Abstract
SR proteins are essential splicing factors that are regulated through multisite phosphorylation of their RS (arginine-serine-rich) domains by two major families of protein kinases. The SRPKs efficiently phosphorylate the arginine-serine dipeptides in the RS domain using a conserved docking groove in the kinase domain. In contrast, CLKs lack a docking groove and phosphorylate both arginine-serine and serine-proline dipeptides, modifications that generate a hyper-phosphorylated state important for unique SR protein-dependent splicing activities. All CLKs contain long, flexible N-terminal extensions (140-300 residues) that resemble the RS domains present in their substrate SR proteins. We showed that the N-terminus in CLK1 contacts both the kinase domain and the RS domain of the SR protein SRSF1. This interaction not only is essential for facilitating hyper-phosphorylation but also induces cooperative binding of SRSF1 to RNA. The N-terminus of CLK1 enhances the total phosphoryl contents of a panel of physiological substrates including SRSF1, SRSF2, SRSF5 and Tra2β1 by 2–3-fold. These findings suggest that CLK1-dependent hyper-phosphorylation is the result of a general mechanism in which the N-terminus acts as a bridge connecting the kinase domain and the RS domain of the SR protein.
Keywords: docking, kinase, kinetics, phosphorylation, splicing
Introduction
As essential regulators of cell function, protein kinases phosphorylate select substrate targets on serine, threonine or tyrosine. This posttranslational modification may alter the conformation and/or the biological activity of the target in complex manners. Protein kinases are now recognized as essential communication molecules and are often arrayed in signaling cascades that stretch from membrane receptors to intracellular targets. How protein kinases recognize specific substrates within these elaborate pathways is an important concern for a general understanding of signal transduction and for the specific development of kinase-directed drugs. Numerous studies have pointed to a classic, two-pronged mechanism for substrate attachment that uses the active site for the recognition of local residues flanking the phosphorylation site (consensus sequence) and a docking groove that recognizes distal residues [1, 2]. In general, the consensus sequence that occupies the active site is about 5-8 residues in length and provides a minimal level of recognition whereas specificity can be generated in distal docking grooves. In all cases studied so far the docking groove is part of a stable, folded domain presenting highly specific donor-acceptor contacts. In this new study, we demonstrate that one family of protein kinases important for RNA splicing incorporates a flexible docking element outside the kinase domain predicted to lack structure that captures similar flexible domains in their target substrates.
RNA splicing is regulated by SR proteins, a family of splicing factors essential for the correct establishment of 5′–3′ splice sites in precursor mRNA [3]. SR proteins derive their name from a C-terminal domain (50–300 residues) rich in Arg-Ser dipeptide repeats (RS domain). While RS domains are vital for SR protein regulation, very little is know about their structure owing to difficulties in expression and poor solubility at high concentrations. Sequence analyses indicate that RS domains lack structure and can be classified as intrinsically disordered [4]. Indeed, recent NMR studies demonstrate that the RS domain in the prototype SR protein SRSF1 is fully disordered in line with these predictions [5]. In contrast, several RNA binding domain structures in SR proteins (RRMs) have been solved and shown to be folded although none contain RS domains [6–8]. The recruitment of SR proteins for splicing reactions is regulated by multisite RS domain phosphorylation. The SR protein kinases [SRPKs] phosphorylate numerous serines in the RS domain, a modification that translocates SR proteins from the cytoplasm to the nucleus for splicing function [9]. SRPK1 binds with high affinity to SRSF1 and readily phosphorylates about 10–12 serines in the RS domain, a reaction that is driven by a docking groove in the large lobe of the kinase domain [10]. The docking groove feeds N-terminal dipeptides in lengthy Arg-Ser repeats into the active site resulting in the net, directional (C-to-N terminal) addition of phosphates onto the RS domain [11]. Thus, SRPK1 uses an active site and a stable docking groove in a folded domain to recognize its substrate, a classic example of the two-prong recognition mechanism.
Once in the nucleus, SR proteins are predominantly hyper-phosphorylated by another family of protein kinases called CLKs. This reaction is thought to mobilize SR proteins from nuclear speckles to the spliceosomal machinery [12, 13]. Similar to SRPKs, CLKs phosphorylate Arg-Ser repeats. Unlike SRPKs, CLKs also phosphorylate serines flanking prolines [14, 15], an activity important for 5′ splice-site selection [16]. Phosphorylation at Ser-Pro dipeptides induces unique substrate conformations since CLK1 treatment of SRSF1 promotes a large gel shift on SDS-PAGE not seen with SRPK1 [17]. CLK1 binds with high affinity to SRSF1, displaying a dissociation constant tighter than that for SRPK1 [14]. CLKs are intriguing because, unlike SRPKs, they possess no docking groove in the kinase domain [18] yet efficiently phosphorylate up to 18 serines in the SRSF1 RS domain [15]. The mechanistic details of how CLKs phosphorylate a narrow polypeptide stretch without a dedicated docking groove are largely unknown.
The CLK family (CLK1-4) possesses lengthy, N-terminal extensions (140-300 residues) that are classified as RS domains owing to the presence of arginine-serine dipeptides [19]. These N-termini are generally rich in many additional disorder-promoting amino acids common to RS domains (e.g., Arg, Ser, Lys, Pro, Gly, etc). Although the N-terminus is not a CLK1 substrate, it has been shown to be important for the recognition of certain RS domain-containing proteins in a yeast two-hybrid screen [19]. What role this N-terminus might play in regulating the SR protein phosphorylation levels has not been explored. To evaluate how CLK1 achieves high-level SR protein phosphorylation, we studied a kinase form lacking the N-terminus [CLK1(ΔN)]. Although CLK1 phosphorylates about 18 serines in SRSF1, CLK1(ΔN) only phosphorylates about 6 serines suggesting that the N-terminus is vital for complete RS domain activation. The N-terminus can make direct contacts with both the unstructured RS domain in SRSF1 and the kinase domain suggesting that this extension is highly dynamic and can act as a bridge between the kinase and substrate. CLK1-dependent phosphorylation induces cooperative binding of SRSF1 to the Ron exonic splicing enhancer (ESE), a phenomenon that is abrogated by deletion of the N-terminus. The CLK1 N-terminus is not only essential for SRSF1 hyper-phosphorylation but is also necessary for high-level phosphorylation of several other SR proteins. These findings suggest that the N-terminal extension serves as a general facilitator of SR protein hyper-phosphorylation by connecting a flexible RS domain to the CLK kinase domain.
