Abstract
Mass spectrometric identification of all types of molecules relies on the observation and interpretation of ion fragmentation patterns. Peptides, proteins, carbohydrates and nucleic acids that are often found as components of complex biological samples represent particularly important challenges. The most common strategies for fragmenting biomolecular ions include low- and high-energy collisional activation, post-source decay, and electron capture or transfer dissociation. Each of these methods has its own idiosyncrasies and advantages but encounters problems with some types of samples. Novel fragmentation methods that can offer improvements are always desirable. One approach that has been under study for years but is not yet incorporated into a commercial instrument is ultraviolet photofragmentation. This review discusses experimental results on various biological molecules that have been generated by several research groups using different light wavelengths and mass analyzers. Work involving short-wavelength vacuum ultraviolet light is particularly emphasized. The characteristics of photofragmentation are examined and its advantages summarized.
Keywords: photodissociation, peptide fragmentation, ultraviolet lasers, MS-MS, tandem MS
I. INTRODUCTION
The fragmentation of biomolecular ions, whether induced by collisions, light, or some kind of charge exchange process, is a fundamentally and intrinsically interesting phenomenon that has been elevated to a practically significant enterprise by the emergence of the field of proteomics. Due to energetic limitations, an individual dissociative event typically involves the cleavage of just one or two bonds. However, it is noteworthy and remarkable that even when thousands of such events are observed only a minority of the numerous chemical bonds that hold a biomolecule together are found to be cleaved. Understanding this selectivity is a familiar and popular goal. Controlling it represents a major challenge. For decades, Reference Mass Spectra have been exploited for the identification of small organic molecules. In these applications it is usually the fragmentation patterns rather than the nominal masses that provide definitive fingerprints and lead to compound identifications. In the last 15 years, the fragmentation of peptide ions to identify the proteins from which they derive has likewise become a routine but indispensable constituent of proteomic technology. A plethora of work on peptides has been reported and most of this article will be devoted to these ions. However, before focusing on their ultraviolet and vacuum ultraviolet photofragmentation, it is instructive to briefly summarize the kinds of results that more conventional methods of ion fragmentation yield. This will provide some perspective and facilitate comparison of the short-comings and idiosyncrasies of various approaches.
II. COLLISIONAL METHODS
Collision-induced fragmentation of peptide ions is by far the most popular approach used in protein identification and it is commonly investigated using ion trap, quadrupole-TOF and tandem-TOF mass analyzers.
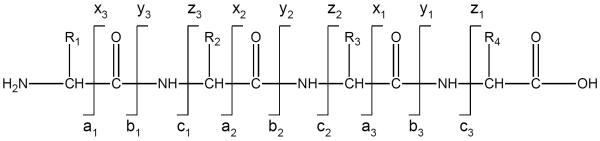
In ion trap experiments activation occurs over a millisecond timescale as a result of a large number of low-energy collisions. A consequence of the introduction of energy through many small steps is that ions typically fragment with internal energies that are only slightly above some threshold value. The latter is related to the energies of the weakest bonds in the molecule. When peptide ions are activated by a series of low-energy collisions, the most commonly observed fragments are produced by cleavage of the peptide bonds that connect one amino acid to the next. As shown in Scheme 1, these are called b- or y-ions depending whether the charge is located on the N-terminal or C-terminal fragment (Biemann, 1990;Roepstorff & Fohlman, 1984). With the exception of proline, the backbone structure of peptides is independent of sequence. Simplemindedly, one might therefore expect to observe similar fragmentation propensities at each peptide bond along the backbone. However, numerous studies have shown that both amino acid side chains and the secondary structure of peptides can significantly affect the propensity for cleaving each bond (Paizs & Surhai, 2005;Tsaprailis et al., 1999). Consequently, fragmentation patterns can be incomplete and difficult to predict.
Scheme 1.
Standard nomenclature for peptide fragmentation
Many of the sequence-specific fragmentation tendencies observed with low-energy CID of peptide ions have been rationalized based on either “mobile” proton processes or charge-remote processes associated with particular amino acid side chains (Wysocki et al., 2000) . A comprehensive review of the wide range of mechanisms proposed for both types of peptide fragmentation was recently published (Paizs & Surhai, 2005). However, since these effects impact some of the photofragmentation studies discussed below, a brief synopsis of the topic is worthwhile. Amino acids with high gas-phase basicity such as arginine (Harrison, 1997) tend to efficiently bind the charge proton. When the number of charges is less than or equal to the number of arginines, protons are effectively sequestered on these basic residues. As a result, fragmentation through charge-remote processes is favored. The most important charge-remote pathways involve the side chains of aspartic and glutamic acid. These chains can fold over so that the acidic proton interacts with the backbone amide on its C-terminal side thereby weakening the peptide bond and promoting its cleavage. When the number of charge protons is greater than the number of basic amino acids, one or more protons can move around the excited ion, inducing “charge-directed” fragmentation processes. The transfer of the charge proton to either the backbone amide or carbonyl groups weakens the adjacent peptide bond. Cleavage N-terminal to proline residues is particularly favored under these conditions, apparently due to the favorability of protonation at proline (Loo et al., 1993). However, when large libraries of peptide fragmentation spectra are examined, the ability of the preferential cleavage rules to predict fragmentation patterns is limited, as the expected fragments are not always observed (Breci et al., 2003;Huang et al., 2005). This may be due to competition between preferential fragmentation sites or the gas-phase structure of the peptide, both of which are extremely difficult to predict. The existence of preferential pathways often results in fragmentation spectra that are dominated by just a few ions and therefore contain limited overall sequence information. Thus, this fundamentally interesting selectivity can be troublesome in practical applications.
Peptide ions in tandem-TOF instruments are commonly activated in two ways: energy transfer during the MALDI process and high-energy collisions with an added gas. MALDI is generally considered to be a “cool” ionization method. Nevertheless, collisions that take place during desorption and ionization provide the ions with significant internal energy. This is particularly evident when the incident MALDI laser fluence is high (Brown & Lennon, 1995;Talbo & Roepstorff, 1993). While most ions are accelerated out of the source intact, a portion will undergo unimolecular decomposition during the 10-50 μs they spend in the field-free region of the mass spectrometer. This process is known as post-source decay (PSD) (Kaufmann et al., 1994;Spengler, 1997). The energy imparted to the ions depends on the MALDI matrix and the laser intensity, but it is not easily controlled. The low fragmentation yield and relatively long timescale of PSD suggest that most of these ions are fragmenting at energies only slightly above threshold. This low-energy and the presence of only a single charge proton in MALDI-generated peptide ions lead to fragmentation being dominated by charge-remote processes. To induce high-energy collisional activation in a MALDI-TOF instrument, a cell containing a collision gas is inserted into the field-free drift region. A collision between a 1000 m/z peptide ion and an air molecule (assumed to be N2) with 2 keV laboratory frame kinetic energy has a center-of-mass collision energy of 54 eV (Sleno & Volmer, 2004). It has been estimated that in this energy range, only about 15%, or 8.1 eV, of the collision energy is converted to internal excitation of the ion (Meroueh & Hase, 1999). This energy is still significantly larger than typical peptide backbone bond strengths of 3-4 eV. Considering that internal energy is also imparted by the MALDI process, it is clear that a single high-energy collision can impart sufficient energy to induce fragmentation. Although the gas pressure in the collision cell is much lower than that used for low-energy CID, it is still detrimental to the performance of the TOF analyzer. Ions can be lost, their velocities modified and their trajectories shifted by collisions with gas molecules anywhere in the mass spectrometer.
High-energy CID yields somewhat different fragmentation patterns than either low-energy CID or PSD. Since the energy is imparted in a single event, excitation is initially localized in one part of the molecule. A wider range of fragment ion types is observed, including most backbone cleavage fragments as well as the d, v and w-type ions that correspond to side-chain losses. A “shattering” mechanism has been proposed to explain how fragmentation occurs promptly near the site of energy deposition (Laskin et al., 2003;Laskin & Futrell, 2003). Spectra obtained by MALDI high-energy CID contain both the typical low-energy b and y fragments and the high-energy fragments consistent with the shattering mechanism (Medzihradszky et al., 2000). In commercial MALDI-TOF-TOF instruments such as those from Applied Biosystems,(Medzihradszky et al., 2000) the recommended CID operating conditions involve low collision gas pressures and spectra are dominated by PSD products rather than those derived from high-energy collisions (Campbell, 2003). There are two major advantages to using these conditions. First, ion transmission and mass resolution are better at low collision gas pressures. Second, a wide array of fragment types including side-chain losses and internal fragments are generated by high-energy CID. To the extent that their appearance is unpredictable, these additional ion types can make fragmentation patterns difficult to interpret compared with PSD spectra that are dominated by b- and y-type sequence fragments.
III. TOP-DOWN PROTEOMICS: ECD AND ETD
The most promising techniques for tandem mass spectrometry of proteins combine novel activation techniques with high-resolution mass spectrometers. Both electron capture (Zubarev et al., 1998) and electron transfer dissociation (Syka et al., 2004) (ECD and ETD), can effectively generate extensive collections of fragment ions from multiply-charged protein ions. With these techniques, fragmentation is induced when low-energy electrons neutralize positive charges on analyte ions. An extensive series of c- and z-type fragments is observed with little sequence specificity. Two mechanisms have been proposed for fragmentation by ECD and ETD. The first involves reactions of a free hydrogen atom generated when the electron is captured by a proton on the analyte ion, (Kruger et al., 1999) and the second involves localization of approximately 6 eV of charge neutralization energy leading to electronic state excitation (Breuker et al., 2004). Implicit in this second mechanism is the notion that excitation of dissociative electronic states can lead to unique, non-ergodic fragmentation even for molecules as large as peptides and proteins. Electronic-to-vibrational relaxation, which results in the loss of any specificity imparted by the excitation technique, is an important competing process that frequently inhibits non-ergodic processes. The large number of fragment ions generated by ECD in conjunction with the accurate masses attainable with FTMS can enable de novo sequencing of small proteins (Horn et al., 2000). Nevertheless, efficient fragmentation is still challenging, since charge repulsion limits the number of electrons that can be injected into the ICR cell. As a result, the acquisition of ECD data can be slow.
Electron transfer dissociation uses a reagent such as anthracene or fluoranthene anion to transfer a low-energy electron to the analyte ion. Since the reagent is an ion rather than a free electron, this technique is compatible with other types of mass spectrometers such as 2D or 3D ion traps. Their lower resolution and mass accuracy initially limited studies to smaller analytes such as peptides. However, the recently developed linear ion trap – orbitrap hybrid instrument (Hu et al., 2005) is extending the capabilities of ETD into “middle down” proteomics, in which proteins are digested into a small number of high-mass pieces (Garcia et al., 2007;McAlister et al., 2008;McAlister et al., 2007). While ECD and ETD probably impart comparable energy to peptide ions, the fragmentation patterns that result are significantly more informative than with high-energy CID. This may be because the energy is deposited more specifically, and that in the CID case variations in kinetic to internal energy conversion yield distributions of internal energies. In summary, the emerging techniques of ECD and ETD have yielded the intriguing conclusion that it is possible to generate much more complete fragmentation patterns from peptide and protein ions than thermal methods provide.
