Abstract
Dysregulation of Forkhead-box (FOX) transcription factors is linked to cancers of numerous tissue types. Here, we report that FOXF2 is frequently silenced in luminal-type and HER2-positive breast cancers, but is overexpressed in basal-like breast cancers; thus, FOXF2 appears to play distinct roles in different breast cancer subtypes. Inactivation of FOXF2 in luminal-type and HER2-positive breast cancers is attributable to epigenetic silencing. Silencing of FOXF2 is associated with poor prognosis in luminal-type breast cancer. Ectopic expression of FOXF2 in luminal and HER2-positive breast cancer cells suppresses their tumorigenic properties in vitro and in vivo via inhibition of the CDK2-RB-E2F cascade. The in vivo function of FOXF2 is to maintain the stringency of DNA replication, and its loss triggers dysregulation of DNA replication, which in turn activates the p53 checkpoint pathway. Besides its role in cell cycle regulation, FOXF2 is functionally required for mobility and epithelial-to-mesenchymal transition (EMT) of normal breast epithelial cells. In basal-like breast cancer cells, the cell-cycle function of FOXF2 is impaired. However, the EMT function of FOXF2 is still required for mobility, invasiveness and anchorage-independent growth of basal-like breast cancer cells. Our gene expression profiling studies demonstrate that FOXF2 regulates the expression of genes implicated in cell cycle and EMT regulation. Moreover, FOXF2 is highly co-expressed with basal- and metastasis-related genes in breast cancer. These findings suggest that FOXF2 has a dual role in breast tumorigenesis and functions as either a tumor suppressor or an oncogene depending on the breast tumor subtype.
Keywords: FOXF2, DNA methylation, epithelial-to-mesenchymal transition, basal-like/triple-negative breast cancer, luminal breast cancer, HER2-positive breast cancer
1. Introduction
The heterogeneity of breast tumors has been attributed as the cause of differential responses of patients to anti-cancer therapy [1,2]. Gene expression profiling and immunohistochemistry studies have been utilized to molecularly classify heterogeneous breast cancers into at least five subtypes, including normal breast-like, luminal A (ER+ and/or PR+/HER2− with a low Ki67 index), luminal B (ER+ and/or PR+/HER2+ or HER2− with a high Ki67 index), HER2-positive (ER−/PR−/HER2+) and basal-like/triple negative (ER−/PR−/HER2−) [3–6]. These distinct subtypes of breast cancer not only display different molecular profiles, but also exhibit varied therapeutic responses and prognosis. For instance, luminal-type breast cancer cells behave like epithelial cells that are more differentiated, have lower metastatic potential, respond to endocrine therapy and show a better prognosis compared to other breast cancer subtypes. In contrast, the basal-like breast cancer subtype, which often shows a triple negative phenotype for ER, PR and HER2 receptors, has a poor prognosis due to a low level of differentiation, a mesenchymal-like stem-cell phenotype, hematogenous dissemination, and resistance to endocrine and anti-HER2 target therapies [4,7,8]. Identification of key molecular regulators functionally contributing to the development of these breast cancer molecular subtypes is important for understanding of the disease formation and for the development of effective therapeutics specific for each breast cancer subtype.
Two major hallmarks of tumors are dysregulation of cell cycle progression and aberrant activation of the epithelial-to-mesenchymal transition (EMT) program. During the cell cycle, DNA re-replication (also called over-replication) results from repeated firing of DNA replication due to loss of DNA replication stringency, mainly due to the deregulation of the replication licensing process [9,10]. DNA re-replication is known to result in gene amplification and chromosomal alterations, in turn promoting cellular transformation and tumorigenesis [9,10]. Several G1-S checkpoint proteins such as ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3 related (ATR) and p53 are involved in monitoring DNA replication stringency and suppressing DNA re-replication for maintaining genomic stability and inhibiting mutagenesis [11]. During tumorigenic progression, cancer cells also acquire an ability to reactivate the EMT program that endows them with increased invasive and migratory abilities, an essential step for cancer cells to become metastatic [12]. Moreover, cancer cells undergoing EMT exhibit cancer-stem-cell phenotypes and are highly resistant to conventional chemotherapy and radiotherapy [13,14]. Identification of transcription factors regulating the expression of genes involved in cell-cycle and EMT regulation and revealing their dysregulation during tumorigenesis are critical for understanding tumorigenic mechanisms and for propelling the development of novel therapies to treat cancer.
Deregulating the subcellular localization, functions and expression of Forkhead box (FOX) transcription factors that are critically involved in embryonic development and multiple biological processes is known to result in the development and progression of diseases, in particular cancer [15–18]. Human FOXF transcription factors, including FOXF1 and FOXF2, are a subfamily of the FOX gene family [19]. Mouse Foxf proteins are well-known mesenchymal factors that modulate extracellular matrix (ECM) synthesis and epithelial-mesenchymal interactions, and play critical roles in regulating embryonic development and organogenesis [20–23]. Our recent findings have revealed that FOXF1 plays a crucial role in maintaining the stringency of DNA replication and functions as a tumor suppressor in breast epithelial cells, which is frequently silenced in breast cancer via epigenetic mechanisms [24]. Moreover, ectopic overexpression of Foxf1 in a mouse mammary epithelial cell line induced EMT, increased invasiveness in vitro and enhanced tumor growth in vivo [25]. These findings suggest that FOXF1 may play a dual role that acts as either a tumor suppressor or an oncogenic factor during tumorigenesis in a context-dependent manner.
Dysregulation of FOXF2 has also been linked to breast tumorigenesis [26–29]. Decreased FOXF2 expression was reported to be associated with early-onset metastasis and poor prognosis in breast cancer [26]. DNA methylation contributes to silencing FOXF2 in cultured breast cancer cell lines in a subtype-specific manner [28]. Although FOXF2 was reported to act as a tumor suppressor by blocking the metastasis of basal-like breast cancer cells via inhibiting EMT [27,29], this paradigm is contradictory to the previously reported mesenchymal roles of FOXF transcription factors that are well-known to promote EMT and mesenchymal phenotypes of stromal and epithelial cells [22,25,30–32]. In this study, we showed that FOXF2 plays a dual role in breast tumorigenesis by functioning either as a tumor suppressor that negatively regulates DNA replication or as an oncogenic factor that promotes the EMT process. Remarkably, we found that FOXF2 loses its cell-cycle function but retains its EMT function in basal-like breast cancer. Our paradigm rationally explicates why FOXF2 is silenced in luminal and HER2-positive breast cancers through epigenetic mechanisms and its restoration exhibits tumor-suppressive features in these two breast cancer subtypes, and why basal-like breast cancers frequently overexpress FOXF2 that turns into an EMT promoter to facilitate tumorigenesis and metastasis of basal-like breast cancer. Our findings have shed light on the biological role of overexpressed FOXF2 in basal-like breast cancer and provide new insights into FOXF2 function in cancer.
2. Materials and methods
2.1. In silico analysis of gene expression
The Oncomine’s Cancer Microarray Database (http://www.oncomine.org) [33] and cBio portal for Cancer Genomics (http://www.cbioportal.org) [34] were used to perform in silico expression analysis of FOXF2 and other genes in normal and cancerous breast tissues.
2.2. Identification of the CpG island of the FOXF2 gene
We obtained the genomic DNA sequence, including the upstream promoter sequence, of the FOXF2 gene from the GenBank Database of the National Center of Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) and also from the Database of Transcriptional Start Sites (DBTSS, http://dbtss.hgc.jp). The extracted 600-bp upstream and 200-bp downstream genomic sequences relative to the transcription start site of the FOXF2 gene were subjected to analysis using the criteria and algorithm of online CpG Island Searcher (http://cpgislands.usc.edu) to identify the CpG island.
2.3. Cell lines and tissue samples
We obtained immortalized and nontumorigenic human mammary epithelial cells (HMEC), including MCF10A and HBL100, and the breast cancer cell lines (listed in Fig. 1A) from ATCC (American Type Culture Collection, Manassas, VA, USA) and cultured them according to the ATCC online instructions. The molecular subtype classification of breast cancer cell lines used in the study was based on the information of two publications [35,36]. Mammary organoids were prepared from reduction mammoplasty specimens of normal women as previously described [24]. The primary HMEC cultures were isolated from digested mammary organoids. We obtained fresh reduction mammoplasty specimens and frozen primary breast cancer samples from the Department of Pathology at the Johns Hopkins Hospital (Baltimore MD). All of human tissue specimens in this study were processed and used with prior approval from the JH Institutional Review Board.
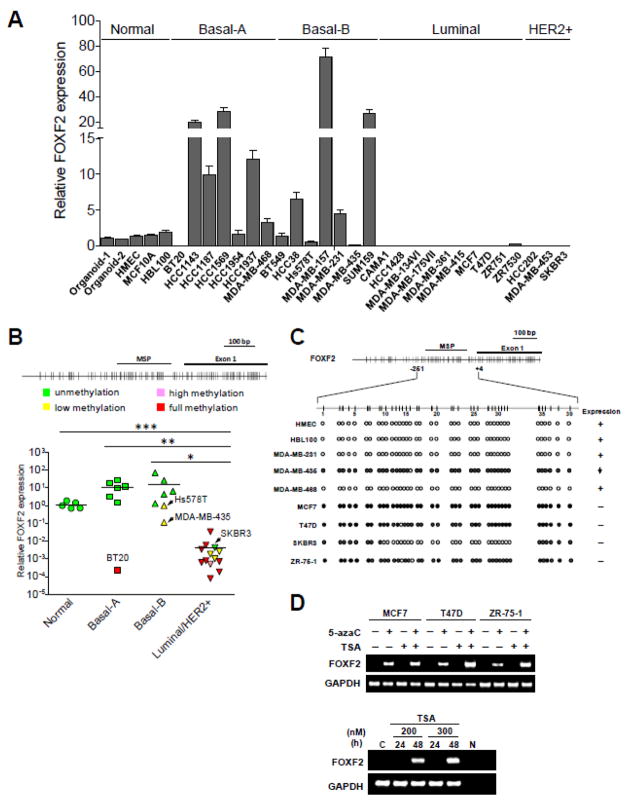
Fig. 1.
The FOXF2 gene is differentially epigenetically silenced in breast cancer molecular subtypes. (A) qRT-PCR analysis of FOXF2 expression in various molecular subtypes of breast cancer cell lines. Total RNAs isolated from two mammary epithelial-enriched organoids, human mammary epithelial cells (HMEC), two immortalized HMEC lines (MCF10A, HBL100), basal-A, basal-B, luminal and HER2-positive breast cancer cell lines were subjected to qRT-PCR analysis of FOXF2 mRNA expression. (B) The relationship between the promoter methylation status and expression of the FOXF2 gene in breast cancer cell lines. Methylation-specific PCR (MSP) analysis of the FOXF2 gene was performed on the same panel of breast cancer cell lines and normal controls shown in (A). The top panel is the map of CpG dinucleotides in exon 1 of the FOXF2 gene and the genomic region upstream of exon 1. The exon 1 and MSP-amplified regions are indicated. (C) Bisulfite sequencing analysis of the FOXF2 promoter in normal breast epithelial and breast cancer cell lines. The amplified FOXF2 DNA region for sequencing analysis encompasses nucleotides from −251 to +4 relative to the first nucleotide (set as the default +1) of exon 1 based on the FOXF2 cDNA sequence (NM_001452). The methylation status of each CpG dinucleotide is indicated by the open circle (unmethylated), partially filled circle (partially methylated), and completely filled circle (completely methylated). (D) RT-PCR analysis of FOXF2 expression in breast cancer cell lines treated with 5-aza-2′-deoxycytidine (5-azaC), trichostatin A (TSA), or a combination (the top panel). SKBR3 cells were treated with TSA at two different doses as indicated for 24 and 48 h (the bottom panel). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression is shown as an internal loading control.
2.4. Methylation-specific PCR and bisulfite sequencing analysis
Bisulfite modification of genomic DNA and methylation-specific PCR (MSP) amplification were performed as previously described [24]. MSP primer sequences for the FOXF2 promoter are: 5′-GGG TTT AGG TTG TGG TTT TAT TTG T-3′ (forward) and 5′-TCA ACT CCT CTA AAA CTT CTA ACA ACA-3′ (reverse) for unmethylated reactions (171 bp), and 5′-GTT TAG GTC GCG GTT TTA TTC-3′ (forward) and 5′-AAC TCC TCT AAA ACT TCT AAC GAC G-3′ (reverse) for methylated reactions (167 bp). Bisulfite sequencing analysis of the FOXF2 promoter was performed as previously described [24] by using a pair of external primers, 5′-GGA G(C/T)G TTT TTG AAG GGA GA-3′ (forward) and 5′-AAC TAC AAC C(A/G)T AAC CAC TCC-3′ (reverse).
