Abstract
High blood pressure is a major risk factor for cardiovascular disease and premature death. However, there is limited knowledge on specific causal genes and pathways. To better understand the genetics of blood pressure, we genotyped 242,296 rare, low-frequency and common genetic variants in up to ~192,000 individuals, and used ~155,063 samples for independent replication. We identified 31 novel blood pressure or hypertension associated genetic regions in the general population, including three rare missense variants in RBM47, COL21A1 and RRAS with larger effects (>1.5mmHg/allele) than common variants. Multiple rare, nonsense and missense variant associations were found in A2ML1 and a low-frequency nonsense variant in ENPEP was identified. Our data extend the spectrum of allelic variation underlying blood pressure traits and hypertension, provide new insights into the pathophysiology of hypertension and indicate new targets for clinical intervention.
Introduction
High blood pressure (BP) or hypertension is a highly prevalent chronic disorder. It is estimated to be responsible for a larger proportion of global disease burden and premature mortality than any other disease risk factor1. Elevated systolic and/or diastolic BP increases the risk of several cardiovascular disorders including stroke, coronary heart disease (CHD), heart failure, peripheral arterial disease and abdominal aortic aneurysms2. BP is a complex, heritable, polygenic phenotype for which genome-wide association studies (GWAS) have identified over 67 genetic regions associated with BP and/or hypertension to date3–11. These variants are common (minor allele frequency, MAF≥0.05), mostly map to intronic or intergenic regions, with the causal alleles and genes not readily identified due to linkage disequilibrium (LD)4,5, and explain only ~2% of trait variance12. Low-frequency (0.01<MAF<0.05) and rare (MAF≤0.01) single nucleotide variants (SNVs), predominantly unexplored by GWAS may have larger phenotypic effects than common SNVs13, and may help to explain the missing heritability, and identify causative genes as demonstrated previously14.
To identify novel coding variants and loci influencing BP traits and hypertension we performed the largest meta-analysis to date that included a total of ~350,000 individuals, directly genotyped with the Exome chip. The Exome chip contains ~240,000 mostly rare and low-frequency variants (Methods). A single-variant discovery analysis was performed, and candidate SNVs were taken forward for validation using independent replication samples. Gene-based tests were used to identify BP associated genes harboring multiple rare variant associations. We next assessed whether the newly identified BP associated SNVs were associated with expression levels of nearby genes, and tested these variants in aggregate for a causal association of BP with other cardiovascular traits and risk factors. Our findings highlight the contribution of rare variants in the aetiology of blood pressure in the general population, and provide new insights into the pathophysiology of hypertension.
Results
Discovery of single variant BP associations
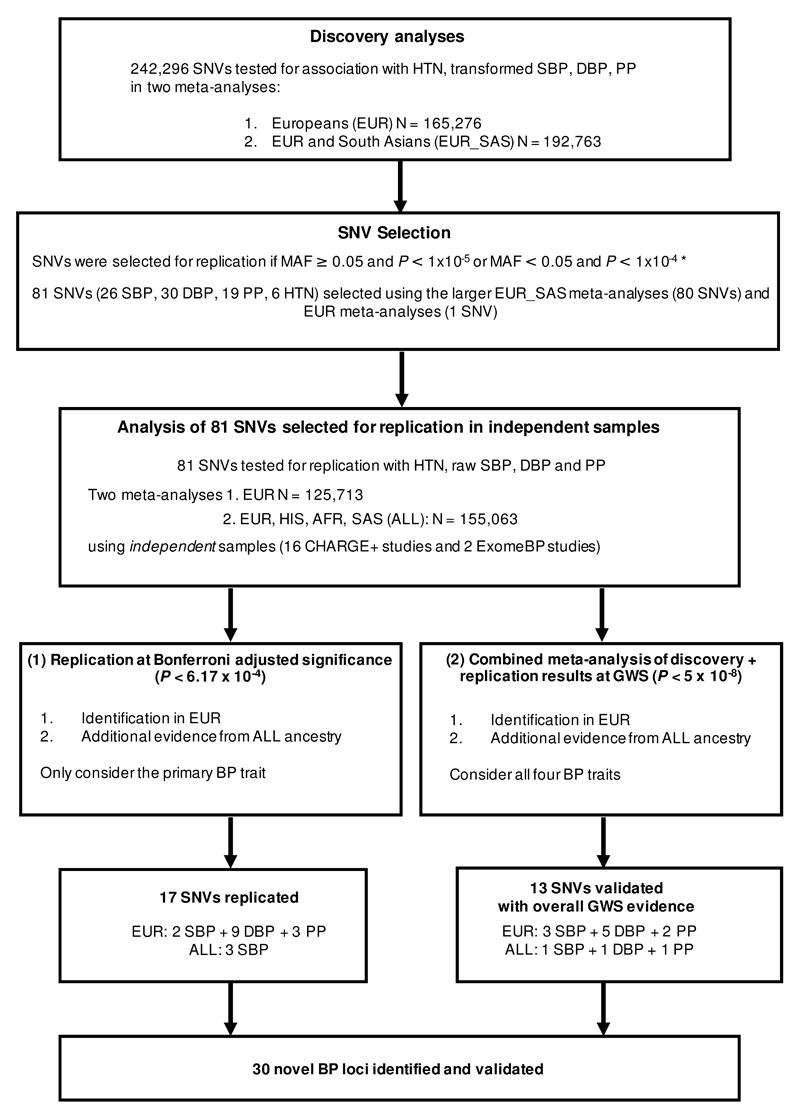
We genotyped 192,763 individuals from 51 studies, and assessed association of 242,296 SNVs with diastolic BP (DBP), systolic BP (SBP), pulse pressure (PP) and hypertension (HTN; Supplementary Tables 1, 2 and 3; Methods). An overview of the SNV discovery study design is given in Figure 1. A fixed effects meta-analysis for each trait was performed using study-level association summary statistics from i) samples of European (EUR) ancestry (up to 165,276 individuals), and ii) a trans-ethnic meta-analysis of the EUR and additional South Asian (SAS) ancestry samples (EUR_SAS; up to 192,763 individuals). Two analyses of DBP, SBP and PP were performed, one in which the trait was inverse normal transformed and a second in which the raw phenotype was analysed. Both sets of results were consistent (Methods), therefore to minimise sensitivity to deviations from normality in the analysis of rare variants, the results from the analyses of the transformed traits were used for discovery. Strong correlations between the BP traits were observed across studies (Methods), hence no adjustment of significance thresholds for independent trait testing was applied.
Figure 1. Study design and work flow diagram of single variant discovery analyses.
EUR=European, SAS=South Asian, HIS=Hispanic, AA=African American, HTN=hypertension, BP=blood pressure, SBP=systolic blood pressure, DBP= diastolic blood pressure, PP=pulse pressure, N=sample size, MAF=minor allele frequency, P=P-value significance threshold, SNV=single-nucleotide variant, GWS=genome-wide significance *Further details of the selection criteria are provided in the methods.
The discovery meta-analyses identified 51 genomic regions with genome-wide significant (GWS) evidence of association with at least one of the four BP traits tested (P<5x10-8; Supplementary Table 4). There were 46 regions associated in the EUR_SAS samples, of which 14 were novel (Supplementary Figure 1). An additional five regions were GWS in the EUR only meta-analyses of which three were novel (Supplementary Figure 2). In total, 17 genomic regions were identified that were GWS for at least one BP trait that have not been previously reported.
Replication of single variant BP associations
Next we sought support for our findings, in an independent replication dataset comprising of 18 studies, 15 of which were from the Cohorts for Heart and Aging Research in Genomic Epidemiology+ (CHARGE+) exome chip blood pressure consortium (Figure 1; Liu et al. Nature Genetics, submitted). Variants were selected for replication first using the larger (transformed) EUR_SAS data, with additional variants from the (transformed) EUR data also selected. SNVs were selected if they mapped outside of known BP genomic regions and had MAF≥0.05 and P<1x10-5 or MAF<0.05 and P<1x10-4 with at least one BP trait, i.e. choosing a lower significance threshold for the selection of rare variants (full details of the selection criteria are provided in the Methods). In total 81 candidate SNVs were selected for replication (Supplementary Table 5). Eighty variants were selected from EUR_SAS (transformed) results and one SNV at the ZNF101 locus from the EUR (transformed) analyses. The results for EUR_SAS and EUR were consistent (association statistics were correlated, ρ=0.9 across ancestries for each of the traits). Of the 81 variants, 30 SNVs were selected for association with DBP as the primary trait, 26 for SBP, 19 for PP and 6 for HTN, with the primary trait defined as the BP trait with the smallest association P-value in the EUR-SAS discovery analyses.
Meta-analyses were performed on results from analyses of untransformed DBP, SBP, PP and HTN (as only results of untransformed traits were available from CHARGE+) in (i) up to 125,713 individuals of EUR descent, and (ii) up to 155,063 individuals of multiple ethnicities (4,632 of Hispanic descent, 22,077 of African American descent, 2,641 SAS samples with the remainder EUR; Figure 1). Given that a large proportion of the ancestries in the trans-ethnic meta-analyses were not included in our discovery samples, we used the EUR meta-analyses as the main data set for replication, but we also report any additional associations identified within the larger trans-ethnic dataset.
Novel BP-SNV associations were identified based on two criteria (Figure 1; Methods). Firstly, replication of the primary BP trait-SNV association was sought at a Bonferroni adjusted P-value threshold in the replication data (P≤6.17x10-4, assuming α=0.05 for 81 SNVs tested and same direction of effect; Methods) without the need for GWS. Secondly, meta-analyses of discovery and replication results across all four (untransformed) BP traits were performed to assess the overall level of support across all samples for the 81 candidate SNVs; those BP-SNV associations that were GWS (with statistical support in the replication studies; P<0.05 and the same direction of effect) were also declared as novel.
Seventeen SNV-BP associations formally replicated with concordant direction of effect at a Bonferroni adjusted significance level for the primary trait. Fourteen were in the EUR meta-analyses, and amongst these was a rare non-synonymous (ns) SNV mapping to COL21A1 (Table 1, Supplementary Table 6). Three associations were in the trans-ethnic meta-analyses, these included two rare nsSNVs in RBM47 and RRAS (Table 1, Supplementary Table 7; Methods).
