small (human NOXA, 54 amino acids) and large (HUWE1, 4374 amino acids) proteins and can be positioned near to or far from the N terminus (e.g., BLID/BRCC2 and mouse Pxt1 vs MCL-1, CHMP5, and HEPB2). Hence, BH3 motifs appear to resemble SLiMs in terms of overall sequence characteristics and positioning.
Evolutionary Plasticity and Taxonomic Distribution
Placed in an evolutionary context, the BH3 motif is plastic because it can arise with very few amino acid substitutions during random or convergent evolution. Thus, distinctions between bona fide BH3 SLiMs and false positives cannot be deduced from sequence alone due to sequence degeneracy [4]. Accurate assignments depend on additional contextual information including primary sequence features, secondary structure (or intrinsic disorder), and validation through some orthogonal (functional, evolutionary) approach. The BH3s of the understudied BCL-2 homologs BCL2A1 and BCL2L10/BCL-B display conserved secondary structure rather than sequence. More challenging is the distinction between BH3s that function in nature versus those that function in experimental settings only. The compiled list of all reported BH3-containing proteins (Figure S1 and Table S1) also contains invertebrates (e.g., C. elegans EGL-1), bacteria [e.g., makes caterpillars floppy (MCF) toxins], viruses (e.g., matrix protein of Newcastle disease virus), and fungi (e.g., YBH3/Ynl305c). While not all reported BH3 motifs may prove to be functional in nature, the existence of seemingly rogue BH3s could suggest that BH3 motifs encompass broad taxonomic diversity and, at least for yeast and bacteria, implies as-yet-unidentified non-BCL-2-binding functions because BCL-2-homologous proteins are restricted to metazoan organisms [3] (Figure 2).
Interaction with Globular Domains
SLiMs are prominently involved in protein–protein interactions where a globular domain serves as a receptor for the SLiM ligand 40, 41 (Box 2). For example, P-X-X-P motifs bind to beta-barrel SH3 domains (PDB code 1SHG), the L-X-X-L-L motif in transcription factor STAT6 binds the PAS-B domain of nuclear coactivator NCoA-1/SRC-1, and the ER-retention motif KDEL found in many proteins apparently binds to the seven-transmembrane domain receptor ERD2. These ligand–receptor interactions are frequently transient and reversible [39] and thus well suited to mediating dynamic processes such as signaling and subcellular targeting. Each type of SLiM typically binds to the members of a single protein family, but due to the loose consensus sequence of SLiMs, specific SLiM-binding proteins usually bind different SLiM motif variations. For instance, SLiMs of class I (X[ST]Xϕ-COOH), class II (XϕXϕ-COOH), or class III (X[ED]Xϕ-COOH) (where X is any residue and ϕ is hydrophobic) bind PDZ domains, which are small globular domains [47]. Binding affinity is specified by the residues of both binding partners, especially SLiM residues flanking the hot-spot residues. Also like BH3s, several SLiMs including NSCaTE [48] and PDZ-binding motifs [49] have an intrinsic propensity for α-helix formation and thus are also designated α-MoRFs 44, 45. The special type of protein–protein interaction mediated by a SLiM and a hydrophobic binding cleft can be druggable [50], as exemplified by the design of small-molecule BH3 mimetics that inactivate antiapoptotic BCL-2 proteins to treat cancer [15] (Box 1).
Analogous to many SLiMs, BH3 motifs bind in the hydrophobic groove of particular BCL-2-homologous proteins. In the quest for clinically useful tools, extensive NMR, surface plasmon resonance, optical spectroscopy, and isothermal titration calorimetry studies have defined the overlapping binding selectivities of peptides derived from BH3-only proteins [15] (see Figure I in Box 1). Because regions adjacent to the BH3 motif influence the mode of binding to BCL-2-homologous proteins, some canonical BH3-only proteins are more promiscuous than others or bind to a defined subset of BCL-2 homologs (giving rise to the sensitizer vs activator subclasses). The combinatorial diversity of BH3 sequences from BH3-only proteins, particularly in the regions flanking the conserved hot-spot residues L-x(3)-G-D and the pairing residues lining the hydrophobic grooves of BCL-2-homologous partners explain the great selectivity observed in these interactions (Figure 1A and Table S1).