2.5. Conventional and real-time quantitative reverse-transcription PCR (RT-PCR) analysis
Total RNA isolated from cancer cell lines or primary tumor tissues was reverse-transcribed and the PCR reaction was carried out as previously described [24]. The primers for FOXF2 RT-PCR amplification are: 5′-GCT CGC GAG GAC CTC TCA GTG-3′ (forward) and 5′-ATC GCT TGG CCT CTT TCC GTT C-3′ (reverse); the primers for GAPDH are the same as previously described [24]. We carried out SYBR Green-based real-time quantitative RT-PCR as described [24]. Data analysis was performed using the 2−ΔΔCT method for relative quantification [37], and all samples were normalized to the expression of the internal control GAPDH.
2.6. Drug treatment
Breast cancer cell lines (MCF7, T47D, ZR-75-1 and SKBR3) were treated with the regimen as previously described [24].
2. 7. Immunohistochemistry assay
The immunohistochemical (IHC) analysis was performed using the avidin-biotin-peroxidase complex (ABC) method. Paraffin-embedded tissue sections (4 uM) were deparaffinized by xylene and rehydrated in serial graded alcohol: 100%, 95%, and 70% (10 min for each). After antigen retrieval of tissue sections using Dako Epitope Retrieval Solution (Dako North America Inc., Carpinteria, CA, USA) and the microwave heating method, the endogenous peroxidase of tissue was blocked by 0.3% H2O2 in methanol. Non-specific staining was blocked with 10% normal goat serum (for polyclonal antibody staining) for 20 min at room temperature. Primary antibodies were then applied at the concentration of 1 μg/ml for anti-FOXF2 antibody (polyclonal, Aviva Systems Biology Corp., San Diego, CA, USA). Tissue sections were incubated with primary antibodies at 4°C overnight. After 1×PBS washing, the biotinylated goat anti-rabbit serum (Vector Laboratory Inc., Burlingame, CA, USA) was added and tissue sections were further incubated at room temperature for 30 min. The avidin-biotin-peroxidase complex (Vector Laboratory Inc.) was then applied for 30 min at room temperature, and the mix of DAB chromogen and substrate was added for color developing. Counterstaining was done by staining tissue sections with DAKO hematoxylin. Scoring of FOXF2 protein expression in human breast cancer samples was scored independently by two individuals based on an H-score, which is derived by multiplying the staining intensity (0–3) with the percentage of epithelial cells with positive IHC staining.
2.8. Clinicopathological characteristics for breast tumors
By the Nottingham combined histologic grading system [38], 15 carcinoma cases were classified as grade 1, 51 cases grade 2, and 51 cases grade 3. According to the TNM stage [39], 34 patients had stage I, 60 patients stage II, and 23 patients stage III. The expression status of hormone receptors, HER2 and p53 for tumor specimens was determined according to the guideline of the American Society of Clinical Oncology/College of American Pathologists [40].
2.9. In silico Kaplan–Meier survival analysis
The Kaplan–Meier plotter for breast cancer (http://kmplot.com/analysis/index.php?p=service&cancer=breast) was used to perform in silico relapse-free survival (RFS) analysis of FOXF2 expression in breast cancer patients.
2.10. Cloning and expression plasmid construction
Full-length cDNA fragments for the FOXF2 gene were obtained by RT-PCR of RNA from normal mammary organoids and cloned into the pGEM-T-Easy vector (Promega Corp., Madison, WI, USA). The sequence confirmed FOXF2 cDNA was subcloned into the pcDNA3.1 (−) vector (Life Technologies Inc., Carlsbad, CA, USA) with the haemagglutinin (HA)-tag coding sequence.
2.11. Transfection and reporter assay
Cells transfected with the vector control or HA-tagged FOXF2 expression plasmid DNA using Lipofectamine 2000 (Life Technologies Inc.) were used for Western blot analysis with antibodies against hemagglutinin (HA) tag (Cell signaling Technology, Danvers, MA, USA), or α-tubulin (Life Technologies Inc.). For reporter assays, cells transfected with reporter-, expression and pCMV-β-galactosidase plasmids were lysed and luciferase activity was measured using a luminometer (BD Biosciences, San Jose, CA, USA). β-galactosidase activities were measured using the reporter assay system (Promega) according to the manufacturer’s instructions. Luciferase activities as measured in cell extracts were normalized by β-galactosidase activities.
2.12. Colony formation assay
At 24 h after transfection, transfected cells were plated in 6-well plates at a density of 6,000–12,000 cells/well in complete medium containing 0.3–0.6 mg/ml geneticin (G418). After 2–3 weeks, colonies were fixed and then stained with 0.1% crystal violet. Unbound dye was washed out with water. Bound dye was then eluted with 10% acetic acid, and quantitated by measuring the absorbance at 590 nm.
2.13. Flow cytometric analysis
2×106 MCF7 cells were grown in 6-well culture plates for 24 h, and cotransfected with 3.5 μg of the control empty vector or HA-tagged FOXF2 expression plasmid DNA and 0.7 μg of the pEGFP-C1 plasmid DNA (Clontech Laboratories Inc., Mountain View, CA, USA). After 72 h, cells were trypsinized, fixed with 2% formaldehyde in PBS at 4°C for 30 min and permeabilized with 70% ethanol overnight at −20°C. Fixed cells were pelleted and resuspended in an isotonic buffered PI-staining solution containing RNase A (0.1 mg/ml) and propidium iodide (20 μg/mL). Cell cycle distribution and the sub-G1 fraction of GFP-positive cells were analyzed by the BD FACSCalibur system (Becton Dickinson, Franklin Lakes, NJ, USA). Apoptosis was scored by assessing the fraction of cells with a sub-G1 DNA content.
2.14. Tumorigenicity assay
T47D cells transfected with either HA-FOXF2 or the empty vector were selected in the G418-containing medium for a week. 2×106 of G418-selected HA-FOXF2-transfected or vector-transfected (as a control) T47D cells were injected into the fourth mammary fat pad of female athymic nude mice with age of 6 weeks (n = 6 for each transfected cells). Nude mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). For xenograft with T47D cells, mice were supplemented with estradiol pellets (0.72 mg, released over 60 days; Innovative Research of America, Sarasota, FL, USA) before tumor cell transplantation. Tumors were measured in two diameters with calibers to permit calculation of tumor volume, V = [(D + d)/2]3, where D and d are the larger and smaller diameters, respectively. Xenograft experiments were performed according to the animal protocol approved by IACUC, which is in accordance with the guidelines established by the USPHS.
2.15. Immunofluorescence assay
Transfected cells on coverslips were fixed with formaldehyde, and then subjected to primary (including anti-HA, anti-CDK2, anti-phospho-CDK2, anti-RB and anti-phospho-RB antibodies) and secondary (Alexa series, Life Technologies Inc.) antibody reactions according to the manufacturer’s instructions. After immunostaining, nuclear DNA of cells was stained with 10μg/ml 4′6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, St. Louis, MO, USA). Immunostained cells on slides were examined under a Nikon Eclipse E800 fluorescence microscope (Nikon, Minato, Tokyo, Japan). The anti-HA, anti-RB, anti-phospho-CDK2 (Thr160) and anti-phospho-RB (Ser807/811) antibodies were obtained from the Cell Signaling Technology Inc. and the goat anti-CDK2 antibody was obtained from the Santa Cruz Biotechnology Inc. (Dallas, TX, USA).
2.16. siRNA transfection
siRNA transfections were performed with 20 nM of each siRNA using Lipofectamine™ RNAiMAX (Life Technologies Inc.) according to the instructions of the manufacturer. The two siRNA sequences for targeting FOXF2 are: 5′-CAACUUCAAUGGGAUUUCU-3′ (FOXF2 siRNA-1) and 5′-GAGCGUCUGUCAGGAUAUU-3′ (FOXF2 siRNA-2). The FOXF2 siRNAs and non-targeting control siRNA (siControl: Catalog No. D-001810-10) were purchased from Dharmacon (Lafayette, CO, USA).
2.17. Western blot analysis
Western blot assays were performed as previously described [24]. Antibodies used in the assays include: anti-HA, anti-phospho-p53 (Ser-15), anti-p53, anti-phospho-CHK2 (Thr-68), anti-phospho-CDK2 (Thr-160), anti-phospho-Rb (Ser-807/811), anti-Rb, anti-phospho-CDC2 (Tyr-15), anti-Smad2, anti-phospho-Smad2 (Ser465/467) (Cell Signaling Technology Inc.); anti-p21WAF1 (Calbiochem Inc., Billerica, MA, USA); anti-CDK2, anti-CDC2 (Santa Cruz Biotechnology Inc.); anti-CDC6, anti-α-tubulin, anti-Vimentin, anti-E-cadherin (Life Technologies Inc.).
2.18. Migration and invasion assays
Wound healing and transwell migration assays were performed to measure the migratory ability of cells. For the wound healing assay, cells were seeded in 6-well plates and cultured until confluent. A (200 μl) pipette tip was used to make a straight scratch, simulating a wound. Cell wound images were taken by a microscope at 0 and 12 (or 24) hours for examining wound healing. For the transwell migration assay, 2×104 cells were seeded in the upper transwell chamber insert. For the invasion assay that measures the invasive ability of cancer cells, 40 μl of matrigel was added into the upper transwell chamber insert to form a thin gel layer in a 37°C incubator for 15–30 minutes and 5×104 cells were then seeded on the top. The lower chamber was filled with the complete culture medium containing 10% serum. Cells were allowed to migrate or invade towards the serum gradient for 24 hours. Migrated or invaded cells were stained with 1% crystal violet and counted using a phase-contrast microscope. Five random fields were counted per experiment.
2.19. Soft agar assay
To perform the soft agar assay, the bottom agar (0.6% agar) was prepared in six-well plates. Once the bottom agar was solidified, 1 ml of the top agar (0.3% agar) was mixed with 2×104 cells and plated into six-well plates. After the top agar became solidified, 2 ml of the culture medium was added and soft agar plates were maintained in a cell culture incubator until spheroid colonies formed. Formed colonies in agar were stained with 0.005% crystal violet and counted (≥ 50 μm) under a dissection microscope.
2.20. Gene expression profiling
48 hours after siRNA transfections, siRNA-transfected MCF10A (or MDA-MB-231) cells were lysed in TRIzol (Life Technologies Inc.) and total RNA was purified according to manufacturer’s instructions. Approximately 20 μg of RNA was treated with RNase-free DNaseI (#2238, Ambion, Ambion Inc., Austin, TX, USA) for 30 minutes at 37°C and subsequently purified using the RNeasy mini kit (#74104, Qiagen, Hilden, Germany) according to manufacturer’s instructions. The integrity of purified DNaseI-treated RNA was evaluated using the Agilent 2100 Bioanalyzer and the RNA nano 6000 kit (Agilent Technologies, Wilmington, DE, USA). For each sample, Biotinylated cRNA was prepared using the Ambion MessageAmplification kit for Illumina arrays (Ambion Inc.) according to the manufacturer’s specifications with an input of 500 ng total RNA. Per sample, 750 ng of the biotinylated cRNA was hybridized onto the Illumina HumanHT-12 v4 Expression BeadChip (Illumina, Inc., San Diego, CA, USA) according to the Illumina Manual “Direct Hybridization Assay Guide”. Hybridized arrays were scanned on an Illumina HiScan microarray scanner. Illumina GenomeStudio was used to transform bead-level data to probe-level intensity values and statistics, which were exported raw data (unfiltered and non-normalized) for bioinformatic analysis. The expression data were quantile normalized using IlluminaGUI in R and log2-transformed, and a rank product analysis was performed using a p-value < 0.05 to identify significant changes of gene expression. The dataset has been deposited in the GEO data repository (http://www.ncbi.nlm.nih.gov/geo, accession number GSE55675).
2.21. Statistical analysis
Comparisons of clinicopathologic characteristics between invasive ductal carcinoma cases with and without the methylated FOXF2 gene were subjected to χ2 analysis using the SPSS software (version 11.5; SPSS Inc., Chicago, IL, USA). The Student’s t-test was used to analyze the significance of the difference between two groups of data using the GraphPad Prism software (version 6.0; GraphPad Software, Inc, La Jolla, CA, USA). P < 0.05 was regarded as statistically significant.