Table 1. Novel blood pressure trait associated loci. Variants with formal replication.
| Variant information | Discovery |
Replication | Combined |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | rsID | Chr:Pos (EA:EAF) | Trait | Pt | PU | N | β | P | N | β | P |
| EUR | |||||||||||
| RNF207 | rs709209 | 1:6.28 (A:0.655) | PP | 4.57 x 10-6 | 1.60 x 10-6 | 122,780 | 0.17 | 5.83 x 10-4 | 284,683 | 0.20 | 9.62 x 10-9 |
| C5orf56 | rs12521868 | 5:131.78 (T:0.373) | DBP | 1.59 x 10-6 | 3.03 x 10-7 | 122,795 | -0.18 | 2.29 x 10-5 | 282,023 | -0.19 | 6.12 x 10-11 |
| PHACTR1 | rs9349379 | 6:12.90 (A:0.566) | SBP | 2.11 x 10-8 | 1.78 x 10-7 | 122,809 | 0.24 | 4.06 x 10-4 | 284,673 | 0.29 | 8.84 x 10-10 |
| COL21A1 | rs200999181† | 6:55.94 (A:0.002) | PP | 3.08 x 10-8 | 2.46 x 10-7 | 121,487 | 2.70 | 1.90 x 10-4 | 242,486 | 3.25 | 6.27 x 10-10 |
| ABO | rs687621 | 9:136.14 (A:0.615) | DBP | 8.80 x 10-8 | 2.55 x 10-7 | 122,798 | 0.16 | 1.96 x 10-4 | 276,014 | 0.19 | 5.45 x 10-10 |
| ADO | rs10995311 | 10:64.56 (C:0.567) | DBP | 1.86 x 10-6 | 1.14 x 10-6 | 122,798 | 0.23 | 8.47 x 10-8 | 266,456 | 0.21 | 1.12 x 10-12 |
| LMO1 | rs110419 | 11:8.25 (A:0.48) | DBP | 9.41 x 10-6 | 2.22 x 10-5 | 122,798 | 0.16 | 1.81 x 10-4 | 279,935 | 0.16 | 3.04 x 10-8 |
| OR5B12 | rs11229457 | 11:58.21 (T:0.236) | SBP | 1.58 x 10-6 | 4.62 x 10-5 | 122,809 | -0.32 | 7.53 x 10-5 | 284,680 | -0.31 | 2.70 x 10-8 |
| CERS5 | rs7302981 | 12:50.54 (A:0.361) | DBP | 1.35 x 10-13 | 4.60 x 10-11 | 122,798 | 0.24 | 2.64 x 10-8 | 284,718 | 0.25 | 1.38 x 10-17 |
| MYH6 | rs452036 | 14:23.87 (A:0.327) | PP | 4.59 x 10-11 | 2.80 x 10-13 | 122,780 | -0.21 | 1.81 x 10-5 | 284,672 | -0.28 | 2.96 x 10-16 |
| DPEP1 | rs1126464 | 16:89.70 (C:0.256) | DBP | 1.19 x 10-9 | 4.35 x 10-11 | 118,677 | 0.24 | 1.68 x 10-6 | 261,564 | 0.28 | 1.02 x 10-15 |
| TBX2 | rs8068318† | 17:59.48 (T:0.698) | DBP | 7.46 x 10-13 | 5.71 x 10-10 | 122,798 | 0.26 | 3.23 x 10-8 | 281,978 | 0.26 | 1.95 x 10-16 |
| RGL3 | rs167479 | 19:11.53 (T:0.486) | DBP | 2.22 x 10-23 | 1.97 x 10-22 | 122,797 | -0.29 | 3.01 x 10-11 | 283,332 | -0.33 | 1.99 x 10-31 |
| PREX1 | rs6095241 | 20:47.31 (A:0.452) | DBP | 5.65 x 10-6 | 2.29 x 10-5 | 122,798 | -0.18 | 2.56 x 10-5 | 281,322 | -0.17 | 4.75 x 10-9 |
| ALL ancestry | |||||||||||
| RBM47 | rs35529250† | 4:40.43 (T:0.01) | SBP | 6.56 x 10-7 | 6.15 x 10-6 | 148,878 | -1.43 | 5.02 x 10-4 | 306,352 | -1.55 | 2.42 x 10-8 |
| OBFC1 | rs4387287 | 10:105.68 (A:0.157) | SBP | 2.23 x 10-8 | 1.32 x 10-7 | 147,791 | 0.28 | 3.37 x 10-4 | 320,494 | 0.36 | 9.12 x 10-10 |
| RRAS | rs61760904† | 19:50.14 (T:0.008) | SBP | 1.96 x 10-6 | 1.90 x 10-5 | 148,878 | 1.38 | 5.70 x 10-4 | 322,664 | 1.50 | 8.45 x 10-8 |
SNV-BP associations are reported for the newly identified BP loci that replicated at P < 6.2 x 10-4 (Bonferroni correction for the 81 variants selected for replication for a primary blood pressure trait; Methods). Loci are categorised into EUR and ALL ancestry based on the meta-analysis used to replicate the variants for the primary BP trait shown in columns labelled ‘Trait’. In the columns that contains the discovery meta-analyses results, Pt represents the P-value for association of the variant with the transformed primary BP trait in the EUR_SAS discovery meta-analyses (which was also used to select the variant for replication) and Pu represents the P-value for association with the untransformed primary BP trait in the ancestry in which the variant replicated. N, β and P, which denote the number of samples, estimated allelic effect and P-value for association with the primary BP trait, are provided for the untransformed primary BP trait in the replication data and also from the combined (discovery and replication) meta-analyses. NB: ALL ancestry corresponds to all ancestries in the combined (discovery + replication) meta-analyses
Locus – Gene or region containing the SNV, rsID - dbSNP rsID. Chr:Pos (EA:EAF) – Chromosome:NCBI Build 37 position in Mb (effect allele:effect allele frequency), Trait – primary blood pressure trait for which the variant was and also replicated, β - effect estimate, N:sample size, EUR - European.
indicates it is a non-synonymous SNV (nsSNV) or is in linkage disequilibrium with a nsSNV (r2 > 0.8) that is predicted to be damaging
In addition to the 17 SNV-BP trait associations that formally replicated, we identified 13 further SNV-associations that were GWS in the combined (discovery and replication) meta-analyses. Ten of these were GWS in the combined EUR analyses, (Table 2; Supplementary Tables 6 and 8a), and three were GWS in the combined trans-ethnic meta-analyses (Table 2; Supplementary Tables 7 and 8b).
Table 2. Novel blood pressure trait associated loci. Variants with GWS evidence of association in combined meta-analyses.
| Variant information | Discovery |
Replication | Combined |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | rsID | Chr:Pos (EA:EAF) | Trait | Pt | PU | N | β | P | N | β | P |
| EUR | |||||||||||
| 2q36.3 | rs2972146 | 2:227.10 (T:0.652) | DBP§ (HTN) |
1.51 x 10-9 | 2.47 x 10-7 | 122,798 | 0.13 | 2.20 x 10-3 | 275,610 | 0.17 | 8.40 x 10-9 |
| ZBTB38 | rs16851397 | 3:141.13 (A:0.953) | DBP§ (SBP) |
6.87 x 10-6 | 3.20 x 10-5 | 122,798 | -0.38 | 1.20 x 10-4 | 284,717 | -0.38 | 3.01 x 10-8 |
| PRDM6 | rs1008058 | 5:122.44 (A:0.135) | SBP | 5.09 x 10-7 | 1.01 x 10-8 | 43,109 | 0.46 | 3.61 x 10-3 | 176,362 | 0.55 | 2.99 x 10-10 |
| GPR20 | rs34591516 | 8:142.37 (T:0.055) | SBP§ (DBP) |
1.54 x 10-6 | 1.01 x 10-7 | 122,807 | 0.51 | 4.20 x 10-4 | 282,009 | 0.64 | 6.10 x 10-10 |
| HOXB7 | rs7406910 | 17:46.69 (T:0.118) | SBP | 6.07 x 10-10 | 2.74 x 10-9 | 122,809 | -0.20 | 4.89 x 10-2 | 284,690 | -0.46 | 3.80 x 10-8 |
| AMH | rs10407022† | 19:2.25 (T:0.82) | PP | 1.63 x 10-7 | 1.73 x 10-7 | 118,656 | -0.19 | 1.62 x 10-3 | 252,525 | -0.26 | 5.94 x 10-9 |
| ZNF101 | rs2304130 | 19:19.79 (A:0.914) | DBP | 1.66 x 10-8 | 1.92 x 10-8 | 122,798 | -0.17 | 1.71 x 10-2 | 284,705 | -0.29 | 1.53 x 10-8 |
| PROCR | rs867186 | 20:33.76 (A:0.873) | DBP | 1.44 x 10-6 | 4.15 x 10-7 | 122,798 | 0.21 | 2.48 x 10-3 | 284,722 | 0.26 | 1.19 x 10-8 |
| RRP1B | rs9306160 | 21:45.11 (T:0.374) | DBP§ (SBP) |
1.04 x 10-8 | 1.90 x 10-6 | 100,489 | -0.16 | 4.30 x 10-4 | 249,817 | -0.18 | 6.80 x 10-9 |
| TNRC6B | rs470113 | 22:40.73 (A:0.804) | PP | 1.48 x 10-10 | 1.31 x 10-9 | 122,780 | -0.14 | 1.37 x 10-2 | 284,683 | -0.25 | 1.67 x 10-9 |
| ALL ancestry | |||||||||||
| 7q32.1 | rs4728142 | 7:128.57 (A:0.433) | SBP | 8.10 x 10-6 | 4.21 x 10-6 | 150,542 | -0.21 | 8.62 x 10-4 | 338,338 | -0.24 | 3.45 x 10-8 |
| PRKAG1 | rs1126930† | 12:49.40 (C:0.036) | PP | 2.12 x 10-6 | 4.62 x 10-7 | 151,481 | 0.36 | 3.74 x 10-3 | 314,894 | 0.50 | 3.34 x 10-8 |
| SBNO1 | rs1060105 | 12:123.81 (T:0.209) | DBP | 6.66 x 10-7 | 1.09 x 10-6 | 150,532 | -0.15 | 2.67 x 10-3 | 336,413 | -0.18 | 3.07 x 10-8 |
SNV-BP associations are reported for the newly identified BP loci that showed genome-wide significant association (P < 5 x 10-8) in the combined discovery and replication meta-analyses. In the columns that contain results from the discovery meta-analyses, Pt represents the P-value for association of the variant with the transformed primary BP trait in the EUR_SAS discovery meta-analyses (used to select the variant for replication) and Pu represents the P-value for association with the untransformed BP trait in the ancestry in which the variant was validated. Loci are categorised into EUR and ALL ancestry based on the ancestry in which the variant showed association with a blood pressure trait at P < 5 x 10-8. N, β and P, which denote the number of samples, estimated allelic effect and P-value for association with the validated BP trait, are provided for the untransformed BP trait in the replication data and also from the combined (discovery and replication) meta-analyses. NB: ALL ancestry corresponds to all ancestries in the combined (discovery + replication) meta-analyses.
