Abstract
A recently discovered class of endogenous mammalian lipids, branched fatty acid esters of hydroxy fatty acids (FAHFAs), possess anti-diabetic and anti-inflammatory activities. Here, we identified and validated carboxyl ester lipase (CEL), a pancreatic enzyme hydrolyzing cholesteryl esters and other dietary lipids, as a FAHFA hydrolase. Variants of CEL have been linked to maturity-onset diabetes of the young, type 8 (MODY8) and to chronic pancreatitis. We tested FAHFA hydrolysis activity of the CEL MODY8 variant and found a modest increase in activity as compared with the normal enzyme. Together, the data suggest that CEL might break down dietary FAHFAs.
Graphical abstract
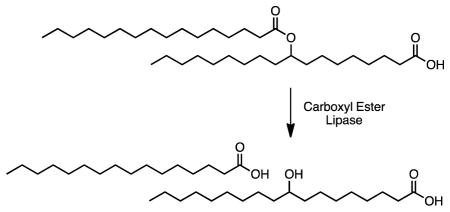
Lipids serve structural, energetic, and signaling roles in biology1. These activities are regulated by the action of lipid-modifying enzymes in the proteome2. The proper characterization of these enzymes, however, depends on knowing the composition of the lipidome, or, the complete set of lipids in a cell, tissue, or organism3–5. The recent discovery of new lipids3, 6, 7 indicates gaps in the current knowledge of lipid biochemistry.
Transgenic mice overexpressing the GLUT4 glucose transporter in adipose tissue have proven to be an important model for understanding the role of adipose tissue in systemic insulin resistance8, 9. These animals are obese, yet have improved glucose tolerance, and increased adipose tissue lipogenesis, which is important in maintaining insulin sensitivity10. The link between adipose lipogenesis and insulin sensitivity in mice and humans led to the hypothesis that the adipose tissue might be producing a metabolically beneficial lipid.
Lipidomic profiling of adipose tissue from the GLUT4 overexpressing mice revealed increased levels of a novel endogenous lipid family7. Structural characterization of these lipids using a combination of mass spectrometry and chemical synthesis identified them, as branched chain fatty acid esters of hydroxy fatty acids (FAHFAs). Subsequent analysis revealed 16 FAHFA families, which differ by the composition of their fatty acid chains. Prominent families include palmitic, oleic, and palmitoleic acid esters of hydroxy stearic acid, referred to as PAHSAs, OAHSAs, and POHSAs, respectively.
Analysis of the PAHSAs also revealed the existence of multiple positional isomers within each family. For instance, 5-PAHSA and 9-PAHSA are two isomers that have the conjoining ester bond at either 5- or 9- position (numbering from the carboxylate). A total of eight PAHSA isomers were identified (13-, 12-, 11-, 10-, 9-, 8-, 7-, and 5-PAHSA) and tissues have different distributions of these isomers7.
Human studies revealed that serum PAHSA concentrations were highly correlated with insulin sensitivity7 and insulin resistant people had low levels. This suggested that raising PAHSA levels in vivo might be beneficial. Indeed, administration of PAHSAs to mice improved glucose tolerance and reduced inflammation, demonstrating that increased FAHFA concentrations are anti-diabetic and anti-inflammatory7.
The discovery of FAHFAs has led to many questions, including what are the enzymes and metabolic pathways responsible for the production and catabolism of these lipids? Efforts towards this goal have already revealed two atypical FAHFA hydrolases11, AIG1 and ADTRP. Here we describe the discovery that the pancreatic enzyme carboxyl ester lipase (CEL), can utilize FAHFAs as substrates.
EXPERIMENTAL SECTION
Chemicals
FAHFAs were purchased from Cayman Chemical and other lipids were purchased from Avanti Polar Lipids or Sigma-Aldrich. FP-rhodamine12, WWL9213, and 9-hydroxyheptadecanoic acid14 were synthesized as reported.
Tissue lysate preparation
Wild-type C57BL/6J mice were purchased from Jackson Laboratories. All animals were housed in groups on a 14-h light, 10-h dark schedule at the Salk. All animal care and experimental procedures were in accordance with the standing committee on the Use of Animals in Research and Teaching at Salk and Institutional Animal Care and Use Committee (IACUC), and the National Institutes of Health Guidelines for the Humane Treatment of Laboratory Animals. Tissues were collected immediately after euthanasia and snap frozen using liquid nitrogen. Tissue lysates were prepared on ice by dounce homogenizing tissues in PBS. Homogenized lysates were centrifuged at 1,000 g for 10 min at 4 °C to pellet cell debris. The supernatant was then used as the total lysate for some assays. To prepare membrane and soluble fractions, the supernatant was separated by ultra-centrifugation at 100,000 × g for 45 min at 4 °C. Protein concentrations were measured using QuickStart Bradford 1× Dye Reagent (Bio-Rad). Aliquots were flash-frozen and stored at −80 °C for further use.