3. Results
3.1. Epigenetic silencing of FOXF2 gene expression in breast cancer is subtype-specific
It has been shown that FOXF2 expression is downregulated in various types of cancer [26,41,42]. Indeed, our in silico meta-analysis of the Oncomine Cancer Microarray Databases (http://www.oncomine.org) [33] also provided the data that FOXF2 was consistently underexpressed in lung, bladder, prostate and testicular cancers (Supplementary Fig. 1, Supplementary Table 1). In contrast, FOXF2 was overexpressed in esophageal and ovarian cancers (Supplementary Table 1). Although DNA methylation-mediated silencing of FOXF2 in cultured breast cancer cell lines was reported elsewhere [28], it is unclear whether this finding can be recapitulated in breast tumors, whether this epigenetic silencing is related to in vivo breast cancer molecular subtypes and whether other epigenetic mechanisms (e.g. histone modifications) are also involved in FOXF2 silencing. To convincingly examine the expression status of FOXF2 in breast cancer molecular subtypes, we first performed qRT-PCR analysis of FOXF2 expression on a panel of breast cancer cell lines consisting of luminal, basal-like (basal-A and basal-B) and HER2+ subtypes. As shown in Fig. 1A, FOXF2 expression was overexpressed in 9 out of 14 (64.3%) of basal-like breast cancer cell lines when compared with normal controls including two organoid samples, human mammary epithelial cells (HMEC) and two immortalized HMEC lines MCF10A and HBL100. In contrast, FOXF2 expression was lost in all of 13 examined luminal and HER2+ breast cancer cell lines. To further validate our finding, we also performed in silico analysis of FOXF2 expression in Neve breast cancer cell line microarray data encompassing three different subtypes (basal-A, n=12; basal-B, n=14; luminal, n=24) [35] using the Oncomine Cancer Microarray database (http://www.oncomine.org) [33]. In line with the result of Fig. 1A, FOXF2 was expressed at a significantly low level in luminal breast cancer cell lines compared with basal-type breast cancer cell lines (Supplementary Fig. 2). Intriguingly, among two basal-subtypes, the basal-B subtype expressed a higher level of FOXF2 than the basal-A subtype (Supplementary Fig. 2). These results, taken together, suggest that dysregulation of FOXF2 expression in breast cancer is subtype-specific, and is able to further segregate subtypes of basal-like breast cancers.
To determine whether loss of FOXF2 expression in luminal and HER2+ breast cancer cells is mediated by epigenetic silencing, we performed methylation-specific PCR (MSP) analysis of the FOXF2 gene. With the exception of SKBR3, MSP detected hypermethylation of the FOXF2 promoter in all of luminal and HER2+ breast cancer cell lines, which correlated with silencing of FOXF2 expression in these cell lines (Fig. 1B). In contrast, only 3 out of 14 basal-type breast cancer cell lines exhibited FOXF2 promoter hypermethylation and two of them showed weak methylation (Fig. 1B). MSP findings were further validated by bisulfite sequencing analysis. As shown in Fig. 1C, the FOXF2 promoter was fully or partially methylated in luminal breast cancer cell lines including MCF7, T47D and ZR-75-1, but not in HER2+ SKBR3 that showed a poorly methylated FOXF2 promoter.
To determine whether epigenetic mechanisms play crucial roles in silencing FOXF2 expression in luminal and HER2+ breast cancer cells, three hypermethylated cell lines were treated with epigenetic modifiers. Treatment of these lines with the demethylating agent (5-aza-2′-deoxycytidine, 5-azaC), the histone deacetylase inhibitor (trichostatin A, TSA) or a combination resulted in the re-expression of FOXF2 mRNA, an effect that was enhanced by co-treatment of some cell lines (T47D and ZR-75-1) with 5-azaC and TSA (the top panel in Fig. 1D). These results indicated that promoter-hypermethylation and histone-deacetylation mechanisms are responsible for silencing FOXF2 expression in luminal breast cancer cell lines. In addition, 48-h TSA treatment revived FOXF2 expression in SKBR3 cells (the bottom panel in Fig. 1D), indicating that in this HER2+ line, histone deacetylation, not DNA methylation, was responsible for silencing FOXF2 expression. This conclusion was further confirmed by the 5-azaC treatment experiment, which showed that the inhibition of DNA methylation was unable to restore FOXF2 expression in SKBR3 cells (Data not shown).
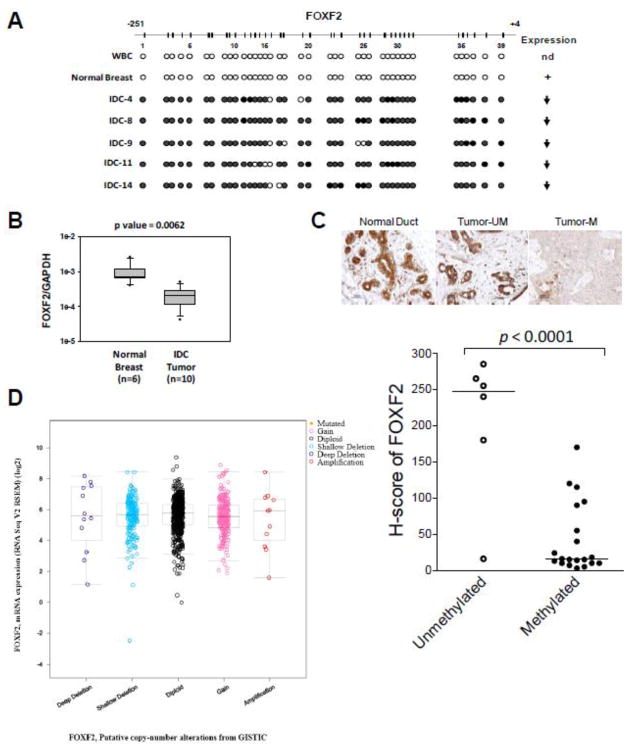
To investigate whether FOXF2 promoter hypermethylation also occurs in primary breast carcinomas, MSP analysis was performed on invasive ductal carcinomas (IDCs; n = 117). The FOXF2 promoter was hypermethylated in 70.9% (83 of 117) of IDC tumors (Table 1), which was in remarkable contrast to normal breast tissue and white blood cells (WBC) that showed the unmethylated FOXF2 promoter (data not shown). Bisulfite sequencing analysis further confirmed that IDC cases (IDC-4, IDC-8, IDC-9, IDC-11, and IDC-14) with positive MSP results had extensive CpG methylation throughout the analyzed genomic region compared with normal breast tissue and WBC DNA samples with the unmethylated CpG island (Fig. 2A). These five IDC samples consistently exhibited downregulation or loss of FOXF2 expression compared with the normal breast tissue (Fig. 2A). Thus, hypermethylation of the FOXF2 promoter occurs frequently in primary breast tumors. Furthermore, statistical analysis of the association between clinicopathological parameters and the FOXF2 methylation status showed that hypermethylation of the FOXF2 gene promoter positively correlates with the tumor size (p = 0.019) and tumor stage (p = 0.022), but are inversely associated with age (p = 0.048) and the triple-negative (TN) status (p = 0.014) (Table 1). It is known that 70–80% of basal-like breast carcinomas are triple-negative [8,43,44]. Therefore, the finding that hypermethylation of the FOXF2 CpG island occurred less frequently in TN breast tumors than in non-TN breast tumors is consistent with the result that basal-like breast cancer cell lines exhibited the low frequency of promoter hypermethylation and expression silencing of the FOXF2 gene when compared to luminal and HER2-positive breast cancer cell lines (Fig. 1).
Table 1.
Correlation between the promoter hypermethylation of the FOXF2 gene and clinicopathologic characteristics of IDC tumors.
| Clinicopathologic characteristics* | Methylation (%)ǂ | p¥ |
|---|---|---|
| Age | 0.048 | |
| < 50 (51) | 41 (80.4) | |
| ≥ 50 (66) | 42 (63.6) | |
| Size | 0.019 | |
| < 2 cm (46) | 27 (58.7) | |
| ≥ 2 cm (71) | 56 (78.9) | |
| Histologic grade | 0.873 | |
| G1 (15) | 9 (60.0) | |
| G2 (51) | 40 (78.4) | |
| G3 (51) | 34 (66.7) | |
| LV invasion | 0.218 | |
| Negative (55) | 36 (65.5) | |
| Positive (62) | 47 (75.8) | |
| Nodal involvement | 0.247 | |
| Negative (66) | 44 (66.7) | |
| Positive (51) | 39 (76.5) | |
| Stage | 0.022 | |
| I (34) | 19 (55.9) | |
| II (60) | 45 (75.0) | |
| III (23) | 19 (82.6) | |
| ER status | 0.559 | |
| Negative (57) | 39 (68.4) | |
| Positive (60) | 44 (73.3) | |
| PR status | 0.247 | |
| Negative (66) | 44 (66.7) | |
| Positive (51) | 39 (76.5) | |
| Her-2/neu status | 0.468 | |
| Negative (95) | 66 (69.5) | |
| Positive (22) | 17 (77.3) | |
| Triple-negative status | 0.014 | |
| Negative (87) | 67 (77.0) | |
| Positive (30) | 16 (53.3) | |
| p53 status | 0.695 | |
| Negative (69) | 48 (69.6) | |
| Positive (48) | 35 (72.9) |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor.
The IDC tumor sample number for each clinicopathological characteristic is shown in parentheses. Classification systems for histologic grades and tumor stages, and methods to determine positivity for ER (estrogen receptor), PR (progesterone receptor), Her-2/neu, and p53 status were described in detail in “Materials and Methods”.
The percentages of IDC cases with the methylated FOXF2 gene are shown in parentheses.
P values with statistical significance are shown in boldface.
Fig. 2.
Epigenetic inactivation of FOXF2 in primary invasive breast tumors. (A) Bisulfite sequencing analysis of the FOXF2 promoter in white blood cells (WBC), normal breast tissue and invasive ductal carcinomas (IDCs). The right panel shows the FOXF2 expression status of these sequenced tissue samples: nd, not determined; (+) expression; (↓), underexpression. (B) qRT-PCR analysis of FOXF2 mRNA expression in normal breast tissues versus primary breast IDCs with the methylated FOXF2 promoter. (C) Immunohistochemistry (IHC) analysis of FOXF2 protein expression in normal breast tissue and primary breast IDC tumors with or without the methylated FOXF2 gene. The representative IHC staining pictures are shown in the top panel. The semi-quantitative H-score result from IHC analysis of FOXF2 in 26 IDC samples (separated into unmethylated and methylated groups) is shown in the bottom panel. The median of each dataset is indicated in the plot. (D) In silico analysis of the relationship between genetic alterations of the FOXF2 gene and the FOXF2 mRNA expression status using the cBioPortal for Cancer Genomics (http://www.cbioportal.org).
To determine the functional consequence of FOXF2 promoter hypermethylation, FOXF2 expression was examined in primary breast carcinomas. Quantitative RT-PCR analysis of 6 normal breast tissues and 10 IDC tumor tissues with the methylated FOXF2 gene showed that FOXF2 mRNA was significantly underexpressed in IDC tumors compared with normal breast tissues (p < 0.05) (Fig. 2B). To further verify the qRT-PCR data, immunohistochemistry (IHC) assays were performed to examine FOXF2 protein expression in paraffin-embedded tissue sections of 26 IDC tumors selected from 117 IDC cases subjected to MSP analysis (Table 1). Normal breast tissue was also examined by IHC and found to display positive immunostaining for the FOXF2 protein in breast epithelial cells, consistent with its status as unmethylation for the FOXF2 gene. We exploited the H-score method to semi-quantify the IHC staining of the FOXF2 protein in tumor tissues. As shown in Fig. 2C, IDC cases (n = 20) with the methylated FOXF2 gene significantly manifested downregulated FOXF2 protein expression when compared with those (n = 6) with the unmethylated FOXF2 gene (p < 0.0001). Collectively, evidence from promoter hypermethylation, mRNA and protein expression studies suggests that FOXF2 promoter hypermethylation correlates significantly with reduction/loss of FOXF2 expression.
To determine whether genetic alterations are involved in regulating FOXF2 expression status in primary breast tumors, we analyzed the correlation between the genetic deletion/amplification of the FOXF2 gene locus and its mRNA expression status in 960 breast tumor cases using the cBio portal for Cancer Genomics (http://www.cbioportal.org) [34]. As shown in Fig. 2D, the frequency of FOXF2 gene deletion and amplification was rare (~1.2%) in this cohort. Furthermore, there was no significant correlation between genetic deletion/amplification at the FOXF2 gene locus and the FOXF2 expression status in genetically altered cases when compared with diploid cases. The results of this bioinformatics-based analysis indicated that dysregulation of FOXF2 expression in breast tumors is unlikely to result from genetic aberrations.