Material & Methods
Materials
Adenosine triphosphate (ATP), 3-(N-morpholino)propanesulphonic acid (Mops), Tris (hydroxymethyl) aminomethane (Tris), MgCl2, NaCl, EDTA, glycerol, sucrose, acetic acid, Lysozyme, DNAse, RNAse, Phenix imaging film, BSA, Whatman P81 grade filter paper, g-agarose, Ni-resin, and liquid scintillant were obtained from Fisher Scientific. [γ-32P] ATP was obtained from NEN Products. Lysobacter enzymogenes endoproteinase Lys-C and protease inhibitor cocktail were obtained from Roche. Anti-His monoclonal antibody was purchased from Biolegend. InstantBlue was purchased from Expedeon, Hybond ECL nitrocellulose blotting membrane was purchased from Amersham, and the KinaseMax™ Kit was purchased form Ambion.
Expression and Purification of Recombinant proteins
Human SRPK1 and SRSF1 and mouse CLK1, CLK1(ΔN) and His-N (CLK1 residues 1-160) were expressed from pET19b vectors containing an N-terminal His Tag. Human Tra2β1, SRSF2, and SRSF5 were expressed from pET28a vectors with a C-terminal His Tag. GST-SRSF1 and GST-N (CLK1 residues 1-160) were expressed from a pGEX vector. The plasmids for SRSF1, SRPK1, Tra2β1 and CLK1 were transformed into BL21(DE3) E. coli strain. SRSF2 and SRSF5 were transformed in plus4RA E. coli strain. Cells were grown at 37°C in LB broth with 100 μg/ml ampicillin [Tra2β1 supplemented with 50 μg/mL Kanamycin], and protein expression was induced with 1 mg/mL IPTG at room temperature for 5 hours for SRSF1, SRSF2, SRSF5, and Tra2β constructs and for 12 hours for SRPK1, and 2.5 mg/mL IPTG for 16 hours for CLK1. All His-tagged CLK1 and SRPK1 proteins were purified by Ni-resin affinity chromatography as previously published [17]. SRSF1, SRSF2, SRSF5, and TRA2β1 were refolded and purified as previously published [15]. GST protein constructs were purified with glutathione-agarose resin according to a published procedure [20].
Phosphorylation Reactions
Substrate phosphorylation was carried out in 100 mM Mops (pH 7.4), 10 mM Mg2+, and 5 mg/mL BSA, at 23 °C according to previously published procedures [11]. Typical time courses were carried out with 30–90 nM enzyme, 20–150 nM SRSF1 and 50–100 μM [γ-32P]ATP (4000–8000 cpm pmol−1). For competition experiments, reactions were carried out using 20 nM CLK1, 20 μM ATP [γ-32P]ATP, 50 nM SR(ΔRRM1) and varying SR protein. All reactions were carried out in a total volume of 10 μL and quenched with 10 μL SDS PAGE loading buffer. Phosphorylated SR proteins were separated from unreacted ATP on 12% SDS-PAGE, cut from the dried SDS PAGE gel and quantitated on the 32P channel in liquid scintillant. The total amount of phosphoproduct was determined from the reaction specific activity (cpm/min).
Pull-down assays
GST-tagged proteins (4 μM) were incubated with His-tagged proteins (0.2 μM) in binding buffer [0.1% NP40, 20 mM Tris (pH 7.5) and 75 mM NaCl] for 30 min at room temperature before incubating with 15 μL glutathione-agarose resin for 30 min. The resin was washed 5X with 200 μL binding buffer, and the bound proteins were eluted with SDS quench buffer and boiling for 5 min. Bound protein was resolved by 12% SDS-PAGE and visualized by Instant Blue Coomassie stain or western blotting with anti-His mouse antibody (Biolegend).
LysC Proteolysis
SR(R214K) (0.25 μM) was phosphorylated using CLK1 and CLK1(ΔN) and 100 μM [γ-32P]ATP (4000–8000 cpm pmol−1) for 60 min in 30 μL total volume. This reaction was split in half, and an equal volume of buffer (50 mM Tris pH 8.5, 2 mM EDTA) with and without 0.35 μg of LysC was added and incubated at 37 °C for 60 min. Phosphorylated SR protein fragments were visualized using 18% SDS-PAGE.
RNA Binding Assays
The binding of a 13mer RNA oligomer based on the Ron ESE (AGGCGGAGGAAGC) to SRSF1 in the absence and presence of CLK1 phosphorylation was assessed using a filter-binding assay. Kinase phosphorylation reactions were carried out in the presence of 100 mM Mops (pH 7.4), 10 mM free Mg2+, 5 mg/mL BSA, 100 μM ATP, 100 nM CLK1 or CLK1(ΔN), and 2 μM SRSF1 at 23 °C for 1 hour. Mock reactions were carried out under the same conditions in the absence of kinase and ATP. RNA labeling was carried out using the KinaseMax™ Kit from Ambion and complete RNA labeling was confirmed by running reactions on a 12% urea PAGE gel. Increasing concentrations of SRSF1 were incubated with 6 pmol of Ron ESE labeled with 32P-ATP at 23 °C for 30 minutes in a final volume of 20 μL in buffer containing 100 mM Mops (pH 7.4), 10 mM free Mg2+, 5 mg/mL BSA, 150 mM NaCl, 10% glycerol, and 5 units of RNAse inhibitor. Samples were then spotted on a Hybond ECL nitrocellulose blotting membrane (0.45 μm, Amersham) using a Bio-Dot microfiltration apparatus (Bio-Rad). A vacuum is applied to capture the SRSF1-RNA complex and each sample washed 4 times with 400 μL wash buffer containing 20 mM Tris-HCL (pH 7.5) and 100 mM NaCl. Membranes were dried and dots corresponding to SRSF1:RNA complex were cut and quantitated on the 32P channel in liquid scintillant. In all cases, the total amounts of 32P-labeled RNA added in the reaction and retained on the filter at high SRSF1 concentrations were identical.