Gel-based ABPP analysis
Tissue and HEK293T cell proteomes (100 μL) were treated with FP-rhodamine (1 μM) for 30 min at 37 °C. Reactions were quenched by the addition of 4× SDS-PAGE loading buffer (40 μL). Competitive gel-based ABPP experiments were performed as previously described15. Samples were visualized in-gel using ChemiDoc MP imaging system (Bio-Rad). The fluorescence from rhodamine is presented in gray scale. 1 s and 5 s exposure times were used for native proteomes and CEL-transfected cell proteomes, respectively.
9-PAHSA and 9-OAHSA substrate hydrolysis assay and analysis
Tissue lysates/proteomes (20 μg) were incubated with lipid substrate (100 μM) in PBS (200 μL) at 37 °C with constant shaking. After 10 minutes, the reaction was stopped by addition of 2:1 mixture of CHCl3:MeOH (400 μL). 9-hydroxyheptadecanoic (9-HHDA) (50 pmol) was premixed into the CHCl3:MeOH mixture as an internal standard. The mixture was vortexed and centrifuged at 2200 × g for 5 min. The bottom organic layer was collected and dried under a stream of nitrogen. The extract was then dissolved in MeOH (200 μL) and a portion of this sample (10 μL) analyzed by LC-MS using a Thermo TSQ Quantiva LC-MS fitted with an Aquity UPLC BEH C18 column (1.7 μM; 2.1 mm × 100 mm; Waters). The LC solvents were as follows: buffer A, H2O + 0.01% NH4OH + 5 mM ammonium acetate; buffer B, 95:5 ACN:H2O + 0.01% NH4OH + 5 mM ammonium acetate. A typical LC run was 15 min long at 0.2 mL/min with a binary gradient consisting of the following steps: 50–90% buffer B over 6 min, 90–100% B over 0.1 min, 100% buffer B for 3 min, 100–50% buffer B over 0.1 min, and 50% buffer B for 6 minutes. MS analyses were performed using electrospray ionization (ESI) in negative ion mode with source parameters of 3kV spray voltage, ion transfer tube temperature of 325 °C, and a vaporizer temperature of 275 °C. Pseudo-MRM (collision energy (CE) of 9 V, RF lens set at 97 V) was used to detect 9-HSA ( m/z 299.3 → 299.3) and 9-HHDA (m/z 285.3 → 285.3). Hydrolysis was quantified by measuring 9-HSA peak area and diving this by 9-HHDA peak area. The ratio of 9-HSA/9-HHDA was then multiplied by 50 pmol (internal standard amount) and then divided by the amount of proteome (20 μg) and reaction time (10 min) to afford hydrolysis rates. Data represents mean values for 2–3 independent replicates.
Broad substrate hydrolysis assay
20 μg of proteome was incubated with 100 μM lipid substrate in a reaction volume of 250 μL PBS at 37 °C with constant shaking. After 30 minutes the reaction was quenched with 400 μL of 2:1 (vol/vol) CHCl3: MeOH doped with internal standard (0.5 nmol C17:1 heptadecenoic acid (C17:1 FFA) and 0.05 nmol 9-hydroxyheptadecanoic acid (9-HHDA)). The mixture was vortexed and centrifuged at 2800 × g for 5 min to separate the aqueous (top) and organic (bottom) phase. The organic phase was collected and dried under a stream of N2, re-solubilized in 100 μL of 2:1 (vol/vol) CHCL3: MeOH, and subjected to LC-MS analysis. A fraction of the organic extract (~15 μL) was injected onto an Agilent 6520 quadrupole-time-of-flight (QTOF) LC-MS and analyzed as described previously16. Data represents mean values for 2–3 replicates.
Western blotting
SDS-PAGE gels were transferred to PVDF membranes by iBlot2 Dry Blotting System (Life Technologies) according to manufacturer’s protocol. Immunoblots were blocked with Odyssey blocking buffer (LI-COR). The primary antibodies and dilutions are as follows: anti-CEL (Rabbit, Proteintech, 15384-1-AP, 1:1,000), anti-V5 (Mouse, Invitrogen, R96025, 1:2,000), and anti-β Actin (Rabbit, LI-COR, 926-42210, 1:1000). Secondary antibodies anti-rabbit IRDye 800CW (goat, 926-32211, LI-COR) and anti-mouse (goat, 925-32210, LI-COR) were applied at a dilution of 1:10,000 in blocking buffer for visualization.