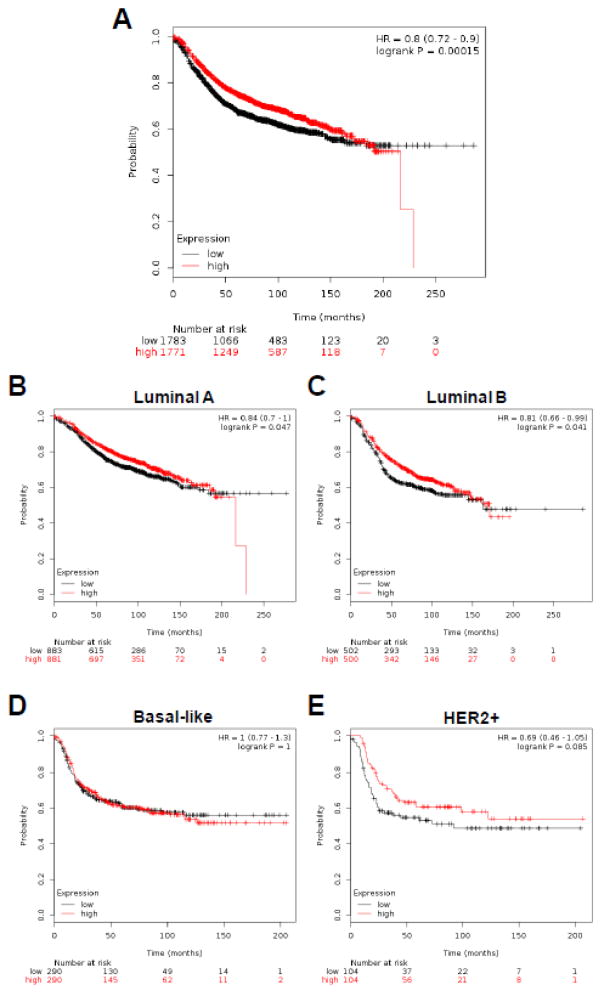
To evaluate the clinical relevance of FOXF2 expression silencing in breast cancer, we analyzed whether the tumor level of FOXF2 expression correlated with patient prognosis. Using expression data derived from a cohort of 3554 patients with breast cancer [45], Kaplan–Meier analysis of overall mortality revealed that breast cancer patients with low FOXF2 expression had a significantly poorer outcome when compared with those with high FOXF2 expression (p = 0.00015, hazard ratio 0.8, 95% confidence interval = 0.72 – 0.90) (Fig. 3A). To reveal whether the FOXF2 expression status can be a prognostic indicator in distinct breast cancer subtypes, we performed Kaplan-Meier survival analysis of FOXF2 in luminal A (1764 cases), luminal B (1002 cases), basal-like (580 cases) and HER2+ (208 cases) breast cancers (Fig. 3B, 3C, 3D and 3E). Low expression of FOXF2 predicted poorer prognosis of luminal A (p = 0.047) and luminal B (p = 0.041) breast cancer patients, but not basal-like breast cancer patients (p = 1.00) (Fig. 3B, 3C, 3D). Although not statistically significant (p = 0.085), HER2+ breast cancer patients with low FOXF2 expression tended to have a poorer outcome (Fig. 3E). The prediction of outcomes based on FOXF2 expression in these four different breast cancer subtypes correlated with our finding that FOXF2 expression is frequently epigenetically silenced in luminal and HER2+ breast cancers compared with basal-like breast cancers. Taken together, the results from multivariate analysis of the FOXF2 promoter hypermethylation taking all clinico-pathological parameters into account and Kaplan-Meier survival analysis of FOXF2 in breast cancer suggest that epigenetic silencing of FOXF2 expression is important for the malignant progression of luminal-type and HER2+ breast tumors, but not for basal-like breast cancer.
Fig. 3.
Kaplan-Meier survival analysis of breast cancer patients is stratified by the FOXF2 expression levels. (A) Kaplan-Meier relapse-free survival (RFS) curves of breast cancer patients (n = 3554) were plotted according to the FOXF2 expression status (high vs. low) using the online survival analysis software “Kaplan-Meier plotter” (http://kmplot.com/analysis). (B, C, D, E) Kaplan-Meier RFS curves of four different molecular subtypes of breast cancer patients were plotted based on the FOXF2 expression levels. The breast cancer subtypes including luminal A (n = 1764), luminal B (n = 1002), basal-like (n = 580) and HER2-positive (n = 208) were analyzed to plot the KM survival curves. The breast cancers were classified into high- or low-expressing groups according to their expression of the FOXF2 gene relative to the median FOXF2 mRNA expression value in the patient cohort. The p value of the log-rank test, hazard ratio, and confidence interval from comparing the two KM RFS curves are shown in each figure.
3.2. Re-expression of FOXF2 inhibits the tumorigenicity of luminal and HER2-positive breast cancer cells
To elucidate the biological significance of epigenetic silencing of FOXF2 in luminal and HER2+ breast cancer cells, we performed a series of functional assays. We examined the impact of ectopic expression of HA-tagged FOXF2 on breast cancer cells with the epigenetically silenced FOXF2 gene. Protein expression and nuclear localization of HA-FOXF2 expressed from the pCMV-HA-tagged FOXF2 expression construct were confirmed by Western blot and immunofluorescence analyses, respectively (Supplementary Fig. 3). To test whether exogenously introduced HA-FOXF2 retains transcription factor functions as the endogenous FOXF2 protein, reporter assays were performed using the reporter plasmid containing a minimal promoter with (p4XFREAC-Luc) or without (pApo-Luc) four tandem copies of the FOXF binding site [46]. Ectopic expression of the HA-FOXF2 protein induced the luciferase activity of the reporter in a highly significant manner in p4XFREAC-Luc-transfected T47D and SKBR3 cells, but not in pApo-Luc-transfected control cells (Fig. 4A).
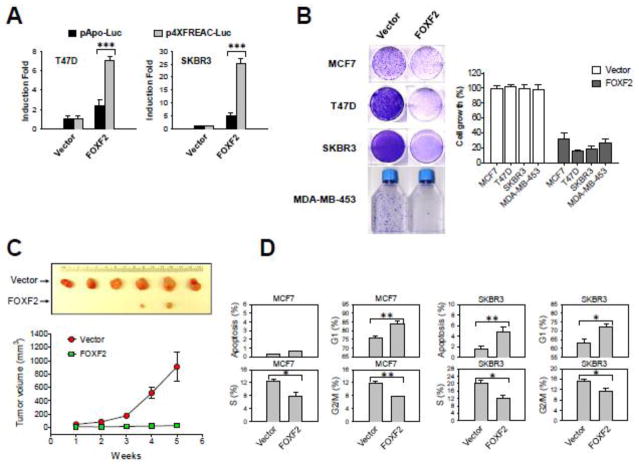
Fig. 4.
FOXF2 re-expression inhibits the growth and tumorigenicity of luminal and HER2-positive breast cancer cells by inducing G1 arrest and/or apoptosis. (A) The transcriptional activity of HA-tagged FOXF2 in T47D and SKBR3 cells was assessed by using the FOXF-responsive reporter plasmid (p4XFREAC-Luc) relative to the empty reporter control pApo-Luc. The error bar represents the standard deviation (SD) from three independent experiments. (B) FOXF2 re-expression suppresses in vitro growth of luminal and HER2-positive breast cancer cell lines. Luminal (MCF7 and T47D) and HER2-positive (SKBR3 and MDA-MB-453) cell lines were transfected with either the empty vector or the FOXF2 expression plasmid and selected in the G418-containing medium to form colonies. G418-resistant colonies were visualized by staining with crystal violet dye (the left panel). Absorbance values at 590 nm of eluted binding dye reflecting overall formed cell colonies were used to plot the bar graph. The error bar represents the SD (n = 3). (C) Restoration of FOXF2 expression inhibits in vivo tumorigenicity of T47D cells. Each point indicates the mean of tumor volume measurements of six xenograph tumors generated from empty vector-transfected or HA-FOXF2-transfected T47D cells. The tumor formation difference between these two distinct transfected cells is statistically significant (p < 0.05). (D) Flow cytometric analysis of cell cycle and apoptotic profiles in FOXF2-transfected MCF7 and SKBR3 cells. The empty control vector or HA-tagged FOXF2 expression plasmid DNA was co-transfected with the green fluorescent protein (GFP) expression plasmid DNA into cells. GFP-positive cells that indicate transfected cells were gated for cell cycle and apoptotic analyses. Bar graphs show mean percentages of GFP-positive cells in sub-G1 (indicating apoptosis), G1, S and G2/M. The error bar represents the SD (n = 3). *p < 0.05, **p < 0.01, *** p < 0.001.
To assess the effect of re-expressed FOXF2 on the growth of luminal and HER2+ breast cancer cells, we transfected the HA-tagged FOXF2 expression plasmid into two cell lines of each breast cancer subtype (MCF7 and T47D for the luminal subtype; SKBR3 and MDA-MB-453 for the HER2+ subtype). As shown in Fig. 4B, FOXF2 re-expression significantly inhibited the growth of all four cell lines examined. To evaluate the effect of FOXF2 restoration on luminal breast tumor formation in vivo, we injected FOXF2-expressing and empty-vector-transfected T47D cells into mammary fat pads of athymic nude mice. The results indicated that FOXF2 re-expression almost completely blocked xenograft tumor formation of T47D cells when compared with vector-transfected T47D cells (Fig. 4C). These findings, taken together, indicate that the FOXF2 transcription factor possesses tumor-suppressor characteristics in luminal and HER2+ breast cancers.
To delineate the mechanism of FOXF2-mediated suppression of cell growth of luminal and HER2-positive breast tumor cells, cell cycle analysis was performed. The FOXF2 expression plasmid was co-transfected with the green fluorescence protein (GFP) expression plasmid (to track the transfected cells) into cells and GFP-positive cells were gated to analyze the effect of FOXF2 re-expression on the cell cycle profile. The results showed that ectopic FOXF2 expression in MCF7 cells led to a significant increase in G1-phase cells and concurrent decreases in both S- and G2/M-phase cells (Fig. 4D), indicating that FOXF2 re-expression induced G1 arrest. In addition to G1 arrest, concurrent apoptosis was observed in FOXF2-expressing, HER2-positive SKBR3 cells (Fig. 4D).
3.3. FOXF2 suppresses breast cancer cell proliferation via impairing the CDK2-RB-E2F cascade
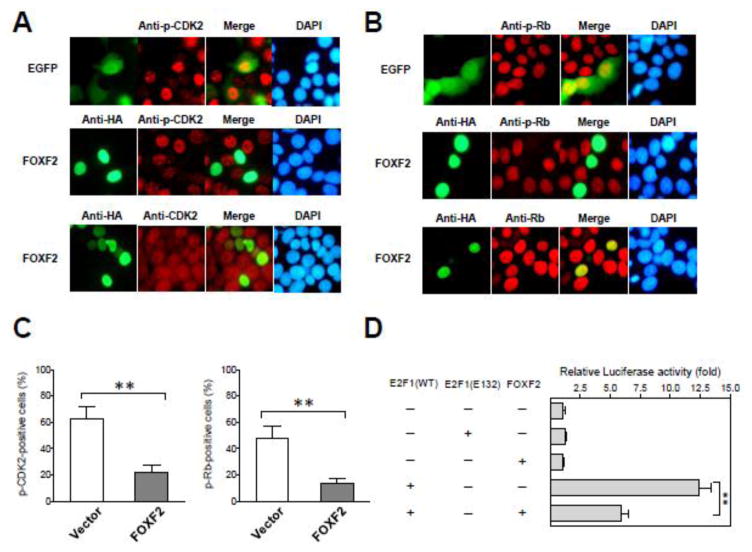
To unravel the underlying molecular mechanisms whereby FOXF2 mediates G1 arrest, protein molecules involved in the CDK2-RB-E2F cascade that controls the G1-S transition were investigated. We first analyzed the phosphorylation of CDK2 at Thr160, which is known to activate CDK2 kinase activity and in turn promote the G1-S transition [47]. Immunofluorescence analysis showed that FOXF2 re-expression in MCF7 cells diminished the phosphorylation of CDK2 at Thr160 (Fig. 5A). We also examined the phosphorylation status of RB at Ser807/811, a phosphorylated target downstream of CDK2, and found that ectopic FOXF2 expression also abrogated RB phosphorylation (Fig. 5B), which is consistent with inactive CDK2 function (Fig. 5A). FOXF2 had no significant effect on total levels of CDK2 (Fig. 5A), indicating that the FOXF2-mediated abrogation of CDK2 activity was not caused by downregulating total protein levels of CDK2. In the immunofluorescence analysis of total RB protein levels, a moderate decrease in protein levels of RB was observed in FOXF2-expressing cells compared with non-transfected cells (Fig. 5B, bottom). This effect, however, could not completely account for the dramatic suppression of RB phosphorylation by FOXF2. To investigate this further, we quantitatively assessed the cell percentage with positive staining for phosphorylated CDK2 or phosphorylated RB among FOXF2-transfected MCF7 cells compared with EGFP-plasmid-transfected control cells. FOXF2-transfected MCF7 cells were counted based on positivity for HA antibody staining that detected HA-tagged FOXF2. GFP-positive cells represented EGFP-plasmid-transfected control cells. The cell percentage positive for phospho-CDK2 or phospho-RB staining was measured among transfected cells. Consistently, FOXF2 re-expression in MCF7 cells significantly reduced cell percentages positive for both phospho-CDK2 and phospho-RB staining when compared with those of EGFP-transfected cells (Fig. 5C). These findings together demonstrate that FOXF2 inhibits CDK2 activation, which in turn suppresses the CDK2-mediated phosphorylation of RB protein.
Fig. 5.