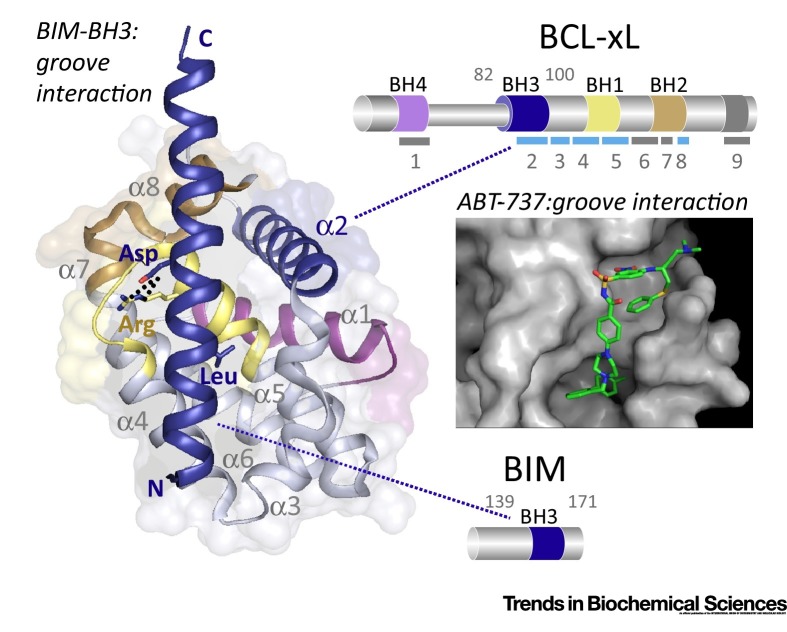
Figure I.
Structure of the Antiapoptotic Protein B Cell Lymphoma (BCL)-xL in Complex with a 33-mer BIM BCL-2 Homology 3 (BH3) Peptide. The ribbon structure (PDB code 1PQ1[94]) shows that BCL-xL is an all-α-helical protein with a central hydrophobic core surrounded by amphipathic helices. This helical bundle arrangement is typical of anti- and proapoptotic BCL-2-homologous proteins. The BIM BH3 peptide (in blue) fits into an extended hydrophobic groove at the surface of BCL-xL. Various interactions (hydrophobic contacts, salt bridges, hydrogen bonds) are formed between BIM-BH3 residues and residues lining the BCL-xL groove. An aspartate residue (Asp) found in all BH3 motifs forms a salt bridge with a conserved arginine (Arg) residue present in the BH1 motif of BCL-xL (broken lines), and an invariant leucine (Leu) in the BIM-BH3 participates in hydrophobic contacts with the BH3-binding groove on the BCL-xL. The BH3 motif of BCL-xL is also colored blue. Diagrams of the two interacting proteins (right side) indicate the amino acid positions of BH3-containing helices and the positions of the nine BCL-xL α helices (numbered bars, light blue bars indicate helices that form the hydrophobic BH3-binding groove). The four BH motifs are shown in magenta (BH4), blue (BH3), yellow (BH1), and brown (BH2) and the C-terminal transmembrane domain (TM) region in gray. BH motifs are depicted using the same color code in the structure and in the motif architecture diagrams. Inset: Crystal structure of Bcl-xL in complex with ABT-737 (PDB code 2YXJ[95]). The figure was prepared with the program PyMOL.
Post-transcriptional and Post-translational Events
SLiMs are often encoded on exons that are alternatively spliced 41, 51, 52, 53. At least for BAX, the BH3 exon is omitted from some splice variants [54]. By contrast, the more conserved BH1 and BH2 motifs of BCL-2 proteins are disrupted by alternative splicing of BCL-2, BCL-X, and MCL1, and this splicing event can generate proapoptotic ‘BH3-only’ splice variants (MCL-1S, BCL-xS). In addition, Ensembl predictions suggest that BH3-containing exons in approximately half of the noncanonical BH3-bearing proteins are candidates for alternative splicing events to either add or remove BH3 motifs (Table S1). SLiMs are reported to be post-translationally regulated, including by phosphorylation 41, 51, 52, 55. Intriguingly, BH3 and BH3-like regions show an abundance of Ser/Thr residues (Figure S1), suggesting that their phosphorylation states might influence the binding of BH3 linear motifs to BCL-2-homologous proteins, as previously reported 56, 57. Therefore, BH3 motifs bear numerous striking similarities with SLiMs, although the next section shows that this is not entirely spot on.
