ABSTRACT
Pre-mRNA splicing in eukaryotes is performed by the spliceosome, a highly complex macromolecular machine. SF3b is a multi-protein complex which recognizes the branch point adenosine of pre-mRNA as part of a larger U2 snRNP or U11/U12 di-snRNP in the dynamic spliceosome machinery. Although a cryo-EM map is available for human SF3b complex, the structure and relative spatial arrangement of all components in the complex are not yet known. We have recognized folds of domains in various proteins in the assembly and generated comparative models. Using an integrative approach involving structural and other experimental data, guided by the available cryo-EM density map, we deciphered a pseudo-atomic model of the closed form of SF3b which is found to be a “fuzzy complex” with highly flexible components and multiplicity of folds. Further, the model provides structural information for 5 proteins (SF3b10, SF3b155, SF3b145, SF3b130 and SF3b14b) and localization information for 4 proteins (SF3b10, SF3b145, SF3b130 and SF3b14b) in the assembly for the first time. Integration of this model with the available U11/U12 di-snRNP cryo-EM map enabled elucidation of an open form. This now provides new insights on the mechanistic features involved in the transition between closed and open forms pivoted by a hinge region in the SF3b155 protein that also harbors cancer causing mutations. Moreover, the open form guided model of the 5′ end of U12 snRNA, which includes the branch point duplex, shows that the architecture of SF3b acts as a scaffold for U12 snRNA: pre-mRNA branch point duplex formation with potential implications for branch point adenosine recognition fidelity.
KEYWORDS: Cryo-EM, integrative structure modeling, spliceosome, SF3b complex, U11/U12 di-snRNP
Introduction
Splicing in eukaryotes is a well orchestrated stepwise process involving a large number of component molecules.1 This process constitutes the removal of non-coding sequences (introns) from pre-mRNAs and subsequent ligation of the coding sequences (exons) to form a mature mRNA.2-4 Several cis (pre-mRNA) and trans (snRNPs, accessory proteins) acting elements coordinate this process with high precision, to ensure fidelity. Higher eukaryotes contain a U2-dependent (major) splicing pathway and a less abundant U12-dependent (minor) pathway, which splices specific pre-mRNA introns (U2-type and U12-type) differing in the splice site and branch point consensus sequences. The U2-dependent pathway involves U1, U2, U5 and U4/U6 snRNPs while the U12-dependent pathway comprises of U11, U12, U5 and U4atac/U6atac snRNPs.5 Multiple intermediary complexes (E, A, B, Bact, B* and C) which are characterized by synergistic protein-protein, protein-RNA and RNA-RNA interactions 2,3 are involved in these pathways. The functional E form is absent in the U12-dependent pathway, where U11 and U12 snRNPs are parts of a stable di-snRNP complex prior to pre-mRNA binding.5-7 All the snRNPs consist of a common set of 7 Sm proteins (B/B′, D3, D2, D1, E, F and G) along with an snRNA and a number of particle-specific proteins.3,5
Splicing Factor SF3b complex is a fundamental component of both the U2 snRNP and U11/U12 di-snRNP.8 It forms a dynamic and integral part of 4 intermediary complexes (A, B, B*and Bact). Previous studies have revealed various roles for SF3b complex including i) recognition of branch point adenosine 9 and promotion of stable interaction for U2 and U11/U12 di-snRNP to pre-mRNA,10 ii) prevention of premature splicing 10 and iii) interaction with snRNAs and pre-mRNA.11,12 The complex is made of 7 proteins, namely, p14, SF3b49, SF3b145, SF3b155, SF3b10, SF3b130 and SF3b14b.12-14 p14, an RRM-domain containing protein, has been shown to recognize the branch point adenosine (BPA) within the bulged branch point duplex (BPD) structure formed between pre-mRNA and branch point recognition sequence (BPRS) of U2/U12 snRNA.9,15 This enables SF3b to present a temporary steric barrier to branch point sequence (BPS) prior to activation.10 In addition, SF3b49 has been shown to interact with the stem-loops at 5′ end of U2 snRNA 11 and is also shown to be cross linked around the BPS along with the other components such as the SF3b145 and SF3b155.12 Further, SF3b49 and SF3b145 are known to participate in multiple protein-protein interactions during transition from A to C in the dynamic splicing pathway.16
The dynamic nature and large size of snRNPs in the spliceosome machinery, have limited the attempts to obtain structural information at the atomic level by X-ray crystallography and NMR. Cryo-EM has emerged as an important technique to characterize the architectures and structures of large macromolecular assemblies. It has the advantage of using small amount of samples and visualizing the complexes in the native cellular conditions.18,19 A U1 snRNP structure was deciphered at a resolution of 10 Å using single particle cryo-Electron Microscopy.17 A cryo-EM map of the human SF3b complex was achieved at a resolution of 9.7 Å in the year 200320 and a cryo-EM map for the U11/U12 di-snRNP was published in the year 2005 at 13.4 Å resolution21 Low resolution cryo-EM maps often do not have sufficient details to understand the mechanistic and structural basis of biological functions of the protein complexes. Hence, integrative methods, where the structures from X-ray crystallography,22 NMR, comparative modeling,23 or de novo protein structure prediction,24 combined with cryo-EM and other experimental data are used to obtain pseudo-atomic models of macromolecular assemblies.25,26
In the previous cryo-EM work on SF3b published in 2003, only 3 of the 7 components of the SF3b complex could be localized into the density map.20 Further, neither crystallographic nor NMR structures of 2 of the components (SF3b49 and p14) or related proteins, were available at that time. Structural details of many SF3b proteins or protein domains have now become available.27 SF3b155 protein was shown to be composed of several N-terminal RWDETP and TPGH repeats (∼450 amino acids) and 22 HEAT repeats at its C-terminus.28 It forms a ladder-like structure folded in S-shape occupying 2 thirds of the density map.29 Subsequently, NMR structures of the 2 RRM domains of SF3b49 (PDB ID: 1X5U and 1X5T; Structural genomics consortium) were also determined. For p14, both crystal (PDB ID: 2F9D, 3LQV)9,30 and NMR structures (PDB ID: 2FHO) 31 became available along with the peptide component of SF3b155. To date, this is the only inter-molecular interaction, defined at atomic level, in the SF3b complex. Further understanding of the detailed structural features of this complex and mechanistic features is desirable as it is a target for many antitumor drugs.32-35
It is more than a decade since the cryo-EM map of the human SF3b complex was obtained and the lack of structural information has left a substantial part of the density unexplained. With the remarkable expansion of the list of known 3D structures of proteins, we now have better opportunities to recognize structures of these components. In addition, given the fact that the number of protein folds is limited 36 and with the development of sensitive fold recognition methods, reliable fold assignments could be made for many of the components.37
In the current work, we provide a description of the structure of human SF3b complex and provide a mechanistic basis of its function by increasing its structural coverage using state-of-the-art integrative structure modeling techniques. The modeling employed here has involved an extensive use of the available X-ray and NMR structures, fold recognition and comparative modeling, as well as substantial conformational and configurational sampling of the interacting protein components. In addition, we have also devised novel methods to integrate both experimental and evolutionary information pertaining to protein folds and dynamics. Our modeling efforts have enabled us to delineate the molecular architecture of the human SF3b complex and obtain a pseudo-atomic model for the entire complex with high structural coverage. Further, the locations of the proteins within the SF3b density map, provided by the pseudo-atomic model, have allowed us to carve out appropriate SF3b density segments to model an open form of the SF3b complex, using an existing U11/U12 cryo-EM map 21 and also fit the component structures into it. Finally, it has enabled us to obtain the structural constraints for modeling of the 5′ end of U12 snRNA and its interaction with pre-mRNA in the branch point duplex (BPD). We discuss the implications for pre-mRNA branch point recognition fidelity, provided by the architecture of SF3b, in the light of proposed pseudo-atomic model. This work adds to the excellent developments made in 2003 20 and 2005 21 and also deepens our understanding of the structure-function relationship of various components of the human SF3b complex.