Data Analysis
The time-dependent production of phosphoproduct was fit to a single exponential function. The initial velocity data were fit to the Michaelis–Menten equation. Vmax values were converted to kcat using the total enzyme from a Bradford assay. The relative initial velocities for the competition data were fit to equation (1)
| (1) |
where vi/vo is the relative initial velocity (ratio in the presence and absence of inhibitor), Eo is the total enzyme concentration, Io is the total substrate inhibitor concentration and appKI is the apparent dissociation constant for the substrate inhibitor. The KI is calculated from appKI using equation (2)
| (2) |
where KI is the true dissociation constant for the substrate inhibitor, Km is the Michaelis constant for the fixed substrate and [S] is the fixed substrate concentration.
Results
CLK1 N-Terminus Regulates SRSF1 Phosphorylation Levels
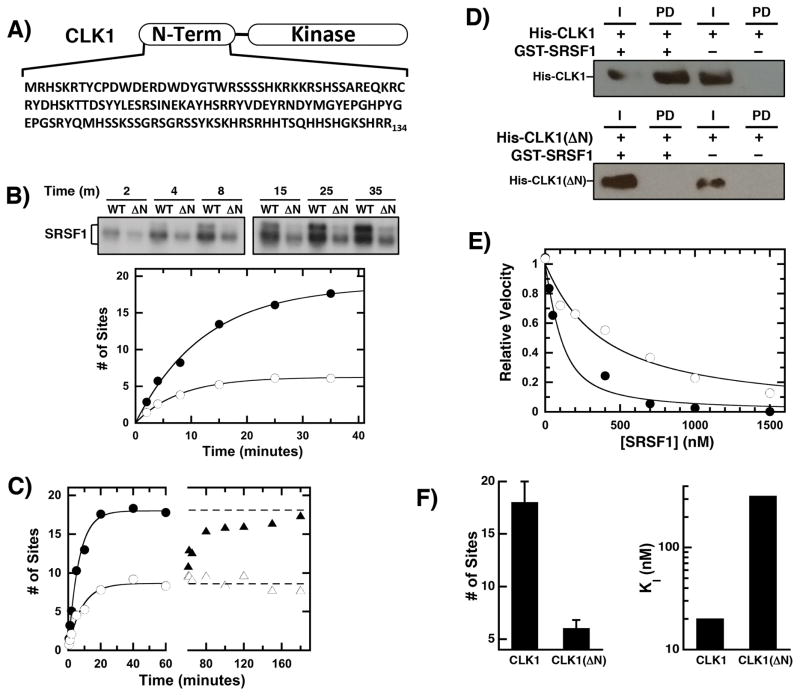
We showed that CLK1 incorporates approximately 18 phosphates into the RS domain of the SR protein SRSF1 [15]. To establish whether the CLK1 N-terminus affects this reaction, we expressed a form lacking N-terminal sequences (Fig. 1A). Unlike the kinase domain, the N-terminus of CLK1 is predicted to be largely disordered and lacking in secondary structure based on computational analyses (Suppl Fig. 1). CLK1(ΔN) incorporated less 32P into a fixed amount of SRSF1 compared to the wild-type enzyme (Fig. 1B). Although CLK1 phosphorylation induces a gel shift resulting from Ser-Pro phosphorylation [17], CLK1(ΔN) did not induce this shift consistent with lower phosphorylation levels in SRSF1 (Fig. 1B). CLK1 incorporated a total of 18 phosphates, whereas CLK1(ΔN) added about 6 phosphates (Fig. 1B). The addition of more CLK1(ΔN) does not improve the net phosphorylation levels indicating that SRSF1 underphosphorylation is not due to an unstable kinase (Fig. 1C). Furthermore, CLK1 doping of the CLK1(ΔN)-phosphorylated SRSF1 enhances total phosphorylation to the same extent as the wild-type control, indicating that lower phosphorylation levels are not the result of poor substrate viability (Fig. 1C). Taken together, these findings indicate that the CLK1 N-terminus regulates high-level SRSF1 phosphorylation.
Figure 1. CLK1 N-terminal sequences regulate RS domain binding & phosphorylation.
A) CLK1 N-terminus. B) Phosphorylation of SRSF1 (0.15 μM) using 32P-ATP (50 μM) and 50 nM CLK1 or CLK1(ΔN) is monitored by SDS-PAGE autoradiography. Time-dependent phosphorylation is fit with an amplitude and rate constant of 18 sites and 0.08 min−1 for CLK1 (●) and 6 sites and 0.13 min−1 for CLK1(ΔN) (○), respectively. C) Enzyme Doping. SRSF1 (60 nM) is phosphorylated using 30 nM CLK1 (●) and CLK1(ΔN) (○) and after 60 minutes, additional CLK1 (▲) or CLK1(ΔN) (△) are added to the CLK1(ΔN) reaction. D) The interaction of His-tagged CLK1 and CLK1(ΔN) with GST-SRSF1 is monitored on g-agarose beads using an anti-His antibody in pull-down assays. I = Input, PD = pull down. E) Competition Experiments. The phosphorylation of 50 nM SR(ΔRRM1) is monitored with varying amounts SRSF1 using CLK1 (●) and CLK1(ΔN) (○). The data are fit to equation (1) to obtain appKI of 50 and 320 nM for CLK1 and CLK1(ΔN). F) Bar Graphs. Total number of phosphorylation sites and true binding affinities of SRSF1 are plotted for CLK1 and CLK1(ΔN). The error bars for the total number of sites were obtained from triplicate measurements.