Special Features of the BH3 SLiMs
BH3 Motifs Are Embedded in Both Structured and Intrinsically Disordered Proteins (IDPs)
Despite their similarities, several features of BH3 motifs suggest that they constitute a new type of SLiM. SLiMs are preferentially found in IDPs or IDP regions (IDRs) 39, 42, whereas BH3s are found in both disordered and structured proteins. Canonical BH3-only proteins lack native structure (BIM, BAD, BMF, HRK, NOXA, PUMA, and worm EGL-1) 58, 59, 60, 61, 62 and the helical folding of their BH3s is coupled with binding to their globular partners 58, 60, 61, analogous to α-MoRFs 44, 45, 63, 64. However, BH3 motifs also occur in the well-folded α-helical bundle domain (outside the unstructured loops) of BCL-2-like, BAX-like, and BID-like proteins 16, 65, 66, 67, 68 as well as in other ordered proteins like the helical protein VIRF1 and the α/β proteins ATG12, RAD9A, and TGM2. In addition, numerous BH3 motifs lie in proteins that are predicted to adopt a tertiary structure (Table S1) (see also [11]). Therefore, in addition to specifying binding partners, inherently structured BH3 SLiMs also have a role in maintaining globular domain structure integrity (and stability [18]). Reciprocally, the hydrophobic cores of structured proteins may facilitate the formation or stabilization of helical structures in BH3 regions, similar to MoRFs or so-called chameleons that change conformation with their surroundings.
BH3 Motifs Escape From Computational SLiM Prediction
This peculiarity that some BH3 SLiMs are embedded in globular proteins explains why BH3s are automatically discarded by SLiM-detection algorithms 38, 55, 69, 70, 71, 72. Similarly, methods for computational identification of SLiMs/MoRFs from structural data exist (e.g., by screening of PDB complexes [73]) but fail in recognizing the BH3 motif because no 3D structures for full-length BH3-only proteins in complex with BCL-2 homologous proteins are available to date. BH3 sequences are also likely to be downgraded by evolutionary filtering approaches for detecting remotely similar motifs 71, 74, 75 because some BH3 instances appear to be conserved among BCL-2-homologous proteins. Conversely, BH3 motifs from canonical and noncanonical BH3-only proteins are overlooked by methods that rely on evolutionary conservation scores to derive sequence signatures (e.g., QuasimotifFinder [76]).
Some BH3 Motifs Are Conditionally Exposed at the Protein Surface
A distinctive trait of the BH3s of globular BCL-2 proteins is their ability to be conditionally exposed on protein surfaces, while SLiMs commonly appear on protein surfaces and exposed unstructured loops [77]. That is, the hydrophobic binding face of the BH3 in BAX and BAK is buried inside the core (in solution) 78, 79, 80. Thus, BH3 motifs buried within structured domains must be exposed through activating events such as conformational changes on membrane association and proteolytic cleavage.
Most BH3 Motifs Have a High Affinity for Their Binding Partners
SLiMs are reported to bind their partners with low affinity (>1 μM) [81]. Although low-affinity interactions have sometimes been measured between prosurvival BCL-2 proteins and BAX BH3 peptides 82, 83, most of the BH3 peptides assessed through biophysical and biochemical assays bind prosurvival BCL-2 homologs with relatively high affinity (nanomolar range K D 84, 85, 86). Factors that increase binding affinity are helical content [14], length of BH3 peptides [87], and, potentially, residues distant from the BH3 core region (i.e., long-range contacts) of BH3-only or BH3-bearing proteins.
Although distinct in some aspects, the BH3 motif fits the definition and essential features of a SLiM involved in binding. Furthermore, the subset of BH3 motifs from IDPs can also be considered MoRFs that change shape on binding [18]. This novel perspective has the major advantage of reconciling the structural versus sequence definitions of BH3 motifs, taking into account their full diversity.
Concluding Remarks
We introduce the concept that the intensely studied, cell death-inducing BH3 motif is a newly recognized SLiM/MoRF that specifies protein-binding interactions with or between globular proteins. The SLiM/MoRF concept allows the field to avoid the easy and comfortable option of relegating most (if not all) of the noncanonical BH3-containing proteins to the rank of ‘false positives’. Validated BH3 motifs are found in IDPs but are also found in structured domains (e.g., globular domains of the BCL-2 type), which is likely to delay the recognition of BH3s, and potentially other motifs, as SLiMs or MoRFs. Unlike most SLiMs, some BH3 motifs are only conditionally available for protein–protein interactions, constituting a distinct class of SLiMs that control the mitochondrial checkpoint of apoptosis. The evidence suggests that some BH3 motifs of both BCL-2-homologous and nonhomologous proteins share common ancestry, supported by conservation at the genomic level despite amino acid sequence divergence.