IV. EARLY IR AND NEAR-UV PHOTODISSOCIATION WORK
Analogous to the electron transfer methods, optical excitation may provide ions with more clearly defined energies. One might hope that by imparting sufficient energy or simply by introducing it in a different way some of the preferential cleavage processes of collisional excitation might be bypassed. Use of laser light also offers the advantage of being able to excite analyte ions without disturbing the operation of the instrument, making it compatible with every type of mass analyzer.
Several groups have previously investigated photodissociation of peptides, and various excitation wavelengths have been utilized from the infrared to the UV. When infrared light is used, the photon energy is low (~0.1 eV) and many photons are required to induce fragmentation, causing this technique to be known as infrared multiphoton dissociation (IRMPD). This series of low-energy activation events creates a slow heating process that can be compared to the many collisions involved in low-energy CID (Little et al., 1994). IRMPD is preferred over CID in certain applications, such as FTICR experiments where the presence of a collision gas would interfere with the operation of the mass spectrometer. It is also effective in the dissociation of large ions for which collisional activation efficiencies are low (Little et al., 1994). Fragmentation of peptide ions activated by blackbody radiation, (Price et al., 1996). and IR multiphoton excitation,(Zimmerman et al., 1991) appears to involve vibrational excitation of precursor ions and yields b and y type product ions.
The higher photon energy of ultraviolet light may in some cases enable single-photon fragmentation processes, similar to the single-collision activation in high-energy CID or single electron that is captured in ECD and ETD. There are two approaches for peptide photodissociation in the UV: excitation through a single localized chromophore or excitation through a general chromophore that appears throughout the molecule. Several groups have photodissociated peptides using the side chains of aromatic amino acids as specific chromophores. Gabryelski and Li, (Gabryelski & Li, 1999) and, more recently, the Dugourd and Kim groups (Joly et al., 2007;Oh et al., 2004;Tabarin et al., 2005) have utilized wavelengths around 266 nm (4.6 eV photon energy) to excite peptide ions through aromatic amino acid chromophores. The Li group photodissociated small (up to five residue) phenylalanine-containing peptides that did not have basic amino acids with 266 nm light in an ion trap-TOF instrument. They observed generally similar distributions of a, b and y fragments with both photodissociation and low-energy CID using broadband activation. A few additional fragments were observed with photodissociation, such as a strong c2 fragment from YPFP (Gabryelski & Li, 1999). Dugourd and coworkers utilized several wavelengths between 220 and 280 nm to photodissociate protonated YGGFL and sodiated Gramicidin A ions in a quadrupole ion trap. They observed a-, b-, y- type fragments, as well as the loss of aromatic side chains and hydrogen atoms to yield odd-electron products. These unusual losses were attributed to direct fragmentation at the UV chromophores (Tabarin et al., 2005). The Kim group photodissociated two tryptophan-containing synthetic peptides with arginine residues at each terminus (RGGWGGGGGR and RGGGGGWGGR) in a curved-field reflectron MALDI-TOF instrument with 266 nm light. They obtained complex fragmentation patterns that included large numbers of a-, y- and z-type fragments as well as a few and b-, c-, and x- type fragments. Significantly more product ions were observed than with post-source decay. They also noted that fragmentation is enhanced adjacent to the tryptophan chromophores, yielding strong a-, c- and y-type ions. These fragments were not favored with PSD (Oh et al., 2004).
Dugord and coworkers have irradiated multiply-charged peptide and DNA negative ions with ultraviolet laser light near 260 nm (Antoine et al., 2007;Gabelica et al., 2007a;Gabelica et al., 2007b;Gabelica et al., 2006;Joly et al., 2008). They observed both electron photodetachment and peptide side chain cleavage. The resulting anion products are sufficiently activated to yield numerous structurally informative fragments. They refer to the process as electron photodetachment dissociation (EPD). It is noteworthy that when performed inside of an ion trap, the radical anion fragments that are created can subsequently be collisionally activated to produce additional higher-order fragment ions.
UV-active tags have been employed by the Russell and Brodbelt groups as an alternative method to introduce a specific chromophore without any sequence restrictions. Ultraviolet light-absorbing dyes (e.g., dinitrophenyl, AlexaFluor 350, 7-amino-4methylcoumarin) were linked to the N-terminus of peptides so that they could be activated at 355 nm (3.5 eV per photon) (Tecklenburg et al., 1989;Wilson & Brodbelt, 2007). Tecklenburg, et al. found that visible and UV light fragmented peptide ions and the chromophore itself in different ways. The photofragmentation spectra of Wilson and Broadbelt contain many of the same b- and y-type ions observed in their CID results. Photodissociation enabled them to avoid the low mass cutoff associated with resonant activation in the trap. In addition, because the chromophore was attached at the N terminus, they could simplify their spectra by using high light fluence to re-fragment all chromophore-containing N-terminal primary fragments without affecting the C-terminal y ions that did not absorb at 355 nm. Since these techniques impart energy through a specific chromophore, extensive fragmentation of the peptide backbone occurs after the energy imparted by the light is redistributed throughout the molecule. Although most of the sequence fragments observed in these experiments result from this energy randomization process, cleavages adjacent to the chromophores in Dugourd and Kim’s experiments suggest that in some cases, fragmentation may occur before the energy is fully randomized. Similarly, the radical side-chain losses they observed may arise from photochemical processes.
Wilson and coworkers (Wilson & Brodbelt, 2007) have recently extended these chromophore-induced photodissociation experiments by synthesizing a novel crown ether molecule that absorbs 355 + 266 nm laser light and also can form non-covalent complexes with peptides (Wilson & Brodbelt, 2007). In principle, this provides a very simple and universal approach to adding a near-UV chromophore to any peptide of interest. They demonstrated that a peptide ion that by itself was not affected by the UV light, could be fragmented into a-, b- and y-type ions once the UV chromophore was incorporated.
V. PHOTODISSOCIATION WITH 193 nm VACUUM ULTRAVIOLET LIGHT
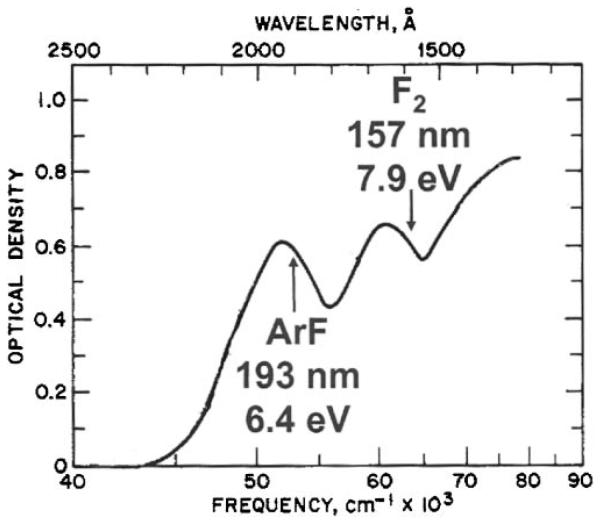
An alternative approach to peptide photodissociation is to select a universal chromophore such as one of the peptide backbone bonds. This should enable the excitation of all peptides without sequence restrictions or the need for derivatization. In addition, since the energy is imparted into the backbone, it may be possible to generate informative sequence fragments without energy redistribution. Bond-selective chemistry has been a goal of photochemists for decades, particularly since the development and proliferation of tunable laser light sources. Nevertheless, in relatively large molecules, this goal has been elusive. Rapid intramolecular vibrational relaxation appears to redistribute energy throughout large molecules on timescales faster than dissociation so that any selectivity that may be injected by an excitation process is lost (Hu et al., 2003). Figure 1 displays the accessible absorption bands of polyalanine that were originally reported by Robin (Robin, 1975). This molecule is an excellent prototype peptide because all of the amino acids have aliphatic side chains and the observed absorption bands are not sequence-dependent. Three major bands appear in Figure 1: one is centered at about 190 nm, another is near 160 nm and a third is at approximately 130 nm. These bands have been associated with the peptide backbone amides (Clark, 1995;Peterson & Simpson, 1957;Woody & Koslowski, 2002). The wavelength region below 200 nm is generally referred to as the vacuum ultraviolet. Since molecules in the air begin to absorb in this region, spectroscopy experiments at these wavelengths are performed with evacuated light paths and spectrometers. Water, in particular, is one of the absorbing species so optical spectra of peptides or proteins at these wavelengths must be recorded with samples that are dry films.
FIGURE 1.
Absorption spectrum of polyalanine in the vacuum ultraviolet (Thompson, 2007).
For photofragmentation studies, laser wavelengths are available that match two of the three absorption bands as indicated in Figure 1: the ArF excimer laser at 193 nm and the F2 laser at 157 nm. The photon energies of these wavelengths, 6.4 eV and 7.9 eV, are significantly higher than peptide bond energies of 3-4 eV. Both lasers generate short pulses of about 10 ns duration, and significant pulse energies up to kHz repetition rates. In many cases the same cavity can be utilized at both wavelengths by simply changing the gas mixture and mirror. ArF is more experimentally convenient than F2, as it lases more efficiently, leading to higher pulse energies. In addition, light at this wavelength is transmitted by fused silica optics and, with only a very small loss, through air. In contrast, lower powers are obtained with F2, and the 157 nm light from this laser requires the use of more expensive MgF2 or CaF2 optics along with an evacuated beamline. As a result of these technological constraints, photodissociation of peptides at 193 nm has been far more prevalent, with several groups employing this wavelength in conjunction with a variety of mass spectrometers (Barbacci & Russell, 1999;Bowers et al., 1984;Harris & Reilly, 1997;Harris & Reilly, 1998;Hunt et al., 1987;Martin et al., 1990;Moon et al., 2005;Thompson et al., 2004;Williams et al., 1990)
The first reported 193 nm peptide photodissociation experiments were performed in 1984 in the McIver laboratory using an FTICR mass spectrometer (Bowers et al., 1984). The MALDI and electrospray ionization techniques were not available at that time, and their chemical ionization source limited their study to the analysis of 2- and 3-residue peptides. They observed an a1 ion and the loss of CO2 from dissociation of protonated Leu-Ala, and a1 and z1 fragments from LGF. While these results are significant as the first demonstration that 193 nm light can be used to induce peptide fragmentation, the limited size of the peptides makes comparison with recent results on larger peptides difficult. The McIver group later photodissociated four- and five-residue peptides ionized in a FAB source and observed primarily b- and y-type fragments (Lebrilla et al., 1989). Hunt and coworkers also investigated 193 nm peptide photodissociation in an FTICR instrument (Hunt et al., 1987;Hunt et al., 1989). Using a liquid SIMS ionization source, they were able to protonate a large peptide and successfully performed de novo sequencing of FQETFEDVFSASPL*R based solely on their photodissociation spectra. A complete y ion series was observed, and they confirmed their interpretation by photodissociating the methyl-esterified form of this peptide. Note that in this sequence the L* could represent either leucine or isoleucine, since in their experiment they could not distinguish these isobaric amino acids (Hunt et al., 1987). They also obtained a y ion series extending from y8 to y28 along with a number of internal fragments from the 29-residue peptide DKPQPVMKLAVGGLLFPPSSLRLPLVPAL (Hunt et al., 1989) The photodissociation of several large peptide antibiotics by FTICR-MS was subsequently reported by the McLafferty group (Williams et al., 1990). Although these molecules were biological in origin, they contained amino acids outside of the standard 20 that are incorporated by ribosomal protein synthesis. They used the same 193 nm laser for both desorption/ionization and dissociation, and obtained nearly 100% destruction of the precursor ion signal. Unlike the prior studies, their ionization process yielded sodiated ions. The cyclic peptide Gramicidin S (-OVPFLOVPFL-, where O = ornithine) was investigated as well as two isomers of Alamethicin (BPBABAQBVBGLBPVBBEQF, where B = 2-aminoisobutyric acid). Because of its cyclic structure, the backbone of Gramicidin S must be cleaved twice to generate an observable fragment, making assignment of exact fragment structures difficult. Two fragment series were observed in the photodissociation spectrum from this molecule, and each contained a full sequence ladder. With CID, significantly fewer fragments were observed. 193 nm photodissociation spectra of the large Alamethicin isomers exhibited relatively poor signal to noise ratios, probably due to the ionization technique used. About a dozen a-type fragments could be identified from each isomer (Williams et al., 1990). The technique of ECD dissociation was also inadvertently discovered by the McLafferty group as a result of photoelectrons generated during a 193 nm photodissociation study (Guan et al., 1996;Zubarev et al., 1998). The Biemann group photodissociated Angiotensin III (RVYIHPF) at 193 nm in a four-sector mass spectrometer (Martin et al., 1990). The combination of the low duty cycle of the laser with a continuous-beam mass analyzer allowed only a small fraction of the precursor ions to be exposed to the laser light. As a result, extremely low photodissociation efficiencies (~0.02%) were obtained in their experiments. This peptide yielded an a-type ion series extending from a2 through a5 as well as d4 and several immonium ions. While a-type fragments are unusual in low-energy CID experiments, the authors pointed out that they can form as a secondary product via the elimination of CO from a b-type ion when higher energy activation is involved.