N-Terminus Controls High-Affinity Binding
To assess the contribution of the N-terminus for substrate binding, we initially performed pull-down assays. While GST-SRSF1 readily pulled-down CLK1, no CLK1(ΔN) was detected under identical conditions (Fig. 1D). This is consistent with an observed 5-fold increase in SRSF1 Km (420 nM) using CLK1(ΔN) compared to CLK1 (90 nM) [17]. Because Km values oftentimes do not reflect true dissociation constants, we used a competition experiment to assess affinities. The phosphorylation rate of an alternate substrate lacking the first RRM1 [SR(ΔRRM1)] was monitored as a function of varying SRSF1 for both CLK1 and CLK1(ΔN) [21] and fit to eqn (1) to obtain an apparent KI (appKI). While SRSF1 readily inhibited CLK1-dependent phosphorylation of SR(ΔRRM1) with an appKI of 50 nM, higher concentrations of SRSF1 were required to inhibit the CLK1(ΔN)-dependent reaction with an appKI of 320 nM (Fig. 1E). Using the Km’s for SR(ΔRRM1) to both kinases [33 nM for CLK1 [17] and 700 nM for CLK1(ΔN)], true KI values of 20 and 300 nM can be estimated for CLK1 and CLK1(ΔN) using eqn (2). Overall, the data indicate that the net effect of the N-terminus of CLK1 is to increase the total phosphoryl content of SRSF1 from 6 to 18 sites and increase the binding affinity of the SR protein by about 15-fold (Fig. 1F).
CLK1 Kinase Domain Broadly Phosphorylates the RS Domain
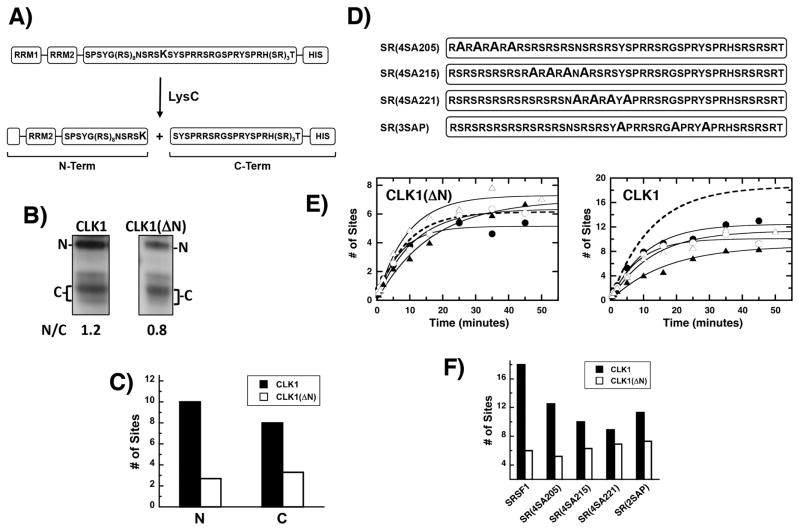
Previous studies showed that unlike SRPK1 that has a preferred consensus sequence, CLK1 phosphorylates most serines in the SRSF1 RS domain with equal efficiency [22]. To investigate whether this phenomenon is induced by the N-terminus, we performed a LysC footprinting experiment [11]. A unique lysine is placed in the center of the RS domain (R224K) so that upon phosphorylation and LysC cleavage, the relative phosphoryl contents of the N- and C-terminal halves of the RS domain can be measured by SDS-PAGE (Fig. 2A). Consistent with previous findings [22], CLK1 phosphorylates the N- and C-terminal portions of the RS domain at levels consistent with the natural dispersion of serines and shows no regiospecificity (Fig. 2B). The ratio of N- and C-terminal fragments (N/C) upon CLK1(ΔN) phosphorylation is similar to the wild-type control (1.2 vs 0.8). Using total phosphoryl contents and N/C ratios, we can calculate the number of phosphates incorporated into the N- and C-termini of the RS domain after CLK1 and CLK1(ΔN) treatment (Fig. 2C). While CLK1(ΔN) showed a slight preference for C- rather than N-terminal serines, the N-terminus of CLK1 did not appear to broadly induce a phosphorylation bias. This supports the idea that the fundamental role of the N-terminus is to increase net phosphate incorporation in the RS domain.
Figure 2. Phosphorylation regiospecificity of the SRSF1 RS domain.
A) Footprinting Strategy. LysC cleavage of SR(R224K) generates two major fragments corresponding to the N- and C-terminal halves of the RS domain. B) LysC cleavage. After complete phosphorylation with CLK1 and CLK1(ΔN) and 32P-ATP, the N- and C-terminal halves of SR(R214K), generated by LysC treatment, are resolved by SDS-PAGE. The relative phosphoryl contents of the N- and C-terminal fragments [N/C] are calculated by a ratio of CPMs in the two bands. C) Phosphoryl contents of N & C are calculated using the total phosphoryl contents of uncleaved SR(R224K) and the N/C ratios. D) Ser-to-Ala Mutants in the SRSF1 RS Domain. E) Phosphorylation kinetics. The time-dependent data for CLK1 are fit to a single exponential function to obtain a rate constant and amplitude of 0.09 min−1 and 12 sites for SR(4SA205) (●), 0.12 min−1 and 10 sites for SR(4SA215) (○), 0.06 min−1 and 9 sites for SR(4SA221) (▲), 0.08 min−1 and 9 sites for SR(4SA221) (△), respectively. For CLK(ΔN), a rate constant and amplitude of 0.14 min−1 and 5.1 sites for SR(4SA205), 0.09 ± 0.01 min−1 and 6.3 sites for SR(4SA215), 0.06 min−1 and 7 sites for SR(4SA221), 0.11 min−1 and 7.2 sites for SR(4SA221), respectively, were obtained. The dashed lines in both plots were taken from Fig. 1B and represent SRSF1 phosphorylation. F) Bar graph showing total number of phosphates added in the presence of CLK1 and CLK(ΔN).