Locus – Gene or region containing the SNV, rsID - dbSNP rsID. Chr:Pos (EA:EAF) – Chromosome:NCBI Build 37 position in Mb (effect allele:effect allele frequency), Trait - blood pressure trait for which association is reported, EUR - European.
At four loci (2q36.3, ZBTB38, GPR20 and RRP1B) the primary trait used to select the variants for replication is given in parentheses because the variant associations were validated in the combined meta-analysis for the listed secondary trait. For these variants, Pt denotes the P-value for association with the primary trait, the other P-values provided are for the secondary trait.
indicates it is a non-synonymous SNV (nsSNV) or is linkage disequilibrium with a nsSNV (r2 > 0.8) that is predicted to be damaging
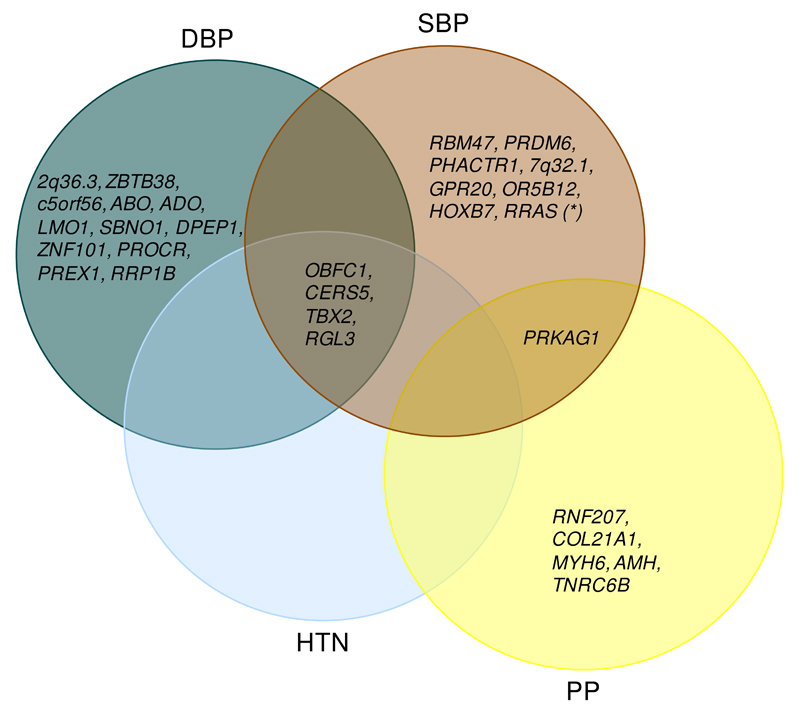
This gives a total of 30 novel SNV-BP associations (15 SNV-DBP, 9 SNV-SBP and 6 SNV-PP; Tables 1 and 2; Supplementary Figures 3 and 4). Five of the SNVs were GWS with more than one BP trait (Figure 2: Tables 1 and 2; Supplementary Table 8). Four loci (CERS5, TBX2, RGL3 and OBFC1) had GWS associations with HTN in addition to GWS associations with DBP and SBP. The PRKAG1 locus had GWS associations with both SBP and PP.
Figure 2. Overlap of the 30 novel loci associations across SBP, DBP, PP and HTN.
The Venn diagram shows which of the 30 newly identified BP loci are associated with multiple BP traits. Only SNV-BP trait associations that were genome-wide significant (P < 5x10-8) in the combined discovery and replication meta-analyses are listed for any given BP trait, within the corresponding ancestry dataset that the given locus was validated for (see Tables 1 and 2). The association of RRAS variant with SBP was replicated in the independent samples, but did not achieve GWS in the combined discovery and replication meta-analysis and is therefore only included for SBP. HTN=hypertension, SBP=systolic blood pressure, DBP= diastolic blood pressure, PP=pulse pressure.
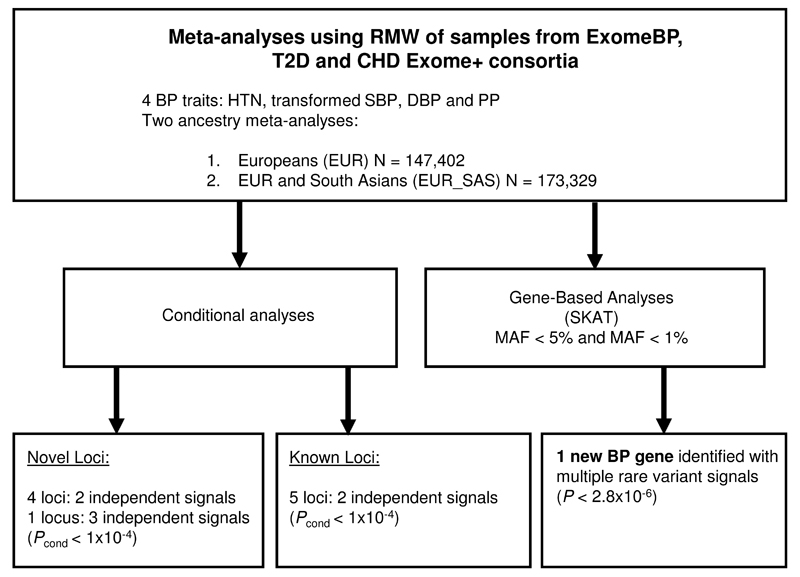
Conditional analyses were performed to identify secondary signals of association within the novel BP loci. The RAREMETALWORKER (RMW) package (Methods)15 allows conditional analyses to be performed using summary level data. Hence, analyses of the transformed primary traits and HTN were re-run in RMW across the discovery studies (Figure 3). The results of the RMW single variant tests were consistent with the initial discovery analyses (Supplementary Information). Given the RMW analyses were based on our discovery samples, the larger EUR-SAS data was used as the main analysis to increase power, but we also report any additional associations with evidence in EUR.
Figure 3. Study design for conditional analyses and rare variant gene-based discovery analyses.
RMW=RareMetalWorker, EUR=European, SAS = South Asian, HTN=hypertension, BP=blood pressure, SBP=systolic blood pressure, DBP= diastolic blood pressure, PP=pulse pressure. N=sample size, MAF=minor allele frequency, P=P-value significance threshold, Pcond=conditional P-value significance threshold
We identified secondary independent signals of association in four loci, PREX1, PRKAG1 and RRP1B within the EUR_SAS analyses and COL21A1 in the EUR analyses (Pconditional<1x10-4, Bonferroni adjusted for ~500 variants within each region; Methods; Supplementary Tables 9 and 10). Three independent association signals were identified in the MYH6 locus in the EUR_SAS analyses (Supplementary Table 11).
Gene-based BP associations
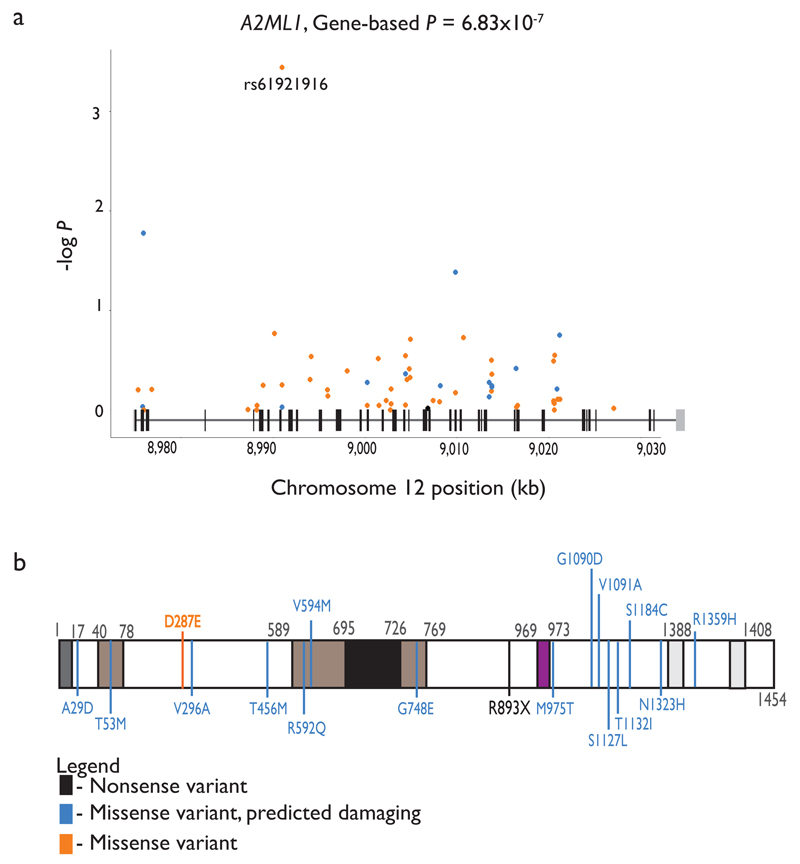
To improve statistical power to detect associations in genes harbouring rare variants, analytical methods that combine effects of variants across a gene into a single test have been devised and are implemented in the RMW package15. We applied the gene-based sequence kernel association test (SKAT)16 and Burden tests17 to the RMW dataset (MAF<0.05 or MAF<0.01; Figure 3; Methods). One previously unidentified BP gene (A2ML1) was associated with HTN (P= 7.73x10-7) in the EUR_SAS studies and also in EUR studies (Supplementary Table 12; Bonferroni-corrected threshold of significance P<2.8x10-6, after adjusting for 17,996 genes tested, Methods). The gene showed residual association with the primary BP trait after conditioning on the most associated SNV in the gene (Pconditional=5.00x10-4; Supplementary Table 12), suggesting that the association is due to multiple rare variants in the gene. One nonsense variant (rs199651558, p.Arg893*, MAF=3.5x10-4) was observed, and there were multiple missense variants (Figure 4). A2ML1 encodes alpha-2-macroglobulin-like 1 protein, and is a member of the alpha macroglobulin superfamily, which comprises protease inhibitors targeting a wide range of substrates. Mutations in this gene are associated with a disorder clinically related to Noonan syndrome, a developmental disorder which involves cardiac abnormalities18. We sought replication in the CHARGE+ studies for this gene, however there was no evidence of association with HTN (P= 0.45). Given the very low frequencies of the variants involved, however, studies in which the variants are polymorphic will be required to replicate the association with HTN. The DBH gene was found to be associated with DBP using the SKAT test (P=2.88x10-6). However, this was not due to multiple rare variants as the association was driven by rs77273740 (Supplementary Table 5) and the SNV was not validated in the replication samples.