The experiments just described demonstrate that 193 nm photodissociation of peptide ions using an ArF laser can be studied with trapping- and beam-type instruments. However, the number of peptides investigated in these studies was far too small and their sequences too diverse for a thorough characterization of the fragmentation that ensues with this activation technique. Additional work in this area was not pursued by the groups who performed these initial studies.
The development of MALDI-TOF-TOF technology led to a resurgence of interest in the VUV photodissociation of peptides in the late 1990s. One reason for this is that the activation of MALDI-generated ions in a time-of-flight instrument is somewhat challenging. High vacuum is required to ensure that collisions with neutral gas molecules do not influence ion velocities or trajectories. This makes efficient collisional activation challenging, and the short (10s of microseconds) timescale of the mass analysis prevents the application of slower activation techniques such as IRMPD with continuous wave lasers. The capabilities and limitations of the two widely used techniques, post-source decay and high-energy collisional activation, were described above. Vacuum ultraviolet excitation is a unique alternative that offers the possibility of activating ions as they travel through a TOF mass analyzer, and light does not otherwise perturb the operation of the instrument. Although ions are not stored in a time-of-flight analyzer, as in an FTICR cell, efficient fragmentation can be achieved by careful synchronization of a single short, intense laser pulse with the ion packet as it travels through the instrument. The destruction of several peptide and small protein ions (bradykinin, mellitin, insulin and lysozyme) by 193 nm light in a MALDI-TOF mass spectrometer was first demonstrated by the Russell group in 1995, although no fragment ions were identified in this study (Gimon-Kinsel et al., 1994).
William Harris investigated peptide photodissociation in a linear MALDI-TOF instrument (Harris & Reilly, 1997;Harris & Reilly, 1998). He employed several different photodissociation wavelengths, including 266 nm (4.6 eV photon energy) 248 nm (5 eV), and 193 nm (6.4 eV). Fragmentation efficiency was found to be best at 193 nm, although it was not clear whether this was because of the higher light absorption cross section or the higher photon energy at the shorter wavelength. The instrument he used was only a single-stage mass spectrometer, and photodissociation was performed within the ion source. Since there was no precursor selection, all ions present in the source were detected. This restricted experiments to the analysis of pure peptides. In addition, the spectra were somewhat difficult to interpret due to the low mass resolution of the linear-TOF and the overlap between background ions and low-mass fragments. The former included matrix clusters and photoions produced by laser ionization of residual gases in the mass spectrometer. Nevertheless, this study demonstrated that UV photodissociation could be successfully performed on the short timescale of a TOF analysis, and certainly motivated our group’s future work.
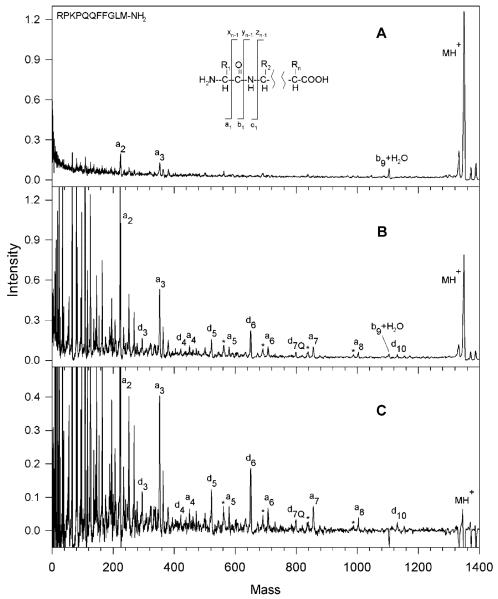
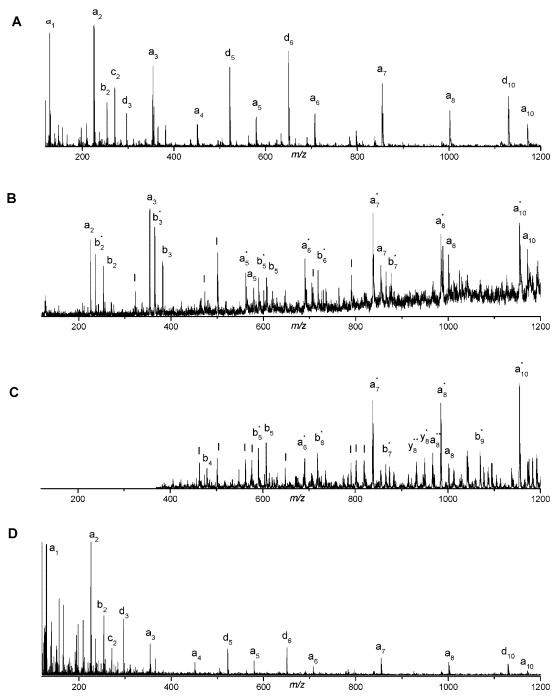
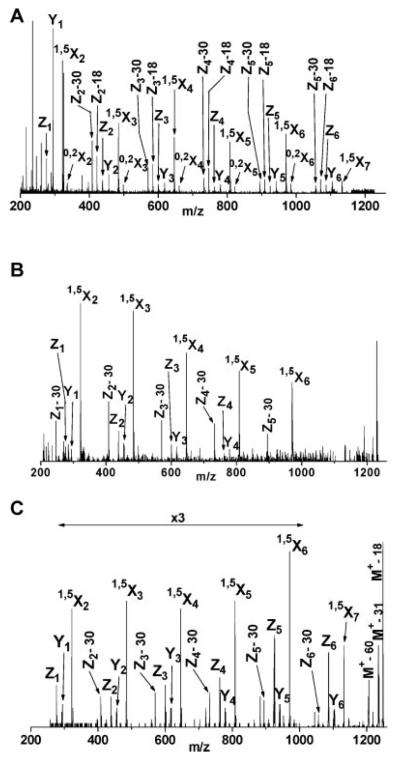
Fragmentation spectra obtained by Harris for Substance P (RPKPQQFFGLM-NH2) are shown in Figure 2. The MALDI-only spectrum (Figure 2A) contains only three weak fragments, apparently the result of in-source fragmentation. Upon adding 193 nm light the spectrum displayed in Figure 2B was recorded. Finally, subtraction of the two to reveal the contribution of the light to the fragmentation process and multiplication of the difference by a factor of two led to the spectrum in Figure 2C. It is evident that light excitation creates a series of a-type ions extending from a2 to a8 accompanied by six d-type ions. This pattern is consistent with the results obtained for a similar molecule having an N-terminal basic group (Angiotensin III) by the Biemann group using 193 nm photodissociation in a four-sector instrument (Martin et al., 1990). However, the peaks are difficult to interpret below 300 m/z due to the overlap between peptide fragments, matrix clusters and other background ions. Harris studied a number of other peptides having arginine residues at or near their N-termini including angiotensin I (DRVYIHPFHL), bombesin (Pyr-QRLGNQWAVGHLM-NH2) and ACTH fragment 18-39 (RPVKVYPNGAEDESAEAFPLEF) and observed similar fragmentation patterns dominated by a- and d-type ions. Bradykinin, which has arginine residues at both termini (RPPGFSPFR) yielded four a-type ions, but, in contrast with the other peptides, three y-type ions also. Since the instrument used in these studies could not select a particular precursor mass for fragmentation, a major consideration when choosing peptides for analysis was the availability of pure samples. Peptides having C-terminal arginines (easily generated by tryptic digestion of proteins) were not specifically targeted. Nevertheless, the observation of d-type fragments led Harris to conclude that peptide ion photodissociation with 193 nm light involves a high-energy, charge-remote process similar to that occurring in high-energy CID. In proteomics applications, the tryptic peptides that contain lysine or arginine at their C-termini are most often encountered. The desire to investigate this class of molecules, to obtain higher quality mass spectra and to analyze mixtures motivated us to develop new instrumentation for peptide photodissociation that is described below.
FIGURE 2.
MALDI mass spectra of substance P in a single-stage linear time-of-flight mass spectrometer. (A) The desorption/ionization laser intensity was set just slightly above that for maximum ionization; (B) 193nm photodissociation light was introduced; (C) the result of subtracting these spectra and amplifying by a factor of two (Harris, 2002).
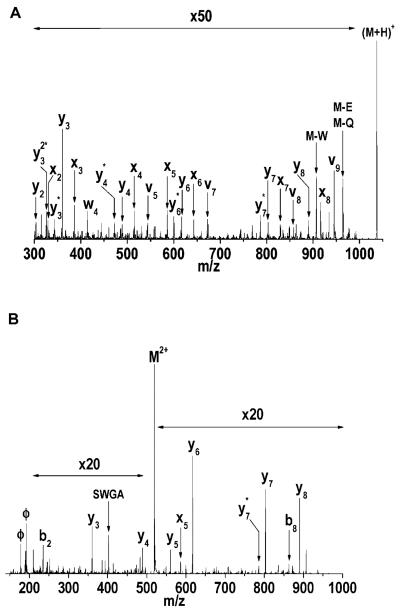
Several other TOF configurations have been utilized in 193 nm peptide photodissociation studies. These include reflectron-TOF instruments with stepped reflector voltages,(Barbacci & Russell, 1999;Hettick et al., 2001) and a curved-field reflectron-TOF(Moon et al., 2005). Results obtained with various tandem-TOF instruments in which fragment ions are reaccelerated following their production are discussed below. While the photodissociation spectra obtained in these studies exhibit some similarity with the results described above, there is also evidence that the analysis timescale of each instrument can influence the observed fragment ion distributions. In the original stepped-reflectron spectrometer employed by the Russell group, the time between photoexcitation and fragment ion separation was about 10 μs,(Barbacci & Russell, 1999) compared to 1.3 μs in Harris’s linear TOF. Their photodissociation spectrum of Substance P (RPKPQQFFGLM-NH2) obtained with the stepped-reflectron instrument contains an a-type ion fragment series extending from a1 to a8, along with a10. This is accompanied by four d ions, and is quite similar to Harris’ results shown in Figure 2. However, in a subsequent study they modified their instrument to include an ion gate, and they selected ions from a peptide mixture (Hettick et al., 2001). With this instrument, Substance P yielded only four a-type ions, while six internal and five b ions not observed in the previous study were identified. Photodissociation of two other peptides was reported in this second publication. Bradykinin (RPPGFSPFR) yielded six a- and six b-type ions, along with one d and one y ion. When the C-terminal arginine was removed (RPPGFSPF), the photodissociation spectrum contained four a, four 4 b, three y, two internal, one d and one w ion. Overall, the photodissociation spectra in this later study appear to be more complex than that presented in the earlier study of the Russell group (Barbacci & Russell, 1999).