In the last decade, electron microscopy studies on spliceosomal complexes, at various stages of the splicing cycle, have provided a glimpse of the overall morphological features of this complex macromolecular machine.38-51 However, recent efforts to determine high resolution cryo-EM density maps, utilizing advances in detector technology 52 coupled with powerful statistical image processing algorithms 53,54 have yielded structures for the U4/U6.U5 tri-snRNP complex from S. cerevisiae 55-57 as well as the U5.U2/U6 intron lariat spliceosome (ILS) complex from S.pombe.58-60 However, in the yeast U5.U2/U6 structure, the U2 snRNP region has an overall resolution of 11 Å and lacks the density for the SF3b sub-complex.58 The only structural data that is currently available for SF3b, although a low resolution cryo-EM density map, is that of human SF3b complex by Golas et. al.20,21 Though such cryo-EM density maps are difficult to interpret, our attempts to derive a reasonable model using multiple methods that involve protein fold recognition, comparative modeling, cryo-EM density fitting as well as an inclusion of currently available experimental datasets is exhaustive. Ultimately, such an integrated approach has been useful to derive a pseudo-atomic model, providing insights into the functioning of SF3b in splicing. Further, SF3b has been found to be a “fuzzy” complex, based on flexibility analysis using normal mode technique and fold-function relationships derived for the individual proteins within the complex. Hence, it is anticipated that the current pseudo-atomic model will provide the necessary impetus and insights for the determination of high resolution structures of the human spliceosomal complexes in the future.
Results
Integrative structure modeling of the human SF3b complex
Human SF3b complex comprises of 7 interacting components, of which only the p14, 2 RRM domains of SF3b49 and an N-terminal SAP motif of SF3b145 have atomic level structural information available (Table S1). We devised a hierarchical modeling protocol to enhance the structural coverage of SF3b complex. This involved an integrative approach that employs a multitude of computational methods summarized in Fig. S1. Briefly, searches for the detection of appropriate templates for comparative modeling of domains or full-length proteins of SF3b14b, SF3b130, SF3b155, SF3b145 and SF3b10 have been performed. This was further augmented with sensitive distant homology detection and fold recognition tools in search databases of known structure. The templates obtained for SF3b14b, SF3b130, SF3b155 and SF3b145 were employed to model these proteins. For those cases with only partial information from homology and domain overlap (SF3b145), we used density and other experimental data (see Materials and Methods) to guide our fold assignments. As we were not able to identify a template with good confidence to model the entire SF3b10 reliably, we used a combined approach that employed de novo structure prediction, SF3b cryo-EM density-guided filtering (Fig. 1A) and scoring using residue-residue contacts, based on the co-evolution principle, to obtain a model. The structural models thus obtained for the SF3b components (Fig. 1B, Fig. S2, Table S1 and see Materials and Methods), were used for further cryo-EM density-based fitting.
Figure 1.
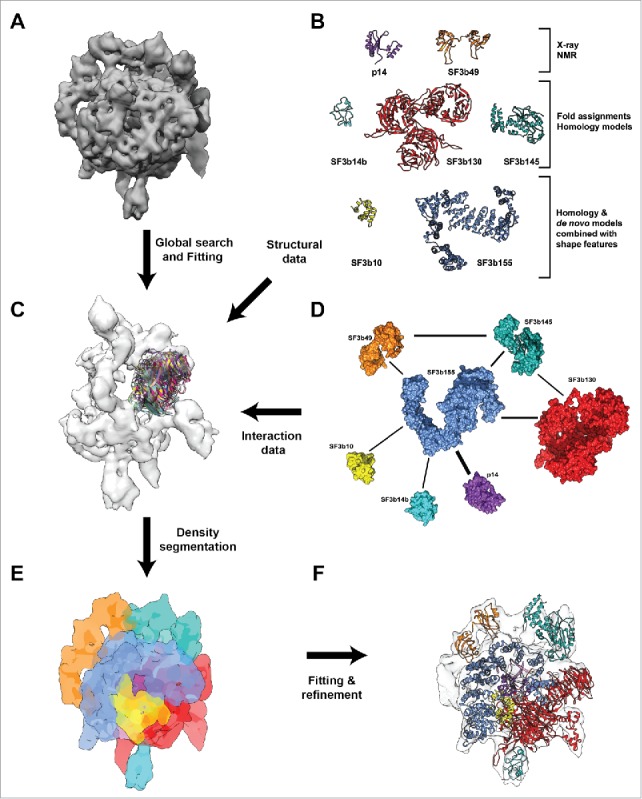
Integrative structural modeling of Human SF3b complex. (A) A 9.7 Å cryo-EM map of SF3b complex displayed at the density threshold level of 0.0158. (B) Structural information for individual components of SF3b complex used to obtain the pseudo atomic model. (C) Global search and fitting for the individual domains of proteins SF3b130, SF3b145 and SF3b14b within the SF3b density region that is currently unoccupied. This was performed across different resolutions (9.7Å, 12Å, 15Å and 18Å) (Fig. S3–S6 and Materials and Methods). (D) Protein-Protein interaction network for the SF3b complex obtained from experimental data. The weights of the edges denote the number of unique experimental methods which showed physical interaction between individual components (Table S2). (E) Density segmentation and localization of individual components into the initial cryo-EM map. This was obtained by integration of several computational methods including fitting techniques, shape features from structural models and experimentally derived interactions among the components. (F) Atomic level representation of SF3b components after flexible fitting into the cryo-EM map, color coded as in B and C.
Spatial locations of the SF3b components in the SF3b density map (EMD-1043) have been identified earlier using the shape features of the respective folds for the proteins p14, SF3b49 and SF3b155.20 In order to locate the other components within the SF3b density, we performed a global search (Fig. 1C), with a fine angular sampling, for the individual domains of proteins SF3b130, SF3b145 and SF3b14b within the SF3b density region that is currently unoccupied. This was performed across different resolutions (9.7Å, 12Å, 15Å and 18Å). Analysis of the top 10 solutions of the SF3b density, based on cross-correlation values and literature-derived SF3b protein-protein interaction information, allowed us to unambiguously locate the proteins within the density map (Fig. S3–S6). We did not perform this procedure for the SF3b10 as we had used an unassigned density segment after sampling to guide its structural modeling. Such a global search ensures that the components are fitted across the un-segmented density map and the best fit is obtained when shape features, derived from structural models, guide local density fitting. Further, the domain boundaries for fitting have been derived after performing flexibility analysis for individual proteins, (see the section on Architecture of the human SF3b complex) to account for the conformational changes as a result of complex formation. It must be noted that the shape features of individual components, based on the structural models, provide us with unique information for localization. This can be seen in the differences in distribution of cross-correlation scores (Fig. S7), as the domains of each component are fitted at each position into a discretized density map of 166 points (Fig. S7). Hence, the final positions of the components in the density map have been obtained by using neighborhood information from the interaction network (Fig. 1D-1F and Table S2), the global search for the components in the density map, local fitting scores (Table S1 and S3) and volume exclusion.
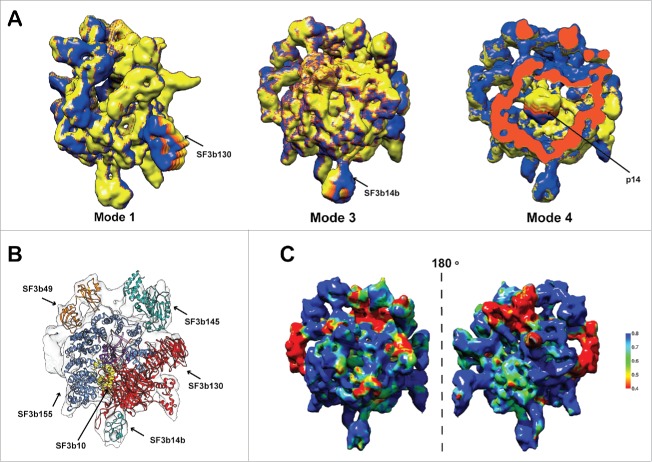
Human SF3b complex is known to undergo large structural changes.21 In order to understand this better, we performed normal mode analysis (NMA) on the density map to identify the regions with high and low conformational flexibility (see Material and Methods). Normal mode analysis on coarse grain representation of cryo-EM maps have been used previously to decipher functionally relevant conformational changes.61,62 NMA results suggested that regions corresponding to proteins SF3b130 (mode 1), SF3b14b (mode 3) and p14 (mode 4) are highly flexible (Fig. 2A). These results contributed toward the localization of the components, as the regions showing flexibility in the low frequency modes are populated with individual compact domains/structures in the pseudo-atomic model (Fig. 2B) (see the section on Architecture of the human SF3b complex).