Cloning and recombinant expression of CEL constructs
Full-length cDNA encoding mouse CEL (GE Dharmacon, in pCMV-SPORT6) was cloned into the pcDNA3.1/V5-His (B) vector. Human CEL wild-type in pcDNA3.1/V5-His (B), Human CEL mutant (1686delT) in pcDNA3.1/V5-His (B), and empty vector pcDNA3.1/V5-His were the same a previously published17, 18. To recombinantly express these CEL constructs, HEK293T cells were grown to 50% confluence in 10 cm tissue culture plates and transiently transfected with 12 μg of the desired construct using Lipofectamine 2000 (ThermoFisher Scientific) as the transfection reagent per the manufacturer’s protocol. “Mock” transfected cells were transfected with 12 μg of empty vector. Thirty-six hours after transfection, cells were washed with PBS (3×), harvested by scraping, and lysed by sonication in PBS. Membrane and soluble fractions were separated by ultra-centrifugation at 100,000 × g for 45 min at 4 °C. Protein concentrations were measured using QuickStart Bradford 1× Dye Reagent (Bio-Rad).
RESULTS and DISCUSSION
We utilized a previously described liquid chromatography-mass spectrometry (LC-MS) assay for measuring FAHFA hydrolysis activity11. Addition of 9-PAHSA to liver lysates resulted in the production 9-hydroxystearic acid (9-HSA), which we use to quantify hydrolysis. Denaturation of the proteome by heat led to loss of 9-PAHSA hydrolysis activity (Supplementary Figure 1).
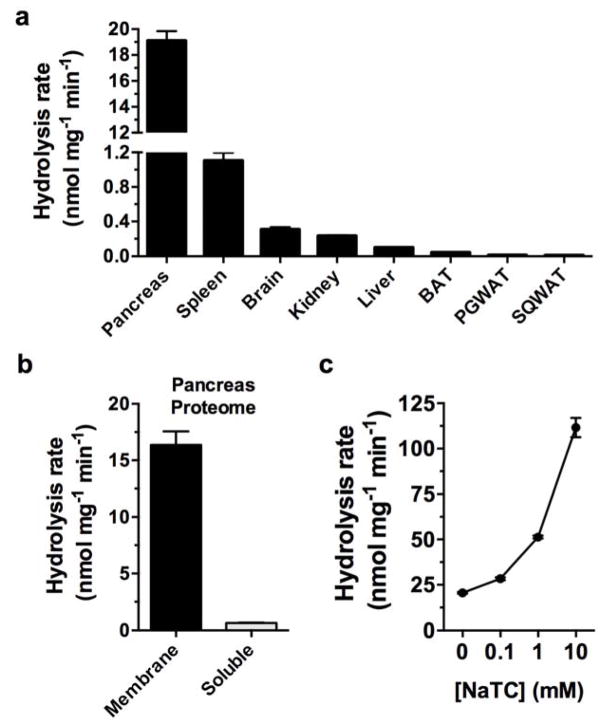
We then profiled the activity of different tissue lysates using the 9-PAHSA hydrolysis assay. The tissue with the greatest amount of 9-PAHSA hydrolysis activity (per mg of lysate) was pancreas (Fig. 1a). The pancreas produces a number of lipases that are secreted into the gastrointestinal (GI) tract to facilitate digestion of lipids from diet19. FAHFAs have been detected in a number of foods7, indicating the need for FAHFA hydrolyzing enzyme(s) in the GI tract. Fractionation of pancreatic tissue into soluble and membrane proteomes revealed that 9-PAHSA hydrolysis activity was specifically associated with the membrane fraction (Fig. 1b). Since the addition of the AIG1 and ADTRP inhibitor KC01 did not affect 9-PAHSA hydrolysis (Supplementary Figure 2a), we hypothesized that the pancreatic FAHFA hydrolase activity originated from another enzyme(s).
Figure 1.
Identification of a bile salt-stimulated pancreatic FAHFA hydrolase. (a) Measurement of 9-PAHSA hydrolysis activity using tissue lysates from various tissues showed varying levels of FAHFA hydrolysis, with the pancreas having the highest activity. (b) Fractionation of the pancreas lysate into membrane and soluble proteomes revealed that the 9-PAHSA hydrolysis activity is largely membrane associated. (c) Addition of the bile salt sodium taurocholate (NaTC) increased 9-PAHSA hydrolysis identifying CEL as the most likely FAHFA hydrolase. Data represents mean ± s.e.m values for three biological replicates.