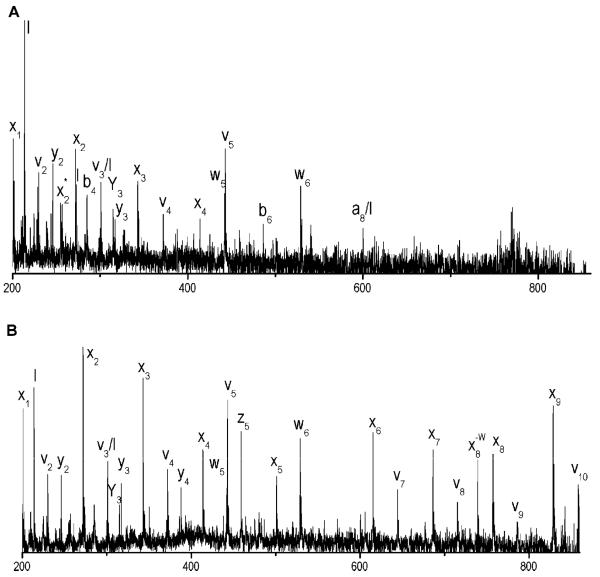
Recently, the Russell group has studied 193 nm peptide photofragmentation using a tandem-TOF instrument (Morgan & Russell, 2006). In this instrument, the precursor ions are separated in the first TOF stage then decelerated prior to entering a field-free region where they are photodissociated. The precursor and fragment ions are then reaccelerated into the second stage reflectron-TOF, where the entire spectrum is acquired. Using peptides having both N-and C-terminal arginines, they compared results obtained with this instrument and their earlier one and found that with the new apparatus they observe fewer b and y fragments. This was attributed to the shorter time available for ion fragmentation (Morgan & Russell, 2006). The critical time between photoexcitation (Thompson et al., 2007) and ion reacceleration in this instrument is about 1 μs. For example, the bradykinin fragment mentioned above (RPPGFSPF) yielded six a, two c and one d and two y ions, along with four low abundance internal fragments in the new apparatus. This was more consistent with their initial results and with those obtained by Harris. They also photodissociated a peptide with a C-terminal arginine residue, bradykinin fragment PPGFSPFR. This molecule yielded primarily C-terminal fragments, including five x, three z, three y and two w ions. The M. S. Kim group has reported 193 nm photodissociation experiments in a curved-field reflectron TOF instrument (Choi et al., 2006;Moon et al., 2005). In this design, no additional electric fields are applied between the MALDI source and the ion reflector. Photodissociation occurs in a field-free region after an ion gate, and the fragments maintain the same velocity as the precursor until they reach the reflector. The fragment ions are separated by the reflector field, and the time between the laser pulse and fragment separation in this apparatus is about 10 μs. In contrast, in our tandem TOF instrument that is discussed below this timescale is only about 1 μs. The Kim group’s 193 nm photodissociation spectra can be considered unusual when compared to previous studies. They observe a near-complete series of high-energy x ions accompanied by v and w ions from several peptides having C-terminal arginines. In one example, photodissociation of the peptide EGVNDNEEGFFSAR at 193 nm yielded an x ion series from x1 to x12 accompanied by eleven 1 v, nine w and eight y ions (Moon et al., 2005). This extensive collection of x-type fragments at 193 nm is noteworthy. Subsequently, we made a detailed comparison of the influence of the wavelength and mass spectrometer on the photodissociation of the these peptides (Thompson, 2007). Once again, mass analysis timescale, in conjunction with light wavelength, may be a critical issue in determining the characteristics of peptide ion photofragmentation spectra. The Kim group has also investigated the photodissociation of peptides having arginine residues at their N-termini or both N- and C-termini. Their results for the bradykinin fragment RPPGFSPF fall between the two reported by the Russell group, with an a ion series extending from a1 to a6 along with two b, one c one y and one internal fragment (Choi et al., 2006). Finally, they have also recently reported the 193 nm photodissociation of larger peptides using a different tandem-TOF apparatus with a shorter time between photodissociation and fragment analysis (Moon et al., 2005). Peptides in the 3 kDa range were found to fragment similar to the smaller peptides investigated in their previous study. For example, the 3188 Da β-casein fragment FPPQSVLSLSQSKVLPVPQKAVPYPQRD generated a large number of x, v, w and y ions. They had difficulty obtaining sufficient fragment ion signal from insulin (5730 m/z) and ubiquitin (8560 m/z), which they suggested was due to the large number of potential fragmentation channels available. Only twelve y and three b ions could be identified in the photodissociation spectrum of ubiquitin, and even fewer fragments were identified from insulin.
VI. PHOTODISSOCIATION WITH 157 nm VACUUM ULTRAVIOLET LIGHT
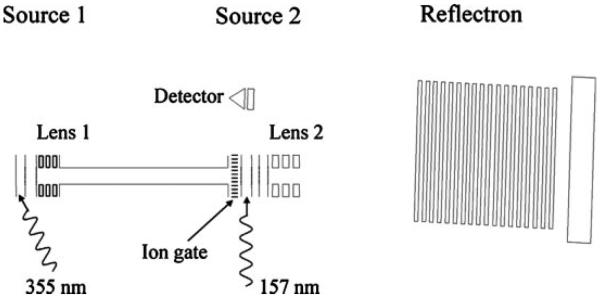
In an attempt to improve on the instrumentation developed in our laboratory by Harris, in 2001 we designed and constructed a tandem photodissociation TOF-TOF instrument that is schematically depicted in Figure 3 (Cui et al., 2005). At the same time, we switched photolysis lasers from a 193 nm ArF excimer to a 157 nm F2 laser in order to investigate whether the higher photon energy and different electronic states associated with the shorter wavelength offer any advantages over other photodissociation wavelengths or conventional activation methods. MALDI-generated peptide ions were mass-separated in a relatively short linear TOF analyzer, isolated by an ion gate, photodissociated then reaccelerated into a second-stage reflectron TOF for product ion mass analysis. An important characteristic of this kind of apparatus is that some of the precursor ions generated by MALDI undergo PSD as they travel through the first stage of the instrument. The resulting fragments continue to travel with the precursor ions until they are all reaccelerated in the second source. As a result, both PSD and photofragments appear in the resulting spectra. In order to distinguish the fragments formed by these two processes the photodissociation laser is triggered on alternate MALDI shots; spectra obtained with and without light activation are recorded in separate memory buffers. In this way, the inevitable slow variations in MALDI ion production are accurately accounted for and the two spectra reflect the PSD contribution and the combined photodissociation + PSD contribution to the tandem mass spectrum. Their subtraction yields the photodissociation spectrum. The only complication to this approach is that because these precursor and PSD product ions travel together through the first drift region, both are exposed to the activating light. It therefore seems inevitable that some of the latter would photofragment. Because of this, spectra in the second buffer reflect a somewhat convoluted combination of PSD and photodissociation effects.
FIGURE 3.
Tandem time-of-flight apparatus used for generating and photodissociating biomolecular ions (Cui et al., 2005).
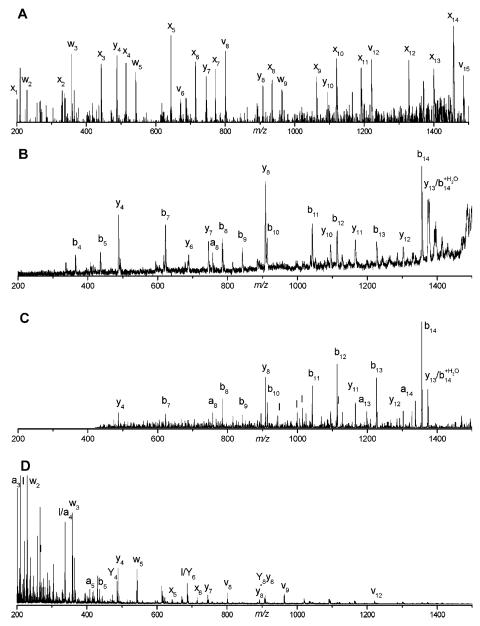
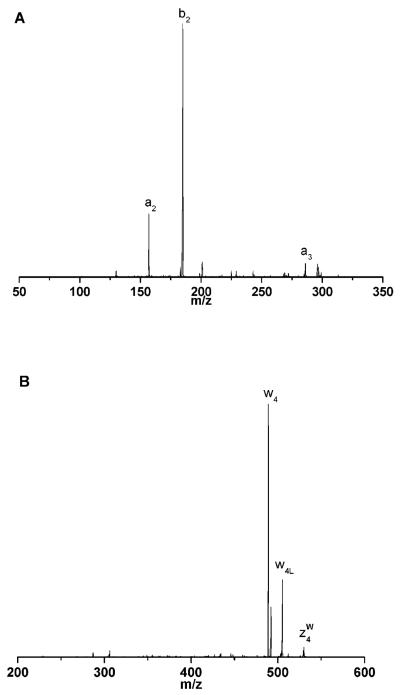
Spectra recorded with this apparatus immediately demonstrated some unique and desirable features. Tryptic peptides were of particular interest, and those containing C-terminal arginines were the focus of our initial experiments. Figure 4 displays a comparison of fragmentation data on peptide VGAHAGEYGAEALER recorded with various instruments and methods of ion activation (Cui et al., 2005). The 157 nm photodissociation spectrum (Figure 4A) is dominated by a series of x-type ions extending from x1 to x14. Also present are several v- and w-type ions. Although these three ion types are indicative of high-energy fragmentation processes, x-type ions appear to be a less dominant product in collision-induced dissociation experiments (Despeyroux et al., 1993;Johnson et al., 1988;Johnson et al., 1987;Medzihradszky et al., 1996;Medzihradszky et al., 2000;Papayannopoulos, 1995;Stimson et al., 1997). Post-source decay and low-energy ion trap CID spectra of the same singly-charged peptide ion are displayed in Figures 4B and 4C. Although these were recorded with different instruments and some mass-dependent effects were instrument-related, the most important characteristic of these spectra is that they are dominated by b- and y-type “thermal” fragment ions. This indicates that energy has sufficient time to be randomized throughout the molecule and the charge proton can be found on either end of the ion. Specific features that stand out in one or both of these spectra include y4, y8, and b14 ions, all of which are formed by cleavage adjacent to an acidic glutamic acid residue. Figure 4D displays the CID spectrum of this peptide recorded with an ABI 4700 TOF-TOF instrument. It contains contributions from both PSD and high-energy collision induced dissociation. The dominance of low-mass ions in the spectrum is probably a consequence of high impact parameter collisions that introduce excessive internal excitation causing the ion to explode into small pieces. Despite that fact that the same types of high-energy fragment ions (x-, v- and w-) appear in this spectrum as in Figure 4A, the overall distribution and the completeness of each ion series is obviously different, indicating a qualitatively different mechanism of formation. Figures 5A-5D display the same comparison of 157 nm photodissociation, post-source decay, ion trap CID and 2 keV TOF-TOF CID data for another arginine-terminated tryptic peptide, SAASLNSR. The results are in qualitative agreement with the previous example. Once again, 157 nm photofragmentation yields a contiguous series of x-type ions along with several other high-energy v- and w- type fragments. PSD and low-energy CID produce spectra that contain both N- and C-terminal fragments. More evident in this figure is the considerable congestion in these spectra that is exacerbated by the occurrence of numerous undesirable and uninformative cleavage processes such as the loss of NH3 and H2O from various fragments. These are labeled with an asterisk and −H2O, respectively. The high-energy TOF-TOF spectrum in Figure 5D again contains a sampling of the ions that appear in the photodissociation spectrum, but not a contiguous distribution. A particularly attractive feature of high-energy peptide fragmentation is that w-type ions make it possible to distinguish leucine from isoleucine (Johnson et al., 1987). w3 ions in Figure 4 and w4 ions in Figure 5 appear in both high-energy CID and photodissociation spectra. We have found that cleavage adjacent to leucine or isoleucine is highly favored and corresponding w-type ions almost always appear in photodissociation spectra of tryptic peptides containing these residues. This is a significant advantage of this high-energy fragmentation method for proteomic applications.