It is unknown whether a BCL-2 family protein can bind to IDPs other than BH3-only proteins. The mainly disordered N-terminal domain of p53 was recently shown to interact with BCL-xL but the surfaces of interaction appear to involve elements of secondary structure on both proteins [88]. It is also unknown whether a BCL-2 family protein can interact with other types of SLiMs or MoRFs in addition to BH3. However, MCL-1 has been reported to interact with proteins sharing a sequence motif distinct from the BH3 in p53 (albeit with the BH3 signature in reverse orientation) 89, 90. Viewing BH3s as SLiMs also raises the possibility that a BH3 motif could interact with globular domains devoid of any sequence and structural similarity to the BCL-2 protein, such as the binding of BAD BH3 to glucokinase [91]. We further hypothesize that non-metazoan species will be found to encode BH3-docking receptors (BCL-2 equivalents) not necessarily sharing a globular 3D structure resembling BCL-2 (see Outstanding Questions and Figure 2). If confirmed, this would suggest that the proapoptotic BH3 motif is also a versatile and evolutionary plastic module associated with binding events in various branches of the tree of life.
Outstanding Questions.
Can we distinguish the functional versus structural definitions of the BH3 motif? Screens for new BH3-containing proteins have generally focused on candidate proteins associated with systems or conditions in which BCL-2-regulated cell death is involved. However, it is possible that BH3 motifs, which are often referred to as α helices, may represent protein–protein interaction modules mediating binding between molecules that have functions related or not related to apoptosis. This idea is consistent with the nonapoptotic roles of BCL-2 proteins that are mediated through the binding of the BH3 of Beclin 1 to regulate autophagy.
Is the BH3 motif the first of many SLiMs with similar features in the proteome universe? Future research is needed to evaluate the proportion of SLiMs that are embedded in both structured proteins and IDPs and that, like BH3 motifs, can achieve high-affinity binding to their receptors.
Do BH3 motifs interact with non-BCL-2 types of globular domain? BH3 motifs may have a broad phylogenetic distribution and could be involved in interactions with structural domains that do not fold like the BCL-2 proteins and that exert functions different from apoptosis regulation. Identification of non-BCL-2-like BH3 receptors across the tree of life offers a new terra incognita for research.
Do BCL-2-homologous proteins interact with other types of SLiM? Because globular domains often bind to multiple linear motif classes, the question of whether BCL-2-homologous proteins can interact with other types of SLiM or MoRF in addition to the BH3 awaits rigorous analysis but is consistent with the finding that the hydrophobic tail of BCL-2 proteins can also occupy the BH3-binding groove.
Acknowledgments
Financial support by the French National Cancer Institute (INCa), the Ligue Nationale Contre le Cancer (A.A.), Odysseus grant G.0029.12 from Research Foundation Flanders (FWO) (to P.T.), and National Institutes of Health grants RO1 NS083372, RO1 NS037402, and RO1 GM077875 (J.M.H.) is gratefully acknowledged.
Glossary
- Futility Theorem
this theorem indicates that searching for bona fide motifs with signatures having weak information content will yield many more random matches than functional hits, making large-scale screens futile.
- Hot-spot residues
refers to those amino acids involved at the interface of protein–protein complexes and that contribute significantly to the binding free energy between the two partners.
- Protein domain
a compact and autonomous folding unit of protein structure, such as the typical ‘helix-bundled’ BCL-2 fold.
- Sequence motif
a conserved amino acid stretch, like the BH1–BH4 regions in BCL-2 family members that distinguish them from other proteins.
- Similarity
the degree of resemblance (the fraction of identical or chemically similar amino acids) between amino acid sequences, which is usually explained through one or more of the following options: shared ancestry (homology), random coincidence (drift), convergence, and parallelism (the three latter options being usually subsumed under the term homoplasy). Although high sequence similarity often portends structural similarity, proteins with low sequence identity and significant structural similarity can occur (e.g., BCL-xL and BID, BCL-xL and diphtheria toxin translocation domain).