The addition of fluorophosphonate rhodamine (FP-Rh), a broad serine hydrolase inhibitor blocked 9-PAHSA hydrolysis, which indicated that the FAHFA hydrolyzing enzyme(s) in the pancreas membrane fraction is a serine lipase (Supplementary Figure 2b). We searched a tissue database of serine hydrolases generated using activity-based protein profiling (ABPP) with a fluorophosphonate biotin (FP-biotin)13. Briefly, a FP-biotin probe was used to label serine hydrolase enzymes, which were thereafter enriched by avidin chromatography, digested by trypsin and the peptides were subsequently subjected to LC-MS/MS analysis13, 20. The similar inhibition activity of FP-Rh and FP-biotin indicated that the enzyme we sought was likely present in the serine hydrolase database13. We identified three candidate lipases within this dataset with preferential expression in the pancreatic membrane: carboxyl ester lipase (CEL), pancreatic lipase (PNLIP), and pancreatic lipase related protein 2 (PNLIPRP2).
CEL is also known as bile salt-stimulated lipase21, and its activity has been reported to increase in the presence of the bile salt taurocholate22, 23. By contrast, PNLIP and PNLIPRP2 are reportedly inhibited by taurocholate24, 25. Therefore, we reasoned that we might be able to distinguish between CEL, PNLIP, or PNLIPRP2 as a candidate FAHFA hydrolase by measuring 9-PAHSA hydrolysis in the presence of taurocholate. Addition of sodium taurocholate (NaTC) to pancreatic membrane lysates increased FAHFA hydrolysis activity in a concentration-dependent manner suggesting that CEL is a putative FAHFA-degrading enzyme (Fig. 1c).
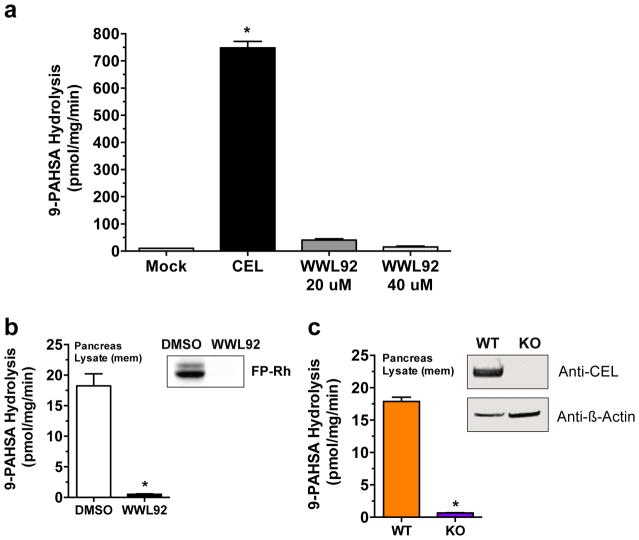
Expression of CEL in HEK293T cells resulted in more than a 50-fold increase in FAHFA hydrolysis activity compared to lysate from control (mock) cells transfected with an empty vector (Fig. 2a), demonstrating that CEL has FAHFA hydrolysis activity. Treatment of CEL-transfected lysates with a CEL inhibitor WWL9213, completely blocked FAHFA hydrolysis activity (Fig. 2a). The addition of WWL92 to pancreatic membrane proteome also abrogated 9-PAHSA activity (Fig. 2b), indicating that CEL is a principal FAHFA hydrolase in the pancreas. Also in support of this conclusion, 9- PAHSA hydrolysis activity was substantially reduced in pancreas tissue membrane from CEL−/− mice compared to control pancreatic tissue from CEL+/+ mice (Fig. 2c). Together, these data provide strong evidence that CEL is a FAHFA hydrolase and the main source of 9-PAHSA hydrolysis activity in the pancreatic lysates.
Figure 2.
Identification of CEL as a FAHFA hydrolase. (a) 9-PAHSA hydrolysis activity for membrane lysates from mock-transfected and CEL-transfected HEK293T cells, and CEL-transfected membrane lysates treated with WWL92 (20 μM and 40 μM). Data represents mean ± s.d for two replicates. ***p < 0.001 by ANOVA. (b) 9-PAHSA hydrolysis and gel-based ABPP (inset) for WT pancreatic membrane lysates treated with DMSO or WWL92 (10 μM). (c) 9-PAHSA hydrolysis and western blot analysis (inset) of of CEL+/+ and CEL−/− pancreatic membrane lysates. Data represents mean ± s.e.m. for three biological replicates. ***p < 0.001 by Student’s t-test. Full images of gel and western blots are provided in Supplementary Figure 3.