Figure 4. Locus plot for A2ML1 and secondary amino acid structure of the gene product.
(a) Locus plot for A2ML1 associated with HTN identified through gene based tests. The variants’ positions along the gene (x axis, based on human genome build 37) and the –log10(P-value of association) (y axis) are indicated. The variants are colour coded: nonsense (black), missense, predicted damaging (blue), and missense (orange). The schematic above the x-axis represents the intron / exon (black vertical bars) structure, the untranslated regions are shown as grey vertical bars.
(b) The white box denotes the full-length amino acid sequence for each of the two gene products. Black numbers denote amino acid residue positions of note. Coloured boxes depict putative functional domains (see below). Coloured vertical lines indicate the amino acid substitutions corresponding to the variants depicted in the locus plots above using the same colour coding. Bold, italic indicates the SNV association with smallest P-value.
Dark grey – signal peptide sequence. Brown – regions of intramolecular disulfide bonds. For simplicity only those regions coinciding with variants described were indicated. Black – bait region described to interact with proteases. Purple – thiol ester sequence region aiding in interaction with proteases. Light grey – alpha helical regions thought to mediate A2ML1 interaction with LRP1, facilitating receptor-mediated endocytosis.
Rare and common variant associations in established BP loci
Of the 67 established BP loci, 35 loci were on the Exome chip (N=43 SNVs or close proxies r2>0.7). All 43 SNVs had at least nominal evidence of association with BP in our discovery samples (P<0.01; Supplementary Table 13). We also assessed if any of the established BP loci contained coding variants that are associated with BP traits and in LD (r2>0.2) with the known BP variants on the Exome chip (Supplementary Table 13), using the 1000G phase 3 release for LD annotation. Focusing on SNVs that were GWS for any BP trait from our transformed discovery data for either ancestry, there were 25 coding variants, of which 6 were predicted to be damaging at loci labelled CDC25A, SLC39A8, HFE, ULK4, ST7L-CAPZA1-MOV10 and CYP1A1-ULK3. Three of these are published variants at loci labelled SLC39A8, HFE and ST7-CAPZA1-MOV10. At CYP1A1-ULK3, the coding variant was in moderate LD with the reported variant, but was less significantly associated with DBP in our EUR_SAS dataset (P=2.24x10-8 compared to P=1.68x10-15 for the published variant). At the ULK4 locus the predicted damaging coding variant had similar association as the published coding variant (predicted to be benign), and prior work has already indicated several associated nsSNVs in strong LD in ULK4 19. The nsSNV within the CDC25A locus (rs11718350 in SPINK8) had similar association with DBP as the intergenic published SNV in our EUR_SAS dataset (P=2.00x10-8 compared to P=2.27x10-8 for the published variant). Overall at least 5 of the known loci are consistent with having a coding causal variant.
Gene-based SKAT tests of all genes that map within 1 Mb of a previously reported SNV association (Supplementary Table 14), indicated no genes with multiple rare or low-frequency variant associations. Single variant conditional analyses showed that rs33966350, a rare nonsense variant in ENPEP (MAF=0.01) was associated with SBP (Pconditional=1.61x10-5) in the EUR_SAS samples (Supplementary Tables 14 and 15; Methods) independently of the known SNV (rs6825911). ENPEP encodes aminopeptidase A (APA) an enzyme of the renin-angiotensin-aldosterone system (RAAS) that converts angiotensin II (AngII) to AngIII.
There were no other established loci with convincing low-frequency or rare SNV associations in the EUR_SAS samples. However, HOXC4, had evidence of a second independent signal with a rare missense SNV in EUR samples (rs78731604; MAF=0.005, Pconditional= 5.76x10-5; Supplementary Table 15). The secondary signal in the HOXC4 region, mapped to CALCOCO1, ~300kb from the known SNV. The gene association (MAF≤0.01, P=2.37x10-5) was below the required significance threshold and attributable to rs78731604, which is not predicted to have detrimental effects on protein structure. Therefore, replication of this association is required. Three loci (ST7L-CAPZA1-MOV10, FIGN-GRB14, and TBX5-TBX3) had evidence of a second independent signal in the region in EUR_SAS samples with a common variant (Pconditional<1x10-4; Supplementary Table 15) that has not been previously reported.
Having identified 30 novel loci associated with BP traits, as well as additional new independent SNVs at four novel loci and five known loci, we calculated the percent of the trait variance explained (Methods). This was 2.08%/2.11%/1.15% for SBP/DBP/PP for the 43 previously reported BP-SNVs covered in our dataset, increasing to 3.38%/3.41%/2.08% respectively with the inclusion of the 30 lead SNVs from novel loci, plus new independent SNV-BP associations identified from novel and known loci.
Effect of BP SNVs on cardiovascular traits & risk factors
Amongst our novel BP-SNV associations, some have previously been reported to be associated with other cardiovascular traits and risk factors (Supplementary Table 16); these include coronary heart disease (CHD: PHACTR1, ABO)20,21, QT interval (RNF207)22, heart rate (MYH6)23, and cholesterol levels (2q36.3, ABO, ZNF101)24.
To test the impact of BP variants on cardiovascular endpoints and risk factors we created three weighted genetic risk scores (GRS) according to SBP/DBP/PP based on the newly identified and previously published BP variants (up to N=125; Methods). The GRS models were used to test the causal effect of BP on the following traits: ischemic stroke (including the subtypes, cardiometabolic, large and small vessel 25), CHD, heart failure,26 left ventricular mass27, left ventricular wall thickness27, high-density lipoprotein cholesterol (HDL-c), low-density lipoprotein (LDL-c), triglycerides, total cholesterol, body mass index (BMI), waist-hip ratio adjusted BMI, height and estimated glomerular filtration rate (eGFR) (Methods). As expected, BP was positively associated with increased CHD risk (OR [95% CI]=1.39[1.22-1.59] per 10mmHg increase in SBP, P=6.07×10-7; 1.62[1.28-2.05] per 10mmHg increase in DBP, P=5.99x10-5; 1.70[1.34-2.16] per 10mmHg increase in PP, P=1.20x10-5; Table 3), and increased risk of ischemic stroke (OR [95% CI]=1.93[1.47-2.55] per 10mmHg increase in DBP, P=2.81×10-6; 1.57[1.35-1.84] per 10mmHg increase in SBP, P=1.16×10-8; 2.12[1.58-2.84] per 10mmHg increase in PP, P=5.35x10-7). The positive association with ischemic stroke was primarily due to large vessel stroke (Table 3). DBP and SBP were also positively associated with left ventricular mass (9.57 [3.98-15.17] gram increase per 10mmHg increase in DBP, P=8.02x10-4 and 5.13 [1.77-8.48] gram increase per 10mmHg increase in SBP, P=0.0027) and left ventricular wall thickness (0.10 [0.06-0.13] cm increase per 10mmHg increase in DBP, P=1.88x10-8 and 0.05 [0.03-0.07] cm increase per 10mmHg increase in SBP, P=5.52x10-6, Table 3). There was no convincing evidence to support the BP associated variants having an effect on lipid levels (P>0.1), BMI (P>0.005), waist hip ratio adjusted BMI (P>0.1), height (P>0.06), eGFR (P>0.02) or heart failure (P>0.04). The causal associations with CHD, stroke, and left ventricular measures augment the results from a previous association analysis using 29 BP variants28. Our data strongly support the previous observations of no causal relationship between BP and eGFR. Lack of evidence of a BP effect with heart failure may only be due to lack of power, as the association was in the expected direction.
Table 3. Results of the genetic risk score analyses across CVD traits and risk factors.
| Outcome | Units | N | DBP (per 10mmHg increase) | SBP (per 10mmHg increase) | PP (per 10mmHg increase) | |||
|---|---|---|---|---|---|---|---|---|
| Effect [95% CI] | P | Effect [95% CI] | P | Effect [95% CI] | P | |||
| CHD | OR | 82,056 | 1.62 [ 1.28, 2.05] | 5.99 x 10-5 | 1.39 [ 1.22, 1.59] | 6.07 x 10-7 | 1.70 [ 1.34, 2.16] | 1.20 x 10-5 |
| Ischemic stroke | OR | 25,799 | 1.93 [ 1.47, 2.55] | 2.81 x 10-6 | 1.57 [ 1.35, 1.84] | 1.16 x 10-8 | 2.12 [ 1.58, 2.84] | 5.35 x 10-7 |
| Cardioembolic stroke | OR | 16,113 | 1.43 [ 0.86, 2.39] | 0.1683 | 1.33 [ 0.99, 1.80] | 0.0584 | 1.73 [ 1.00, 3.02] | 0.0518 |
| Large vessel stroke | OR | 13,903 | 2.26 [ 1.25, 4.08] | 0.0068 | 1.85 [ 1.32, 2.59] | 3.61 x 10-4 | 3.05 [ 1.64, 5.68] | 4.37 x 10-4 |
| Small vessel stroke | OR | 15,617 | 1.96 [ 1.13, 3.41] | 0.0168 | 1.56 [ 1.13, 2.16] | 0.0064 | 1.98 [ 1.09, 3.61] | 0.0248 |
| Heart failure | OR | 13,282 | 1.48 [ 1.02, 2.17] | 0.0409 | 1.25 [ 1.00, 1.57] | 0.0512 | 1.33 [ 0.88, 2.02] | 0.1757 |
| Left ventricular mass | g | 11,273 | 9.57 [ 3.98,15.17] | 8.02 x 10-4 | 5.13 [ 1.77, 8.48] | 0.0027 | 5.97 [-0.38,12.31] | 0.0653 |
| Left ventricular wall thickness | cm | 11,311 | 0.10 [ 0.06, 0.13] | 1.88 x 10-8 | 0.05 [ 0.03, 0.07] | 5.52 x 10-6 | 0.05 [ 0.01, 0.09] | 0.0187 |
| HDL | mg/dl | 80,395 | 0.25 [-1.00, 1.51] | 0.6930 | 0.21 [-0.50, 0.92] | 0.5622 | 0.47 [-0.79, 1.73] | 0.4668 |
| LDL | mg/dl | 77,021 | -1.57 [-5.20, 2.06] | 0.3972 | 0.07 [-2.03, 2.16] | 0.9498 | 1.87 [-1.86, 5.59] | 0.3255 |
| Total cholesterol | mg/dl | 80,455 | -1.34 [-5.90, 3.22] | 0.5639 | 0.70 [-1.93, 3.32] | 0.6029 | 3.68 [-0.97, 8.33] | 0.1209 |
| Triglycerides | mg/dl | 77,779 | 0.02 [-0.03, 0.08] | 0.3859 | 0.02 [-0.01, 0.05] | 0.2697 | 0.03 [-0.03, 0.08] | 0.3025 |
| BMI | INVT | 526,508 | -0.10 [-0.18,-0.01] | 0.0342 | -0.07 [-0.13,-0.02] | 0.0058 | -0.12 [-0.23,-0.02] | 0.0165 |
| WHRadjBMI | INVT | 344,369 | 0.03 [-0.04, 0.11] | 0.4025 | 0.03 [-0.02, 0.08] | 0.2170 | 0.06 [-0.03, 0.15] | 0.1885 |
| Height | INVT | 458,927 | 0.02 [-0.15, 0.18] | 0.8592 | -0.04 [-0.15, 0.06] | 0.4170 | -0.18 [-0.37, 0.01] | 0.0683 |
| eGFR | INVT | 51,039 | -0.02 [-0.15, 0.11] | 0.7810 | -0.03 [-0.10, 0.04] | 0.4080 | -0.07 [-0.20, 0.06] | 0.2741 |
CHD, coronary heart disease; HDL, high density lipoprotein; LDL, low density lipoprotein; eGFR, estimated glomerular filtration rate; DBP, diastolic blood pressure; SBP systolic blood pressure; PP, pulse pressure; OR, odds ratio; g, grams; INVT, inverse normally transformed (hence no units); N, sample size; P, P-value of association of BP with the trait listed; CI, confidence interval. Results are considered significant if P < 0.0038, which corresponds to a Bonferroni correction for 13 phenotypes tested.