Restoration of FOXF2 expression in FOXF2-deficient luminal breast cancer cells inhibits the CDK2-RB-E2F cascade pathway. (A) Immunofluorescence (IF) analysis was performed on GFP-transfected or HA-FOXF2-transfected MCF7 cells using the anti-HA (green) or anti-phospho-CDK2 (Thr160) (red) antibody (shown in the top and middle panels). The bottom panel is IF analysis of total CDK2 protein levels. DNA was labeled with DAPI (blue). (B) IF analysis of the transfected cells same as (A) using the anti-phospho-RB (Ser807/811) (the top and middle panels) or anti-RB (the bottom panel) antibody. (C) Quantitative analysis of the effect of ectopic expression of FOXF2 on the staining positivity of phospho-CDK2 and phospho-RB proteins in MCF7 cells. Empty vector-transfected and FOXF2-transfected MCF7 subjected to the immunostaining of phospho-CDK2 and phospho-RB as described in (A) and (B) were examined by a fluorescent microscopy to quantitate phospho-CDK2-positive and phospho-RB-positive percentages among one hundred GFP-positive (as an empty vector control) or FOXF2-positive MCF7 cells. The bar graphs (mean ± SD) from three independent experiments are shown. (D) FOXF2 attenuates E2F1 transcriptional activity. MCF7 cells were transfected with the E2F-responsive luciferase reporter plasmid DNA plus different expression plasmid DNA combinations as indicated. The β-galactosidase expression plasmid was used in assays to serve as a transfection efficiency control. The luciferase reporter activity was normalized by the β-galactosidase activity and then converted into a fold relative to the empty vector control (set as default 1). The error bar represents the SD of three independent experiments. **p < 0.01.
Aforementioned immunofluorescence and quantitative data indicate that FOXF2 re-expression suppresses CDK2 activity and consequently leads to dephosphorylation of RB, suggesting that RB function is activated to inhibit its downstream E2F1 transcriptional activity in FOXF2-re-expressing breast cancer cells. To test this possibility, we next examined the activity of the E2F1 transcription factor in FOXF2-re-expressing MCF7 cells. The effect of ectopic FOXF2 re-expression on E2F1 transcriptional activity was evaluated by the reporter assay using a reporter plasmid containing the E2F-responsive DNA polymerase-α promoter upstream of the luciferase reporter gene [48]. The luciferase activity of this E2F-responsive reporter was induced by wild-type E2F1 (almost 12.5-fold induction), but not mutant E2F1 (E132) [49], when compared with the vector control (Fig. 5D). Co-transfection of the FOXF2 expression plasmid with the wild-type E2F1 expression plasmid significantly suppressed the E2F1-mediated transactivation (~50% reduction) of E2F reporter activity (Fig. 5D). This suppression was E2F1-specific since FOXF2 had no significant effect on the reporter activity in the absence of E2F1 (Fig. 5D). The in vitro reporter assay data were reflective of the immunofluorescence data showing a consistent, moderate decrease in endogenous total RB protein levels in FOXF2-transfected cells because RB is one of known target genes downstream of E2F transcription factors. These results, taken together, suggest that FOXF2-induced inhibition of the CDK2-RB-E2F signaling cascade contributes to a blockage of the G1-S transition of the cell cycle.
3.4. FOXF2-dependent DNA replication regulation is lost in basal-like breast cancer
Since FOXF2 is a negative regulator of the G1-S transition of the cell cycle, it raises an interesting question of how basal-like breast cancer cells can tolerate high expression levels of FOXF2. To address this question, we performed siRNA knockdown experiments to examine the effect of loss of endogenous FOXF2 function on cell cycle progression. We tested the knockdown efficiencies of four different FOXF2 siRNAs in MCF10A cells and identified siRNA-1 and siRNA-3 as most effective for FOXF2 knockdown (Supplementary Fig. 4). To evaluate the effect of FOXF2 inactivation on cell cycle progression, we performed FOXF2 knockdown using these two most effective FOXF2 siRNAs on human mammary epithelial cells (HMECs) isolated from reduction mammoplasty tissues. Flow cytometric analysis was performed on siRNA-knockdown HMECs stained with propidium iodide for generating cell cycle profiles. As shown in Fig. 6A, the cell cycle progression of HMECs was arrested in the G1 phase of the cell cycle after treatment with these two distinct siRNAs, indicating that FOXF2 is functionally required for the normal cell cycle progression of HMECs. Given the outcomes from these two different siRNA knockdowns were consistent with each other, the most effective FOXF2 siRNA-3 was utilized throughout the following experiments. To examine the FOXF2-knockdown effect on basal-like breast cancer cells, we performed BrdU incorporation analysis with the co-staining of propidium iodide on MCF10A (the normal control), HBL-100 (the immortalized HMEC line with inactive p53), basal-like MDA-MB-231 and MDA-MB-468 cell lines transfected with either the non-targeting control or FOXF2 siRNA. The knockdown efficiencies of FOXF2 siRNA-3 in these four cell lines were confirmed by qRT-PCR (Supplementary Fig. 5). Similar to HMECs, FOXF2 knockdown strongly abolished DNA replication and induced G1 arrest in MCF10A cells (Fig. 6B). In contrast, FOXF2 knockdown had no effect on the DNA replication, cell cycle progression and polyploid status of HBL-100, MDA-MB-231 and MDA-MB-468 cells (Fig. 6B).
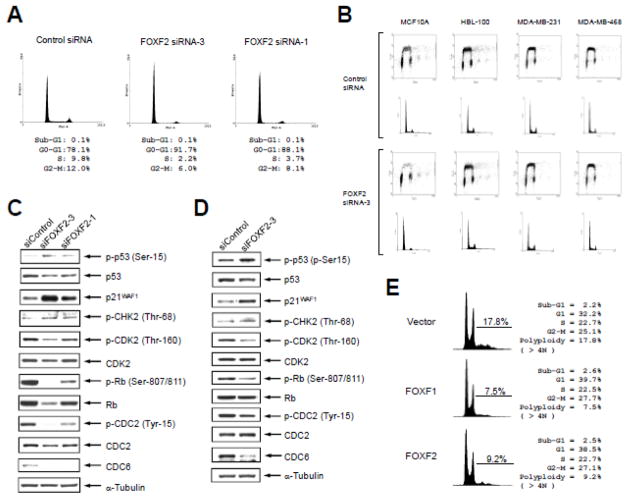
Fig. 6.
FOXF2 functions as a negative regulator of DNA replication and its inhibition leads to activation of the CHK2-p53 signal pathway. (A) FOXF2 knockdown induces G1 arrest in HMEC cells. HMEC cells were transfected with two different FOXF2 siRNAs and their cell cycle profiles were analyzed by flow cytometry 48 hrs after transfections. (B) The effect of FOXF2 knockdown on the DNA replication and cell cycle progression correlates with the functional status of p53. BrdU incorporation assays were performed on four breast cell lines transfected with either control or FOXF2 siRNA as indicated. For each siRNA transfection group, the 2D dot plot profiles of the incorporated BrdU fluorescent intensity vs. DNA content (indicated by prodium iodide staining intensity) are shown in the top panel and the DNA content histograms are shown in the bottom panel. (C, D) FOXF2 knockdown activates the CHK2-p53-p21WAF1 signaling axis in normal breast epithelial cells. Primary HMEC cultures isolated from breast tissue were transfected with two distinct FOXF2 siRNAs (or the control siRNA). Cells were harvested 48 h post-transfection for Western blot analysis of proteins as indicated in (C). Immortalized HMECs (MCF10A) were transfected with siFOXF2-3 (or the control siRNA) and were then subjected to the same protein analysis as mentioned in (C). WB results of MCF10A cells are shown in (D). (E) Ectopic expression of FOXF2 suppresses DNA re-replication of Hs578T cells. Hs578T cells were transfected with the empty vector, FOXF1 (a positive DNA re-replication inhibitor) or FOXF2 expression plasmid DNA. Cells were harvested 48 h post-transfection for propidium iodide staining and flow cytometric analysis. Polyploid cells with the DNA content over 4N were gated for measuring the percentage of cells with DNA re-replication.
It is known that DNA over-replication (re-replication) elicits ATM-CHK-dependent DNA damage signaling that activates the p53 pathway, which leads to cell cycle arrest and/or apoptosis for maintaining genomic stability [9,10,50]. Given that FOXF2 inhibits DNA replication by blocking the CDK2-RB-E2F cascade, we postulated that abrogation of FOXF2 function results in aberrant DNA replication (re-replication) and consequently triggers the activation of ATM-CHK-p53 signaling to arrest cell cycle progression for prevention of re-replication occurrence. To test this possibility, we performed Western blot analysis of DNA damage signaling components and key molecules involved in ATM-CHK-p53 signaling and G1-S as well as G2-M progression on FOXF2-knockdown HMECs by either FOXF2 siRNA-1 or siRNA-3 compared with control siRNA-transfected cells. As shown in Fig. 6C, FOXF2 knockdown activated CHK2 (indicated by increased phosphorylation at Thr-68), a DNA-damage-dependent kinase that is downstream of ATM and activates p53 signaling. Consistently, we observed p53 activation (indicated by increased phosphorylation at Ser-15) and the increased expression of the p53-downstream target p21WAF1 (Fig. 6C). Therefore, the activation of the CHK2-p53-p21 signaling axis by FOXF2 knockdown is likely a main mechanism that suppresses the CDK2-RB cascade (indicated by decreased phospho-CDK2 and phospho-RB protein levels), which is critically involved in initiating the G1-S transition of the cell cycle (Fig. 6C). In contrast, the kinase activity of CDC2 was stimulated by FOXF2 knockdown (indicated by its decreased phosphorylation at Tyr-15, an inhibitory phosphorylation site) (Fig. 6C). Inhibition of CDK2 and activation of CDC2 are consistent with the G1 arrest phenotype in FOXF2-knockdown HMECs. In addition, we found that the protein level of the licensing factor CDC6, a key molecule involved in DNA replication initiation [50], was substantially decreased in FOXF2-depleted HMECs. This effect also contributed to the inhibition of DNA replication in FOXF2-knockdown cells (Fig. 6A). Since activation of p53 by DNA damage has been shown to promote CDC6 degradation by the anaphase-promoting complex [51], downregulation of CDC6 protein levels in FOXF2-knockdown HMECs was possibly attributable to activated p53. Given that both FOXF2 siRNA-1 and siRNA-3 gave rise to the same phenotype, these outcomes were not caused by the non-specific targeting effect of the siRNA. These results were recapitulated in MCF10A cells with FOXF2 knockdown by siRNA-3 (Fig. 6D). Since both MDA-MB-231 and MDA-MB-468 are p53 mutant cell lines and HBL-100 harbors the large T antigen to inactivate p53 protein, FOXF2 knockdown was unable to activate the p53-p21 pathway in these cell lines, which possibly explains why FOXF2 abrogation had no impact on the cell cycle progression of these three cell models (Fig. 6B).
To address whether FOXF2 is able to negatively regulate DNA re-replication as we postulated, we transfected the FOXF2 expression plasmid into the basal-like Hs578T breast cancer cell line that contains a substantial polyploid cell population (17.8%). Given that polyploid cells harbor re-replicated DNA, the percentage of the polyploid cell population indicates the extent of DNA re-replication (over-replication) [50]. Therefore, Hs578T cells had re-replicated DNA due to defects in DNA replication stringency. Hs578T is one of few basal-like breast cancer cell lines with moderate silencing of FOXF2 expression compared with normal breast epithelial cells (Fig. 1A) and also carries the epigenetically silenced FOXF1 [24] and FOXF2 (Fig. 1B) genes, correlating with the polyploid phenotype of this cell line. As shown in Fig. 6E, ectopic FOXF2 overexpression significantly decreased polyploid cells by 48.3% compared to vector-transfected control cells, which was similar to FOXF1 (a DNA re-replication inhibitor control reported by us [24,52]) that reduced polyploid cells by 57.9%. These data together demonstrate that FOXF2 is a negative regulator of DNA re-replication and thus plays a critical role in maintaining genomic stability. This function is lost in p53-inactivated, FOXF2-expressing breast cells (e.g. HBL-100, MDA-MB-231, MDA-MB-468) since FOXF2 knockdown in these cell lines failed to induce an increase in the polyploid cell population (re-replication) (Fig. 6B).
3.5. FOXF2 is required for the invasive and tumorigenic ability of basal-like breast cancer cells
It has been shown that FOXF2 is involved in the epithelial-to-mesenchymal transition (EMT) and mediates invasion and metastasis in lung cancers [32]. These findings raise the possibility that FOXF2 function is required for the invasive ability of FOXF2-expressing basal-like breast cancer cells. To test this possibility, we first examined whether FOXF2 is functionally required for cell mobility of normal mammary epithelial cells. To assess the role of FOXF2 in cell mobility, we performed wound healing assays on HMECs transfected with either the control or FOXF2 siRNAs. As shown in Fig. 7A (the left panel), FOXF2 knockdown by siRNA-3 manifested the substantial inhibitory effect on cell mobility of HMECs (~75% inhibition) in comparison with siRNA-1 showing a lesser inhibitory effect (~40% inhibition), consistent with the knockdown efficiencies of these two FOXF2 siRNAs (Supplementary Fig. 4). We observed the same inhibitory effect when FOXF2 knockdown was performed on MCF10A cells (Fig. 7A, the right panel). Consistently, we observed that FOXF2 knockdown in MCF10A cells altered their cell morphology from the more mesenchymal-like, spindle shape to the more epithelial-like, polygonal shape (Fig. 7B), implying a mesenchymal-to-epithelial transition (MET) after FOXF2 knockdown. These data demonstrate that FOXF2 function is vital for cell mobility of normal mammary epithelial cells.