Figure 2.
Flexibility and molecular architecture of Human SF3b complex. (A) Normal mode analysis of the density map and the associated conformational changes. The arrows show the regions of maximum flexibility observed in the corresponding low frequency modes where the proteins SF3b130 (mode 1), SF3b14b (mode 3) and p14 (mode 4) are located. The variations in the colors represent the different degrees of displacements in that particular mode. (B) Pseudo-atomic model of SF3b complex shown with the locations of its components. (C) Confidence assessment of atomic models by local correlation. The local cross-correlation was calculated for each voxel between the simulated density and the experimental density map with a grid size of 5×5 ×5 Å.
Structural models (Fig. 1B) were localized into the cryo-EM map using neighborhood information from the interaction network (Fig. 1D), exhaustive map search procedures using the shape features and considerations of flexibility (Fig. 2A). Subsequently, local fitting of all the proteins into the density map, using refined density segmentation (Fig. 1E), was performed. This was followed by a final round of flexible fitting. The resulting pseudo-atomic model provides structural information for 2682 residues (64.5%) of the human SF3b complex (Fig. 1F). Thus, for the first time, we report the relative spatial positions and interactions of all the components of the human SF3b complex (Fig. 2B and Video S1). The final pseudo-atomic model has an overall coverage of 81.3% (Volume = 333.3×103 Å3) of the map at a density threshold of 0.0108. It is important to note that the SF3b complex is dynamic and harbors considerable amount of intrinsically disordered regions (Fig. S8).27,63 Generation of a reasonable structural model for the complex with high agreement to the cryo-EM data (cross-correlation scores ranging from 0.61–0.88 for the components) (Fig. 2C) required an effective protocol involving diverse and powerful computational approaches.
Architecture of the human SF3b complex
From the pseudo-atomic model derived in this study, we provide structural information for 5 proteins (SF3b10, SF3b155, SF3b145, SF3b130 and SF3b14b) and localization information for 4 proteins (SF3b10, SF3b145, SF3b130 and SF3b14b) for the first time. For the remaining proteins in the SF3b complex, this information is already available from earlier work.20 Learning on the 5 proteins obtained using our integrative structure modeling protocol (Fig. 2B and Fig. S9–S11, S13–S14, S19–S20) is described below in further detail.
SF3b14b – a triquetra knotted structure
The SF3b14b structure has been modeled based on the NMR structure of a yeast homolog of Rds3p, which shares nearly 56% sequence identity (Table S1). A triquetra knotted-like structure containing 3 zinc clusters at each of the 3 vertices, as in Rds3p,64 is conserved in the SF3b14b (Fig. S11). This model has been localized to a peripheral region of the density in the cryo-EM map (Fig. 2B and Fig. S6, S11). We find that such a fitting resulted in the least deviation of mass expected from the density region (11.4 kDa) and the theoretical mass of the modeled protein (12.4 kDa). The yeast homolog of SF3b14b, Rds3p, is known to lack nucleic acid binding capacity.64 However, a point mutation in the Rds3p protein weakens Rse1p (yeast homolog of SF3b130) association with yeast U2 SF3b,65 showing that it is likely to mediate protein-protein interactions, as was also proposed earlier.64 A similar role can be envisaged for SF3b14b as its location is proximal to SF3b130.
SF3b130 – a highly flexible structure with 3 7-bladed WD40 β propellers and a C–terminal helical domain
The structure of SF3b130 has been modeled using a DNA Damage-binding protein 1 (Table S1 and see Materials and Methods) as template. To understand the conformational variability of the SF3b130 that leads to differences in the shape features of the localized density and the modeled structure, a comparative structural analysis was performed. This analysis involved comparisons of 31 structures of the DNA Damage-binding protein 1, often in complex with several proteins (Table S4). Further, clustering of these template structures, using an RMSD cutoff of 5 Å, resulted in 5 clusters showing high conformational variability with a maximum variation of about 21.4 Å among 2 inter-cluster proteins (Fig. S12, Video S2). We then modeled the SF3b130 sequence on these structures and performed principal component analysis (PCA) and normal mode analysis. The results suggested that one of the 7-bladed β-propeller WD40 domains undergoes large-scale motions coupled with the conformational changes in the rest of the structure (Fig. S12). Therefore, the combination of structural and normal mode analysis on the density map (mode 1) renders support to the localization of this protein (Fig. 2B) in the cryo-EM density map. Additionally, it has allowed us to confidently perform the fitting based on the conformation observed in the density, without over fitting to cryo-EM data (Fig. 2B and Fig. S13). Structures with large deviations, when compared to the density maps, are not uncommon and result from differences in the functional states of the entire complex.66,67 Since the template for SF3b130 interacts with nucleic acids as well as proteins,68 it can be expected that SF3b130 might play a similar role in the SF3b complex, when it is a part of U2 snRNP or U11/U12 di-snRNP.
SF3b155 – a highly curved HEAT repeat protein
SF3b155 is the longest protein (1304 amino acids) among the SF3b components and interacts with other proteins in the assembly (Fig. 1D and Table S1). We have modeled only the C-terminal HEAT repeat region (469–1204 amino acids) of this protein,28 as much of the N-terminal region is predicted to be intrinsically disordered (Fig. S15). Previously, localization of this protein in the SF3b cryo-EM map was achieved based on the observation of a repeating structure.20 Initially, we noted that the curvature of the SF3b155 model is different from the curvature of its density in the cryo-EM map. Hence, we performed a fragment-based fitting protocol to localize the HEAT repeats of this protein into the cryo-EM map followed by flexible fitting (see Materials and Methods and Fig. S14). Further, we observed that many templates corresponding to proteins with HEAT repeats have wide range of curvatures for their HEAT repeat region (Fig. S15 and Table S5). Normal mode analysis on these templates showed that they have varying degrees of flexibility with movements among the segments of the HEAT repeats (Fig. S16).
The observation of movements in the HEAT repeats of SF3b155 templates prompted us to perform a hybrid fragment-based comparative modeling, (Fig. S17) to explore if we could capture the S-like curvature of SF3b155, which was proposed earlier on the basis of the shape of cryo-EM density.29 This involved generation of multiple SF3b155 models of various curvatures, by combining the short-length alignments with diverse template structures (Table S5) and multi-template comparative modeling (see Materials and Methods and Fig. S17). This provided us with a decoy of about 1000 structures and on comparing the SF3b155 density curvature with that of the models generated the S-shaped curvature (in terms of dihedral angles) was not observed in the models, although the models showed a wide range of curvatures (Fig. S17). Large scale changes in curvature are not uncommon for HEAT repeat structures, as has been shown in a number of molecular dynamics studies.69,70 Thus, it is likely that the partly disordered SF3b155 region (HEAT repeats) and its interactions with all other SF3b proteins (Fig. 1D) constrains the HEAT repeat structure, thereby, forcing it to take such a peculiar curvature.71,72 Bias in the fitting of the SF3b155 model to the cryo-EM data is low due to the reasons mentioned above. Hence, our approach of fragment-based fitting and refinement by flexible fitting, has allowed us to assign the HEAT repeats in the density with reasonable confidence and obtain insights into the SF3b155 structure (Fig. S14).
SF3b145 – a stem loop binding protein containing an RNase H-like fold
SF3b145 contains a SAP domain at its N-terminus (sequence region 24–68) as shown by an available NMR structure (PDB ID: 2DO5 - to be published) for this region. Further, this protein contains intrinsically disordered regions interspersed with folded regions, as arrived at from disorder predictions (Fig. S18). The SF3b145 region (361–556 amino acids) is relatively less disordered and it has been modeled using the template 3′hExo (3′-5′ exoribonuclease) (PDB ID: 4L8R Chain B), which has an RNase H-like fold (see Materials and Methods). The SF3b145 model fits well to a density region (cross-correlation - 0.61) (Fig. S3, S19) and this position in the cryo-EM map is in agreement with an earlier experimental observation of its interaction with SF3b49 (Fig. 1D).73
Earlier, it has been reported that SF3b145 can bind to stem-loop IIa of U2 snRNA.74 Hence, we modeled the SF3b145-stem loop RNA complex using the 3′hExo template, which has been solved in a stem-loop RNA bound form (see Material and Methods). It has been shown that the Glu181 to Lys mutation in Cus1 (yeast homolog of SF3b145) suppresses mutations in stem-loop IIa of yeast U2 snRNA.75 The equivalent position of this mutation in SF3b145 is Lys487. It has been proposed earlier that Lys487, being in an α-helical region of SF3b145 (from secondary structure predictions), could interact with the stem-loop RNA.75 Remarkably, we observe the same interaction in the SF3b145-stem-loop RNA complex model (Fig. S19). We have also observed a positively charged surface around the stem-loop binding region in the SF3b145 model (Fig. S19). Further, 3′hExo (template of SF3b145) is believed to bind to the stem-loop RNA by recognizing its shape rather than sequence.76 Hence, it can be speculated that the structure of SF3b145 helps it to bind to both stem-loop IIa of U2 snRNA and U12 snRNA, without the requirement of RNA sequence specificity.