CEL is expressed in the acinar cells of the exocrine pancreas26. Upon feeding, CEL, along with a variety of other lipases, are released into the gut to break down complex lipids and aid with their absorption. Several lipids, including triglycerides, retinyl esters, phospholipids, and cholesteryl esters are reported as CEL substrates based on in vitro studies. In vivo, however, CEL has no impact on triglyceride27 or retinyl ester absorption28, but is reported to influence cholesteryl ester hydrolysis29.
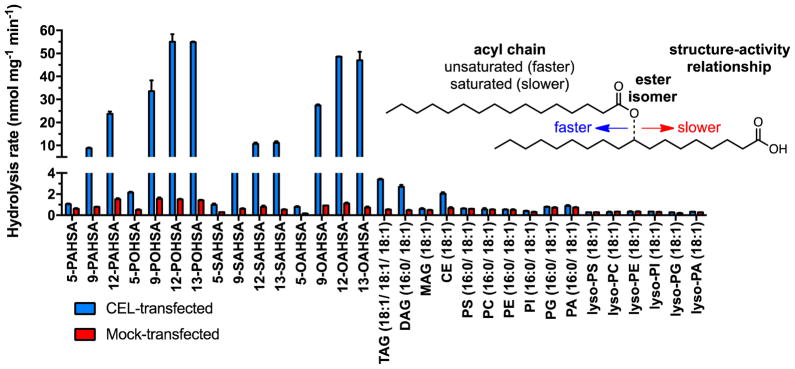
To ascertain the relative activity of CEL against different lipids, including FAHFAs, we performed a broad substrate analysis (Fig. 3). This assay used a total of 31 total lipids, including 15 FAHFAs, a triacylglycerol (TAG), a diacylglycerol (DAG), a cholesteryl ester (CE), along with a variety of phospholipids and lysophospholipids. Activity was monitored by the release of fatty acids from the different substrates upon hydrolysis by CEL. The substrate hydrolysis activity profile was generated using HEK293T membrane lysates overexpressing mouse CEL using a previously established LC-MS assay11. As reported previously, we found that CEL can hydrolyze TAGs, DAGs, and CEs (Fig. 3). However, CEL hydrolyzed FAHFAs at much greater rates than these reported substrates under the reaction conditions employed.
Figure 3.
In vitro lipid substrate hydrolysis assay for cellular lysates of CEL- and mock-transfected HEK293T cells. Data represents mean ± s.d. for two replicates.
Analysis of the hydrolysis data also revealed that CEL had preferences among different FAHFAs. FAHFAs are divided into families and isomers. Families are comprised of different acyl chains (i.e. oleic acid or palmitic acid) and isomers describe the same acyl chain combination with the ester bond at different positions (Fig. 3). Two clear preferences emerged from the substrate hydrolysis activity profile with different FAHFAs. First, and most dramatically, CEL preferentially hydrolyzed FAHFAs with the ester bond further away from the carboxylate (12-PAHSA > 9-PAHSA ≫ 5-PAHSA). A similar trend was observed for all the FAHFA families tested. Second, unsaturated FAHFAs were hydrolyzed more quickly than saturated FAHFAs (POHSAs/OAHSAs > PAHSAs/SAHSAs). The physiological significance of these trends is unclear, but they do suggest that individual FAHFA isomers can be metabolized at different rates.