Footnotes
Supplementary information associated with this article can be found online http://dx.doi.org/10.1016/j.tibs.2015.09.007.
Supplementary Information
References
- 1.Czabotar P.E. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 2.Hardwick J.M., Soane L. Multiple functions of BCL-2 family proteins. Cold Spring Harb. Perspect. Biol. 2013;5:a008722. doi: 10.1101/cshperspect.a008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aouacheria A. Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-only, and BNip families of apoptotic regulators. Mol. Biol. Evol. 2005;22:2395–2416. doi: 10.1093/molbev/msi234. [DOI] [PubMed] [Google Scholar]
- 4.Aouacheria A. Evolution of Bcl-2 homology motifs: homology versus homoplasy. Trends Cell Biol. 2013;23:103–111. doi: 10.1016/j.tcb.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kvansakul M., Hinds M.G. The structural biology of BH3-only proteins. Methods Enzymol. 2014;544:49–74. doi: 10.1016/B978-0-12-417158-9.00003-0. [DOI] [PubMed] [Google Scholar]
- 6.Shamas-Din A. BH3-only proteins: orchestrators of apoptosis. Biochim. Biophys. Acta. 2011;1813:508–520. doi: 10.1016/j.bbamcr.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Letai A. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 8.Kim H. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 9.Moldoveanu T. BID-induced structural changes in BAK promote apoptosis. Nat. Struct. Mol. Biol. 2013;20:589–597. doi: 10.1038/nsmb.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llambi F. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBartolo J. Genome-wide prediction and validation of peptides that bind human prosurvival Bcl-2 proteins. PLoS Comput. Biol. 2014;10:e1003693. doi: 10.1371/journal.pcbi.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moldoveanu T. Many players in BCL-2 family affairs. Trends Biochem. Sci. 2014;39:101–111. doi: 10.1016/j.tibs.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westphal D. Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ. 2014;21:196–205. doi: 10.1038/cdd.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petros A.M. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davids M.S., Letai A. Targeting the B-cell lymphoma/leukemia 2 family in cancer. J. Clin. Oncol. 2012;30:3127–3135. doi: 10.1200/JCO.2011.37.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czabotar P.E. Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis. J. Biol. Chem. 2011;286:7123–7131. doi: 10.1074/jbc.M110.161281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeBartolo J. Predictive Bcl-2 family binding models rooted in experiment or structure. J. Mol. Biol. 2012;422:124–144. doi: 10.1016/j.jmb.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee E.F. The functional differences between pro-survival and pro-apoptotic B cell lymphoma 2 (Bcl-2) proteins depend on structural differences in their Bcl-2 homology 3 (BH3) domains. J. Biol. Chem. 2014;289:36001–36017. doi: 10.1074/jbc.M114.610758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunelle J.K., Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tait S.W., Green D.R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 21.Youle R.J., Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 22.Hawley R.G. An integrated bioinformatics and computational biology approach identifies new BH3-only protein candidates. Open Biol. J. 2012;5:6–16. doi: 10.2174/1874196701205010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaczmarek K. Overexpression of peroxisomal testis-specific 1 protein induces germ cell apoptosis and leads to infertility in male mice. Mol. Biol. Cell. 2011;22:1766–1779. doi: 10.1091/mbc.E09-12-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D.H. Interaction of a putative BH3 domain of clusterin with anti-apoptotic Bcl-2 family proteins as revealed by NMR spectroscopy. Biochem. Biophys. Res. Commun. 2011;408:541–547. doi: 10.1016/j.bbrc.2011.04.054. [DOI] [PubMed] [Google Scholar]
- 25.Molouki A. The matrix (M) protein of Newcastle disease virus binds to human bax through its BH3 domain. Virol. J. 2011;8:385. doi: 10.1186/1743-422X-8-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodolfo C. Tissue transglutaminase is a multifunctional BH3-only protein. J. Biol. Chem. 2004;279:54783–54792. doi: 10.1074/jbc.M410938200. [DOI] [PubMed] [Google Scholar]
- 27.Tan K.O. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J. Biol. Chem. 2001;276:2802–2807. doi: 10.1074/jbc.M008955200. [DOI] [PubMed] [Google Scholar]
- 28.Czabotar P.E. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Wasserman W.W., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 30.Schad E. Exon-phase symmetry and intrinsic structural disorder promote modular evolution in the human genome. Nucleic Acids Res. 2013;41:4409–4422. doi: 10.1093/nar/gkt110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedorov A. Influence of exon duplication on intron and exon phase distribution. J. Mol. Evol. 1998;46:263–271. doi: 10.1007/pl00006302. [DOI] [PubMed] [Google Scholar]