FIGURE 4.
Tandem mass spectra of the singly charged hemoglobin tryptic peptide VGAHAGEYGAEALER obtained by (A) 157 nm photodissociation, (B) post-source decay, (C) ion trap CID, (D) 2 keV TOF-TOF CID (Cui et al., 2005).
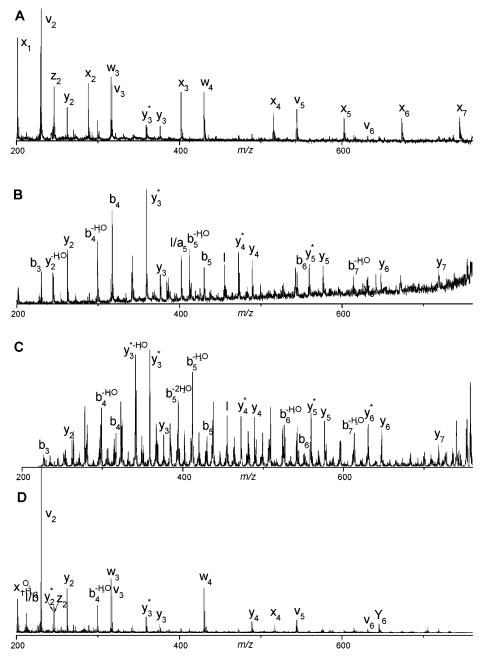
FIGURE 5.
Tandem mass spectra of the singly charged trypsinogen tryptic peptide SAASLNSR obtained by (A) 157 nm photodissociation, (B) post-source decay, (C) ion trap CID, (D) 2 keV TOF-TOF CID. Ions labeled by * have lost NH3. Peak labeled by I is an internal ion fragment (Cui et al., 2005).
Peptides containing arginine at their N-termini yield spectra that are comparably predictable. Results for Substance P (RPKPQQFFGLM-NH2) are displayed in Figure 6. As expected, photodissociation (Figure 6A) exhibits a strong propensity for forming N-terminal fragments. a-type ions result from cleavage of the same α carbon-carbonyl carbon bond that is cleaved to form x-type ions, except that the charge now remains with the N-terminal fragment. d-type ions are analogous to the w-type ions just discussed in that their observation also enables leucine and isoleucine to be distinguished. Post-source decay and low-energy collision induced dissociation (Figures 6B and 6C) again yield more congested spectra. Many fragments lose NH3 and both a- and b-type ions are readily formed. The photodissociation spectrum contains at least comparable useful information with significantly less congestion so its interpretation is more facile. The TOF-TOF CID spectrum in Figure 6D is most similar to photodissociation providing further confirmation that the latter is a high-energy process. Loss of ammonia is much more prevalent in the low-energy spectra, apparently due to the difference in fragmentation timescale.
FIGURE 6.
Tandem mass spectra of singly charged substance P (RPKPQQFFGLM-NH2) obtained by (A) 157 nm photodissociation, (B) post-source decay, (C) ion trap CID, (D) 2 keV TOF-TOF CID. (Cui et al., 2005).
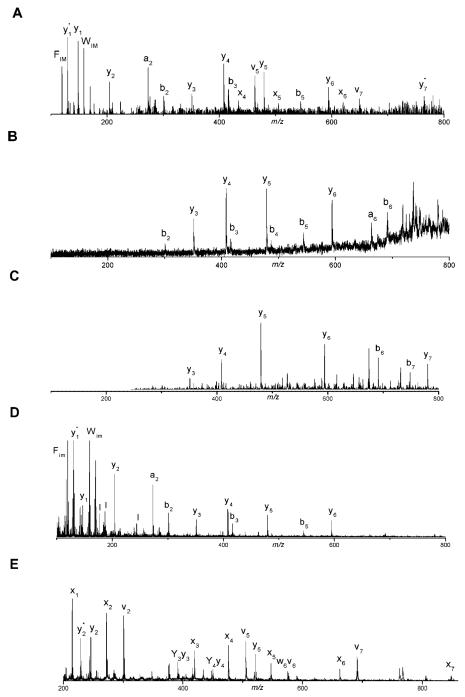
Photofragmentation of tryptic peptides that are terminated in lysine yields results that are different from those just discussed. Spectra in Figure 7 were recorded with peptide NWDAGFGK. The 157 nm photodissociation spectrum (Figure 7A) contains a discontinuous series of x-, y-, a- and b-type ions indicating that the charging proton shows little preference to remain on one end of the peptide. In fact, this spectrum looks like a composite of the three spectra below it that were recorded by post-source decay (Figure 7B), low-energy CID (Figure 7C), and high-energy TOF-TOF CID (Figure 7D). While at first glance this result might seem surprising, one must remember that in this experiment precursor ions are formed by MALDI. Through this process, they receive enough internal excitation that many ions undergo postsource decay fragmentation. One must assume that other ions are at least somewhat internally excited, even if they remain intact. This internal excitation is apparently sufficient to mobilize protons that might otherwise reside on lysines. Thus by the time a peptide ion is irradiated with VUV light in the TOF-TOF instrument, its charging proton could be anywhere. Guanidination of the lysine creates a highly basic C-terminal homoarginine group that is capable of sequestering a single charge (Beardsley et al., 2000). For such peptides, one would expect results that are comparable to what we observed with arginine-terminated peptides. Photodissociating the guanidinated peptide leads to the spectrum displayed in Figure 7E. As expected, thermal fragment ion peaks are either reduced in intensity or eliminated and they are replaced by a distribution of high-energy C-terminal fragment ions comparable to those obtained with arginine-containing peptides. Most significantly, the contiguous ion series found in these mass spectra enable the direct determination of sequence information. This capability should facilitate the identification of mutations, genome sequencing errors and post-translational modifications.
FIGURE 7.
Tandem mass spectra of the singly charged Carboxypeptidase A tryptic peptide NWDAGFGK obtained by (A) 157 nm photodissociation, (B) post-source decay, (C) ion trap CID, (D) 2 keV TOF-TOF CID, (E) 157 nm photodissociation following peptide guanidination (Cui et al., 2005).
We have performed 157 nm photofragmentation experiments on platforms other than the TOF-TOF instrument. Linear ion traps are particularly compatible with this activation approach (Kim et al., 2005) . With the ThermoElectron LTQ ion trap there is a hole in the back lens of the trap that is exploited in certain commercial configurations to deliver ions to another mass analyzer. The hole provides convenient access for laser light, facilitating excellent overlap between the trapped cloud of ions and the light beam. The LTQ detectors are mounted off to the sides of the trap so they do not block the path of the incoming axial light beam. Photodissociation experiments with the ion trap provide a number of new capabilities and advantages compared with the TOF-TOF analyzer. First, ions are generated continuously, by electrospray, in contrast with the pulsed MALDI source of the TOF-TOF. The resulting ions should have significantly less internal energy before they are excited by the light. However, the timescale for mass analysis after photoactivation is significantly longer than with the TOF-TOF instrument (tens of milliseconds in contrast with microseconds) and this can also affect the resulting spectra. Figure 8A displays the 157 nm photodissociation spectrum of peptide FSWGAEGQR recorded with the ion trap. While a substantial number of high-energy x-, v- and w-type ions again appear in the spectrum, there are more thermal y and y-NH3 ions than we typically observe with our TOF-TOF apparatus. It seems likely that these are products of slow unimolecular dissociation of initially formed high-energy photofragment ions.
FIGURE 8.
157 nm ion trap photodissociation spectra of (A) singly-charged and (B) doubly-charged peptide FSWGAEGQR (Kim et al., 2005).
The electrospray source also allowed us to investigate the effect of precursor ion charge state on the resulting fragmentation pattern. Figure 8B displays the 157 nm photodissociation spectrum of doubly-charged FSWGAEGQR peptide. In this case, the high-energy fragment ions are almost entirely lost from the spectrum: only x5 is intense enough to be selected for labeling in the figure. Although in this type of experiment it is not possible to determine whether high-energy fragment ions are formed with considerable internal excitation and undergo rapid unimolecular decomposition to thermal b- and y-type ions or whether they are simply not formed, but the former would be our hypothesis.