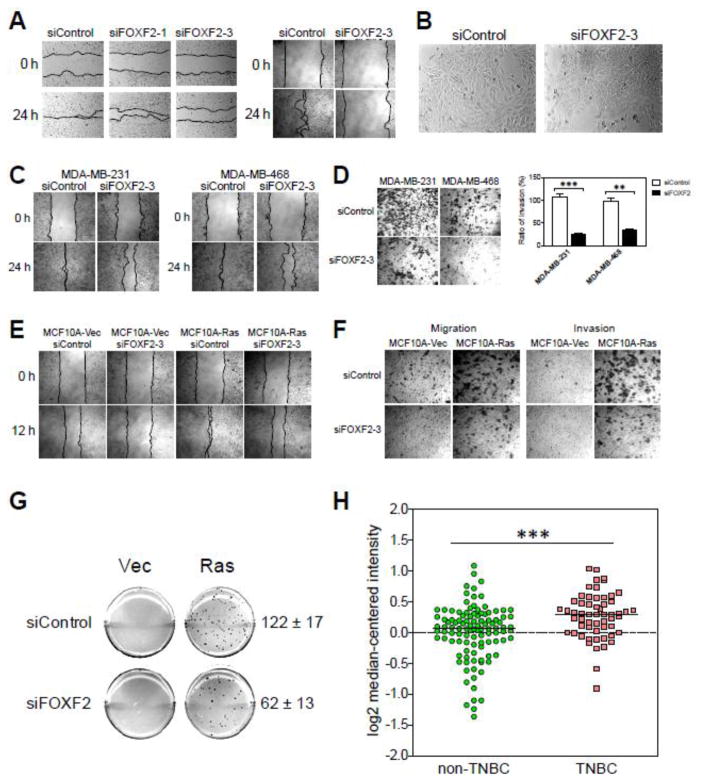
Fig. 7.
FOXF2 is functionally required for the mobility of breast epithelial cells and for the invasiveness as well as anchorage-independent growth of basal-like breast cancer cells, and is overexpressed in triple-negative breast tumors. (A) Inactivation of FOXF2 inhibits cell mobility of HMECs. Primary and immortalized (MCF10A) HMECs were transfected with the control or FOXF2 siRNA and then subjected to wound healing assays 24 h post-transfection. Transfected cells were allowed to migrate for 24 h before pictures were taken. (B) FOXF2 inhibition alters the morphology of MCF10A cells from the spindle-like shape to the polygonal shape. (C) Abrogation of FOXF2 attenuates cell mobility of basal-like breast cancer cells. At 24 h post-transfection, control or FOXF2 siRNA-transfected MDA-MB-231 and -486 cells were subjected to would healing assays. Migration pictures were taken at 24 hr of assays. (D) FOXF2 is functionally required for invasiveness of basal-like breast cancer cells. Control or FOXF2 siRNA-transfected MDA-MB-231 and -468 cells were subjected to invasion assays for 24 h and then stained with crystal violet for visualization (the left panel) and quantitation (the right panel). The error bar represents the SD (n = 3). (E) Abrogation of FOXF2 attenuates the mobility of Ras-transformed MCF10A cells. MCF10A-Vec and MCF10A-Ras cells transfected with either the control or FOXF2 siRNA were subjected to wound healing analysis for assessing their migration ability. (F) FOXF2 inactivation suppresses the migratory and invasive ability of Ras-transformed MCF10A cells. siRNA-transfected cells aforementioned in (E) were subjected to assays using transwell and invasion chambers for measuring their migratory and invasive ability. (G) FOXF2 knockdown inhibits anchorage-independent growth of Ras-transformed MCF10A cells. siRNA-transfected cells aforementioned in (E) were subjected to soft agar assays for examining their anchorage-independent growth. Three independent experiments were performed and the difference between the numbers of colonies formed from siControl- and siFOXF2-transfected MCF10A-Ras cells is statistically significant (p < 0.01). (H) In silico expression analysis of FOXF2 in invasive ductal breast carcinomas (IDC). FOXF2 expression in IDC tumors (n = 163) was plotted based on previously published Tabchy’s Breast Cancer microarray expression dataset deposited in the Oncomine microarray database. IDC tumors subjected to in silico expression analysis were classified into triple-negative breast cancers (TNBC) (n = 57) and non-TNBC (n = 106) according to ER, PR and ERBB2 status. The median of each dataset is indicated in the plot. ** p < 0.01; *** p < 0.001.
To decipher whether FOXF2 is functionally required for the cell migration and invasion ability of basal-like breast cancer cells, we performed wound healing (Fig. 7C) and invasion (Fig. 7D) assays on FOXF2-knockdown MDA-MB-231 and MDA-MB-468 cells. The results showed that inhibition of FOXF2 significantly impaired cell migration (~45% inhibition in MDA-MB-231 cells; ~33% inhibition in MDA-MB-468 cells) and invasion (~73% inhibition in MDA-MB-231 cells; ~59% inhibition in MDA-MB-468 cells) of these two cell lines (Fig. 7C, 7D). Moreover, we tested the effect of FOXF2 knockdown on Ras-transformed MCF10A cells, a basal-like pre-malignant breast cell model manifesting accelerating cell migration and acquired invasive ability. Consistently, abrogation of FOXF2 significantly inhibited cell migration (~45% inhibition) and invasion (~63% inhibition) ability of MCF10A-Ras cells (Fig. 7E, 7F). We also examined whether FOXF2 is pivotal for the anchorage-independent growth of MCF10A-Ras cells. FOXF2 knockdown resulted in the 50% inhibition of the anchorage-independent growth of MCF10A-Ras cells (Fig. 7G). These findings, taken together, demonstrate that FOXF2 function is required for the malignant features of basal-like breast cancer cells, including cell migration, invasion and anchorage-independent growth. To unravel whether FOXF2 expression status is associated with basal-like breast cancers, we performed in silico expression analysis of FOXF2 in triple-negative breast cancers (TNBC) that encompass 70–80% of basal-like breast cancers compared with non-TNBC. We analyzed Tabchy’s Breast Cancer Microarray Dataset (including 163 IDC cases) [53] deposited in the Oncomine database [33] and found that FOXF2 was overexpressed in TNBC (n = 57) when compared with non-TNBC (n = 106) (p < 0.001) (Fig. 7H). This finding indicates that FOXF2 tends to be overexpressed in TN/basal-like breast cancers in contrast to its overall underexpression trend in non-TNBC, consistent with the aforementioned result that the DNA methylation status of the FOXF2 promoter was negatively associated with the TN status of IDC (Table 1). This suggests that aberrant overexpression of FOXF2 in TN/basal-like breast cancers may contribute to the development of this breast cancer subtype.
3.6. FOXF2 has a dual role in regulating cell growth and EMT during breast tumorigenesis
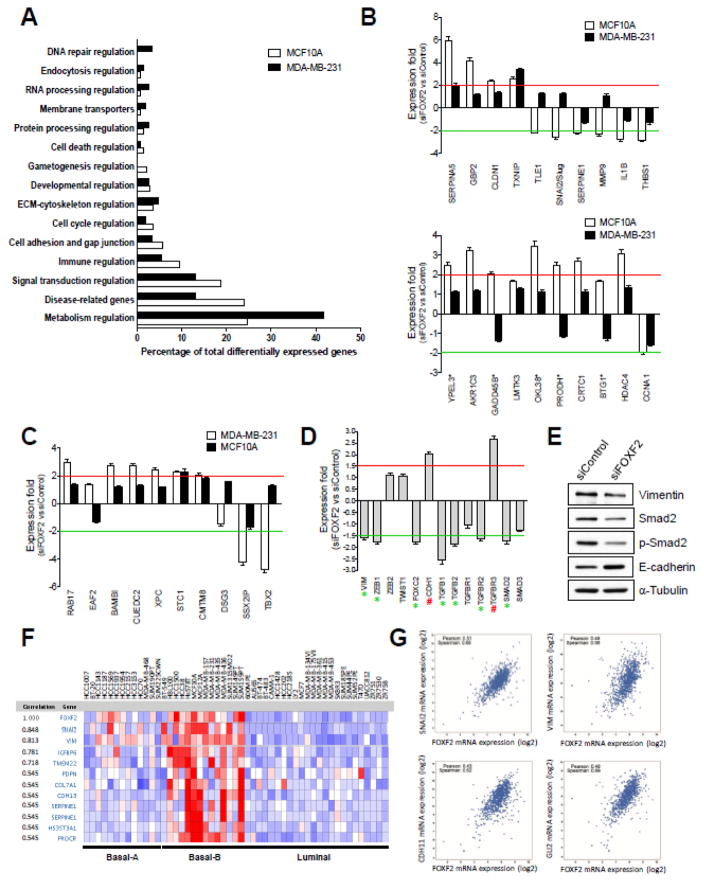
To unveil the biological roles of FOXF2 in normal breast epithelial and metastatic basal-like breast cancer cells, we performed global gene expression profiling analyses of FOXF2-knockdown MCF10A and MDA-MB-231 cells compared with their control siRNA-transfected cells using microarrays. We identified 199 and 309 differentially expressed genes (≥ 2 fold) in FOXF2-knockdown MCF10A and MDA-MB-231 cells, respectively (Supplementary Tables 2 and 3). Intriguingly, only 15 genes (highlighted by red color in Supplementary Tables 2 and 3) overlap between these two differentially expressed gene sets, indicating FOXF2-dependent gene regulation was significantly altered in this triple-negative metastatic breast cancer cell line. Based on gene ontology enrichment analysis, 138 and 144 differentially expressed genes from FOXF2-knockdown MCF10A and MDA-MB-231 cells, respectively, were classified into 15 different functional categories. In comparison to MCF10A, metastatic MDA-MB-231 cells expressed more FOXF2-regulated genes involved in regulating metabolism, ECM-cytoskeleton, protein processing, membrane transporters, RNA processing, endocytosis and DNA repair, and exhibited less FOXF2-regulated genes implicated in regulating disease-related mechanisms, signal transduction, immunological responses, cell adhesion and gap junction, cell cycle, gametogenesis and cell death (Fig. 8A).
Fig. 8.
The FOXF2 transcription factor regulates genes involved in controlling cell cycle progression and EMT, and FOXF2 is co-expressed with EMT-programming and metastasis-related genes in breast tumors. (A) Functional ontology of genes differentially expressed in FOXF2-knockdown MCF10A and MDA-MB-231 cells. (B) Expression of EMT- (the top graph) and cell-cycle-related (the bottom graph) genes selected from the microarray data of FOXF2-knockdown MCF10A cells was validated by using qRT-PCR analysis. The results from qRT-PCR analysis of the same set of genes in FOXF2-knockdown MDA-MB-231 cells are also included in bar graphs for comparison. The expression fold was calculated by the gene expression value of siFOXF2 relative to that of siControl. The p53 target genes are indicated by asterisks. (C) Expression of EMT- and metastasis-related genes selected from the microarray data of FOXF2-knockdown MDA-MB-231 cells was validated by using qRT-PCR analysis. The results from qRT-PCR analysis of the same set of genes in FOXF2-knockdown MCF10A cells are also included in the bar graph for comparison. The expression fold was calculated as described in (B). (D) The effect of FOXF2 inhibition on the expression of genes involved in EMT and TGFβ signaling. Quantitative RT-PCR analysis of 13 genes functionally implicated in EMT and TGFβ signaling was performed on control and FOXF2 siRNA-transfected MCF10A cells. Upregulated and downregulated genes (≥1.5 fold) in FOXF2 siRNA-transfected cells are indicated by the pound (#) and asterisk (*) signs, respectively. (E) The effect of FOXF2 knockdown on the protein expression of Vimentin, Smad2, phospho-Smad2 and E-cadherin. Western blot analysis of Vimentin, Smad2, phospho-Smad2 (Ser465/467), E-cadherin and α-tubulin was performed on control and FOXF2 siRNA-transfected MCF10A cells. α-tubulin protein levels were used as a loading control. (F) In silico co-expression analysis of FOXF2 with EMT/basal- and stem-cell-related genes in 51 breast cancer cell lines. The co-expression results were from in silico analysis of the Oncomine cancer microarray database. (G) In silico co-expression analysis of FOXF2 with EMT- and metastasis-related genes in breast tumors. Co-expression analysis of FOXF2 against three EMT-related genes (SNAI2, VIM, CDH11) and one metastasis-related gene (GLI2) was derived from in silico analysis of their expression in 960 breast tumors using the cBioPortal for Cancer Genomics.