- 32.Ren R. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 33.Semenza J.C. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- 34.Heery D.M. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 35.Puntervoll P. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balla S. Minimotif Miner: a tool for investigating protein function. Nat. Methods. 2006;3:175–177. doi: 10.1038/nmeth856. [DOI] [PubMed] [Google Scholar]
- 37.Linding R. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neduva V. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davey N.E. Attributes of short linear motifs. Mol. Biosyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 40.Tompa P. A million peptide motifs for the molecular biologist. Mol. Cell. 2014;55:161–169. doi: 10.1016/j.molcel.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 41.Van Roey K. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 2014;114:6733–6778. doi: 10.1021/cr400585q. [DOI] [PubMed] [Google Scholar]
- 42.Fuxreiter M. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 43.Weatheritt R.J., Gibson T.J. Linear motifs: lost in (pre)translation. Trends Biochem. Sci. 2012;37:333–341. doi: 10.1016/j.tibs.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Cumberworth A. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. Biochem. J. 2013;454:361–369. doi: 10.1042/BJ20130545. [DOI] [PubMed] [Google Scholar]
- 45.Mohan A. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 46.Tan K.O. A novel BH3-like domain in BID is required for intramolecular interaction and autoinhibition of pro-apoptotic activity. J. Biol. Chem. 1999;274:23687–23690. doi: 10.1074/jbc.274.34.23687. [DOI] [PubMed] [Google Scholar]
- 47.Tonikian R. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taiakina V. The calmodulin-binding, short linear motif, NSCaTE is conserved in L-type channel ancestors of vertebrate Cav1.2 and Cav1.3 channels. PLoS ONE. 2013;8:e61765. doi: 10.1371/journal.pone.0061765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popovic M. Flexibility of the PDZ-binding motif in the micelle-bound form of Jagged-1 cytoplasmic tail. Biochim. Biophys. Acta. 2012;1818:1706–1716. doi: 10.1016/j.bbamem.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 50.Cheng Y. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Akiva E. A dynamic view of domain–motif interactions. PLoS Comput. Biol. 2012;8:e1002341. doi: 10.1371/journal.pcbi.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Roey K. The switches.ELM resource: a compendium of conditional regulatory interaction interfaces. Sci. Signal. 2013;6:rs7. doi: 10.1126/scisignal.2003345. [DOI] [PubMed] [Google Scholar]
- 53.Weatheritt R.J. Linear motifs confer functional diversity onto splice variants. Nucleic Acids Res. 2012;40:7123–7131. doi: 10.1093/nar/gks442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maia S. Gene expression profiling identifies BAX-δ as a novel tumor antigen in acute lymphoblastic leukemia. Cancer Res. 2005;65:10050–10058. doi: 10.1158/0008-5472.CAN-05-1574. [DOI] [PubMed] [Google Scholar]
- 55.Dinkel H. ELM – the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datta S.R. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 57.Zalckvar E. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrera-Vilarmau S. Intrinsic order and disorder in the bcl-2 member harakiri: insights into its proapoptotic activity. PLoS ONE. 2011;6:e21413. doi: 10.1371/journal.pone.0021413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Craxton A. NOXA, a sensor of proteasome integrity, is degraded by 26S proteasomes by an ubiquitin-independent pathway that is blocked by MCL-1. Cell Death Differ. 2012;19:1424–1434. doi: 10.1038/cdd.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinds M.G. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–136. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- 61.Rogers J.M. Folding and binding of an intrinsically disordered protein: fast, but not ‘diffusion-limited’. J. Am. Chem. Soc. 2013;135:1415–1422. doi: 10.1021/ja309527h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan N. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol. Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 63.Hsu W.L. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22:258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Lee R. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petros A.M. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 66.Dewson G. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 2012;19:661–670. doi: 10.1038/cdd.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gahl R.F. Conformational rearrangements in the pro-apoptotic protein, Bax, as it inserts into mitochondria: a cellular death switch. J. Biol. Chem. 2014;289:32871–32882. doi: 10.1074/jbc.M114.593897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding J. After embedding in membranes antiapoptotic Bcl-XL protein binds both Bcl-2 homology region 3 and helix 1 of proapoptotic Bax protein to inhibit apoptotic mitochondrial permeabilization. J. Biol. Chem. 2014;289:11873–11896. doi: 10.1074/jbc.M114.552562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davey N.E. SLiMSearch 2.0: biological context for short linear motifs in proteins. Nucleic Acids Res. 2011;39:W56–W60. doi: 10.1093/nar/gkr402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edwards R.J. Interactome-wide prediction of short, disordered protein interaction motifs in humans. Mol. Biosyst. 2012;8:282–295. doi: 10.1039/c1mb05212h. [DOI] [PubMed] [Google Scholar]