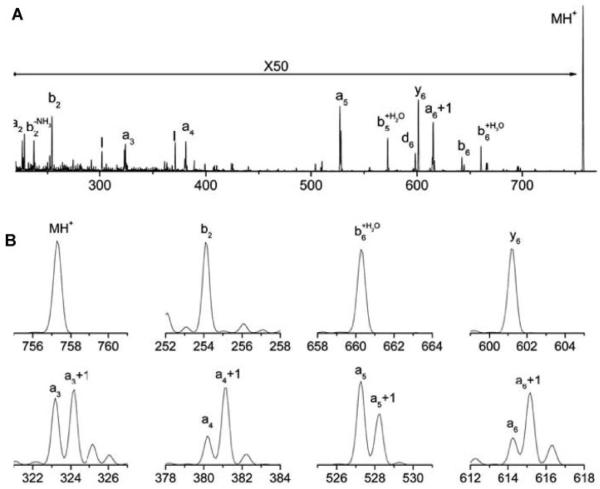
The ion trap has two other distinct characteristics that make it particularly attractive for photodissociation studies. The first is its ability to isolate precursor ions with superior resolution. While the our TOF-TOF spectrometer (like its commercial equivalent) has an ion gate that transmits a 10-15 Da wide batch of ions for subsequent dissociation, the ion trap has little difficulty selecting a single nominal mass for fragmentation. This significant advantage eliminates complications from 13C isotope effects that might otherwise confuse the interpretation of certain mechanistic experiments. For example, Figure 9 displays the 157 nm photodissociation spectrum of peptide RPPGFSP. Because the precursor ion could be isolated with unit mass resolution, fragment ion spectra contain no 13C contributions. Indeed, as evident in Figure 9B, expanded views of the precursor ion and fragment ion peaks associated with three different thermal fragments, b2, b6+H2O, and y6, display no 13C contribution adjacent to each of the fragment ion peaks. However, high-energy fragments a3, a4, a5 and a6 all have partners one Da higher in mass that are of significant intensity, indicating that an+1 radical ions are formed in the photodissociation process. Observation of these intermediate species supports a homolytic cleavage mechanism for the photofragmentation process (Zhang et al., 2006). Another example of a mechanistic study involved the use of deuterium substitution to determine whether the double bond in an a-type peptide fragment ion is created along the backbone or in the side chain of the C-terminal residue. Because hydrogen atoms bound to nitrogens are easily exchangeable but those bound to carbon atoms are not, deuterium substitution can be used to establish whether or not the putative double bond in an a-type ion connects to a backbone nitrogen atom (Zhang et al., 2006). Note that this experiment relies on the capability of observing subtle one-Da differences caused by single H-D substitutions. The conclusion of this experiment was somewhat more complicated than had been expected: For some residues, only the backbone double bond forms. For others, only the side chain double bond forms. For still others, both bonds form and the probability for producing each depends on the conjugative properties of the side chain.
FIGURE 9.
157 nm photodissociation spectrum of singly-charged RPPGFSP obtained in the linear ion trap (A) the entire spectrum, (B) expanded parts of the spectrum. In this experiment, monoisotopic precursor ions were isolated and irradiated (Zhang et al., 2006).
Perhaps the most significant advantage of an ion trap over a tandem-TOF instrument is the capability of performing MSn experiments that can substantiate the identification of ambiguously-assigned species. An excellent example of this arose in a 157 nm photofragmentation study of singly-charged ALELFR ions. Two unanticipated product ions, at m/z 305 and 548, were observed and tentatively assigned as b3 and z4. The former was unexpected because of the presence of the C-terminal arginine and the latter is one of only a handful of z-type ions that we have ever tentatively identified in our photodissociation work. However, isolation of these two photofragment ions followed by their collisional dissociation led to the MS3 spectra displayed in Figure 10. The fragment ions that were observed convincingly demonstrated that the two proposed assignments were correct.
FIGURE 10.
CID spectra of putative (A) b3 and (B) z4 ions generated by 157nm photodissociation of singly-charged ALELFR precursor ions inside of a linear ion trap (Kim et al., 2005).
In another ion trap experiment, Zubarev and coworkers (Fung et al., 2005) have demonstrated that disulfide linkages are effectively cleaved by 157 nm light excitation. They photodissociated peptide and small protein ions inside of their LTQ linear ion trap. Interestingly, they showed that collisional activation of photofragments leads to numerous smaller ion fragments that convey abundant structural and mechanistic information.
As discussed above, most of the earlier peptide photodissociation work had been performed with 193 nm ArF excimer laser light. Since this wavelength has some advantages in terms of ease of light generation and transmission through air and optics, it would be preferred if it could activate ions as effectively as 157 nm light. In our own tandem-TOF instrument we have directly compared 157 nm and 193 nm photodissociation and generally found that the longer wavelength leads to inferior photofragmentation spectra. As an example of this, Figures 11A and 11B compare data recorded for peptide AAAANSAAAR with 193 and 157 nm, respectively. The longer wavelength produces fewer high-mass ions, for reasons that are not entirely clear. The shorter wavelength provides about 1.5 eV more activation energy per photon and the absorption spectrum displayed in Figure 1 suggests that the two wavelengths excite different electronic states. However, it is not obvious why either of these factors should favor the production of smaller, rather than larger, high-energy fragments. Surprisingly, when the photofragmentation experiments are performed in the linear ion trap, spectra recorded at the two wavelengths appear very similar (Thompson, 2007). (As noted above, more thermal fragment ions are observed in ion trap spectra than in tandem TOF data). Although there are a number of differences in the experiment performed in the two instruments, a key factor may be the orders-of-magnitude time difference between photoactivation and product ion drawout in the two instruments that was pointed out above.
FIGURE 11.
Tandem-TOF photodissociation of singly-charged AAAANSAAAR peptide ions using (A) 193 and (B) 157 nm light. W refers to loss of water (Thompson et al., 2007).
Along similar lines, the Kim group has investigated the 193 nm photodissociation of a few peptide ions that we had studied at 157nm using our TOF-TOF apparatus (Moon et al., 2005). Remarkably their data exhibited many of the characteristics that we were only able to observe using the shorter light wavelength. Once again the different fragment ion observation timescales in the two experiments may be the source of this discrepancy.
VII. VUV PHOTOFRAGMENTATION OF OTHER BIOMOLECULAR IONS
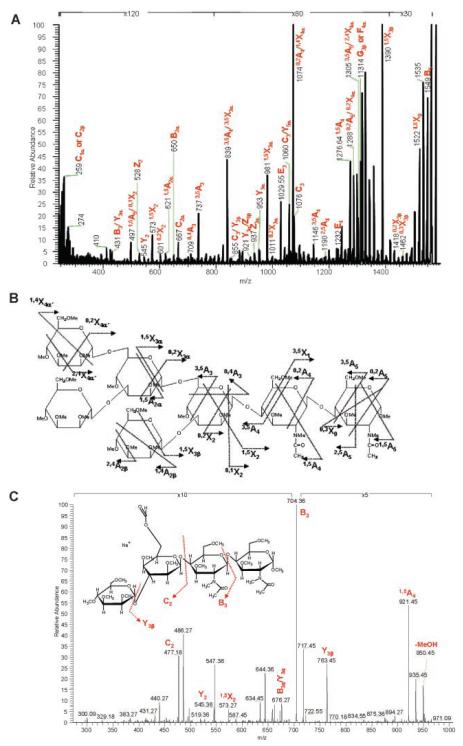
One of the most complex and ubiquitous of all post-translational modifications is glycosylation (Bertozzi & Kiessling, 2001;Varki, 1993). Glycoproteins and carbohydrates play roles in inter- and intra-cellular activities (Helenius & Aebi, 2001), the function of the immune system (Rudd et al., 2001), protein regulation and interactions (Bianco et al., 2006) and therapeutics (Shriver et al., 2004). As their importance becomes better appreciated, interest in their analysis and identification grows. Nevertheless, compared with peptides, glycans are particularly challenging to the mass spectrometrist. They do not contain as many basic sites as peptides so they are more difficult to ionize. More significantly, their branching, or antennary structure, enables glycans to serve their biological functions, and it must be completely characterized for each molecule of interest. Glycan ions fragment in two principal ways: glycosidic cleavages between adjacent sugar residues and cross-ring cleavages within the same sugar unit (Domon & Costello, 1988). The former are observed in low-energy fragmentation experiments while the latter require high-energy activation. Based on our experiments with peptides we were optimistic that 157 nm light could provide sufficient excitation for all of these processes. However, lack of vacuum ultraviolet spectroscopic data on glycans made it difficult to predict what chromophore might be excited and therefore whether photodissociation would provide an abundance of useful fragmentation information. Nevertheless, these experiments proved to be successful. One of the first experiments involved a comparison of different ways of fragmenting Girard’s T-derivatized maltoheptaose ions (Devakumar et al., 2005). (In this case, the derivatization label provided a fixed charge to the carbohydrate, thereby improving the observed ion signal). Figure 12 compares the high-energy CID fragmentation of these ions in an ABI 4700 TOF-TOF instrument with 157 nm photodissociation results obtained with our tandem-TOF and linear ion trap instruments. Extensive cross-ring fragmentation is evident in all three spectra including a contiguous series of 1,5Xn ions from n= 2-6. Although the simplicity of these photodissociation data is attractive, for practical glycan analysis the observation of different types of cross-ring fragments is really needed to elucidate complex glycan branching patterns. We have investigated a number of these more complex systems (Devakumar et al., 2008b;Devakumar et al., 2007). Figure 13A presents an example of a 157 nm photofragmentation spectrum of a branched permethylated glycan whose structure appears in Figure 13B. In addition to the B-, C- and Z- type ions that result from glycosidic cleavages, we observed several different types of A- and X-type ions indicative of various kinds of cross-ring fragmentation processes that provide definitive structural information. For example, the 3,5A3 and 0,4A3 ions enable the composition of each antenna to be determined. As noted previously, a unique feature of the ion trap as a photodissociation platform is its capability of performing higher order MSn experiments. This can be invaluable for confirming the assignments of peaks in fairly congested spectra such as those of glycans. One photofragment ion that appears at m/z 981.5 in Figure 13A is tentatively assigned as 1,5X3α. When ions of this mass are collisionally activated inside the trap, the MS3 spectrum in Figure 13C results. The simplicity of this spectrum facilitates its interpretation and the assignment of the structure of the m/z 981.5 precursor ion. It is hoped that these kinds of MSn spectra, that are generated by combining stages of photo- and collision-induced fragmentation, can be straightforwardly interpreted using automated, computer-based algorithms. In some cases this has already been achieved in low-energy CID MSn experiments on glycans (Ashline et al., 2005;Ashline et al., 2007;Lapadula et al., 2005;Zhang et al., 2005). However, it is likely that the ability to induce cross-ring fragments using VUV light will facilitate the elucidation of glycan branching structure in fewer MSn stages.
FIGURE 12.
MS2 spectra of singly-charged Girard’s T derivatized Maltoheptaose obtained by (A) high-energy CID and (B) 157 nm photodissociation in a MALDI-TOF-TOF instrument (c) 157 nm photodissociation in a linear ion trap (Devakumar et al., 2005).
FIGURE 13.
(A) Ion trap photodissociation mass spectrum of permethylated Man5 N-glycan (m/z 1580) ions from ribonuclease B (B) precursor structure and cross-ring fragments (Devakumar et al., 2007); (C) structure and CID spectrum of m/z 981 photofragment ion.
The fragmentation of lipids using both157 nm light and low-energy CID have been compared using the linear ion trap apparatus discussed above (Devakumar et al., 2008a). It was demonstrated that leukotriene C4 isomers could be distinguished with either approach, but the high-energy nature of the photofragmentation process significantly enhanced the production of informative product ions that facilitated this. The ion trap again found use in hybrid MSn experiments in which photofragments were detected, isolated, collisionally dissociated and thereby definitively identified.
VIII. SUMMARY AND CONCLUSIONS
Photofragmentation of biomolecular ions with vacuum ultraviolet light has been shown to be both an interesting phenomenon and an effective analytical tool. It can be performed on a number of mass spectrometer platforms and the resulting mass spectra vary somewhat from one instrument to another, apparently because the fragment analysis time following photoexcitation varies from one instrument to another. In some experiments, excitation wavelengths of 193 and 157 nm appear to yield strikingly different results, while in other apparati they produce comparable fragment distributions. It has nevertheless been ubiquitously observed that VUV photodissociation is a high-energy process. The capability of inducing high-energy fragmentation in any type of mass analyzer independent of vacuum requirements is perhaps one of its clearest virtues. The unique fragment distributions and high information content in spectra generated by photodissociation guarantee that the method will be helpful for solving specific mass spectrometric problems. It remains to be demonstrated whether it is compatible with truly high throughput experiments that will dominate the field of proteomics.