Stabilizing Phosphate Addition Through the N-Terminus
We made block mutations of 3-4 residues in the SRSF1 RS domain (Fig. 2D) to determine whether the N-terminus induces site-specific phosphorylation. We found that serine-to-alanine mutations did not affect the ability of CLK1(ΔN) to phosphorylate the RS domain (Fig. 2E). These findings suggest that the kinase domain phosphorylates a defined number of serines without site specificity (Fig. 2E). Thus, the kinase domain has high flexibility in the groups of serines modified but is limited in the total number of phosphate additions allowed. Surprisingly, the block mutations had large, negative effects on phosphoryl content that exceeded the total number of sites mutated in all cases for CLK1 (Fig 2E, F). These findings suggest that the N-terminus not only enhances total phosphorylation but also helps stabilize phosphate additions in the RS domain during multisite catalysis.
N-Terminus Makes Direct Contacts With SRSF1
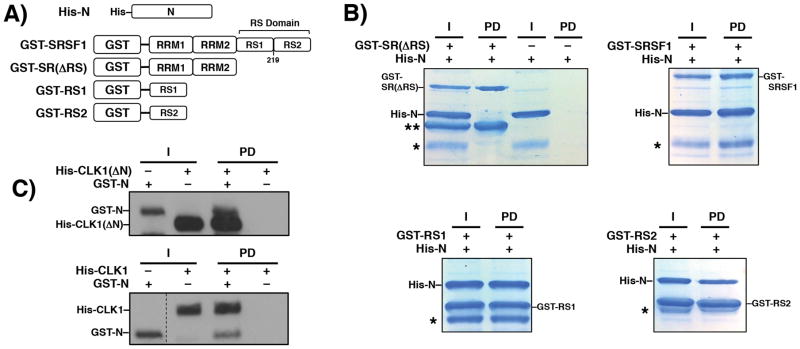
Although we showed that the N-terminus of CLK1 significantly enhances SRSF1 binding and phosphorylation (Fig. 1), the mechanism underlying this phenomenon warranted investigation. To address whether the N-terminus could make direct contacts with the substrate, we expressed and purified a His-tagged form of the N-terminus (His-N) and determined whether it could interact with several GST-tagged forms of SRSF1 in pull-down assays (Fig. 3A). We found that GST-SRSF1 efficiently pulled down His-N whereas SRSF1 lacking the RS domain [GST-SR(ΔRS)] did not pull down His-N (Fig. 3B). Both GST-RS1 and GST-RS2 pulled down His-N (Fig. 3B), indicating that the CLK1 N-terminus can interact with the N- and C-terminal halves of the RS domain. A smaller impurity in the His-N preparation that is likely a proteolytic fragment of the N-terminus was also pulled down with substrate forms containing an RS domain. To determine whether the N-terminus can also interact with the kinase domain, we expressed a GST-tagged form (GST-N) and found that it pulled down CLK1(ΔN) (Fig. 3C). GST-N contains a His tag to confirm its presence on the g-agarose beads using the anti-His antibody. GST-N also interacts with CLK1, suggesting that the N-terminus contacts the RS and kinase domains. In keeping with a prior report [19] we found that His-N cannot be phosphorylated by the kinase suggesting that the N-terminus is not likely to be autophosphorylated in the recombinant CLK1 (data not shown). Taken together, these findings indicate that the CLK1 N-terminus can directly interact with SRSF1 exclusively through the RS domain.
Figure 3. CLK1 N-Terminus interacts with the SRSF1 RS domain.
A) GST-tagged Constructs of SRSF1. B) Pull-down assays using His-N and GST-tagged constructs on g-agarose resin. An asterisk denotes an impurity in the His-N preparation and a double asterisk denotes an impurity in the GST-SR(ΔRS) preparation. C) Pull-down assays using GST-N and His-tagged kinases and g-agarose resin. Kinases are probed using an anti-His antibody. I = Input, PD = pull down. Vertical dashed line indicates where lane from the same gel is spliced.
CLK1 Interacts With Phosphorylated SRSF1
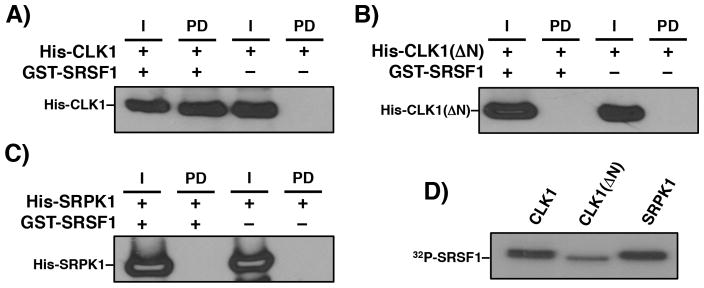
In previous studies we and others showed that SRSF1 interacts with SRPK1 in the absence but not in the presence of ATP suggesting that RS domain phosphorylation disrupts the kinase-substrate complex [23, 24]. We performed a pull-down experiment in the presence of ATP to determine whether CLK1 also discriminates between phosphorylated and unphosphorylated SRSF1 and found that phosphorylated GST-SRSF1 interacts strongly with CLK1 (Fig. 4A). We showed that CLK1 incorporated 32P into GST-SRSF1 indicating that the RS domain is, indeed, phosphorylated in these studies (Fig. 4D). As expected, phosphorylated GST-SRSF1 did not pull down CLK1(ΔN) or SRPK1, indicating that the CLK1 N-terminus is necessary for interactions with the phosphorylated RS domain (Fig. 4B, C). In both cases, we confirmed that these kinases phosphorylated GST-SRSF1 (Fig. 4D). Overall, these experiments indicate that the N-terminus of CLK1 flexibly interacts with the RS domain irrespective of its phosphorylation state.
Figure 4. Phosphorylation and SRSF1 binding to CLK1.
GST-SRSF1, bound to g-agarose resin, is used to pull down His-CLK1 (A), His-CLK1(ΔN) (B), and His-SRPK1 (C) in the presence of pre-phosphorylation with ATP. The His-tagged kinases were detected using an anti-His antibody. I = Input, PD = pull down. D) Autoradiogram showing phosphorylation of GST-SRSF1 by the kinases.