To understand the functional roles of FOXF2 in the regulation of the cell cycle and EMT, the differentially expressed genes with relevant roles in these two biological processes were selected (Supplementary Tables 4 and 5). Consistent with the aforementioned FOXF2-knockdown phenotypes in MCF10A cells, most of upregulated EMT-related genes function either as EMT and metastasis suppressors (e.g. SERPINA5, TXNIP, RPS6KA6, STC1, PRSS8), or as cell adhesion and gap junction molecules (e.g. CLDN1, COL8A1, PANX2, GJD3); whereas, all downregulated EMT-related genes (TLE1, SNAI2/Slug, SERPINE1, CSF3, MMP9, CCL20, TMSL8, IL1B, THBS1) are involved in promoting EMT and metastatic processes (Supplementary Table 4). Among 18 upregulated cell-cycle-related genes, five of them are p53 target genes (YPEL3, GADD45B, OKL38, PRODH, BTG1) (Supplementary Table 4), which was in line with the activated p53 pathway in FOXF2-knockdown MCF10A cells (Fig. 6C, 6D). In contrast, they were not activated in FOXF2-knockdown MDA-MB-231 cells, consistent with the p53-mutated status of this cell line. With regard to the rest of upregulated genes, the majority of them play oncogenic roles and are involved in regulating cell cycle progression and facilitating cell growth via multiple mechanisms, including AKR1C3, LMTK3, SERPINA3, GABARAPL1, MIF4GD, CRTC1, HDAC4, PINK1, IFITM3, and SPIN1 (Supplementary Table 4). Interestingly, we observed that FOXF2 may be required for Cyclin A1 expression in MCF10A cells as its expression was downregulated by FOXF2 knockdown (Supplementary Table 4). In FOXF2-knockdown MDA-MB-231 cells, we identified 11 upregulated and 3 downregulated genes implicated in the regulation of EMT and/or cell growth (Supplementary Table 5). Although only three genes (TXNIP, EFNA1, STC1) show overlap between these two sets of differentially expressed genes listed in Supplementary Table 4 and Table 5, FOXF2-regulated genes in MDA-MB-231 show the similar functional trend as those genes found in MCF10A cells. For upregulated genes in FOXF2-knockdown MDA-MB-231 cells, they function as the suppressors to inhibit cell migration, invasion and proliferation via various mechanisms including inactivation of TGFβ (BAMBI), NFκB (CUEDC2) and c-Met/ERK (CMTM8) signaling pathways and activation of E-cadherin expression (XPC) (Supplementary Table 5). In contrast, three downregulated genes (DSG3, SSX2IP, TBX2) function as the promoters to enhance EMT and metastasis (Supplementary Table 5). These aforementioned comparative analyses of two expression microarray datasets (Supplementary Table 4, 5) indicate that MDA-MB-231 cells largely lost the normal EMT-regulatory function of FOXF2 which was found in MCF10A cells and developed altered FOXF2 function that regulates EMT and metastasis.
To validate the microarray-based gene expression profiling results, we performed quantitative RT-PCR assays to analyze the expression of these identified genes involved in the regulation of the cell cycle and EMT. We validated 10 EMT-related as well as 10 cell-cycle-related genes identified from MCF10A cells and 10 genes from MDA-MB-231 cells. As shown in Fig. 8B and 8C, expression of over 80% of these analyzed genes could be validated in qRT-PCR analysis. Owing to the limited sensitivity and accuracy of microarray analysis in gene expression detection, we performed additional qRT-PCR analysis of 13 genes involved in EMT and TGFβ signaling, a critical pathway to induce EMT. As shown in Fig. 8D, FOXF2 knockdown resulted in downregulating (≥1.5 fold downregulation) the expression of EMT-programming genes (Vimentin (VIM), ZEB1, FOXC2) and TGFβ pathway genes (TGFB1, TGFB2, TGFBR2, SMAD2) in MCF10A cells. Conversely, the expression of the cell adhesion protein E-cadherin was upregulated (> 2.0 fold) in MCF10A cells by FOXF2 knockdown (Fig. 8D). Interestingly, the expression of TGFBR3, a membrane receptor that inhibits TGFβ signaling, was significantly enhanced (> 2.0 fold) in MCF10A cells by FOXF2 inhibition (Fig. 8D). Western blot analysis of protein levels of Vimentin, Smad2 and E-cadherin in FOXF2-knockdown MCF10A cells confirmed the qRT-PCR result (Fig. 8E). We also found that FOXF2 knockdown attenuated TGFβ signaling, indicated by decreased levels of phospho-Smad2 (Ser465/467) (Fig. 8E). These findings are consistent with the results of functional (Fig. 7) and global gene expression profiling (Supplementary Table 4) studies, indicating that FOXF2 knockdown leads to inhibition of EMT. These gene expression analysis results together suggest that FOXF2 has a dual functional role in regulating EMT and cell-cycle-regulatory genes to promote EMT and inhibit cell growth, respectively. In addition, our findings also show that loss or downregulation of FOXF2 function indirectly activates the p53 checkpoint pathway via deregulating DNA replication and cell cycle progression.
To determine whether FOXF2 is co-expressed with EMT/basal-related genes in breast cancer, we performed in silico analysis of cancer microarray databases using Oncomine (http://www.oncomine.org) [33]. As shown in Fig. 8F, FOXF2 was significantly co-expressed with EMT/basal-related genes in 51 breast cancer cell lines. Remarkably, FOXF2 was highly expressed with these EMT/basal genes in most of basal-B subtype breast cancer cell lines (Fig. 8F). It is noteworthy that FOXF2 was highly co-expressed with two well-known EMT genes SNAI2/Slug and Vimentin (VIM) in basal-A and basal-B breast cancer cell lines (Fig. 8F). Our expression profiling studies also identified that SNAI2/Slug is positively regulated by FOXF2 in MCF10A cells (Fig. 8B, Supplementary Table 4). Through in silico analysis of FOXF2 expression in 960 primary breast tumors using the cBioPortal for Cancer Genomics (http://www.cbioportal.org) [34], we found that FOXF2 was significantly co-expressed with three EMT/basal genes (SNAI2/Slug, VIM, CDH11) in breast carcinomas (Fig. 8G). CDH11 has been reported as a TGFβ-inducible gene whose function is required for TGFβ-induced EMT [54]. Interestingly, we also found that FOXF2 was highly co-expressed with GLI2 in breast cancer, a well-known breast cancer metastasis-promoting gene critically involved in the hedgehog signaling pathway (Fig. 8G) [55,56]. These in silico analysis findings together with aforementioned gene expression profiling results strongly suggest that FOXF2 is a basal-specific gene critically implicated in promoting EMT and metastasis of triple-negative/basal subtype breast cancer.
4. Discussion
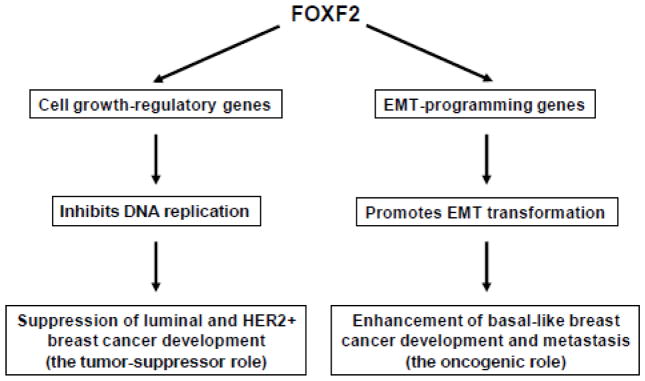
In this article, we report that FOXF2 plays a dual role in regulating DNA replication and EMT, which enables this gene to function either as a tumor suppressor or as an oncogene in a context-dependent manner (Fig. 9). We found that FOXF2 is frequently silenced in luminal and HER2-positive breast cancers via epigenetic mechanisms, which is predominantly attributable to the hypermethylation of the FOXF2 CpG-island-containing promoter. Hypermethylation of the FOXF2 promoter is positively correlated with the tumor size and stage, but inversely associated with patients’ age and triple-negative status. The low expression of FOXF2 is associated with the poor prognosis of luminal-type breast cancers. Restoration of FOXF2 expression in luminal and HER2+ breast cancer cells inhibits their tumorigenicity. FOXF2-mediated tumor suppression involves a blockage of cell cycle progression at the G1-S transition and induction of apoptosis. FOXF2 restoration inhibits DNA replication in FOXF2-deficient breast cancer cells via suppressing the CDK2-RB-E2F cascade. In contrast, FOXF2 is frequently overexpressed in triple-negative (TN)/basal-like breast cancers, in particular the more malignant basal-B subgroup. The tumor-suppressive function of FOXF2 in regulating DNA replication is lost in basal-like breast cancer, explaining why this subtype of breast cancer can exhibit the overexpressed levels of FOXF2. FOXF2 is required for malignant characteristics of basal-like breast cancer, including cell migration, invasion and anchorage-independent growth. Moreover, we found that FOXF2 expression in breast cancer is associated with basal-signature and stem-cell-related genes. The dual role of FOXF2 in breast carcinogenesis is further supported by genome-wide gene expression analyses using microarray, qRT-PCR and Western blot methods, showing that FOXF2 can regulate genes involved in controlling cell cycle progression, EMT and TGFβ signaling.
Fig. 9.
The paradigm for the dual role of FOXF2 that functions as either a tumor suppressor or an oncogene in breast cancers.
Our new findings presented here have revealed that luminal and HER2-positive breast cancer cells adopt the epigenetic mechanism to silence FOXF2 expression. The DNA methyltransferase-mediated DNA methylation of the CpG-island-containing FOXF2 promoter is the predominant mechanism for epigenetically silencing FOXF2 expression. In addition, our studies have also shown that histone deacetylation is involved in epigenetically silencing FOXF2 expression in an independent manner or in collaboration with DNA methylation. It has recently been shown that FOXF2 is the target of oncogenic and tumor-suppressive microRNAs, which are another epigenetic mechanism to silence FOXF2 expression. FOXF2 was reported to be directly targeted and downregulated by oncogenic miR-182-5p that is involved in promoting cell invasion and proliferation of prostate and colorectal cancers [57,58]. Later FOXF2 expression was also found to be negatively regulated by miR-301 that plays an oncogenic role in human breast cancer and is involved in facilitating cell proliferation, clonogenicity, migration, invasion, tamoxifen resistance, tumor growth, and microvessel density [59]. Moreover, miR-519a is a most recently reported oncogenic miRNA that promotes proliferation and inhibits apoptosis of hepatocellular carcinoma cells by targeting FOXF2 [60]. Therefore, downregulation of FOXF2 expression by oncogenic microRNAs supports the tumor-suppressive role of FOXF2 in breast, colorectal, prostate and liver cancers. Besides silencing by oncogenic microRNAs, Kundu et al. recently reported that the miR-200 family and the miR-183 ~ 96 ~ 182 cluster target and downregulate FOXF2 to suppress invasion and metastasis in lung cancers, indicating that FOXF2 functions as a metastatic promoter [32]. These five lines of miRNA-related evidence together suggest that FOXF2 is capable of functioning as either a tumor suppressor or an oncogene in a tissue- or stage-specific manner, which is consistent with the conclusion from our studies. It is noteworthy that the miR-200 family is selectively silenced in basal-like breast cancer, not in luminal-type breast cancer [61]. Therefore, basal-specific silencing of the miR-200 family potentially contributes to the FOXF2-overexpressing phenotype in basal-like breast cancer.
Multiple lines of evidence presented in this paper have revealed that FOXF2 plays differential roles in distinct breast cancer molecular subtypes. Our studies showed that ectopic expression of FOXF2 in FOXF2-deficient luminal and HER2+ breast cancer cells led to a blockage in the CDK2-Rb-E2F signal axis, triggered G1 arrest with or without induced apoptosis and suppressed xenograft tumor formation in vivo. Therefore, in luminal-type and HER2-positive breast cancers, FOXF2 functions as a tumor suppressor to stringently control DNA replication and restrict cell growth. Moreover, these findings correlate with the survival analysis showing that low expression of FOXF2 is statistically associated with the poor prognosis of luminal breast cancer patients. In normal breast epithelial cells, abrogation of FOXF2 function by siRNA knockdown resulted in the elevated expression of growth-promoting genes. These aberrant alterations are the potential causes that drive activation of the p53 checkpoint pathway, which in turn transactivates p53 target genes involved in growth suppression and induces cell cycle arrest at the G1 phase. What surveillance mechanism is induced in FOXF2-deficient cells to activate the ATM-CHK-p53-p21 checkpoint cascade is unknown. However, it may be the same mechanism that elicits activation of the ATM-CHK-p53-p21 signaling axis upon overexpression of DNA replication initiation factors (e.g. Cdt1 and Cdc6) or Cyclin E [50,62]. In contrast, FOXF2 inhibition in p53-inactivated breast epithelial cells (e.g. HBL-100) and p53-mutated basal-like breast cancer cells (e.g. MDA-MB-231 and MDA-MB-468) had no effect on DNA replication, cell cycle progression and polyploid (re-replication) status, suggesting that FOXF2 likely loses its DNA-replication-regulatory function during or after p53 function is abrogated. Our gene expression profiling findings that showed substantial alterations in FOXF2 transcriptional function in basal-like breast cancer cells support this paradigm. This aberration likely allows FOXF2-overexpressing basal-like breast cancer cells to escape from the FOXF2-mediated tumor suppressive effect and to take advantage of the oncogenic effect of FOXF2 in facilitating EMT transformation. This proposed paradigm (Fig. 9) is based on multiple lines of evidence from our functional studies, global gene expression profiling and in silico analyses of multiple public cancer gene expression databases, including: (1) abrogation of FOXF2 by siRNA knockdown has no impact on the cell cycle and polyploidy, but attenuates the invasive capability of basal-like breast cancer cells; (2) FOXF2 tends to be overexpressed in TN/basal-like breast cancers (especially basal-B); (3) expression of EMT inhibitor and metastatic suppressor genes is upregulated by FOXF2 knockdown in basal-like breast cancer cells, whereas expression of EMT and metastatic promoter genes is downregulated under the same FOXF2 knockdown condition. These results suggest that FOXF2 is an intrinsic EMT and metastatic promoter; (4) FOXF2 is co-expressed with EMT inducer (e.g. SNAI2/Slug, Vimentin, CDH11) and metastatic promoter (GLI2) genes in breast carcinomas.