Possible functional variants at BP loci and candidate genes
Twenty-six of our newly discovered BP associated SNVs had MAF≥0.05 and therefore due to extensive LD with other SNVs not genotyped on the Exome array, identifying the causal genes requires additional information. If a SNV is associated with increased or decreased expression of a particular gene, i.e. it is an expression quantitative trait locus (eQTL) this suggests the gene on which the SNV acts could be in the causal pathway. To help identify potential candidate causal genes in the novel BP loci (Supplementary Table 9), information from publicly available eQTL databases was investigated (MuTHER for LCL, adipose and skin and GTEx for nine tissues including the heart and tibial artery; Methods).
The DBP increasing allele of the nsSNV, rs7302981-A, was associated with increased expression of CERS5 in: LCLs (PMuTHER=3.13x10-72) skin (PMuTHER=2.40x10-58) adipose (PMuTHER=2.87x10-54) and nerve (PGTEx=4.5x10-12) (Supplementary Figure 5). Additional testing (Methods) provided no evidence against colocalisation of the eQTL and DBP association signals, implicating CERS5 as a candidate causal gene for this DBP locus. CERS5 (LAG1 homolog, ceramide synthase 5) is involved in the synthesis of ceramide, a lipid molecule involved in several cellular signaling pathways. Cers5 knockdown has been shown to reduce cardiomyocyte hypertrophy in mouse models29. However, it is unclear whether the blood pressure raising effects at this locus are the cause or result of any potential effects on cardiac hypertrophy. Future studies investigating this locus in relation to parameters of cardiac hypertrophy and function (e.g. ventricular wall thickness) should help address this question.
The DBP raising allele of the nsSNV (rs867186-A) was associated with increased expression of PROCR in adipose tissue (PMuTHER=3.24x10-15) and skin (PMuTHER=1.01x10-11) (Supplementary Figure 5). There was no evidence against colocalisation of the eQTL and DBP association thus supporting PROCR as a candidate causal gene. PROCR encodes the Endothelial Protein C receptor, a serine protease involved in the blood coagulation pathway, and rs867186 has previously been associated with coagulation and haematological factors.30,31 The PP decreasing allele of, rs10407022-T, which is predicted to have detrimental effects on protein structure (Methods) was associated with increased expression of AMH in muscle (PGTEx=9.95x10-15), thyroid (PGTEx=8.54x10-7), nerve (PGTEx=7.15x10-8), tibial artery (PGTEx=6.46x10-9), adipose (PGTEx=4.69x10-7), and skin (PGTEx=5.88x10-8) (Supplementary Figure 5). There was no evidence against colocalisation of the eQTL and PP association, which supports AMH as a candidate causal gene for PP. Low AMH levels have been previously associated with hypertensive status in women with the protein acting as a marker of ovarian reserve32. The intergenic SBP raising allele of rs4728142-A was associated with reduced expression of IRF5 in skin (PMuTHER=5.24x10-31) and LCLs (PMuTHER=1.39x10-34), whole blood (PGTEx=3.12x10-7) and tibial artery (PGTEx=1.71x10-7).
Three novel rare nsSNVs were identified that map to RBM47, RRAS (both associated with SBP) and COL21A1 (associated with PP). They had larger effect sizes than common variant associations (>1.5mmHg per allele; Supplementary Figure 6) and were predicted to have detrimental effects on protein structure (Supplementary Table 16; Methods). In RBM47, rs35529250 (p.Gly538Arg) is located in a highly conserved region of the gene and was most strongly associated with SBP (MAF=0.008; +1.59 mmHg per T allele; P=5.90x10-9). RBM47 encodes the RNA binding motif protein 47 and is responsible for post-transcriptional regulation of RNA, through its direct and selective binding with the molecule.33 In RRAS, rs61760904 (p.Asp133Asn) was most strongly associated with SBP (MAF=0.007; +1.51 mmHg per T allele; P=8.45x10-8). RRAS encodes a small GTPase belonging to the Ras subfamily of proteins H-RAS, N-RAS, and K-RAS and has been implicated in actin cytoskeleton remodelling, and controlling cell proliferation, migration and cycle processes34. The nsSNV in COL21A1 (rs200999181, p.Gly665Val) was most strongly associated with PP (MAF=0.001; +3.14 mmHg per A allele; P=1.93x10-9). COL21A1 encodes the collagen alpha-1 chain precursor of type XXI collagen, a member of the FACIT (fibril-associated collagens with an interrupted triple helix) family of proteins35. The gene is detected in many tissues, including the heart and aorta. Based on our results, these three genes represent good candidates for functional follow-up. However, due to the incomplete coverage of all SNVs across the region on the Exome chip, it is possible that other non-genotyped SNVs may better explain some of these associations. We therefore checked for variants in LD (r2>0.3) with these three rare nsSNVs in the UK10K + 1000G dataset36 to ascertain if there are other candidate SNVs at these loci (Supplementary Table 17). There were no SNVs within 1Mb of the RBM47 locus in LD with the BP associated SNV. At the COL21A1 locus there were only SNVs in moderate LD, and these were annotated as intronic, intergenic or in the 5’UTR. At the RRAS locus, there were two SNVs in strong LD with the BP associated SNV, which both mapped to introns of SCAF1 and are not predicted to be damaging. All SNVs in LD at both loci were rare as expected (Supplementary Table 17) supporting a role for rare variants. Hence, the rare BP associated nsSNVs at RBM47, COL21A1 and RRAS remain the best causal candidates.
Pathway and network analyses
To identify connected gene sets and pathways implicated by the BP associated genes we used Meta-Analysis Gene-set Enrichment of variant Associations (MAGENTA)37 and GeneGo MetaCore (Thomson Reuters, UK). MAGENTA tests for over-representation of BP associated genes in pre-annotated pathways (gene sets) (Methods and Supplementary Table 18a). GeneGo Metacore identifies potential gene networks. The MAGENTA analysis was used for hypothesis generation and results were compared with the GeneGo Metacore outputs to cross-validate findings.
Using MAGENTA there was an enrichment (P<0.01 and FDR<5% in either the EUR_SAS or the EUR participants) of six gene sets with DBP, three gene sets with HTN and two gene sets for SBP (Supplementary Table 18b). The RNA polymerase I promoter clearance (chromatin modification) pathway showed the most evidence of enrichment with genes associated with DBP (PReactome=8.4x10-5, FDR=2.48%). NOTCH signalling was the most associated pathway with SBP (PReactome = 3.00x10-4, FDR = 5%) driven by associations at the FURIN gene. The inorganic cation anion solute carrier (SLC) transporter pathway had the most evidence of enrichment by HTN associated genes (PReactome=8.00x10-6, FDR=2.13%).
Using GeneGo MetaCore, five network processes were enriched (FDR<5%; Methods; Supplementary Tables 19 and 20). These included several networks with genes known to influence vascular tone and BP: inflammation signalling, P=1.14x10-4 and blood vessel development P=2.34x10-4. The transcription and chromatin modification network (P=2.85x10-4) was also enriched, a pathway that was also highlighted in the MAGENTA analysis, with overlap of the same histone genes (HIST1H4C, HIST1H2AC, HIST1H2BC, HIST1H1T) and has also been recently reported in an integrative network analysis of published BP loci and whole blood expression profiling38. Two cardiac development pathways were enriched: the oxidative stress-driven (ROS/NADPH) (P=4.12x10-4) and the Wnt/β-catenin/integrin-driven (P=0.0010). Both these cardiac development pathways include the MYH6, MYH7, and TBX2 genes, revealing a potential overlap with cardiomyopathies and hypertension, and suggesting some similarity in the underlying biological mechanisms.
Discussion
By conducting the largest ever genetic study of BP, we identified further novel common variants with small effects on BP traits, similar to what has been observed for obesity and height39,40. More importantly, our study identified some of the first rare coding variants of strong effect (>1.5mmHg) that are robustly associated with BP traits in the general population, complementing and extending the previous discovery and characterisation of variants underlying rare Mendelian disorders of blood pressure regulation 41. Using SNV associations in 17 genes reported to be associated with monogenic disorders of blood pressure (Methods) we found no convincing evidence of enrichment (Penrichment=0.044). This suggests that BP control in the general population may occur through different pathways to monogenic disorders of BP re-enforcing the importance of our study findings. The identification of 30 novel BP loci plus further new independent secondary signals within four novel and five known loci (Methods) has augmented the trait variance explained by 1.3%, 1.2% and 0.93% for SBP, DBP and PP respectively within our data-set. This suggests that with substantially larger sample sizes, for example through UK BioBank42, we expect to identify 1000s more loci associated with BP traits, and replicate more of our discovery SNV associations that are not yet validated in the current report.