CLK1 N-Terminus Induces Cooperative Binding of SRSF1 to the Ron ESE
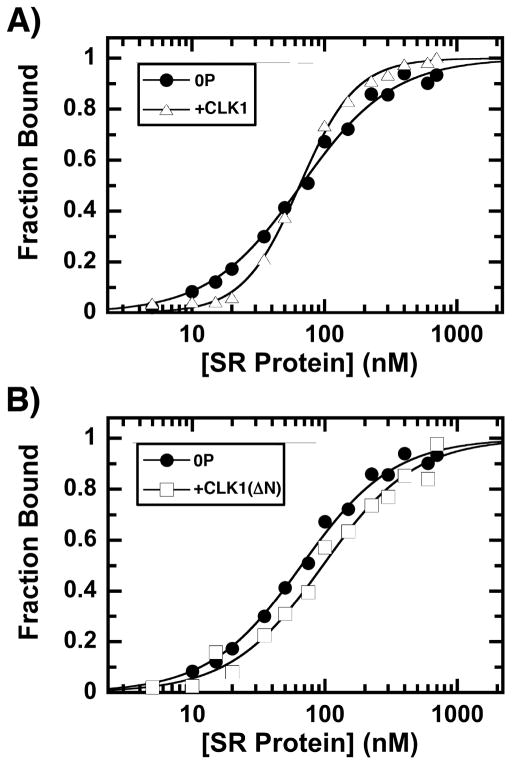
Since the N-terminus generates a hyper-phosphorylated form of SRSF1, we wished to determine whether this form displays any unique RNA binding properties. For these studies we used a filter-binding assay [16] to measure the association of SRSF1 to the Ron ESE, an RNA oligomer based on the SRSF1 exonic enhancer sequence in the Ron proto-oncogene pre-mRNA, a receptor tyrosine kinase. Using a fixed amount of 32P-labeled Ron ESE, increasing SRSF1 was added and the fraction of RNA bound on the filter was assessed as a function of total SRSF1 (Fig. 5A). The data were fitted to a Hill coefficient (N) of 1.2 and a half maximal saturation (K0.5) of 66 nM. Although pre-phosphorylation of SRSF1 with low, catalytic amounts of CLK1 had no effect on overall affinity (K0.5=65 nM), the cooperativity of the binding increased significantly (N=2: Fig. 2A). These findings suggest that CLK1 phosphorylation of the RS domain impacts the mechanism of SRSF1 binding to RNA. Interestingly, although pre-phosphorylation of SRSF1 caused a small reduction in RNA binding affinity (K0.5 = 96M), more importantly we observed no phosphorylation-dependent increase in cooperativity upon CLK1(ΔN) pre-treatment (N= 1.2; Fig. 5B). These finding indicate that the N-terminus of CLK1 plays an important role in regulating the sensitivity of SRSF1 to RNA binding through hyper-phosphorylation.
Figure 5. Phosphorylation-dependent binding of SRSF1 to the Ron ESE.
A) CLK1 phosphorylation induces cooperative binding of SRSF1 to the Ron ESE. Fraction bound is plotted against the total concentration of unphosphorylated (●) and CLK1-phosphorylated SRSF1 (△). The values of N and K0.5 are 1.2 ± 0.1 and 66 ± 2 nM for unphosphorylated SRSF1 and 2.0 ± 0.10 and 65 ± 2 nM for CLK1-phosphorylated SRSF1. B) CLK1(ΔN) phosphorylation does not induce cooperative binding of SRSF1 to the Ron ESE. Fraction bound is plotted against the total concentration of unphosphorylated (from panel A) and CLK1(ΔN)-phosphorylated SRSF1 (□). The values of N and K0.5 are 1.2 ± 0.1 and 96 ± 5 nM for CLK1(ΔN)-phosphorylated SRSF1.
CLK1 N-Terminus Is A General Activator of RS Domain Phosphorylation
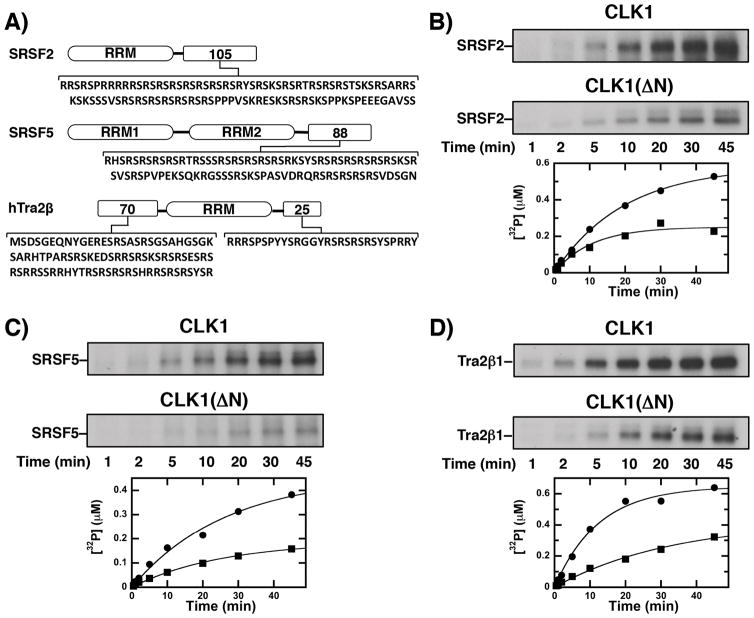
Because it induces high-level phosphorylation of SRSF1 (Fig 1), we assessed whether the CLK1 N-terminus also serves a similar role for other SR proteins. We expressed additional CLK1 substrates including the SR proteins SRSF2 (aka SC35) and SRSF5 (aka SRp40) along with the SR-like protein Tra2β1 (Fig. 6A). Each protein contains a distinctive C-terminal RS domain that differs in length and Arg-Ser content. Interestingly, Tra2β1 contains an additional N-terminal RS domain. We added CLK1 or CLK1(ΔN) to a fixed amount of each substrate and measured 32P incorporation. Overall, we found that CLK1(ΔN) under-phosphorylated all SR and SR-like proteins compared to the wild-type kinase (Fig. 6B–D). Thus, CLK1 phosphorylated approximately twice as many sites in these substrates as did CLK1(ΔN). Taken together, these findings suggest that the CLK1 N-terminus serves a general function in regulating high-level RS domain phosphorylation in SR proteins.