Our findings support FOXF2 functions as a promoter for EMT and metastasis, which is contradictory to findings from Feng’s group [27,29]. This discrepancy between our and Feng’s findings may be due to differences in the experimental approach, such as the FOXF2 knockdown strategies, the FOXF2 expression plasmid and cell line properties (e.g. FOXF2 expression status). However, the conclusions from Feng’s studies are inconsistent with the intrinsic functional roles of FOXF transcription factors in inducing and maintaining the mesenchymal/EMT phenotypes of stromal and basal-type cancer cells reported elsewhere [22,25,30–32]. In contrast, our findings from studies of FOXF1 [24,52] and FOXF2 support a model that the tumor-suppressor roles of FOXF factors are derived from their functions as the negative regulators of DNA replication, but not from their EMT functions. Based on our model, the paradigm proposed by Feng and colleagues [27,29] is unable to explain how basal-like breast cancer cells can tolerate overexpressed levels of FOXF2. Recently Kundu et al. reported that FOXF2 expression can be induced by TGFβ treatment, suggesting FOXF2 mediates the TGFβ signaling effect [32]. In addition, they also found that the miR-200 family, functioning as EMT suppressors, downregulate FOXF2 expression [32]. Importantly, our studies showed that FOXF2 functions as the activator of TGFβ signaling by regulating the expression of TGFβ pathway components (Fig. 8D, 8E). Therefore, FOXF2 forms a positive feedback regulation loop with TGFβ signaling. These findings together support our proposed paradigm because FOXF2, as the upstream activator and downstream mediator of TGFβ signaling, may mediate the dual role of TGFβ signaling in suppressing cell cycle progression (the tumor-suppressor function) and in promoting the EMT and metastatic processes (the oncogenic function).
In conclusion, our studies for the first time decipher that FOXF2 possesses a dual function to act as either a tumor suppressor that negatively regulates DNA replication or an oncogenic factor that drives EMT and metastatic progression in a context-dependent manner. This paradigm provides a rational explanation for the contradictory phenotypes of downregulated FOXF2 expression in luminal-type and HER2-positive breast cancers but upregulated expression in basal-like breast cancers. Our findings reported here also suggest a hypothesis that during the development of basal-like breast cancer, the transcriptional regulation by FOXF2 is altered to change FOXF2 function from a tumor suppressor to an oncogenic factor. Elucidation of the molecular mechanism underlying this alteration will be clinically relevant as this aberrant mechanism can be potentially targeted for therapeutically restoring the tumor-suppressive function of FOXF2 in basal-like breast cancer cells.
Supplementary Material
Highlights.
FOXF2 plays a dual role in regulation of DNA replication and EMT.
FOXF2 is epigenetically silenced in luminal and HER2-positive breast cancers.
FOXF2 functions as a tumor suppressor in luminal and HER2-positive breast cancers.
FOXF2 is frequently overexpressed in triple-negative/basal-like breast cancers.
FOXF2 is an oncogenic factor required for basal-like breast cancer development.
Acknowledgments
The work was supported by the National Institutes of Health [grant number: SPORE-1 P50 CA088843-08] and the Department of Defense [grant number: Center of Excellence-W81XWH-04-1-0595] to SS. We thank Dr. W. Kaelin, Dana-Farber Cancer Institute at Boston, for kindly providing the promoter-reporter plasmid of DNA polymerase α gene and E2F1 expression plasmids (wild-type and E132 mutant); and Dr. P. Carlsson, Göteborg University, Sweden, for kindly providing reporter plasmids pApo-Luc and p4XFREAC-Luc.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 2.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Rakha EA, Richardson AL, Schmitt FC, Tan PH, Tse GM, Weigelt B, Ellis IO, Reis-Filho JS. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 9.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 12.Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 16.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H. Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia (Review) Oncol Rep. 2014;32:1327–1334. doi: 10.3892/or.2014.3357. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016;144:194–201. doi: 10.1016/j.lfs.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genomics. 2010;4:345–352. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 21.Mahlapuu M, Enerback S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development. 2001;128:2397–2406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- 22.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, Jiang R. A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development. PLoS Genet. 2016;12:e1005769. doi: 10.1371/journal.pgen.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ, Sadik H, Argani P, Pandita TK, Sukumar S. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70:6047–6058. doi: 10.1158/0008-5472.CAN-10-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson J, Helou K, Kovacs A, Bendahl PO, Bjursell G, Ferno M, Carlsson P, Kannius-Janson M. Nuclear Janus-activated kinase 2/nuclear factor 1-C2 suppresses tumorigenesis and epithelial-to-mesenchymal transition by repressing Forkhead box F1. Cancer Res. 2010;70:2020–2029. doi: 10.1158/0008-5472.CAN-09-1677. [DOI] [PubMed] [Google Scholar]
- 26.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS One. 2013;8:e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai J, Tian AX, Wang QS, Kong PZ, Du X, Li XQ, Feng YM. FOXF2 suppresses the FOXC2-mediated epithelial-mesenchymal transition and multidrug resistance of basal-like breast cancer. Cancer Lett. 2015;367:129–137. doi: 10.1016/j.canlet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Tian HP, Lun SM, Huang HJ, He R, Kong PZ, Wang QS, Li XQ, Feng YM. DNA Methylation Affects the SP1-regulated Transcription of FOXF2 in Breast Cancer Cells. J Biol Chem. 2015;290:19173–19183. doi: 10.1074/jbc.M114.636126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang QS, Kong PZ, Li XQ, Yang F, Feng YM. FOXF2 deficiency promotes epithelial-mesenchymal transition and metastasis of basal-like breast cancer. Breast Cancer Res. 2015;17:30. doi: 10.1186/s13058-015-0531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito RA, Micke P, Paulsson J, Augsten M, Pena C, Jonsson P, Botling J, Edlund K, Johansson L, Carlsson P, Jirstrom K, Miyazono K, Ostman A. Forkhead box F1 regulates tumor-promoting properties of cancer-associated fibroblasts in lung cancer. Cancer Res. 2010;70:2644–2654. doi: 10.1158/0008-5472.CAN-09-3644. [DOI] [PubMed] [Google Scholar]
- 31.Malin D, Kim IM, Boetticher E, Kalin TV, Ramakrishna S, Meliton L, Ustiyan V, Zhu X, Kalinichenko VV. Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-beta3 expression. Mol Cell Biol. 2007;27:2486–2498. doi: 10.1128/MCB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, Tong P, Creighton CJ, Gibbons DL. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35:173–186. doi: 10.1038/onc.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riaz M, van Jaarsveld MT, Hollestelle A, Prager-van der Smissen WJ, Heine AA, Boersma AW, Liu J, Helmijr J, Ozturk B, Smid M, Wiemer EA, Foekens JA, Martens JW. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013;15:R33. doi: 10.1186/bcr3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Elston CW, Ellis IO. Pathology and breast screening. Histopathology. 1990;16:109–118. doi: 10.1111/j.1365-2559.1990.tb01078.x. [DOI] [PubMed] [Google Scholar]
- 39.Singletary SE, Allred C, Ashley P, Bassett LW, Berry D, Bland KI, Borgen PI, Clark GM, Edge SB, Hayes DF, Hughes LL, Hutter RV, Morrow M, Page DL, Recht A, Theriault RL, Thor A, Weaver DL, Wieand HS, Greene FL. Staging system for breast cancer: revisions for the 6th edition of the AJCC Cancer Staging Manual. Surg Clin North Am. 2003;83:803–819. doi: 10.1016/S0039-6109(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 40.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010;134:907–922. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Heul-Nieuwenhuijsen L, Dits N, Van Ijcken W, de Lange D, Jenster G. The FOXF2 pathway in the human prostate stroma. Prostate. 2009;69:1538–1547. doi: 10.1002/pros.20996. [DOI] [PubMed] [Google Scholar]
- 42.Zheng YZ, Wen J, Cao X, Yang H, Luo KJ, Liu QW, Huang QY, Chen JY, Fu JH. Decreased mRNA expression of transcription factor forkhead box F2 is an indicator of poor prognosis in patients with resected esophageal squamous cell carcinoma. Mol Clin Oncol. 2015;3:713–719. doi: 10.3892/mco.2015.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D. How basal are triple-negative breast cancers? Int J Cancer. 2008;123:236–240. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- 44.Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA, Evans AJ, Blamey R, Reis-Filho JS, Foulkes WD, Ellis IO. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res. 2009;15:2302–2310. doi: 10.1158/1078-0432.CCR-08-2132. [DOI] [PubMed] [Google Scholar]
- 45.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 46.Hellqvist M, Mahlapuu M, Samuelsson L, Enerback S, Carlsson P. Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. J Biol Chem. 1996;271:4482–4490. doi: 10.1074/jbc.271.8.4482. [DOI] [PubMed] [Google Scholar]
- 47.Fesquet D, Labbe JC, Derancourt J, Capony JP, Galas S, Girard F, Lorca T, Shuttleworth J, Doree M, Cavadore JC. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. Embo J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellers WR, Rodgers JW, Kaelin WG., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci U S A. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG., Jr Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 50.Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 51.Duursma A, Agami R. p53-Dependent regulation of Cdc6 protein stability controls cellular proliferation. Mol Cell Biol. 2005;25:6937–6947. doi: 10.1128/MCB.25.16.6937-6947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lo PK, Lee JS, Sukumar S. The p53-p21WAF1 checkpoint pathway plays a protective role in preventing DNA rereplication induced by abrogation of FOXF1 function. Cell Signal. 2012;24:316–324. doi: 10.1016/j.cellsig.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabchy A, Valero V, Vidaurre T, Lluch A, Gomez H, Martin M, Qi Y, Barajas-Figueroa LJ, Souchon E, Coutant C, Doimi FD, Ibrahim NK, Gong Y, Hortobagyi GN, Hess KR, Symmans WF, Pusztai L. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin Cancer Res. 2010;16:5351–5361. doi: 10.1158/1078-0432.CCR-10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider DJ, Wu M, Le TT, Cho SH, Brenner MB, Blackburn MR, Agarwal SK. Cadherin-11 contributes to pulmonary fibrosis: potential role in TGF-beta production and epithelial to mesenchymal transition. Faseb J. 2012;26:503–512. doi: 10.1096/fj.11-186098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sterling JA, Oyajobi BO, Grubbs B, Padalecki SS, Munoz SA, Gupta A, Story B, Zhao M, Mundy GR. The hedgehog signaling molecule Gli2 induces parathyroid hormone-related peptide expression and osteolysis in metastatic human breast cancer cells. Cancer Res. 2006;66:7548–7553. doi: 10.1158/0008-5472.CAN-06-0452. [DOI] [PubMed] [Google Scholar]
- 56.Johnson RW, Nguyen MP, Padalecki SS, Grubbs BG, Merkel AR, Oyajobi BO, Matrisian LM, Mundy GR, Sterling JA. TGF-beta promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer Res. 2011;71:822–831. doi: 10.1158/0008-5472.CAN-10-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS One. 2013;8:e55502. doi: 10.1371/journal.pone.0055502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Wang X, Wang Z, Tang H, Fan H, Guo Q. miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep. 2015;33:2592–2598. doi: 10.3892/or.2015.3833. [DOI] [PubMed] [Google Scholar]
- 59.Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, Hui AB, Sykes J, P’ng C, Miller N, McCready D, Fyles A, Liu FF. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926–2937. doi: 10.1158/0008-5472.CAN-10-3369. [DOI] [PubMed] [Google Scholar]
- 60.Shao J, Cao J, Liu Y, Mei H, Zhang Y, Xu W. MicroRNA-519a promotes proliferation and inhibits apoptosis of hepatocellular carcinoma cells by targeting FOXF2. FEBS Open Bio. 2015;5:893–899. doi: 10.1016/j.fob.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Hilmarsdottir B, Briem E, Bergthorsson JT, Magnusson MK, Gudjonsson T. Functional Role of the microRNA-200 Family in Breast Morphogenesis and Neoplasia. Genes (Basel) 2014;5:804–820. doi: 10.3390/genes5030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minella AC, Swanger J, Bryant E, Welcker M, Hwang H, Clurman BE. p53 and p21 form an inducible barrier that protects cells against cyclin E-cdk2 deregulation. Curr Biol. 2002;12:1817–1827. doi: 10.1016/s0960-9822(02)01225-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.