ACKNOWLEDGEMENTS
Our VUV photofragmentation work has been supported by the National Science Foundation grants CHE 0518324 and CHE 0431991 and the NIH/NCRR National Center for Glycomics and Glycoproteomics (NCGG) grant number RR018942. I wish to acknowledge the students and postdoctoral researchers in our laboratory whose contributions to our understanding of biomolecule photofragmentation are summarized in this review. In particular, the efforts of Matthew Thompson, Weidong Cui, Arugadoss Devakumar, Liangyi Zhang, Tae-Young Kim and William Harris were crucial to this work.
Biographical Sketch
Dr. James Reilly obtained his Ph.D. from the University of California, Berkeley in 1977 while working on ultrasensitive intracavity dye laser spectroscopy. Subsequently he did postdoctoral research at the Max Planck Institute for Laserforschung in Garching, Germany where he helped to open up the new field of multiphoton ionization mass spectroscopy. Starting at Indiana University in 1979, he initiated studies of the kinetic energy distribution of the photoelectrons ejected by laser ionizing diatomic and small aromatic molecules and thereby established the field of laser photoelectron spectroscopy. During the 1980’s his group developed an interest in high resolution time-of-flight mass spectrometry. In the early 1990’s, this TOF experience enabled them to develop space-velocity correlation focusing (often referred to as delayed extraction) as a method to improve the resolution of MALDI-TOF mass spectra of biological molecules. Over the last fifteen years, his group has applied MALDI and electrospray ionization in the analysis of biomolecules and in various bacterial proteomics experiments. A dominant theme in the group’s recent work has been the photofragmentation of biomolecular ions, which is the subject of this review article.

X. REFERENCES
- Antoine R, Joly L, Tabarin T, Broyer M, Dugourd P, Lemoine J. Photo-induced formation of radical anion peptides. Electron photo-detachment dissociation experiments. Rapid Commun. Mass Spectrom. 2007;21:265–268. doi: 10.1002/rcm.2810. [DOI] [PubMed] [Google Scholar]
- Ashline D, Singh S, Hanneman A, Reinhold V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal. Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashline DJ, Lapadula AJ, Liu Y-H, Lin M, Grace M, Pramanik B, Reinhold VN. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal. Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci DC, Russell DH. Sequence and side-chain specific photofragment (193 nm) ions from protonated substance P by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 1999;10:1038–1040. [Google Scholar]
- Beardsley RL, Karty JA, Reilly JP. Enhancing the intensities of lysine-terminated tryptic peptide ions in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2000;14:2147–2153. doi: 10.1002/1097-0231(20001215)14:23<2147::AID-RCM145>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Bertozzi CR, Kiessling LL. Chemical Glycobiology. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- Bianco GA, Toscano MA, Ilarregui JM, Rabinovich GA. Impact of protein-glycan interactions in the regulation of autoimmunity and chronic inflammation. Autoimmun. Rev. 2006;5:349–356. doi: 10.1016/j.autrev.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Biemann K. Sequencing of peptides by tandem mass spectrometry and high-energy collision-induced dissociation. Methods Enzymol. 1990;193:455–479. doi: 10.1016/0076-6879(90)93433-l. [DOI] [PubMed] [Google Scholar]
- Bowers WD, Delbert SS, Hunter RL, McIver RT., Jr. Fragmentation of oligopeptide ions using ultraviolet laser radiation and Fourier transform mass spectrometry. J. Am. Chem. Society. 1984;106:7288–7289. [Google Scholar]
- Breci LA, Tabb DL, Yates JR, III, Wysocki VH. Cleavage N-Terminal to Proline: Analysis of a Database of Peptide Tandem Mass Spectra. Anal. Chem. 2003;75:1963–1971. doi: 10.1021/ac026359i. [DOI] [PubMed] [Google Scholar]
- Breuker K, Oh H, Lin C, Carpenter BK, McLafferty FW. Nonergodic and conformational control of the electron capture dissociation of protein cations Proc. Natl. Acad. Sci. U. S. A. 2004;101:14011–14016. doi: 10.1073/pnas.0406095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RS, Lennon JJ. Mass resolution improvement by incorporation of pulsed ion extraction in a matrix-assisted laser desorption/ionization linear time-of-flight mass spectrometer. Anal. Chem. 1995;67:1998–2003. doi: 10.1021/ac00109a015. [DOI] [PubMed] [Google Scholar]
- Campbell JM. 51st Annual Conference on Mass Spectrometry and Allied Topics; Montreal, Canada. 2003. [Google Scholar]
- Choi KM, Yoon SH, Sun M, Oh JY, Moon JH, Kim MS. Characteristics of photodissociation at 193 nm of singly protonated peptides generated by matrix-assisted laser desorption ionization (MALDI) J. Am. Soc. Mass Spectrom. 2006;17:1643–1653. doi: 10.1016/j.jasms.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Clark LB. Polarization Assignments in the Vacuum UV Spectra of the Primary Amide, Carboxyl, and Peptide Groups. J. Am. Chem. Soc. 1995;117:7974–7986. [Google Scholar]
- Cui W, Thompson MS, Reilly JP. Pathways of peptide ion fragmentation induced by vacuum ultraviolet light. J. Am. Soc. Mass Spectrom. 2005;16:1384–1398. doi: 10.1016/j.jasms.2005.03.050. [DOI] [PubMed] [Google Scholar]
- Despeyroux D, Wright AD, Jennings KR. Comparison of collision-induced dissociation and surface induced dissociation mass spectra of peptides obtained using a four-sector mass spectrometer. Int. J. Mass Spectrom. Ion Proc. 1993;126:95–99. [Google Scholar]
- Devakumar A, O'Dell DK, Walker JM, Reilly JP. Structural analysis of leukotriene C4 isomers using collisional activation and VUV photodissociation. J. Am. Soc. Mass Spectrom. 2008a;19:14–26. doi: 10.1016/j.jasms.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Devakumar A, Mechref Y, Kang P, Novotny MV, Reilly JP. Laser-induced photofragmentation of neutral and acidic glycans inside an ion-trap mass spectrometer. Rapid Commun. Mass Spectrom. 2007;21:1452–1460. doi: 10.1002/rcm.2981. [DOI] [PubMed] [Google Scholar]
- Devakumar A, Mechref Y, Kang P, Novotny MV, Reilly JP. Identification of Isomeric N-Glycan Structures by Mass Spectrometry with 157 nm Laser-Induced Photofragmentation. J. Am. Soc. Mass Spectrom. 2008b;19:1027–1040. doi: 10.1016/j.jasms.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devakumar A, Thompson MS, Reilly JP. Fragmentation of Oligosaccharide Ions with 157 nm Vacuum Ultraviolet Light. Rapid Commun. Mass Spectrom. 2005;19:2313–2320. doi: 10.1002/rcm.2058. [DOI] [PubMed] [Google Scholar]
- Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate. 1988;5:397–809. [Google Scholar]
- Fung YME, Kjeldsen F, Silivra OA, Chan TWD, Zubarev RA. Facile Disulfide bond Cleavage in Gaseous Peptide and Protein Cations by Ultraviolet photodissociation at 157 nm. Angew. Chem. 2005;44:6399–6403. doi: 10.1002/anie.200501533. [DOI] [PubMed] [Google Scholar]
- Gabelica V, Rosu F, De Pauw E, Antoine R, Tabarin T, Broyer M, Dugourd P. Electron photodetachment dissociation of DNA anions with covalently or noncovalently bound chromophores. J. Am. Soc. Mass Spectrom. 2007a:1990–2000. doi: 10.1016/j.jasms.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Gabelica V, Rosu F, Tabarin T, Kinet C, Antoine R, Broyer M, De Pauw E, Dugourd P. Base-Dependent Electron Photodetachment from Negatively Charged DNA Strands upon 260-nm Laser Irradiation. J. Am. Chem. Soc. 2007b;129:4706–4713. doi: 10.1021/ja068440z. [DOI] [PubMed] [Google Scholar]
- Gabelica V, Tabarin T, Antoine R, Rosu F, Compagnon I, Broyer M, De Pauw E, Dugourd P. Electron photodetachment dissociation of DNA polyanions in a quadrupole ion trap mass spectrometer. Anal. Chem. 2006;78:6564–6572. doi: 10.1021/ac060753p. [DOI] [PubMed] [Google Scholar]
- Gabryelski W, Li L. Photo-induced dissociation of electrospray generated ions in an ion trap/time-of-flight mass spectrometer. Rev. Sci. Instrum. 1999;70:4192–4199. [Google Scholar]
- Garcia BA, Siuti N, Thomas CE, Mizzen CA, Kelleher NL. Characterization of neurohistone variants and post-translational modifications by electron capture dissociation mass spectrometry. International Journal of Mass Spectrometry. 2007;259:184–196. [Google Scholar]
- Gimon-Kinsel ME, Kinsel GR, Edmondson RD, Russell DH. Photodissociation of high molecular weight peptides and proteins in a two-stage linear time-of-flight mass spectrometer. J. Am. Soc. Mass Spectrom. 1994;6:578–587. doi: 10.1016/1044-0305(95)00247-B. [DOI] [PubMed] [Google Scholar]
- Guan Z, Kelleher NL, O'Connor PB, Aaserud DJ, Lettle DP, McLafferty FW. 93 nm phototdissociation of larger multiply-charged biomolecules. Int. J. Mass Spectrom. Ion Proc. 1996;157/158:357–364. [Google Scholar]
- Harris W, Reilly JP. 45th ASMS Conference on Mass Spectrometry; Palm Springs, CA. 1997. [Google Scholar]
- Harris WA, Reilly JP. 46th ASMS Conference on Mass Spectrometry; Orlando, FL. 1998. [Google Scholar]
- Harrison AG. The gas-phase basicities and proton affinities of amino acids and peptides. Mass Spectrom. Rev. 1997;16:201–217. [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. 291(5512), 2364-2369. [DOI] [PubMed] [Google Scholar]
- Hettick JM, McCurdy KL, Barbacci DC, Russell DH. Optimization of sample preparation for peptide sequencing by MALDI-TOF photofragment mass spectrometry. Anal. Chem. 2001;73:5378–5386. doi: 10.1021/ac0102157. [DOI] [PubMed] [Google Scholar]
- Horn DM, Zubarev RA, McLafferty FW. Automated de novo sequencing of proteins by tandem high-resolution mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10313–10317. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Cooks RG. The Orbitrap: a new mass spectrometer. J. Am. Soc. Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- Hu Y, Hadas B, Davidovitz M, Balta B, Lifshitz C. Does IVR take place prior to peptide ion dissociation? J. Phys. Chem. A. 2003;107:6507–6514. [Google Scholar]
- Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH. Statistical Characterization of the Charge State and Residue Dependence of Low-Energy CID Peptide Dissociation Patterns. Anal. Chem. 2005;77:5800–5813. doi: 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DF, Shabanowitz J, Yates JR, III, Griffin PR, Zhu NZ. Tandem quadrupole Fourier-transform mass spectrometry. Anal. Chim. Acta. 1989;225:1–10. [Google Scholar]