Figure 6. Effects of CLK1 N-Terminus on the phosphorylation of several SR proteins.
A) RS domain sequences. B–D) CLK1 and CLK1(ΔN) are incubated with SRSF2 (B), SRSF5 (C), Tra2β1 (C) and 32P-ATP and the reaction is monitored by SDS-PAGE autoradiography. The amount of 32P incorporated in the presence of CLK1 (●) and CLK1(ΔN) (■) are plotted as a function of time. For SRSF2, amplitudes and rate constants of 0.58 μM and 0.050 min−1 for CLK1 and 0.25 μM and 0.095 min−1 for CLK1(ΔN). are obtained For SRSF5, amplitudes and rate constants of 0.45 μM and 0.038 min−1 for CLK1 and 0.19 μM and 0.039 min−1 for CLK1(ΔN) are obtained. For Tra2β1, amplitudes and rate constants of 0.65 μM and 0.081 min−1 for CLK1 and 0.42 μM and 0.030 min−1 for CLK1(ΔN) are obtained.
Discussion
The last several decades have revealed that protein kinases often use a two-pronged mechanism for substrate recognition involving the active-site pocket and a distal docking groove. Both prongs are set in folded domains that provide a classical mechanism for targeted protein substrate binding. Many examples of this mechanism have been discerned within the protein kinase family. For instance, the MAP kinases possess a “D motif” docking groove near the β7-β8 reverse turn in the kinase domain that recognizes 13-16 residue spans in the target substrate [2]. The phosphoinositide-dependent kinase PDK1 possesses a hydrophobic pocket (PIF pocket) in the N-terminal lobe of the kinase domain that recognizes a hydrophobic motif in its AGC kinase substrates [25]. In the cyclin-dependent kinases (CDKs), a tightly bound regulatory subunit (cyclin) interacts with a short peptide from the protein target, thereby, tethering the substrate to the kinase domain [26, 27]. In many cases, docking grooves enormously increase phosphorylation efficiency. For example, MAPK phosphorylates the transcription factor ATF2 1300-fold more efficiently than a 14-residue peptide derived from the substrate that only can interact with the active site [28]. Overall, while the traditional view provides a workable model for understanding many kinase-substrate pairs, our data suggest that a docking element need not possess intrinsic structure but could use inherent flexibility to induce high-affinity interactions and robust phosphorylation of its target substrate.
The recent discovery that CLKs lack a conserved docking groove similar to SRPKs [18] raises the important question of how they bind RS domains with such high affinity and attain hyper-phosphorylation of SR proteins. In the present study we showed that these special attributes of one member of the CLK family (CLK1) are not the result of a classic docking groove in a well-folded domain but rather are the result of a flexible N-terminus that curiously bears similarity to its substrate targets. For SRSF1, we showed that the N-terminus increases net phosphorylation levels by an astonishing 12 phosphates and increases binding affinity by more than an order of magnitude. This is not a substrate-specific phenomenon but rather appears to reflect a general mechanism exploited with other known physiological substrates including SRSF2, SRSF5 and Tra2β1. Interestingly, although the related SR-directed kinase SRPK1 also contains an N-terminal extension, this segment is not an RS domain, does not affect the phosphorylation level or specificity in SRSF1 and has very little effect on binding affinity [29]. Thus, CLKs appear to possess unique structural features compared to other SR-specific kinases that are vital for attracting and hyper-phosphorylating SR proteins.
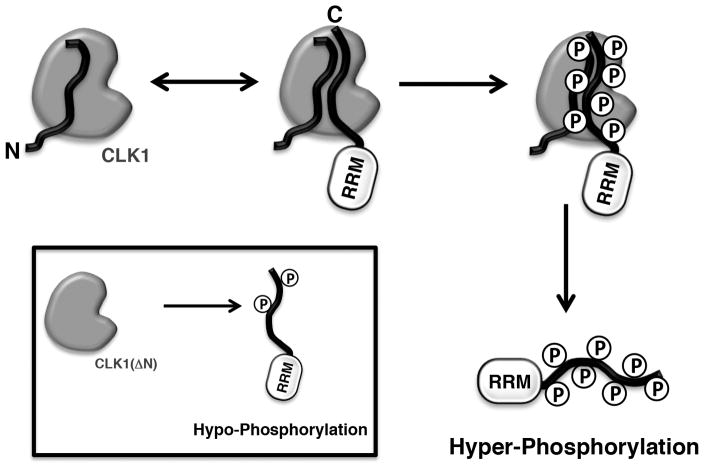
The data presented herein can now be assembled into a working model that describes SR protein hyper-phosphorylation by CLK1. With its ability to interact with both SRSF1 and CLK1, the N-terminus may act as a bridge that links the RS domain to the kinase domain (Fig. 6). The CLK1 N-terminus is classified as an RS domain [19], a structure that is intrinsically disordered [4, 5] and readily interacts with other RS domains. The nature of this association event is not well understood but it has been proposed that RS domains might adopt beta-like structures and form intermolecular “polar zippers” [30]. We also showed that the N-terminus can interact with phosphorylated RS domains suggesting that charge-charge interactions may strongly stabilize the hyper-phosphorylated state of the SR protein and work to transition intermediate forms of the phospho-RS domain in the active site. In the current model, we propose that the N-terminus is physically attached to the kinase domain acting as a bridge that connects the RS domain to the active site. It is also possible that the N-terminus may bind the RS domain independently drawing the substrate into the active site. This proposal awaits a detailed structural analysis of CLK1 with its N-terminus attached, a form that has been resistant to high-level expression and crystallization studies. Regardless, the present studies reveal a unique mechanism for attaining phosphorylated SR proteins that abandons the classical docking groove in place of a flexible N-terminus that bridges the kinase domain and the substrate RS domain. Why CLKs use a different mechanism than their splicing kinase cousins, the SRPKs, is not understood but the data clearly demonstrate that such a mechanistic variation is critical for attaining SR protein hyper-phosphorylation and influencing RNA binding interactions which may be important for spliceosome assembly and alternative gene splicing.