SF3b10 – structural similarity to RNA-binding domain of VP35 protein
SF3b10 is the smallest component of the SF3b complex (86 amino acids). We have used a density-guided de novo structure prediction strategy to model its structure. This was required to help localize its position in the cryo-EM map without depending on the shape information from its structure. Our integrative structure modeling protocol resulted in density regions with no structural information in the cryo-EM map. We chose a density region between SF3b155 and SF3b130 as the location for this protein (Fig. S20), based on the neighborhood information from the SF3b protein-protein interaction network (Fig. 1D). We also observed that this part of the density map formed a separate region upon density segmentation (Fig. 1E). Consequently, from our modeling exercise, which involved using shape information from this density region, we applied our robust scoring scheme (see Materials and Methods) to select the top-scoring model from a decoy of 100,000 models (Fig. S20).
We further searched in the PDB 77,78 using DALI,79 to explore if the fold of the top-scoring model resembled any existing structure. It showed structural similarity to a DnaD domain-like protein (PDB ID: 2I5U Chain A),80 with an RMSD of 3.1Å (Dali Z-score 4.2). Based on the inter-residue contact analysis of the structurally aligned PDB structure and the model, many inter-helix contacts were found to be conserved at structurally equivalent positions (Fig. S21). We also found structural similarity to an RNA-binding domain of the VP35 protein from Reston ebolavirus 81 (PDB ID: 3KS8 Chain A) with an RMSD of 3.0 Å (Dali Z-score 4.2). This structure has been solved in dsRNA-bound form. It was observed that the RNA interacting residues Gln263 and Lys271 of VP35 are structurally equivalent to Arg63 and Lys71 of the SF3b10 model (Fig. S21). Additional analysis of the electrostatic surface potential of the SF3b10 model showed the existence of a positively charged surface around the residues Arg63 and Lys71 (Fig. S21). Hence, this region could form the RNA-binding surface of SF3b when it integrates into U2 snRNP or U11/U12 di-snRNP. Interestingly, it has also been reported that residues at positions 33 to 85 are important for the functioning of Rcp10p (yeast homolog of SF3b10).82
Hence, our structural models show that the human SF3b complex comprises of a multiplicity of folds (Table S1 and Fig. S2), including previously known RRM-binding proteins (p14 and SF3b49). This information is crucial in understanding the functional and mechanistic roles of individual components. Our model also serves to provide structural insights and suggests a mechanistic hypothesis for the available experimental information on the functioning of SF3b.
Opening of the SF3b complex in the context of U11/U12 di-snRNP
SF3b forms an active and crucial component of the U2 snRNP and the U11/U12 di-snRNP in the U2-dependent and U12-dependent pathways respectively, in metazoans. The U11/U12 di-snRNP cryo-EM map, determined by Golas et al.,21 showed that the integration of human SF3b complex into the larger di-snRNP, involves a large structural rearrangement resulting in a relatively more open conformation. In the previous work, localization information at the protein level was restricted to only the RRM-domain containing proteins of U11/U12 di-snRNP.21
Knowledge of the molecular architecture of the closed SF3b complex has enabled us to successfully model the components of the SF3b into the U11/U12 di-snRNP density map (Fig. 3). We used iterative segmentation of the SF3b density map (closed form) to obtain a set of parsimonious density fragments (13 fragments; Fig. S22). These fragments were then localized and fitted into the U11/U12 di-snRNP map (13.4 Å at FSC0.5) based on the relative positions of the components, as in the closed SF3b complex (Fig. 3A). This enabled us to assess the local cross-correlation (Fig. 3B) across the densities between the fragmented SF3b and U11/U12 di-snRNP. Further, it encouraged us to fit the structural models for all the 7 components of the SF3b (Fig. 3C and Fig. S23). This model provided us with structural details for the open form of SF3b complex (Video S3).
Figure 3.
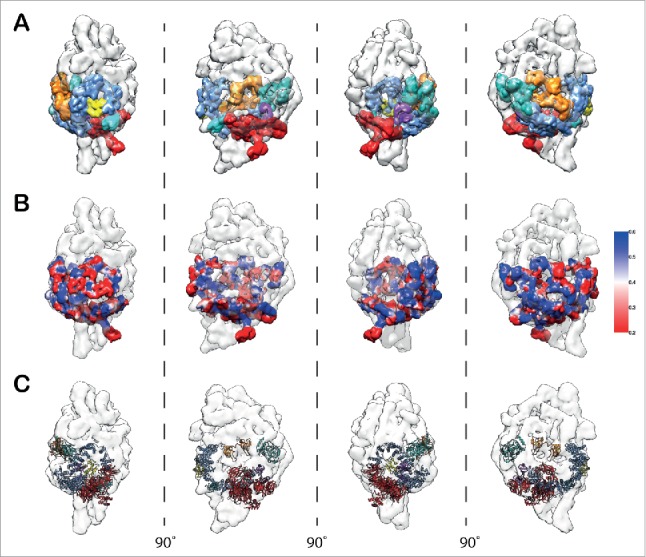
Modeling of SF3b components in the context of U11/U12 di-snRNP. (A) Segmented densities derived from SF3b complex (closed form) fitted into the 13.4 Å U11/U12 di-snRNP cryo-EM map (Fig. S22 and Supplementary Materials and Methods). The various colors represent the 7 SF3b components. (B) Confidence assessment of segmented SF3b densities by local cross-correlation. The local cross-correlation was calculated for each voxel between the fitted density of SF3b (9.7 Å) and the experimental density map of U11/U12 di-snRNP (13.4 Å) with a grid size of 5×5 ×5 Å. (C) Atomic level representation of SF3b components after fitting into the U11/U12 cryo-EM map (Fig. S23 and Video S3).
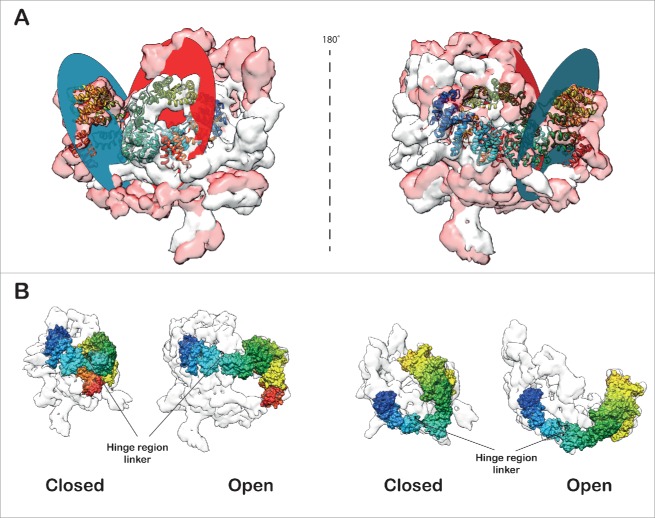
Comparison of the open and closed forms (Video S4) of the SF3b density revealed large structural changes (cross-correlation: 0.46). This structural rearrangement is caused primarily by a large segment of the density comprising mainly of the protein SF3b155. The localization and relative orientation of this segment in the U11/U12 di-snRNP appears to demarcate the open from the closed form (Fig. 4). We have also identified a flap, primarily comprised of SF3b155 HEAT repeats and SF3b10 (see Materials and methods). We find that the curvature of a large segment of the HEAT repeat around SF3b10 enables it to make extensive contacts with this protein. We speculate that this interaction network maintains the integrity of the flap and stabilizes it (Fig. S24). Indeed, this corroborates with earlier reports that show the yeast homolog of SF3b10, Rcp10p interacts with Hsh155p (yeast homolog of SF3b155) and depletion of Rcp10p affects the stability of yeast SF3b complex.82 Further, modeling of the closed and open forms of SF3b pseudo-atomic model has allowed us to determine the scale of the flap motion, in terms of the conformational changes in SF3b155 HEAT repeat structure. The flap undergoes roughly 54Å shift (Centroids of the planes in closed and open conformation of the SF3b155 HEAT repeat structure), which involves movement of the plane of the flap by 65˚ (Fig. 4A; Video S5 and S6). This movement is caused by a hinge motion at the linker region (Fig. 4B) in between the HEAT repeats 6 and 7 of SF3b155. This motion exposes p14 and increases the volume thereby, facilitating the accommodation of snRNA and other proteins in the interior of the U11/U12 di-snRNP complex.