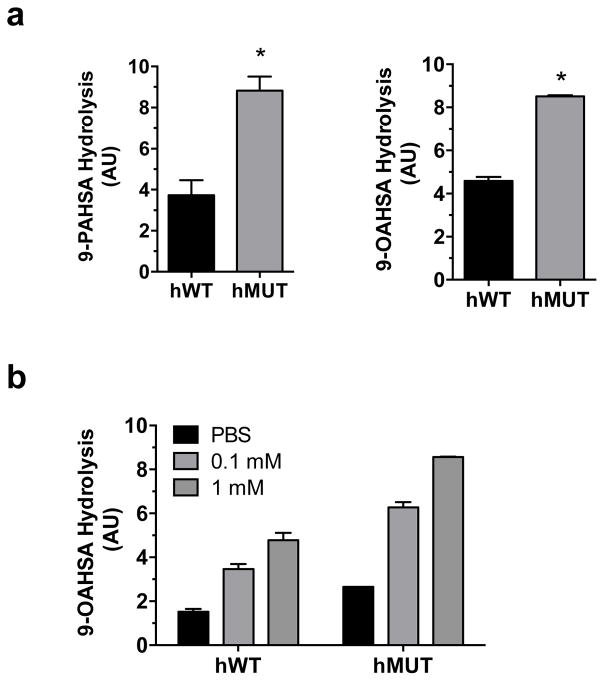
CEL has been linked to diabetes, exocrine pancreatic dysfunction and chronic pancreatitis30, 31. As such, rare variants of the CEL gene can cause maturity-onset diabetes of the young (MODY), a hereditary, autosomal dominant form of diabetes32. The MODY type caused by CEL mutations is denoted MODY8. Considering further the metabolic and anti-inflammatory activities of FAHFAs, we tested the effect of a MODY8-associated human CEL mutant (1686delT) on 9-PAHSA and 9-OAHSA hydrolysis activity. The human CEL mutant exhibited a 2-fold increase in FAHFA hydrolysis compared with human CEL protein (Figure 4a). We also tested the effect of NaTC on these two normal and disease-associated variants and both were bile-salt sensitive, with mutant CEL having slightly higher 9-PAHSA hydrolysis activity (Figure 4b). Current evidence indicates that MODY8 mutations result in CEL variants that are prone to protein aggregation in the pancreas, which could induce tissue damage leading to the symptoms of pancreatic dysfunction17, 18. While there is no dramatic enzymatic difference between normal CEL and the MODY8 mutant, aggregation of the latter could nevertheless lead to physiological changes in CEL activity, which again might affect endogenous FAHFA levels.
Figure 4.
(a) In vitro 9-PAHSA and 9-OAHSA hydrolysis activity comparing human normal wild type (hWT) CEL-transfected HEK293T cells and human MODY8 mutant (hMUT) CEL-transfected HEK293T cells. (b) The effect of NaTC on 9-PAHSA hydrolysis activity for hWT CEL and hMUT CEL-transfected membrane lysates. Hydrolysis rate was normalized to western blot expression of each construct, which was monitored by immunostaining for their V5 tag. AU (arbitrary units). Data represents mean ± s.d. for two biological replicates. *p < 0.05 by two-sided Student’s t-test for hMUT
CONCLUSION
Knowing the substrates of an enzyme is a critical piece of information needed to understand its endogenous functions of the protein. In this study, we have utilized an unbiased approach that combines biochemistry and ABPP to characterize CEL as a FAHFA hydrolase. The complementary tissue distribution of CEL compared to other recently discovered FAHFA hydrolases – AIG1 and ADTRP – indicates the metabolism of FAHFAs may be differentially regulated across the body. Future studies with knockout mice and in vivo-active inhibitors of individual FAHFA hydrolases should enrich our understanding of the respective roles that these enzymes and their lipid substrates play in mammalian biology and disease. In this regard, our results provide new hypothesis about the biochemical function of CEL that extends beyond previously established TAG/DAG and CE substrates to include the regulation of bioactive FAHFAs. Our findings may help to explain some of the endogenous phenotypes seen in CEL−/− mice, such as how CEL influences lipoprotein assembly and secretion33 and how loss of CEL promotes atherosclerosis34.
Supplementary Material
Acknowledgments
Funding Sources
No competing financial interests have been declared.
This work was supported by the US National Institutes of Health (DA033760, DK909810), The Leona M. and Harry B. Helmsley Charitable Trust (grant no. 2012-PG-MED002 to A.S.), National Cancer Institute Cancer Center Support grant P30 (CA014195 MASS core, A.S.), Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.), DK098002 (B.B.K.), a grant from the JPB foundation (B.B.K.), DK106210 (A.S. and B.B.K.) a Hewitt Foundation for Medical Research Fellowship (to W.H.P.), a Chapman Charitable Trust Fellowship (to M.J.K.), UCSD Medical Scientist Training Program funding (T32 GM007198 to M.J.K.) and an Irving S. Sigal postdoctoral fellowship from the American Chemical Society (to S.S.K.), and grants from Western Norway Regional Health Authority (A.M.), Novo Nordisk Foundation (A.M.) and KG Jebsen Foundation (K.F.).
We thank Dr. Philip Howles for the CEL KO mice.