Supplementary Material
Figure 7. Model for induction of SR protein hyper-phosphorylated state by the CLK1 N-terminus.
N-terminus of CLK1 can interact with RS domain in both unphosphorylated and phosphorylated states, thereby inducing the hyper-phosphorylated states of SR proteins. In the absence of the N-terminus [CLK1(ΔN)], only hypo-phosphorylated state is possible.
Summary Statement.
The N-terminus of the protein kinase CLK1 induces hyper-phosphorylation of SR proteins.
Acknowledgments
We thank Drs. Patricia Jennings and Michael Jamros for careful reading and criticism of the manuscript
Funding Sources
This work was supported by an NIH grant [GM98528] to J.A.A. R.M.P. was supported by the Ruth L. Kirschstein National Research Service Award [GM090484].
Abbreviations
- CLK1
Cdc2-like kinase 1
- RRM
RNA recognition motif
- RS domain
domain rich in arginine-serine repeats
- SR protein
splicing factor containing a C-terminal RS domain
- SRPK1
SR-specific protein kinase 1
- SRSF1
SR protein splicing factor 1 (aka ASF/SF2)
Footnotes
Author Contribution
Brandon Aubol and Malik Keshwani performed all the experiments in the study. Ryan Plocinik, Maria McGlone and Jonathan Hagopian made the DNA constructs. Gourisankar Ghosh and Xiang-Dong Fu participated in discussions and critical analysis of data. Joseph Adams planned the experiments and wrote the paper.
References
- 1.Adams JA. Kinetic and Catalytic Mechanisms of Protein Kinases. Chemical Reviews. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 2.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 4.Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic acids research. 2006;34:305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang S, Gapsys V, Kim HY, Bessonov S, Hsiao HH, Mohlmann S, Klaukien V, Ficner R, Becker S, Urlaub H, Luhrmann R, de Groot B, Zweckstetter M. Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure. 2013;21:2162–2174. doi: 10.1016/j.str.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Tintaru AM, Hautbergue GM, Hounslow AM, Hung ML, Lian LY, Craven CJ, Wilson SA. Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 2007;8:756–762. doi: 10.1038/sj.embor.7401031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phelan MM, Goult BT, Clayton JC, Hautbergue GM, Wilson SA, Lian LY. The structure and selectivity of the SR protein SRSF2 RRM domain with RNA. Nucleic acids research. 2012;40:3232–3244. doi: 10.1093/nar/gkr1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clery A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nature structural & molecular biology. 2011;18:443–450. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. The FEBS journal. 2011;278:587–597. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Molecular cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu XD, Adams JA. Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. Journal of molecular biology. 2008;376:55–68. doi: 10.1016/j.jmb.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Molecular cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 13.Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, Riederer P, Moller HJ, Andreadis A, Henkel K, Stamm S. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer’s disease. Journal of neurochemistry. 2006;96:635–644. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- 14.Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD, Adams JA. Partitioning RS Domain Phosphorylation in an SR Protein through the CLK and SRPK Protein Kinases. Journal of molecular biology. 2013 doi: 10.1016/j.jmb.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velazquez-Dones A, Hagopian JC, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD, Adams JA. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. The Journal of biological chemistry. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- 16.Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD, Adams JA. Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. Journal of molecular biology. 2013;425:2894–2909. doi: 10.1016/j.jmb.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullock AN, Das S, Debreczeni JE, Rellos P, Fedorov O, Niesen FH, Guo K, Papagrigoriou E, Amos AL, Cho S, Turk BE, Ghosh G, Knapp S. Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation. Structure. 2009;17:352–362. doi: 10.1016/j.str.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. The EMBO journal. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh N, Ma CT, Giang N, Hagopian J, Ngo J, Adams J, Ghosh G. Allosteric interactions direct binding and phosphorylation of ASF/SF2 by SRPK1. Biochemistry. 2009;48:11432–11440. doi: 10.1021/bi901107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagopian JC, Ma CT, Meade BR, Albuquerque CP, Ngo JC, Ghosh G, Jennings PA, Fu XD, Adams JA. Adaptable molecular interactions guide phosphorylation of the SR protein ASF/SF2 by SRPK1. Journal of molecular biology. 2008;382:894–909. doi: 10.1016/j.jmb.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma CT, Ghosh G, Fu XD, Adams JA. Mechanism of dephosphorylation of the SR protein ASF/SF2 by protein phosphatase 1. Journal of molecular biology. 2010;403:386–404. doi: 10.1016/j.jmb.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubol BE, Adams JA. Applying the brakes to multisite SR protein phosphorylation: substrate-induced effects on the splicing kinase SRPK1. Biochemistry. 2011;50:6888–6900. doi: 10.1021/bi2007993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) The Journal of biological chemistry. 1999;274:11125–11131. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- 25.Biondi RM, Komander D, Thomas CC, Lizcano JM, Deak M, Alessi DR, van Aalten DM. High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. The EMBO journal. 2002;21:4219–4228. doi: 10.1093/emboj/cdf437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 28.Hawkins J, Zheng S, Frantz B, LoGrasso P. p38 map kinase substrate specificity differs greatly for protein and peptide substrates. Arch Biochem Biophys. 2000;382:310–313. doi: 10.1006/abbi.2000.2005. [DOI] [PubMed] [Google Scholar]
- 29.Plocinik RM, Li S, Liu T, Hailey KL, Whitesides J, Ma CT, Fu XD, Gosh G, Woods VL, Jr, Jennings PA, Adams JA. Regulating SR protein phosphorylation through regions outside the kinase domain of SRPK1. Journal of molecular biology. 2011;410:131–145. doi: 10.1016/j.jmb.2011.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perutz MF, Staden R, Moens L, De Baere I. Polar zippers. Curr Biol. 1993;3:249–253. doi: 10.1016/0960-9822(93)90174-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.