Figure 4.
Open and closed forms of SF3b complex. (A) The open and closed forms of SF3b complex differ remarkably from each other. The main distinction is observed in the SF3b155 protein (rainbow-colored) which forms the flap. The flap opening (∼65˚ rotation) is dictated by the 12 residue linker (-LMGCAILPHLRS-) segment (black loop) in between HEAT repeats 6 and 7 which acts as a hinge (674–680; -LMGCAIL-) enabling substantial conformational change (Flap is translated by 12.8 Å). The closed and open forms were aligned (Video S4) to maximize the overlap (Cross correlation: 0.46) and (B) translated by 200 Å along X-axis to show the difference in flap conformations. The circular discs define the plane of the flap in the open and closed forms of SF3b (Video S5 and S6).
Earlier studies have identified multiple cancer causing mutations on SF3b155.32,34,83 A mapping of these mutations on the pseudo-atomic model showed that they cluster around 2 regions in the SF3b structure (Fig. S25 and Video S7). These mutations have the potential to affect assembly of either U2 or U11/U12 di-snRNPs. Few of the mutation sites are concentrated near the linker region of SF3b155 and hence can affect the flap opening. This is consistent with the previously proposed bivalve shell opening mechanism.21 The opening of the SF3b complex exposes both sides of the flap to other trans-acting factors, including RNA, thus enabling U2 snRNP remodeling.33 This also brings into focus, the RNA binding capacity of U2 or U11/U12 di-snRNP.11
U12 snRNA and branch point duplex binding to SF3b open form
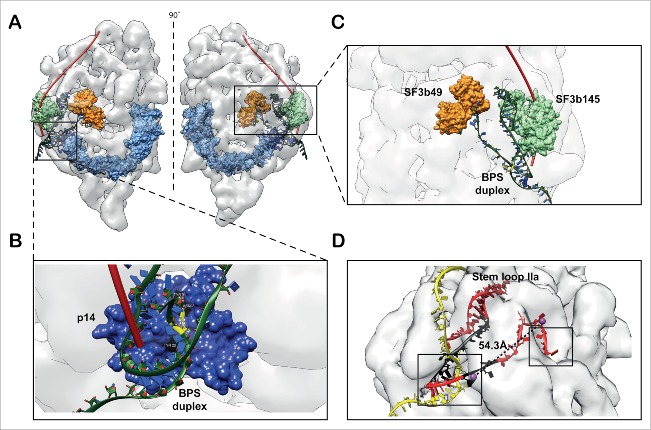
It is well known that the closed form of SF3b is generally stable and does not contain RNA molecules. SF3b integration into U11/U12 di-snRNP involves a complex sequence of rearrangements.21 Knowledge of the open form structure of SF3b in the context of U11/U12 di-snRNP sheds light on the interpretation of a wealth of biochemical data involving RNA binding. p14 is exposed in the open form and interacts with the branch point adenosine (BPA) of the branch point sequence (BPS) in pre-mRNA. This sequence forms a duplex structure with the branch point recognition sequence (BPRS) of U2 snRNA or the U12 snRNA. Hence, we modeled the branch point duplex (BPD) containing U12 snRNA and pre-mRNA, including the BPA, using an available crystal structure of BPD (PDB ID: 1I9X) 84 and prior knowledge of U12 snRNA: pre-mRNA base pairing.85 The information on the recognition of BPA by p14 was collated from multiple sources 9,30 and the p14-BPD complex structure was modeled using a restrained docking procedure (see Materials and Methods). Further, we found that the p14-BPD docked complex structures fitted snugly into the groove above p14 density surface, when fitted in the U11/U12 di-snRNP cryo-EM map. The 5′ end of pre-mRNA also traced a path similar to the one proposed along the longitudinal groove.21 Additionally, we have learnt from cross linking experiments 12 that the 5′ end of the pre-mRNA interacts with SF3b49 (orange) and SF3b145 (green) along the length of pre-mRNA and the duplex is formed by only a few nucleotides along the 3′ direction (Fig. 5). This information also substantiates the directionality of the pre-mRNA obtained from the current model (Fig. 5A).
Figure 5.
Model for RNA binding. (A) The pre-mRNA binds to the SF3b complex in the context of U11/U12 complex. The pre-mRNA (red tube) traces a path on the surface of the U11/U12 di-snRNP to form a duplex with U11/U12 di-snRNA near the base, where the branch point adenosine is recognized by the (B) exposed p14 (open form of SF3b). (C) The U12 snRNA occupies substantial mass and fills the density of U11/U12 cryo-EM map. Here the U12 snRNA is stabilized by interactions with SF3b145 and SF3b49. The 5′ end of this RNA also forms the part of the branch point duplex. The models of the RNA duplex and stem loops were derived from existing crystal structures. (D) The 5′ end of U12 snRNA (red) and pre-mRNA (yellow) is shown. The black colored regions in RNA correspond to the spacers in the earlier deletion analysis.87 The spheres correspond to the centroids of the nucleotides 4 and 16 of U12 snRNA. The dotted lines show the spacing between the nucleotides with a distance of 54.3 Å and a close agreement with earlier biochemical studies.85 The boxes show the exposed U12 snRNA regions where it interacts with 5′ splice site and branch point adenosine of pre-mRNA. The pre-mRNA (yellow strand) that interacts with U12 snRNA and forms a duplex (boxed region) is modeled on a U2 snRNA: pre-mRNA crystal structure duplex, the rest of the strand is modeled to show a speculative path.
Agreement with biochemical data encouraged us to model the U12 snRNA and examine the role of SF3b components in BPD formation. Earlier, cross-linking and mutational experiments have shown that SF3b49 and SF3b145 interact with stem-loop I (SLI) and stem-loop IIa (SLIIa) of U2 snRNA, respectively.11,75,86 The 5′ end of U2 and U12 snRNAs folds into similar secondary structures, due to the presence of intramolecular stem-loops SLI, SLIIa and SLIIb.87 Thus, we modeled the U12 snRNA and its interactions with the SF3b49 and the SF3b145 proteins, using fragments of RNA, from known homologous protein-RNA complexes (see Materials and Methods). The spatial positioning of SF3b49, SF3b145 and p14-BPD complex in the U11/U12 cryo-EM map guided the RNA modeling (Fig. S26) which was difficult due to the uncertainties in the demarcation of RNA and protein regions in the density map. Our model is purely guided by the architectural organization of the SF3b open form, resulting in a model of the SF3b open form-RNA complex structure. The structure of the RNA until stem-loop IIa of U12 snRNA, including BPD with the pre-mRNA, could be successfully modeled. This model provided structural insights into protein-RNA interactions (Fig. 5 and Fig. S27). Furthermore, the availability of this model has also allowed us to localize the potential Sm complex positions of U12 snRNP into the U11/U12 di-snRNP density. This localization was again based on a global fitting process and the use of biochemical data on Sm complex-RNA interactions, which showed that the Sm site is present downstream of the stem-loop IIa of U12 snRNA (Fig. S28).87,88
Discussion
Our integrative structure modeling efforts has shed light on the detailed molecular architecture of the human SF3b complex. This work has also enhanced the structural coverage of the U11/U12 complex and helps us to understand the flap opening mechanism of SF3b 21 to accommodate RNA and other trans-acting factors. Our model is built from an integration of a vast amount of existing experimental data coupled with state-of-the-art structural modeling and cryo-EM density fitting methods. It has allowed us to establish a substantial amount of structure-function relationships of the SF3b complex, including branch point adenosine recognition of pre-mRNA, U12 snRNA binding and localization of several other proteins. Our model also allowed us to interpret several pieces of experimental data.