ABBREVIATIONS
- FAHFAs
Fatty acid esters of hydroxy fatty acids
- PAHSAs
palmitic acid esters of hydroxy stearic acid
- OAHSA
oleic acid esters of hydroxy stearic acid
- SAHSAs
stearic acid esters of hydroxy stearic acids
- POHSAs
palmitoleic acid esters of hydroxystearic acids
- CEL
carboxyl ester lipase
- TAGs
triacylglycerols
- DAGs
diacylglycerols
- CEs
cholesterol esters
Footnotes
Author Contributions
M.J.K., E.A.H, B.B.K and A.S. conceived the project. M.J.K., S.S.K., W.H.P., E.A.H, T.M., A.S., and B.F.C. designed experiments. M.J.K., S.S.K., W.H.P., E.A.H, T.M. performed the experiments. M.J.K. and S.S.K. performed substrate assays and biochemical experiments. O.D.P. and I.S. performed all the animal work and provided all of the tissues used in this study. A.M. and K.F. prepared the plasmids with human CEL MODY8 mutants. M.J.K., S.S.K., W.H.P., E.A.H, T.M., B.B.K., B.F.C., and A.S. analyzed and interpreted the data. M.J.K. and A.S. wrote the paper. S.S.K, W.H.P., K.F., A.M., B.B.K. and B.F.C. edited the paper.
The Supporting Information is available free of charge on the ACS Publications website.
A detailed description of the materials and methods, and additional data (PDF).
References
- 1.Berg JM, Tymoczko JL, Stryer L. Biochemistry. W. H. Freeman; 2010. [Google Scholar]
- 2.Vance JE, Vance DE. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier Science; 2008. [Google Scholar]
- 3.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 4.Wenk MR. The emerging field of lipidomics. Nature Reviews Drug Discovery. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 5.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nature reviews Molecular cell biology. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 6.Saghatelian A, McKinney MK, Bandell M, Patapoutian A, Cravatt BF. A FAAH-regulated class of N-acyl taurines that activates TRP ion channels. Biochemistry. 2006;45:9007–9015. doi: 10.1021/bi0608008. [DOI] [PubMed] [Google Scholar]
- 7.Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, Dhaneshwar AS, Hammarstedt A, Smith U, McGraw TE, Saghatelian A, Kahn BB. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho E, Kotani K, Peroni OD, Kahn BB. Adipose-specific overexpression of GLUT4 reverses insulin resistance and diabetes in mice lacking GLUT4 selectively in muscle. Am J Physiol Endocrinol Metab. 2005;289:E551–561. doi: 10.1152/ajpendo.00116.2005. [DOI] [PubMed] [Google Scholar]
- 9.Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- 10.Herman MA, Peroni OD, Villoria J, Schon MR, Abumrad NA, Bluher M, Klein S, Kahn BB. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons WH, Kolar MJ, Kamat SS, AB, Hulce JJ, Saez E, Kahn BB, Saghatelian A, Cravatt BF. AIG1 and ADTRP are atypical integral membrane hydrolases that degrade bioactive FAHFAs. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn BB, Herman MA, Saghatelian A, Homan EA. USTPO, editor. Lipids That Increase Insulin Sensitivity And Methods Of Using The Same. Harvard College, Beth Israel Hospital Association, Beth Israel Deaconess Medical Center; USA: 2015. [Google Scholar]
- 15.Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd , Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 16.Kamat SS, Camara K, Parsons WH, Chen DH, Dix MM, Bird TD, Howell AR, Cravatt BF. Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay. Nat Chem Biol. 2015;11:164–171. doi: 10.1038/nchembio.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson BB, Torsvik J, Bjorkhaug L, Vesterhus M, Ragvin A, Tjora E, Fjeld K, Hoem D, Johansson S, Raeder H, Lindquist S, Hernell O, Cnop M, Saraste J, Flatmark T, Molven A, Njolstad PR. Diabetes and pancreatic exocrine dysfunction due to mutations in the carboxyl ester lipase gene-maturity onset diabetes of the young (CEL-MODY): a protein misfolding disease. J Biol Chem. 2011;286:34593–34605. doi: 10.1074/jbc.M111.222679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torsvik J, Johansson BB, Dalva M, Marie M, Fjeld K, Johansson S, Bjorkoy G, Saraste J, Njolstad PR, Molven A. Endocytosis of secreted carboxyl ester lipase in a syndrome of diabetes and pancreatic exocrine dysfunction. J Biol Chem. 2014;289:29097–29111. doi: 10.1074/jbc.M114.574244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carey MC, Small DM, Bliss CM. Lipid digestion and absorption. Annual Review of Physiology. 1983;45:651–677. doi: 10.1146/annurev.ph.45.030183.003251. [DOI] [PubMed] [Google Scholar]