The discovery of rare missense variants has implicated several interesting candidate genes, which are often difficult to identify from common variant GWAS, and should therefore lead to more rapidly actionable biology. A2ML1, COL21A1, RRAS and RBM47 all warrant further follow-up studies to define the role of these genes in regulation of BP traits, as well as functional studies to understand their mechanisms of action. COL21A1 and RRAS warrant particular interest since both are involved in blood vessel remodelling, a pathway of known aetiological relevance to hypertension.
We observed a rare nonsense SBP associated variant in ENPEP (rs33966350; p.Trp317* ): this overlaps a highly conserved region of both the gene and protein and is predicted to result in either a truncated protein with reduced catalytic function or is subject to nonsense mediated RNA decay. ENPEP converts angiotensin II (AngII) to Ang-III. AngII activates the angiotensin 1 (AT1) receptor resulting in vasoconstriction, while AngIII activates the angiotensin 2 (AT2) receptor that promotes vasodilation and protects against hypertension.43 The predicted truncated protein may lead to predominant AngII signaling in the body, and increases in BP. This new observation could potentially inform therapeutic strategies. Of note, angiotensin-converting-enzyme (ACE) inhibitors are commonly used in the treatment of hypertension. However, patients who suffer from adverse reactions to ACE inhibitors, such as dry cough and skin rash, would benefit from alternative drugs that target RAAS. Murine studies have shown that in the brain, AngIII is the preferred AT1 agonist that promotes vasoconstriction and increases blood pressure, as opposed to AngII in the peripheral system. These results have motivated the development of brain specific APA inhibitors to treat hypertension44. Our results confirm APAs, such as ENPEP, as a valid target to modify blood pressure, but suggest that long-term systemic reduction in APA activity may lead to an increase in blood pressure. Future studies are needed to examine the effects of the p.Trp317* variant on the RAAS system, specifically in the brain and peripheral vasculature, in order to test the benefits of the proposed therapeutic strategy in humans.
In addition to highlighting new genes in pathways of established relevance to BP and hypertension, and identifying new pathways, we have also identified multiple signals at new loci. For example, there are three distinct signals at the locus containing the MYH6/MYH7 genes, and we note that TBX2 maps to one of the novel regions. These genes are related to cardiac development and/or cardiomyopathies, and provide an insight into the shared inheritance of multiple complex traits. Unravelling the causal networks within these polygenic pathways may provide opportunities for novel therapies to treat or prevent both hypertension and cardiomyopathies.
Online Methods
Overview of discovery studies
The cohorts contributing to the discovery meta-analyses comprise studies from three consortia (CHD Exome+, ExomeBP, and GoT2D/T2D-GENES) with a total number of 192,763 unique samples. All participants provided written informed consent and the studies were approved by their local Research Ethics Committees and/or Institutional Review Boards.
The CHD Exome+ consortium comprised 77,385 samples: eight studies (49,898 samples) of European (EUR) ancestry, two studies (27,487 samples) of South Asian (SAS) ancestry (Supplementary Table 1). The ExomeBP consortium included 25 studies (75,620 samples) of EUR ancestry (Supplementary Table 1). The GoT2D consortium comprised 14 studies (39,758 samples) of Northern EUR ancestry from Denmark, Finland, and Sweden (Supplementary Table 1). The participating studies and their characteristics including BP phenotypes are detailed in Supplementary Tables 1 and 2. Note, any studies contributing to multiple consortia were only included once in all meta-analyses.
Phenotypes
Four blood pressure (BP) traits were analysed: systolic blood pressure (SBP), diastolic blood pressure (DBP), pulse pressure (PP) and hypertension (HTN). For individuals known to be taking BP lowering medication, 15/10 mmHg was added to the raw SBP/DBP values, respectively, to obtain medication-adjusted SBP/DBP values45. PP was defined as SBP minus DBP, post-adjustment. For HTN, individuals were classified as hypertensive cases if they satisfied at least one of: (i) SBP≥140 mmHg, (ii) DBP≥90 mmHg, (iii) taking anti-hypertensive or BP lowering medication. All other individuals were included as controls. The four BP traits were correlated (SBP:DBP correlations were between 0.6 and 0.8, and SBP:PP correlations were ~0.8). However, they measure partly distinct physiological features including, cardiac output, vascular resistance, and arterial stiffness, all measures for determining a cardiovascular risk profile. Therefore the genetic architecture of the individual phenotypes are of interest, and a multi-phenotype mapping approach was not adopted.
Genotyping
All samples were genotyped using one of the Illumina HumanExome Beadchip arrays (Supplementary Table 3). An Exome chip quality control Standard Operating Procedure (SOP) developed by Anubha Mahajan, Neil Robertson and Will Rayner at the Wellcome Trust Centre for Human Genetics, University of Oxford was used by most studies for genotype calling and QC46 (Supplementary Table 3). All genotypes were aligned to the plus strand of the human genome reference sequence (Build37) prior to any analyses and any unresolved mappings were removed. Genotype cluster plots were reviewed for all the novel rare variants (both lead and secondary signals) and for rare variants that contributed to the gene-based testing.
Meta-analyses
Meta-analyses were performed using METAL47, for both discovery and replication analyses, using inverse variance weighted fixed effect meta-analysis for the continuous traits (SBP, DBP and PP) and sample size weighted meta-analysis for the binary trait (HTN).
Discovery SNV analyses
Analyses of both untransformed and inverse normal transformed SBP, DBP and PP were conducted within each contributing study. The analyses of the transformed traits were performed in order to minimise sensitivity to deviations from normality in the analysis of rare variants and for discovery of new SNV-BP associations. The residuals from the null model obtained after regressing the medication-adjusted trait on the covariates (age, age2, sex, BMI, and disease status for CHD) within a linear regression model, were ranked and inverse normalised. These normalised residuals were used to test trait-SNV associations. All SNVs that passed QC were analysed for association, without any further filtering by MAF, but a minor allele count of 10 was used for the analysis of HTN. An additive allelic effects model was assumed.
Two meta-analyses were performed for each trait, one with EUR and SAS ancestries combined (EUR_SAS) and another for EUR ancestry alone. Contributing studies used principal components (PCs) to adjust for population stratification. Consequently minimal inflation in the association test statistics, λ, was observed (λ=1.07 for SBP, 1.10 for DBP, 1.04 for PP and <1 for HTN in the transformed discovery meta-analysis in EUR_SAS; λ= 1.06 for SBP, 1.09 for DBP, 1.05 for PP and <1 for HTN in the transformed discovery meta-analysis in EUR; Supplementary Figure 7). The meta-analyses were performed independently in two centres and results were found to be concordant between centres. Given the studies contributing to the discovery analyses were ascertained on CHD or T2D, we tested potential systematic bias in calculated effect estimates amongst these studies. No evidence of bias in the overall effect estimates was obtained.
The results for the transformed traits were taken forward and used to select candidate SNVs for replication. Results (P-values) from the transformed and untransformed analyses were strongly correlated (r2>0.9).
Replication SNV analyses
SNVs associated with any of the transformed traits (SBP, DBP, PP) or HTN were annotated using the Illumina SNV annotation file, humanexome-12v1_a_gene_annotation.txt, independently across two centres. Given the difference in power to detect common versus low frequency and rare variant associations, two different significance thresholds were chosen for SNV selection. For SNVs with MAF≥0.05, P≤1x10-5 was selected, while, P≤1x10-4 was used for SNVs with MAF < 0.05. By choosing a significance threshold of P<1x10-4 we maximized the opportunity to follow-up rare variants (making the assumption that any true signals at this threshold could replicate at Bonferroni adjusted significance, P≤6.17x10-4, assuming α=0.05 for 81 SNVs). All previously published BP associated SNVs and any variants in LD with them (r2>0.2), were removed from the list of associated SNVs as we aimed to replicate new findings only. SNVs for which only one study contributed to the association result or showed evidence of heterogeneity (Phet<0.0001) were removed from the list as they were likely to be an artefact. Where SNVs were associated with multiple traits, to minimise the number of tests performed, only the trait with the smallest P-value was selected as the primary trait in which replication was sought. Where multiple SNVs fitted these selection criteria for a single region, only the SNV with the smallest P-value was selected. In total, 81 SNVs were selected for validation in independent samples. These 81 SNVs had concordant association results for both transformed and non-transformed traits. Eighty SNVs were selected from EUR_SAS results (with consistent support in EUR), and one SNV from EUR results only. In the next step, we looked up the 81 SNV-BP associations using data from a separate consortium, the CHARGE+ exome chip blood pressure consortium (who had analysed untransformed SBP, DBP, PP and HTN), and UHP and Lolipop (ExomeBP consortium; Supplementary Tables 2 and 3). The analysed residuals from CHARGE+ were approximately normally distributed in their largest studies (Supplementary Figure 8).
Two meta-analyses of the replication datasets were performed: one of EUR samples, and a second of EUR, African American, Hispanics and SAS ancestries (“ALL”). Replication was confirmed if P (1-tailed) < 0.05/81=6.17x10-4 and the effect (beta) was in the direction observed in discovery meta-analyses for the selected trait. A combined meta-analysis was performed of discovery (untransformed results as only untransformed data was available from CHARGE+ exome chip blood pressure consortium) and replication results across the four traits to assess the overall support for each locus. For the combined meta-analyses, a GWS threshold of, P≤5x10-8, was used to declare a SNV as novel rather than a less stringent experiment wide threshold, as GWS is used to declare significance in GWAS and we wish to minimise the possibility of false positive associations. (Note that GWS is equivalent to an exome-wide threshold of P≤2x10-7 adjusted for four traits).
Note: all validated BP-associated variants were associated at P<10-5 in the discovery dataset (for the primary trait). Hence, we could have used the same inclusion criteria for both common and rare SNVs. Therefore the optimal threshold to choose for future experiments may need further consideration.
Conditional analyses and gene-based tests
The RAREMETALWORKER (RMW) tool15 (version 4.13.3) that does not require individual level data to perform conditional analyses and gene-based tests was used for conditional analyses. All studies that contributed to the SNV discovery analyses were re-contacted and asked to run RMW. Only FENLAND, GoDARTS, HELIC-MANOLIS, UKHLS and EPIC-InterAct were unable to run RMW, while two new studies were included, INCIPE and NFBC1966 (Supplementary Table 1 and 2). In total, 43 studies (147,402 samples) were included in the EUR analyses and 45 studies (173,329 samples) in the EUR_SAS analyses (Supplementary Tables 2 and 3). Comparison of discovery and RMW study level results were made (Supplementary Information).