SF3b – a fuzzy complex
SF3b contains proteins with highly flexible folded regions along with intrinsically disordered regions (IDR) (Table S1 and Fig. S2, S8), hence it is a “fuzzy” complex.89 Such fuzziness in multi-protein complexes has also been attributed to the accommodation of a diverse array of heterogeneous folds, as is the case in the SF3b.90 As a consequence, though we were able to assign much of the cryo-EM density to folded regions, there are additional density regions which lacked any structural data to begin with. In this work, we speculated that the region of density located between the proteins SF3b49, SF3b145 and SF3b155 corresponds to the IDR of these SF3b components. Thus, our analysis shows that majority of the intrinsically disordered regions of SF3b components are concentrated to one side of the SF3b cryo-EM map (Fig. 6).
Figure 6.
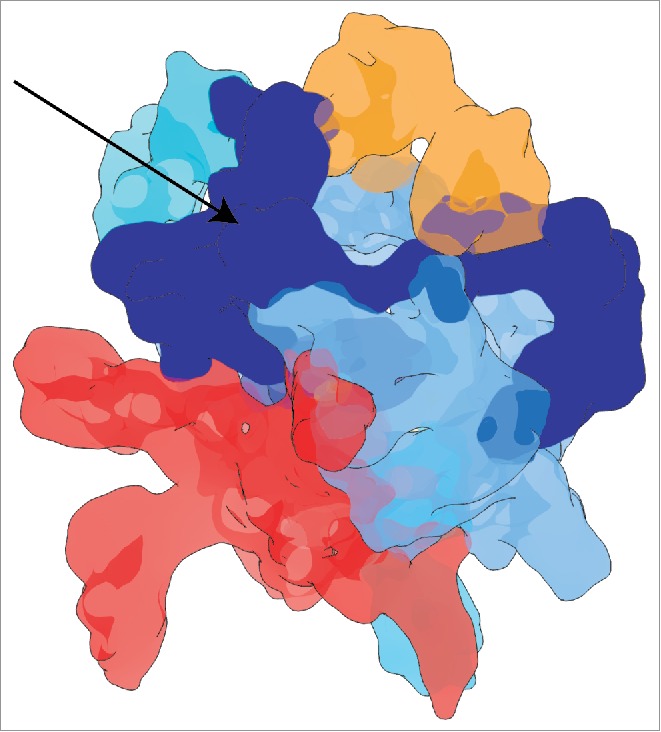
Intrinsically disordered region in the SF3b cryo-EM map. The blue colored density region and the arrow shows the mapping of the intrinsically disordered region to one side of the SF3b cryo-EM density map. As SF3b components harbor intrinsically disordered regions, this density region has been obtained after fitting most of the structured regions into the density map.
We performed an ensemble structural modeling of the SF3b49 IDR (211–424 amino acids), as it has a continuous disordered region and is predicted to interact with other proteins (Fig. S29). This involved generating 500,000 sterically plausible decoys by performing extensive conformational sampling in a probabilistic Ramachandran angle phase space (see Materials and Methods). SF3b49 IDR is proline-rich (83 out of 213 residues) and polyproline II (PPII) helices are known to possess molecular recognition features (MoRFs) 91 and nucleic acid binding properties.92 Hence, we calculated the conformational propensities for these generated conformers in the PPII region of the Ramachandran map. Some of the residues (85 out of 213 residues) have high propensity for PPII conformation with a significant proportion being proline, (38 out of 85 residues) indicating the importance of this IDR region in either protein or nucleic acid binding (Fig. S30). Further, we used a density region C-terminus to the SF3b49 RRM-domains to identify spatial orientation of the model which matched the shape of the identified density. Interestingly the top hits obtained based on cross-correlation values (0.76–0.79) enclosed the density volume quite well despite being intrinsically unstructured (Fig. S30).
Further, it is interesting to note that the yeast homolog of SF3b49, Hsh49p does not contain a proline-rich region (Fig. S29) and it is likely that SF3b49 has evolved a proline rich region to help it to recruit many binding partners possibly via a “fly-casting” mechanism or through motif-based interactions, as generally seen in proteins containing IDRs.93 Similarly, SF3b145 also contains a higher proportion of IDRs when compared to the yeast homolog Cus1p (Fig. S18). In a recent study, both these proteins were shown to be involved in interactions with multiple proteins in the human spliceosome.16 Also, the SF3b complex in the human (30.5%) has more intrinsically disordered content than in the yeast homolog (15.54%), implying more fuzziness for the human SF3b (Fig. S3). We believe that this fuzziness of the human SF3b has direct consequences in the recruitment of several new factors. This also explains the difference in the number of essential factors involved in yeast and human splicing machinery.
The mapping of IDR to one side of the complex can serve as a potential guide for future crystallization trials focusing on individual sub-complexes or protocols, with the possible use of in situ limited proteolysis strategy, as seen in a U1 snRNP structure determination.94 Besides, the strategy of large scale conformational sampling and density-guided filtering can also be used to obtain predictive atomic resolution descriptions of the intrinsically disordered regions for other spliceosomal proteins.95 Therefore, the compositional bias provided by the highly disordered and partly disordered regions (SF3b155 HEAT repeats) and highly flexible components provides the necessary fuzziness to human SF3b. This seems to facilitate both for its integration into U11/U12 di-snRNP 21 and also in the rewiring of the protein-protein interaction networks in the spliceosome.16
SF3b open form – a scaffold for 5′end of U12 snRNA
SF3b exists in an open form in the U11/U12 di-snRNP which binds to pre-mRNA stably; hence it comes in contact with both U12 snRNA and pre-mRNA. From our model of the SF3b open form-RNA complex, we provide a structural view point to some of the observations made in earlier biochemical studies. Previously, a combination of deletion analysis and RNA cross-linking studies have reported that the U12 snRNA interacts with both the 5′ splice site and branch point sequence, within an estimated distance of about 40 to 50 Å between nucleotides 4 and 16 of U12 snRNA.7,85 We measured the distance between these nucleotides in our U12 snRNA structure model. Remarkably, it has a close agreement (∼54.3 Å) with the earlier observation (Fig. 5D). This prompts us to speculate that the SF3b open form acts as a “scaffold” for the 5′ end of U12 snRNA, which is topologically consistent with the biochemical data.85 Furthermore, it is important to note that the U12 snRNA: pre-mRNA structure was modeled purely guided by the structural constraints provided by the spatial positioning of proteins SF3b49, SF3b145 as well as p14 (interaction with branch point duplex). Therefore, these proteins, especially SF3b49 and SF3b145, can possibly act as RNA chaperones 96,97 for the U12 snRNA to take up the right tertiary structure, analogous to that observed in the case of ribosomal proteins for rRNA.98-100
Earlier, extensive mutational and deletion analysis of the SLI (14–17 nucleotides) and single stranded linker region (24–31 nucleotides) of U12 snRNA have shown that they affect splicing. So these regions have been implicated to be spacer elements or distance constraints in the functional form of U12 snRNA, such as during its interaction with pre-mRNA.87 We note a similar role for these nucleotides from our RNA model, as they seem to connect the branch point duplex to SLI and SLII regions of U12 snRNA (Fig. 5D). Loss of splicing activity was also reported upon deletion of the stem loop IIa of U12 snRNA.87 From our SF3b open form-RNA model, we observe that this is primarily due to the loss of U12 snRNA interaction with SF3b145, thus causing some changes in the U12 snRNA structure (Fig. 5B). Further, the stem loops of the U12 snRNA might help in binding of SF3b proteins such as SF3b145.101
The three RNA-binding proteins p14, SF3b49 and SF3b145 face the inner layer of SF3b155 HEAT repeats in the SF3b complex. Hence, it is likely that the binding of the U12 snRNA, coupled with the phosphorylation of the partly disordered protein SF3b155,28 acts as a trigger for the SF3b to undergo such a huge conformational change during its integration into the U11/U12 complex. Moreover, as seen in some of the HEAT repeat containing homologues of SF3b155 (Table S5, Figs. S15 and S16) and from the repertoire of curvatures observed in our hybrid fragment-based comparative modeling (Fig. S17), this protein has the ability to act as a “molecular sling shot” in the SF3b complex, utilizing its stored elastic potential energy as a wound spring during the transformation of SF3b from closed to the open state.69,72
SF3b open form – potential implications for branch point adenosine recognition fidelity
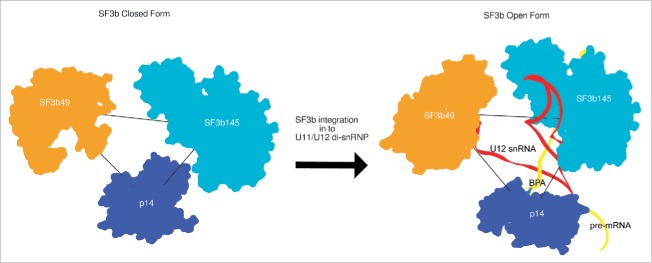