- 20.Speers AE, Cravatt BF. Activity-Based Protein Profiling (ABPP) and Click Chemistry (CC)–ABPP by MudPIT Mass Spectrometry. Current protocols in chemical biology. 2009:29–41. doi: 10.1002/9780470559277.ch090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackberg L, Hernell O. The bile-salt-stimulated lipase in human milk. Purification and characterization. Eur J Biochem. 1981;116:221–225. doi: 10.1111/j.1432-1033.1981.tb05322.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Sternby B, Nilsson A. Hydrolysis of triacylglycerol arachidonic and linoleic acid ester bonds by human pancreatic lipase and carboxyl ester lipase. Biochim Biophys Acta. 1989;1004:372–385. doi: 10.1016/0005-2760(89)90086-6. [DOI] [PubMed] [Google Scholar]
- 23.Hernell O. Human milk lipases. III. Physiological implications of the bile salt-stimulated lipase. Eur J Clin Invest. 1975;5:267–272. doi: 10.1111/j.1365-2362.1975.tb02294.x. [DOI] [PubMed] [Google Scholar]
- 24.Roussel A, Yang Y, Ferrato F, Verger R, Cambillau C, Lowe M. Structure and activity of rat pancreatic lipase-related protein 2. Journal of Biological Chemistry. 1998;273:32121–32128. doi: 10.1074/jbc.273.48.32121. [DOI] [PubMed] [Google Scholar]
- 25.Morgan R, Huffman N. The interacion of lipase, lipase cofactor and bile salts in triglyceride hydrolysis. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism. 1971;248:143–148. doi: 10.1016/0005-2760(71)90086-5. [DOI] [PubMed] [Google Scholar]
- 26.Aho HJ, Sternby B, Kallajoki M, Nevalainen TJ. Carboxyl ester lipase in human tissues and in acute pancreatitis. International Journal of Pancreatology. 1989;5:123–134. doi: 10.1007/BF02924413. [DOI] [PubMed] [Google Scholar]
- 27.Vesterhus M, Raeder H, Kurpad AJ, Kawamori D, Molven A, Kulkarni RN, Kahn CR, Njolstad PR. Pancreatic function in carboxyl-ester lipase knockout mice. Pancreatology. 2010;10:467–476. doi: 10.1159/000266284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weng W, Li L, van Bennekum AM, Potter SH, Harrison EH, Blaner WS, Breslow JL, Fisher EA. Intestinal absorption of dietary cholesteryl ester is decreased but retinyl ester absorption is normal in carboxyl ester lipase knockout mice. Biochemistry. 1999;38:4143–4149. doi: 10.1021/bi981679a. [DOI] [PubMed] [Google Scholar]
- 29.Howles PN, Carter CP, Hui DY. Dietary free and esterified cholesterol absorption in cholesterol esterase (bile salt-stimulated lipase) gene-targeted mice. J Biol Chem. 1996;271:7196–7202. doi: 10.1074/jbc.271.12.7196. [DOI] [PubMed] [Google Scholar]
- 30.Fjeld K, Weiss FU, Lasher D, Rosendahl J, Chen JM, Johansson BB, Kirsten H, Ruffert C, Masson E, Steine SJ, Bugert P, Cnop M, Grutzmann R, Mayerle J, Mossner J, Ringdal M, Schulz HU, Sendler M, Simon P, Sztromwasser P, Torsvik J, Scholz M, Tjora E, Ferec C, Witt H, Lerch MM, Njolstad PR, Johansson S, Molven A. A recombined allele of the lipase gene CEL and its pseudogene CELP confers susceptibility to chronic pancreatitis. Nat Genet. 2015;47:518–522. doi: 10.1038/ng.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raeder H, Johansson S, Holm PI, Haldorsen IS, Mas E, Sbarra V, Nermoen I, Eide SA, Grevle L, Bjorkhaug L, Sagen JV, Aksnes L, Sovik O, Lombardo D, Molven A, Njolstad PR. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nat Genet. 2006;38:54–62. doi: 10.1038/ng1708. [DOI] [PubMed] [Google Scholar]
- 32.Molven A, Njølstad PR. Role of molecular genetics in transforming diagnosis of diabetes mellitus. Expert review of molecular diagnostics. 2011;11:313–320. doi: 10.1586/erm.10.123. [DOI] [PubMed] [Google Scholar]
- 33.Kirby RJ, Zheng S, Tso P, Howles PN, Hui DY. Bile salt-stimulated carboxyl ester lipase influences lipoprotein assembly and secretion in intestine: a process mediated via ceramide hydrolysis. J Biol Chem. 2002;277:4104–4109. doi: 10.1074/jbc.M107549200. [DOI] [PubMed] [Google Scholar]
- 34.Kodvawala A, Ghering AB, Davidson WS, Hui DY. Carboxyl ester lipase expression in macrophages increases cholesteryl ester accumulation and promotes atherosclerosis. J Biol Chem. 2005;280:38592–38598. doi: 10.1074/jbc.M502266200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.