For each novel locus, the genomic coordinates and size of the region were defined according to recombination rates (Supplementary Table 9) around the lead variant. For known loci, a 1 Mb window was used (Supplementary Table 14). Conditional analyses were performed across each region, in both EUR and EUR_SAS samples, for the transformed phenotype corresponding to the validated BP trait for novel loci and the published BP trait for known loci.
Gene based tests were performed in both the EUR and EUR_SAS datasets using the Sequence Kernel Association Test (SKAT)16 method implemented in RMW as it allows for the SNVs to have different directions and magnitudes of effect. Burden tests were also performed but are not presented as only SKAT provided significant results. The variants in the gene-based tests using SKAT were weighted using the default settings, i.e. a beta distribution density function to up-weight rare variants, Beta(MAFj,1,25) where MAFj represents the pooled MAF for variant j across all studies. Analyses were restricted to coding SNVs with MAF<5% and <1%. Genes were deemed to be associated if P <2.8x10-6 (Bonferroni adjusted for 17,996 genes). To confirm the gene associations were not attributable to a solitary SNV, a gene-based test conditional on the most associated SNV was performed (Pconditional< 0.001). The QC of all SNVs contributing to the gene based tests including the number of samples and studies were checked prior to claiming association. We sought replication of associated genes in the CHARGE+ exome chip blood pressure consortium.
Pathway analyses with MAGENTA
We tested seven databases in MAGENTA37 (BioCarta, Kyoto Encyclopedia of Genes and Genomes, Ingenuity, Panther, Panther Biological Processes, Panther Molecular Functions and Reactome) for overrepresentation of the SNV discovery results from both EUR and EUR_SAS ancestries. Each of the four BP phenotypes were tested. Pathways exhibiting P<0.01 and FDR<5% were considered statistically significant.
GeneGo MetaCore Network analyses
A set of BP genes based on previously published studies and our current results (locus defined as r2>0.4 and 500kb on either side of the lead SNV; Supplementary Table 19) were tested for enrichment using the THOMSON REUTERS MetaCore™ Single Experiment Analysis workflow tool. The data were mapped onto selected MetaCore ontology databases: pathway maps, process networks, GO processes and diseases / biomarkers, for which functional information is derived from experimental literature. Outputs were sorted based on P- and FDR-values. A gene set was considered enriched for a particular process if P<0.05 and FDR<5%.
Genetic Risk Score
To assess the effect of BP on CHD, ischemic stroke (and subtypes: large vessel, small vessel and cardioembolic stroke) left ventricular mass, left ventricular wall thickness, heart failure, HDL-c, LDL-c, total cholesterol, triglycerides and eGFR, we performed a weighted generalized linear regression of the genetic associations with each outcome variable on the genetic associations with BP.
When genetic variants are uncorrelated, the estimates from such a weighted linear regression analysis using summarized data, and a genetic risk score analysis using individual-level data, are equal48. We refer to the analysis as a genetic risk score (also known as a polygenic risk score) analysis as this is likely to be more familiar to applied readers. As some of the genetic variants in our analysis are correlated, a generalized weighted linear regression model is fitted that accounts for the correlations between variants, as follows: If βX are the genetic associations (beta-coefficients) with the risk factor (here, BP) and βY are the genetic associations with the outcome, then the causal estimate from a weighted generalized linear regression is (βXTΏ-1βX)-1 βXTΏ-1βY, with standard error,
where T is a matrix transpose, σ̂ is the estimate of the residual standard error from the regression model, and the weighting matrix Ώ has terms
, where σYj is the standard error of the genetic association with the outcome for the jth SNV, and ρj1j2 is the correlation between the j1th and j2 th SNVs. The presence of the estimated residual standard error allows for heterogeneity between the causal estimates from the individual SNVs as overdispersion in the regression model (in the case of underdispersion, the residual standard error estimate is set to unity). This is equivalent to combining the causal estimates from each SNV using a multiplicative random-effects model49.
For each of SBP, DBP and PP, the score was created using both the novel and known BP SNVs or a close proxy (r2>0.8). Both the sentinel SNV association and any secondary SNV associations that remained after adjusting for the sentinel SNV were included in the genetic risk score. For the 30 validated novel SNV-BP associations, βs were taken from the independent replication analyses (Table 1 and 2) to weight the SNV in the genetic risk score. For the secondary SNVs from the seven novel loci and five known loci, βs were taken from the discovery analyses (Supplementary Tables 10 and 15). For the 82 known SNVs, 43 were either genotyped or had proxies on the Exome chip and the βs were taken from discovery results (Supplementary Table 13), the remaining βs were taken from published effect estimates. This strategy for selecting betas for use in the GRS was taken to minimize the influence of winner’s curse. The associations between the BP variants with CHD, HDL-c, LDL-c, total cholesterol, log(triglycerides) and log(eGFR) were obtained using the CHD Exome+ Consortium studies, the associations with BMI, waist-hip ratio adjusted BMI and height from the GIANT consortium (unpublished data), ischemic stroke from METASTROKE25, and left ventricular mass, left ventricular wall thickness and heart failure from EchoGen27 and CHARGE-HF26. A causal interpretation of the association of GRS with the outcome as the effect of BP on the outcome assumes that the effects of genetic variants on the outcome are mediated via blood pressure and not via alternate causal pathways, for example via LV thickness. There are also limitations of the Mendelian randomization approach in distinguishing between the causal effects of different measures of blood pressure, due to the paucity of genetic variants associated with only one measure of blood pressure.
eQTL analyses
The MuTHER dataset contains gene expression data from 850 UK twins for 23,596 probes and 2,029,988 (HapMap 2 imputed) SNVs. All cis–associated SNVs with FDR<1%, within each of the 30 novel regions (IMPUTE info score >0.8) were extracted from the MuTHER project dataset for, LCL (n=777), adipose (n=776) and skin (n=667) 50. The pilot phase of the GTEx Project (dbGaP Accession phs000424.v3.p1) provides expression data from up to 156 individuals for 52,576 genes and 6,820,472 genotyped SNVs (imputed to 1000 Genomes project, MAF≥5%)51. The eQTL analysis was focused on subcutaneous adipose tissue (n=94), tibial artery (n=112), heart (left ventricle) (n=83), lung (n=119), skeletal muscle (n=138), tibial nerve (n=88), skin (sun exposed, lower leg) (n=96), thyroid (n=105) and whole blood (n=156) which have >80 samples and genes expressed at least 0.1 RPKM in 10 or more individuals in a given tissue. All transcripts with a transcription start site (TSS) within one of the 30 new BP loci and for which there was a cis-associated SNV (IMPUTE info score >0.4) within 1Mb of the TSS at FDR<5%, were identified. Kidney was not evaluated because the sample size was too small (n=8). From each resource, we report eQTL signals, which reach the resource-specific thresholds for significance described above, for SNVs that are in LD (r2>0.8) with our sentinel SNV.
For identified eQTLs, we tested whether they colocalised with the BP associated SNV52. Colocalisation analyses were considered to be significant if the posterior probability of colocalisation was greater than 0.95.
Annotation of variants
In silico prediction of the functional effect of associated variants was based on the annotation from dbSNP, the Ensembl Variant Effect Predictor tool and the Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA.
Trait variance explained
The percentage trait variance explained for SBP, DBP, PP was assessed with 5,861 individuals with complete information for all phenotypes and covariates from the population-based cohort, 1958BC.
Two genetic models were investigated: one containing the 43 previously known BP associated SNVs covered on the Exome chip; the other additionally including the 30 novel lead SNVs and 9 conditionally independent SNVs from both novel and known loci. These nine conditionally independent SNVs were taken from the EUR results, as 1958BC is EUR. They included four from novel loci (PREX1, COL21A1, PRKAG1 and MYH6 (there was only 1 in EUR); Supplementary Table 10) and five from known loci (ST7L-CAPZA1-MOV10, FIGN-GRB14, ENPEP, TBX5-TBX3 and HOXC4; Supplementary Table 15).
The residual trait was obtained by adjusting each of the BP traits in a regression model with sex and BMI variables (not age or age2 as all 1958BC individuals were aged 44 years). The residual trait was regressed on all SNVs within the corresponding model and adjusted for the first ten PCs. The R2 calculated from this regression model was used as the percentage trait variance explained.
Monogenic Enrichment analyses
To determine if sub-significant signals of association were present in a set of genes associated with monogenic forms of disease, we performed an enrichment analysis of the discovery single variant meta-analyses association results for all four traits, both for EUR and EUR_SAS datasets.
The monogenic gene set included: WNK1, WNK4, KLHL3, CUL3, PPARG, NR3C2, CYP11B1, CYP11B2, CYP17A1, HSD11B2, SCNN1A, SCNN1B, SCNN1G, CLCNKB, KCNJ1, SLC12A1, SLC12A33. The association results of coding SNVs in these genes were extracted and the number of tests with P<0.001 observed. In order to determine how often such an observation would be observed by chance, we constructed 1,000 matched gene sets. The matching criteria for each monogenic gene was the intersection of all genes in the same exon length quintile and all genes in the same coding variant count decile. Within the matched sets, the number of variants with P<0.001 was observed. The empirical P-value was calculated as the fraction of matched sets with an equal or larger number of variants less than 0.001.
Supplementary Material
Acknowledgements
Footnotes
URLs
Exome chip design information: http://genome.sph.umich.edu/wiki/Exome_Chip_Design
RareMetalWorker information: http://genome.sph.umich.edu/wiki/RAREMETALWORKER
Summary SNV association results: http://www.phenoscanner.medschl.cam.ac.uk
Databases used for variant annotation: http://www.ncbi.nlm.nih.gov/SNP/ http://www.ensembl.org/info/docs/tools/index.html and http://evs.gs.washington.edu/EVS/
UCSC reference file used for annotation of variants with gene and exon information: http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/refFlat.txt.gz
Databases used for pathway analysis: MAGENTA (https://www.broadinstitute.org/mpg/magenta/) and THOMSON REUTERS MetaCore™ Single Experiment Analysis workflow tool (http://thomsonreuters.com/en/products-services/pharma-life-sciences/pharmaceutical-research/metacore.html).