In metazoans, the spliceosome machinery faces an overwhelming task to achieve fidelity given the degeneracy of splice sites, branch point sequence and varying length of introns.102,103 Therefore, in the light of our SF3b open form - RNA complex model we provide insights into the branch point adenosine recognition fidelity, facilitated by its structural organization. As the SF3b open form likely acts as a structural scaffold for 5′ end of U12 snRNA, we believe that it helps in presenting the branch point recognition sequence (BPRS) of U12 snRNA to pre-mRNA branch point sequence (BPS). In addition, from our model and the U11/U12 di-snRNP cryo-EM map, we observe a cleft above p14 and near SF3b49, which can potentially help the U12 snRNA to form RNA-RNA interactions, both with the branch point and 5′ splice site (Fig. 5D). Besides, the thermodynamic stability of this interaction 104 is vital for proper splicing of U12-type pre-mRNAs in the absence of a polypyrimidine (Py) tract and increased distance between the degenerate 3′ splice sites from the branch point.105 Therefore, it can be speculated that the proteins SF3b49, SF3b145 as well as p14 act as “molecular rulers” to improve the fidelity of branch point adenosine selection both by stabilizing and proof-reading the U12 snRNA: pre-mRNA interaction (Fig. 7). We compared the centroid positions of these proteins in both the SF3b closed form and SF3b open form-RNA complex and observed changes both in terms of orientation and distances (Fig. 7). These changes can be attributed to the malleability provided by the mapped IDR region in the SF3b cryo-EM density (Fig. 6) and also from a higher degree of fluctuation in the vicinity of p14 (mode 4), from normal mode analysis of the density map (Fig. 2A).
Figure 7.
Molecular rulers of U12 snRNA-pre-mRNA interaction in closed and open forms. The three spatially juxtaposed proteins SF3b49, SF3b145 and p14 in SF3b complex are shown. The closed and open forms of these proteins were aligned with respect to the SF3b145 position. On comparing the distance between the centroid positions for these proteins in SF3b closed and open forms we observe that for SF3b49-SF3b145 it reduced from 74.8 Å to 70.5 Å, p14-SF3b49 it increased from 62.4 Å to 73.1 Å and for SF3b145-p14 it reduced from 61.9 Å to 50.3 Å. This shows significant differences in the arrangement of these proteins on U12 snRNA binding also stabilizing its interaction with pre-mRNA.
SF3b proteins interact with the 5′ part of U2 snRNA and also help in recruiting U2 snRNP to the highly degenerate BPS.11,74 The role of SF3b in the remodeling of U2 snRNP and the presentation of BPRS has been well studied using the anti-tumor drugs E7107.33 More specifically, Spliceostatin A (SSA), by binding to SF3b, was shown to inhibit the transition of the pre-spliceosome to B complex 106 and reduces the fidelity of BPA recognition.107 We believe that the architectural design of SF3b has a crucial role to play in improving the fidelity of BPA recognition even in the U2 splicing pathway (Fig. S27), analogous to its role in the U11/U12 di-snRNP. SSA is also known to bind to SF3b130 and SF3b155 34 and based on our SF3b pseudo-atomic model (Fig. 2B), we conjecture that these interactions might lock the SF3b130-SF3b155 interaction (Fig. S27), thereby disabling the proper exposure of SF3b proteins (SF3b49, SF3b145 and p14) for interaction with U2 snRNA and pre-mRNA. As BPS in U2-type introns is known to be degenerate,108 any small perturbation in the SF3b integration into U2 snRNP might cause non-specific base pairing of U2 snRNA with pre-mRNA by preventing proper exposure of BPRS.107 We believe that this locking mechanism causes the non-specific base-pairing.
It is also interesting to note that the ortholog of p14 is absent in yeast (S. cerevisiae) as well as in the parasitic organism G. lambia which has very few introns.43,109 Given the fact that yeast has a strong consensus sequence around the branch point adenosine when compared to that of metazoan introns, there is a special requirement for specific factors (p14) to maintain specificity for degenerate BPA recognition in metazoans. Moreover, p14 alone has been shown to lack specificity to bind the branch-point duplex (BPD).110 This information enables us to conjecture that the snRNA-binding proteins SF3b49, SF3b145 and their geometrical arrangement relative to p14 has a vital role in p14 recognizing BPA. This spatial arrangement of the proteins (Fig. 7) might also enable the spliceosome to achieve the right catalytic orientation for splicing, 85,111 as observed in the recent near atomic structure of the yeast spliceosome where much of the catalytic center is shaped by protein components.59 Finally, this arrangement might also aid in the attack of BPA at the 5′ splice site in a diffusion limited regime by limiting pre-mRNA motions in the local vicinity of the spliceosomal proteins.112 Our model shows that SF3b components are imperative for spliceosome to achieve branch point adenosine recognition fidelity.
In conclusion, using an integrative approach, primarily guided by the cryo-EM map, we have deciphered the molecular architecture of the human SF3b machinery in the closed and open forms as well as its contribution for the branch point adenosine recognition. To our knowledge, this is the first such work in the field of splicing where the structure of a “fuzzy” complex such as the SF3b has been obtained using an integrative structure modeling approach. We have tested the limits of many modeling techniques and show convincingly that such a strategy can guide structural modeling of other dynamic snRNPs involved in the spliceosome machinery. Our work also shows the difficulties in obtaining structural information for the spliceosome. Clearly, existing structural data guided by evolutionary principles and experimental restraints can help in discerning the structural organization of this highly dynamic and malleable machine.113 Our work has the potential to initiate and design single molecule experiments on SF3b.1 This would not only provide credence to our views on structural insights into the assembly of the U2 snRNP or U11/U12 di-snRNP, especially with respect to integration of the SF3b into these complexes, but also establish structure-function relationships of this complex machine.
Materials and methods
We have used a diverse array of computational methods for our integrative structure modeling procedure. Detailed procedures are provided in the Materials and Methods section. Here, we summarize the entire methodology. All the components of the SF3b complex lacking X-ray or NMR structures were modeled and fitted into the cryo-EM density map along with the proteins of known structure. For the proteins with no known structure, we employed diverse relationship detection methods ranging from simple sequence search to sophisticated fold recognition methods. Once a convincing template was identified, we used comparative modeling to model the structure (Table S1). In the absence of related proteins of known structure, de novo modeling using Rosetta approach,114,115 followed by rigorous assessment of the model was performed. To obtain neighborhood information for the components a consensus interaction network was computed by collating experimental interaction data from multiple resources deposited in the BIOGRID 116 (Table S2). To locate the individual domains of SF3b130, SF3b145 and SF3b14b within a region of the SF3b cryo-EM density (EMD-1043) that is unoccupied, a global search was performed with a fine angular sampling of 3°, using the colores program in the SITUS package (see Materials and Methods). The neighborhood information from the Protein-Protein Interaction (PPI) data and normal mode analysis of the density map as well as components was utilized for obtaining the final localization information for all the components. Segger tool 117,118 of UCSF Chimera 119,120 was used for both density discretization as well as interactive density segmentation of the SF3b cryo-EM density at a threshold of 0.0158. Further, local fitting of the component proteins was performed into their corresponding map segments based on the localization information with a 6-dimensional search and an angular step of 3° using the colores program in SITUS package.121,122 The fits were further optimized by incorporating other sources of experimental information (Table S3) or domain orientation information from homologues of known structure with the help of Fit in Map module of UCSF Chimera (Table S5). This was followed by a round of flexible fitting using MDFF 123,124 and refinement into the density for SF3b155, SF3130, SF3b49, SF3b145 and SF3b10. To avoid over fitting of the highly flexible SF3b components (SF3b155 and SF3b130) into the density, additional domain restraints were incorporated into the default MDFF protocol.