- Hunt DF, Shabanowitz J, Yates JR., III Peptide sequence analysis by laser photodissociation Fourier transform mass spectrometry. J. Chem. Soc., Chem. Commun. 1987;8:548–550. [Google Scholar]
- Johnson RS, Martin SA, Biemann K, Stults JT, Watson JT. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: Differentiation of leucine and isoleucine. Anal. Chem. 1987;59:2621–2625. doi: 10.1021/ac00148a019. [DOI] [PubMed] [Google Scholar]
- Johnson RS, Martin SA, Biemann K. Collision-induced fragmentation of (M + H)+ ions of peptides. Side chain specific sequence ions. International. Journal of Mass Spectrometry and Ion Processes. 1988;86:137–154. [Google Scholar]
- Joly L, Antoine R, Broyer M, Lemoine J, Dugourd P. Electron Photodetachment from Gas Phase Peptide Dianions, Relation with Optical Absorption Properties. J. Phys. Chem. 2008;112:898–903. doi: 10.1021/jp0752365. [DOI] [PubMed] [Google Scholar]
- Joly L, Antoine R, Broyer M, Dugourd P, Lemoine J. Specific UV photodissociation of tyrosyl-containing peptides in multistage mass spectrometry. J. Mass Spectrometry. 2007;42:818–824. doi: 10.1002/jms.1222. [DOI] [PubMed] [Google Scholar]
- Kaufmann R, Kirsch D, Spengler B. Sequencing of peptides in a time-of-flight mass spectrometer: evaluation of postsource decay following matrix-assisted laser desorption ionization (MALDI) Int. J. Mass Spectrom. Ion Proc. 1994;131:355–385. [Google Scholar]
- Kim T-Y, Thompson MS, Reilly JP. 157 nm photodissociation of peptides in a linear ion trap mass spectrometer. Rapid Commun. Mass Spectrom. 2005;19:1657–1665. doi: 10.1002/rcm.1969. [DOI] [PubMed] [Google Scholar]
- Kruger NA, Zubarev RA, Carpenter BK, Kelleher NL, Horn DM, McLafferty FW. Electron capture versus energetic dissociation of protein ions. International Journal of Mass Spectrometry. 1999;182/183:1–5. 182/183 1-5. [Google Scholar]
- Lapadula AJ, Hatcher PJ, Hanneman A, Ashline DJ, Zhang H, Reinhold VN. Congruent strategies for carbohydrate sequencing. 3. OSCAR: An algorithm for assigning oligosaccharide topology from MSn data. Anal. Chem. 2005;77:6271–6279. doi: 10.1021/ac050726j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskin J, Bailey TH, Futrell JH. Shattering of peptide ions on self-assembled monolayer surfaces. J. Am. Chem. Soc. 2003;125:1625–1632. doi: 10.1021/ja027915t. [DOI] [PubMed] [Google Scholar]
- Laskin J, Futrell JH. Surface-induced dissociation of peptide ions: kinetics and dynamics. J. Am. Soc. Mass Spectrom. 2003;14:1340–1347. doi: 10.1016/j.jasms.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Lebrilla CB, Wang DTS, Mizoguchi TJ, McIver RT., Jr. Comparison of the fragmentation produced by fast atom bombardment and photodissociation of peptides. J. Am. Chem. Soc. 1989;111:8593–8598. [Google Scholar]
- Little DP, Speir JP, Senko MW, O'Connor PB, McLafferty FW. Infrared Multiphoton Dissociation of Large Multiply Charged Ions for Biomolecule Sequencing. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- Loo JA, Edmonds CG, Smith RD. Tandem mass spectrometry of very large molecules. 2. Dissociation of multiply charged proline-containing proteins from electrospray ionization. Anal. Chem. 1993;65:425–438. doi: 10.1021/ac00052a020. [DOI] [PubMed] [Google Scholar]
- Martin SA, Hill JA, Kittrell C, Biemann K. Photon-induced dissociation with a four-sector tandem mass spectrometer. J. Am. Soc. Mass Spectrom. 1990;1:107–109. doi: 10.1016/1044-0305(90)80013-D. [DOI] [PubMed] [Google Scholar]
- McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of Electron-Transfer Dissociation on a Hybrid Linear Ion Trap -- Orbitrap Mass Spectrometer. Anal. Chem. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Berggren WT, Griep-Raming J, Horning S, Makarov A, Phanstiel D, Stafford G, Swaney DL, Syka JEP, Zabrouskov V, Coon JJ. A Proteomics Grade Electron Transfer Dissociation-Enabled Hybrid Linear Ion Trap-Orbitrap Mass Spectrometer. J. Proteome Res. 2008 doi: 10.1021/pr800264t. x:published online 7/10/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzihradszky KF, Campbell JM, Baldwin MA, Falick AM, Juhasz P, Vestal ML, Burlingame AL. The characteristics of peptide collision-induced dissociation using a high-performance MALDI-TOF/TOF tandem mass spectrometer. Anal. Chem. 2000;72:552–558. doi: 10.1021/ac990809y. [DOI] [PubMed] [Google Scholar]
- Medzihradszky KF, Adams GW, Burlingame AL, Bateman RH, Green MR. Peptide sequence determination by matrix-assisted laser desorption ionization employing a tandem double focusing magnetic-orthogonal acceleration time-of-flight mass spectrometer. J. Am. Soc. Mass Spectrom. 1996;7:1–10. doi: 10.1016/1044-0305(95)00621-4. [DOI] [PubMed] [Google Scholar]
- Meroueh O, Hase WL. Collisional Activation of Small Peptides. J. Phys. Chem. A. 1999;103:3981–3990. [Google Scholar]
- Moon JH, Yoon SH, Kim MS. Photodissociation of singly protonated peptides at 193 nm investigated with tandem time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:3248–3252. doi: 10.1002/rcm.2184. [DOI] [PubMed] [Google Scholar]
- Morgan JW, Russell DH. Comparative studies of 193-nm photodissociation and TOF-TOFMS analysis of bradykinin analogues: The effects of charge site(s) and fragmentation timescales. J. Am. Soc. Mass Spectrom. 2006;17:721–729. doi: 10.1016/j.jasms.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Oh JY, Moon JH, Kim MS. Sequence- and site-specific photodissociation at 266 nm of protonated synthetic polypeptides containing a tryptophanyl residue. Rapid Commun. Mass Spectrom. 2004;18 doi: 10.1002/rcm.1679. [DOI] [PubMed] [Google Scholar]
- Paizs B, Surhai S. Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 2005;24:508–528. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos IA. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom. Rev. 1995;14:49–73. [Google Scholar]
- Peterson DL, Simpson WT. Polarized electronic absorption spectrum of amides with assignments of transitions. J. Am. Chem. Society. 1957;79:2375–2382. [Google Scholar]
- Price WD, Schnier PD, Williams ER. Tandem Mass Spectrometry of Large Biomolecule Ions by Blackbody Infrared Radiative Dissociation. Anal. Chem. 1996;68:859–866. doi: 10.1021/ac951038a. [DOI] [PubMed] [Google Scholar]
- Robin M. Higher excited states of polyatomic molecules. Academic Press; Orlando: 1975. [Google Scholar]
- Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomedical Mass Spectrometry. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science (Washington, DC, United States) 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- Shriver Z, Raguram S, Sasisekharan R. Glycomics: a pathway to a class of new and improved therapeutics. Nature Reviews Drug Discovery. 2004;3:863–873. doi: 10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. Journal of Mass Spectrometry and Ion Processes. 2004;39:1091–1112. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- Spengler B. Post-source decay analysis in matrix-assisted laser desorption/ionization mass spectrometry of biomolecules. J. Mass Spectrometry. 1997;32:1019–1036. [Google Scholar]
- Stimson E, Truong O, Richter WJ, Waterfield MD, Burlingame AL. Enhancement of charge remote fragmentation in protonated peptides by high-energy CID MALDI-TOF-MS using "cold" matrices. International Journal of Mass Spectrometry. 1997;169/170:231–240. [Google Scholar]
- Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabarin T, Antoine R, Broyer M, Dugourd P. Specific photodissociation of peptides with multi-stage mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:2883–2892. doi: 10.1002/rcm.2124. [DOI] [PubMed] [Google Scholar]
- Talbo G, Roepstorff P. Determination of sulfated peptides via prompt fragmentation by UV matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1993;7:201–204. [Google Scholar]
- Tecklenburg RE, Jr., Miller MN, Russell DH. Laser ion beam photodissociation studies of model amino acids and peptides. J. Am. Chem. Soc. 1989;111:1161–1171. [Google Scholar]
- Thompson MS, Cui W, Reilly JP. Fragmentation of singly-charged peptide ions by photodissociation at = 157 nm. Angewantde Chemie. 2004;116:4895–4898. doi: 10.1002/anie.200460788. [DOI] [PubMed] [Google Scholar]
- Thompson MS. Doctoral. Indiana University; 2007. Vacuum Ultraviolet Photofragmentation of Peptide Ions Department of Chemistry. [DOI] [PubMed] [Google Scholar]
- Thompson MS, Cui W, Reilly JP. Factors that impact the vacuum ultraviolet photofragmentation of peptide ions. J. Am. Soc. Mass Spectrom. 2007;18:1439–1452. doi: 10.1016/j.jasms.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Tsaprailis G, Nair H, Somogyi A, Wysocki VH, Zhong W, Futrell JH, Summerfield SG, Gaskell SJ. Influence of Secondary Structure on the Fragmentation of Protonated Peptides. Journal of the American Chemical Society. 1999;121:5142–5154. [Google Scholar]
- Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ER, Henry KD, McLafferty FW, Shabanowitz J, Hunt DF. Surface-induced dissociation of peptide ions in Fourier-transform mass spectrometry. J. Am. Soc. Mass Spectrom. 1990;1:413–416. doi: 10.1016/1044-0305(90)85022-E. [DOI] [PubMed] [Google Scholar]
- Wilson JJ, Brodbelt JS. MS/MS Simplification by 355 nm Ultraviolet Photodissociation of Chromophore-Derivatized Peptides in a Quadrupole Ion Trap. Anal. Chem. 2007;79:7883–7892. doi: 10.1021/ac071241t. [DOI] [PubMed] [Google Scholar]
- Woody RW, Koslowski A. Recent developments in the electronic spectroscopy of amides and a-helical polypeptides. Biophysical Chemistry. 2002;101:102–535. doi: 10.1016/s0301-4622(02)00187-4. [DOI] [PubMed] [Google Scholar]
- Wysocki VH, Tsaprailis G, Smith LL, Breci LA. Mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectrometry. 2000;35:1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Zhang H, Singh S, Reinhold VN. Congruent strategies for carbohydrate sequencing. 2. FragLib: An MSn spectral library. Anal. Chem. 2005;77:6263–6270. doi: 10.1021/ac050725r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cui W, Thompson MS, Reilly JP. Structures of alpha-type ions formed in the 157 nm photodissociation of singly-charged peptide ions. J. Am. Soc. Mass Spectrom. 2006;17:1315–1321. doi: 10.1016/j.jasms.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Zimmerman JA, Watson CH, Eyler JR. Multiphoton ionization of laser-desorbed neutral molecules in a Fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 1991;63:361–365. [Google Scholar]
- Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. Journal of the American Chemical Society. 1998;120:3265–3266. [Google Scholar]