Author contributions
Supervision and management of the project: JMHH and PBM. The following authors contributed to the drafting of the manuscript: JMMH, PBM, PSu, HW, ASB, FD, JPC, DRB, KW, MT, FWA, LVW, NJS, JD AKM, HY, CMM, NG, XS, TaT, DFF, MHs, OG, TF, VT. All authors critically reviewed and approved the final version of the manuscript. Statistical analysis review: JMMH, PSu, FD, HW, JPC, RY, NM, PBM, LVW, HY, TF, EMi, ADM, AM, AM, EE, ASB, FWA, MJC, CF, TF, SEH, ASH, JEH, JL, GM, JM, NM, APM, APo, NJS, RAS, LS, KE, MT, VT, TVV, NV, KW, AMY, WZg, NG, CML, AKM, XS, TT. Central Data QC: JMMH, ASB, PSu, RY, FD, HW, JPC, TF, LVW, PBM, EMi, NM, CML, NG, XS, AKM. Central Data analysis: JMMH, PSu, FD, HW, JPC, NG, CML, AKM, XS. Pathway analysis and literature review: JMMH, DRB, PBM, MT, KW, VT, OG, AT, FWA. GWAS lookups, eQTL analysis, GRS, variant annotation and enrichment analyses: JMMH, ASB, DRB, JRS, DFF, FD, MHr, PBM, FWA, TT, CML, AKM, SBu. Study Investigators in alphabetical order by consortium (CHD Exome+, ExomeBP and GoT2D): DSA, PA, EA, DA, ASB, RC, JD, JF, IF, PF, JWJ, FKe, ASM, SFN, BGN, DS, NSa, JV, FWA, PIWB, MJB, MJC, JCC, JMC, IJD, GD, AFD, PE, TE, PWF, GG, PH, CH, KH, EI, MJ, FKa, SK, JSK, LLi, MIM, OM, AMe, ADM, APM, PBM, MEN, SP, CP, OPo, DP, SR, OR, IR, VS, NJS, PSe, TDS, JMS, NJW, CJW, EZ, MB, IB, FSC, LG, TH, EKH, PJ, JKu, ML, TAL, AL, KLM, HO, OPe, RR, JT, MU. Study Phenotyping in alphabetical order by consortium (CHD Exome+, ExomeBP and GoT2D): PA, DA, SBl, MC, JF, JWJ, FKe, KK, SFN, BGN, CJP, AR, MS, NSa, JV, WZo, RAB, MJB, MJC, JCC, JMC, AFD, ASFD, LAD, TE, AF, GG, GH, PH, AS H, OLH, EI, MJ, FK, JSK, LLi, LLa, GM, AMc, PM, AMe, RMg, MJN, MEN, OPo, NP, FR, VS, NJS, TDS, AVS, JMS, MT, AV, NV, NJW, TiT, CC, LLH, MEJ, AK, PK, JL DPS, SM, ERBP, AS, TS, HMS, BT. Study Data QC and analysis in alphabetical order by consortium (CHD Exome+, ExomeBP and GoT2D): ASB, AJMC, JMMH, JK, SFN, BGN, MMN, SP, MP, PSu, ST, GV, SMW, RY, FWA, JPC, FD, AF, TF, CH, AMc, AMj, APM, PBM, CP, WR, FR, NJS, MT, VT, HW, HY, NG, AKM, XS. Exome chip data QC in alphabetical order by consortium (CHD Exome+, ExomeBP and GoT2D): ASB, JMMH, SFN, BGN, PSu, RY, FWA, PIWB, AIFB, JCC, JPC, PD, LAD, FD, EE, CF, TF, SEH, PH, SSH, KH, JEH, EK, AMj, GM, JM, NM, EMi, AMo, APM, PBM, CPN, MJN, CP, AP, WR, NRR, RAS, NS, LS, KES, MDT, VT, TVV, TVV, NV, HW, HY, AMY, EZ, WZg, NG, CML, AKM, XS. Exome chip Data analysis in alphabetical order by consortium (CHD Exome+, ExomeBP and GoT2D): JMMH, PSu, RY, FWA, PIWB, AIFB, RAB, MJC, JCC, JPC, PD, LAD, PE, EE, CF, TF, PWF, SF, CG, SEH, PH, ASH, CH, OLH, JEH, EI, MJ, FKa, JSK, DCML, LLi, JL, GM, RMr, JM, NM, MIM, PM, OM, CM, EMi, AMo, APM, RMg, PBM, CPN, MJN, TO, APo, APa, WR, NRR, NJS, RAS, NS, LS, TDS, KES, MDT, ET, VT, TVV, NV, LVW, NJW, HW, HY, AMY, EZ, HZ, WZg, LLB, APG, NG, MHs, JRH, AUJ, JBJ, CML, AKM, NN, XS, AS, AJS. GRS lookups: AEJ, EMa, HFM, HL, HMH, JFF, MTr, RSV, WL.
Conflict of interests
N. P. has received financial support from several pharmaceutical companies that manufacture either blood pressure lowering or lipid lowering agents, or both, and consultancy fees.
S. K. has received Research Grant-Merck, Bayer, Aegerion; SAB-Catabasis, Regeneron Genetics Center, Merck, Celera; Equity-San Therapeutics, Catabasis; Consulting-Novartis, Aegerion, Bristol Myers-Squibb, Sanofi, AstraZeneca, Alnylam.
P. Sever has received research awards from Pfizer Inc.
A. Malarstig and M. Uria-Nickelsen are full time employees of Pfizer.
D. Reily and M. Hoek are full time employees of Merck and co Inc.
M.J. Caulfield is Chief Scientist for Genomics England a UK Government company.
The authors declare no competing financial interest.
References
- 1.Lim SS, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–60. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rapsomaniki E, et al. Blood pressure and incidence of twelve cardiovascular diseases: lifetime risks, healthy life-years lost, and age-specific associations in 1.25 million people. Lancet. 2014;383:1899–911. doi: 10.1016/S0140-6736(14)60685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munroe PB, Barnes MR, Caulfield MJ. Advances in blood pressure genomics. Circ Res. 2013;112:1365–79. doi: 10.1161/CIRCRESAHA.112.300387. [DOI] [PubMed] [Google Scholar]
- 4.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wain LV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nature Genetics. 2011 doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson T, et al. Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomaszewski M, et al. Genetic architecture of ambulatory blood pressure in the general population: insights from cardiovascular gene-centric array. Hypertension. 2010;56:1069–76. doi: 10.1161/HYPERTENSIONAHA.110.155721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tragante V, et al. Gene-centric meta-analysis in 87,736 individuals of European ancestry identifies multiple blood-pressure-related loci. Am J Hum Genet. 2014;94:349–60. doi: 10.1016/j.ajhg.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganesh SK, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013 doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simino J, et al. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am J Hum Genet. 2014;95:24–38. doi: 10.1016/j.ajhg.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, et al. Meta-analysis of Correlated Traits via Summary Statistics from GWASs with an Application in Hypertension. Am J Hum Genet. 2015;96:21–36. doi: 10.1016/j.ajhg.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salfati E, Morrison AC, Boerwinkle E, Chakravarti A. Direct Estimates of the Genomic Contributions to Blood Pressure Heritability within a Population-Based Cohort (ARIC) PLoS One. 2015;10:e0133031. doi: 10.1371/journal.pone.0133031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–9. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu DJ, et al. Meta-analysis of gene-level tests for rare variant association. Nat Genet. 2014;46:200–4. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vissers LE, et al. Heterozygous germline mutations in A2ML1 are associated with a disorder clinically related to Noonan syndrome. Eur J Hum Genet. 2015;23:317–24. doi: 10.1038/ejhg.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Consortium for Blood Pressure Genome-Wide Association, S et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coronary Artery Disease Genetics Consortium C4D A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–44. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 21.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arking DE, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826–36. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.den Hoed M, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–31. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Global Lipids Genetics Consortium et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traylor M, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–62. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith NL, et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010;3:256–66. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasan RS, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–78. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo SB, et al. Ceramide synthase 5 mediates lipid-induced autophagy and hypertrophy in cardiomyocytes. J Clin Invest. 2012;122:3919–30. doi: 10.1172/JCI63888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oudot-Mellakh T, et al. Genome wide association study for plasma levels of natural anticoagulant inhibitors and protein C anticoagulant pathway: the MARTHA project. Br J Haematol. 2012;157:230–9. doi: 10.1111/j.1365-2141.2011.09025.x. [DOI] [PubMed] [Google Scholar]
- 31.Smith NL, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121:1382–92. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bleil ME, Gregorich SE, McConnell D, Rosen MP, Cedars MI. Does accelerated reproductive aging underlie premenopausal risk for cardiovascular disease? Menopause. 2013;20:1139–46. doi: 10.1097/GME.0b013e31828950fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan R, et al. rbm47, a novel RNA binding protein, regulates zebrafish head development. Dev Dyn. 2013;242:1395–404. doi: 10.1002/dvdy.24039. [DOI] [PubMed] [Google Scholar]
- 34.Wozniak MA, Kwong L, Chodniewicz D, Klemke RL, Keely PJ. R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol Biol Cell. 2005;16:84–96. doi: 10.1091/mbc.E04-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuckwell D. Identification and analysis of collagen alpha 1(XXI), a novel member of the FACIT collagen family. Matrix Biol. 2002;21:63–6. doi: 10.1016/s0945-053x(01)00176-7. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segre AV, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huan T, et al. Integrative network analysis reveals molecular mechanisms of blood pressure regulation. Mol Syst Biol. 2015;11:799. doi: 10.15252/msb.20145399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park HW, et al. Serine-threonine kinase with-no-lysine 4 (WNK4) controls blood pressure via transient receptor potential canonical 3 (TRPC3) in the vasculature. Proc Natl Acad Sci U S A. 2011;108:10750–5. doi: 10.1073/pnas.1104271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Te Riet L, van Esch JH, Roks AJ, van den Meiracker AH, Danser AH. Hypertension: Renin-Angiotensin-aldosterone system alterations. Circ Res. 2015;116:960–75. doi: 10.1161/CIRCRESAHA.116.303587. [DOI] [PubMed] [Google Scholar]
- 44.Gao J, et al. A new strategy for treating hypertension by blocking the activity of the brain renin-angiotensin system with aminopeptidase A inhibitors. Clin Sci (Lond) 2014;127:135–48. doi: 10.1042/CS20130396. [DOI] [PubMed] [Google Scholar]
- 45.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–35. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 46.Mahajan A, et al. Identification and Functional Characterization of G6PC2 Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the G6PC2-ABCB11 Locus. PLoS Genet. 2015;11:e1004876. doi: 10.1371/journal.pgen.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–60. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson SG, Sharp SJ. Explaining heterogeneity in meta-analysis: a comparison of methods. Stat Med. 1999;18:2693–708. doi: 10.1002/(sici)1097-0258(19991030)18:20<2693::aid-sim235>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 50.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.GTEx, Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.