- 71.Neduva V., Russell R.B. DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res. 2006;34:W350–W355. doi: 10.1093/nar/gkl159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen Ba A.N. Proteome-wide discovery of evolutionary conserved sequences in disordered regions. Sci. Signal. 2012;5:rs1. doi: 10.1126/scisignal.2002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varadi M. pE-DB: a database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Res. 2014;42:D326–D335. doi: 10.1093/nar/gkt960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davey N.E. The SLiMDisc server: short, linear motif discovery in proteins. Nucleic Acids Res. 2007;35:W455–W459. doi: 10.1093/nar/gkm400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davey N.E. SLiMFinder: a web server to find novel, significantly over-represented, short protein motifs. Nucleic Acids Res. 2010;38:W534–W539. doi: 10.1093/nar/gkq440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gutman R. QuasiMotiFinder: protein annotation by searching for evolutionarily conserved motif-like patterns. Nucleic Acids Res. 2005;33:W255–W261. doi: 10.1093/nar/gki496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Via A. A structure filter for the Eukaryotic Linear Motif Resource. BMC Bioinformatics. 2009;10:351. doi: 10.1186/1471-2105-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dewson G. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell. 2008;30:369–380. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 79.Gavathiotis E. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell. 2010;40:481–492. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim H. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diella F. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 82.Liu D. A chemical strategy to promote helical peptide–protein interactions involved in apoptosis. Bioorg. Med. Chem. Lett. 2005;15:4467–4469. doi: 10.1016/j.bmcl.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 83.Zhai D. Differential regulation of Bax and Bak by anti-apoptotic Bcl-2 family proteins Bcl-B and Mcl-1. J. Biol. Chem. 2008;283:9580–9586. doi: 10.1074/jbc.M708426200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen L. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 85.Dai H. Evaluation of the BH3-only protein Puma as a direct Bak activator. J. Biol. Chem. 2014;289:89–99. doi: 10.1074/jbc.M113.505701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ku B. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21:627–641. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kelekar A., Thompson C.B. Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 88.Follis A.V. The DNA-binding domain mediates both nuclear and cytosolic functions of p53. Nat. Struct. Mol. Biol. 2014;21:535–543. doi: 10.1038/nsmb.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Placzek W.J. Identification of a novel Mcl-1 protein binding motif. J. Biol. Chem. 2011;286:39829–39835. doi: 10.1074/jbc.M111.305326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao H. Anti-apoptosis proteins Mcl-1 and Bcl-xL have different p53-binding profiles. Biochemistry. 2013;52:6324–6334. doi: 10.1021/bi400690m. [DOI] [PubMed] [Google Scholar]
- 91.Szlyk B. A phospho-BAD BH3 helix activates glucokinase by a mechanism distinct from that of allosteric activators. Nat. Struct. Mol. Biol. 2014;21:36–42. doi: 10.1038/nsmb.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holinger E.P. Bak BH3 peptides antagonize Bcl-xL function and induce apoptosis through cytochrome c-independent activation of caspases. J. Biol. Chem. 1999;274:13298–13304. doi: 10.1074/jbc.274.19.13298. [DOI] [PubMed] [Google Scholar]
- 93.Roberts A.W. Phase 1 study of the safety, pharmacokinetics, and antitumour activity of the BCL2 inhibitor navitoclax in combination with rituximab in patients with relapsed or refractory CD20+ lymphoid malignancies. Br. J. Haematol. 2015;170:669–678. doi: 10.1111/bjh.13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu X. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 95.Lee E.F. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 2007;14:1711–1713. doi: 10.1038/sj.cdd.4402178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.