For modeling the SF3b open form in the context of U11/U12 di-snRNP, we used EMD-1096. After iterative and parsimonious segmentation of SF3b closed map, we performed fitting of the individual fragments into the larger U11/U12 density to obtain the open form of SF3b complex, using the Fit in Map module of UCSF Chimera. This was followed by assessments and atomic level fitting of the individual components into the density. The open and closed forms were compared with each other after density alignments and atomic level fitting of SF3b155. The flap of SF3b complex was characterized by fitting a plane to Cα atoms in the large segment of SF3b155 spanning the region 675–1204 in both the closed and open forms using UCSF Chimera. The inter-planar angles and the centroids were also computed for the planes defining the open and closed forms. Modeling of RNA interactions to SF3b components was performed for the open form in the context of U11/U12 di-snRNP using a combination of approaches including existing structural data on protein-RNA complexes and available experimental data.11,12,21
Supplementary Material
Acknowledgments
We thank Tom Goddard of UCSF Chimera, Kwok Yan Chan of MDFF, Johannes Soeding of HHSuite, Alexandre M. J. J. Bonvin of HADDOCK and Fabrice Jossinet of Assemble2 for help with the respective softwares. We also thank Dr. Sankaran Sandhya for her critical help in the improvement of the manuscript. We are grateful to the anonymous reviewer for his/her painstaking efforts in reviewing this paper and for some excellent suggestions. This work is supported by DBT/COE grant (BT/01/COE/09/01), Indian Institute of Science - Department of Biotechnology partnership program, Mathematical Biology initiative sponsored by Department of Science and Technology and CEFEIPRA Indo-French collaborative grants. NS is a J C Bose National Fellow.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authors Contributions
R.R., A.P.J. and N.S. designed the research; R.R. designed and performed the computational studies and analyzed the data; R.R. interpreted the data with help from R.M.B; R.R., R.M.B. and N.S. wrote the manuscript. All authors have read and approved the manuscript as submitted.
References
- 1.Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science 2011; 331:1289-95; PMID:21393538; http://dx.doi.org/ 10.1126/science.1198830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sperling J, Azubel M, Sperling R. Structure and function of the Pre-mRNA splicing machine. Structure 2008; 16:1605-15; PMID:19000813; http://dx.doi.org/ 10.1016/j.str.2008.08.011 [DOI] [PubMed] [Google Scholar]
- 3.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3:1-24; PMID:21441581; http://dx.doi.org/ 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet 2002; 36:333-60; PMID:12429696; http://dx.doi.org/ 10.1146/annurev.genet.36.043002.091635 [DOI] [PubMed] [Google Scholar]
- 5.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol 2003; 4:960-70; PMID:14685174; http://dx.doi.org/ 10.1038/nrm1259 [DOI] [PubMed] [Google Scholar]
- 6.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 1996; 84:801-11; PMID:8625417; http://dx.doi.org/ 10.1016/S0092-8674(00)81057-0 [DOI] [PubMed] [Google Scholar]
- 7.Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev 1999; 13:851-63; PMID:10197985; http://dx.doi.org/ 10.1101/gad.13.7.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Will CL, Schneider C, Reed R, Luhrmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 1999; 284:2003-5; PMID:10373121; http://dx.doi.org/ 10.1126/science.284.5422.2003 [DOI] [PubMed] [Google Scholar]
- 9.Schellenberg MJ, Dul EL, MacMillan AM. Structural model of the p14/SF3b155 . branch duplex complex. RNA 2011; 17:155-65; PMID:21062891; http://dx.doi.org/20089683 10.1261/rna.2224411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lardelli RM, Thompson JX, Yates JR 3rd, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 2010; 16:516-28; PMID:20089683; http://dx.doi.org/ 10.1261/rna.2030510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Luhrmann R. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol 2006; 26:2803-16; PMID:16537922; http://dx.doi.org/ 10.1128/MCB.26.7.2803-2816.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev 1996; 10:233-43; PMID:8566756; http://dx.doi.org/ 10.1101/gad.10.2.233 [DOI] [PubMed] [Google Scholar]
- 13.Das BK, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol 1999; 19:6796-802; PMID:10490618; http://dx.doi.org/ 10.1128/MCB.19.10.6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J 2002; 21:4978-88; PMID:12234937; http://dx.doi.org/ 10.1093/emboj/cdf480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacMillan AM, Query CC, Allerson CR, Chen S, Verdine GL, Sharp PA. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev 1994; 8:3008-20; PMID:8001820; http://dx.doi.org/ 10.1101/gad.8.24.3008 [DOI] [PubMed] [Google Scholar]
- 16.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell 2012; 45:567-80; PMID:22365833; http://dx.doi.org/ 10.1016/j.molcel.2011.12.034 [DOI] [PubMed] [Google Scholar]
- 17.Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 2001; 409:539-42; PMID:11206553; http://dx.doi.org/ 10.1038/35054102 [DOI] [PubMed] [Google Scholar]
- 18.Chiu W, Baker ML, Jiang W, Dougherty M, Schmid MF. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure 2005; 13:363-72; PMID:15766537; http://dx.doi.org/ 10.1016/j.str.2004.12.016 [DOI] [PubMed] [Google Scholar]
- 19.Orlova EV, Saibil HR. Structural analysis of macromolecular assemblies by electron microscopy. Chem Rev 2011; 111:7710-48; PMID:21919528; http://dx.doi.org/ 10.1021/cr100353t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science 2003; 300:980-4; PMID:12738865; http://dx.doi.org/ 10.1126/science.1084155 [DOI] [PubMed] [Google Scholar]
- 21.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol Cell 2005; 17:869-83; PMID:15780942; http://dx.doi.org/ 10.1016/j.molcel.2005.02.016 [DOI] [PubMed] [Google Scholar]
- 22.Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-ray crystallography and electron microscopy. Structure 2005; 13:355-62; PMID:15766536; http://dx.doi.org/ 10.1016/j.str.2005.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J Struct Biol 2005; 149:191-203; PMID:15681235; http://dx.doi.org/ 10.1016/j.jsb.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 24.Baker ML, Jiang W, Wedemeyer WJ, Rixon FJ, Baker D, Chiu W. Ab initio modeling of the herpesvirus VP26 core domain assessed by CryoEM density. PLoS Comput Biol 2006; 2:e146; PMID:17069457; http://dx.doi.org/ 10.1371/journal.pcbi.0020146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindert S, Stewart PL, Meiler J. Hybrid approaches: applying computational methods in cryo-electron microscopy. Curr Opin Struct Biol 2009; 19:218-25; PMID:19339173; http://dx.doi.org/ 10.1016/j.sbi.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem 2008; 77:443-77; PMID:18318657; http://dx.doi.org/ 10.1146/annurev.biochem.77.060407.135530 [DOI] [PubMed] [Google Scholar]
- 27.Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res 2012; 40:7046-65; PMID:22573172; http://dx.doi.org/ 10.1093/nar/gks347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev 1998; 12:1409-14; PMID:9585501; http://dx.doi.org/ 10.1101/gad.12.10.1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark H, Luhrmann R. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct 2006; 35:435-57; PMID:16689644; http://dx.doi.org/ 10.1146/annurev.biophys.35.040405.101953 [DOI] [PubMed] [Google Scholar]
- 30.Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Lührmann R, Glover JN, MacMillan AM. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci U S A 2006; 103:1266-71; PMID:16432215; http://dx.doi.org/ 10.1073/pnas.0508048103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwasako K, Dohmae N, Inoue M, Shirouzu M, Taguchi S, Guntert P, Séraphin B, Muto Y, Yokoyama S. Complex assembly mechanism and an RNA-binding mode of the human p14-SF3b155 spliceosomal protein complex identified by NMR solution structure and functional analyses. Proteins 2008; 71:1617-36; PMID:18076038; http://dx.doi.org/ 10.1002/prot.21839 [DOI] [PubMed] [Google Scholar]
- 32.Webb TR, Joyner AS, Potter PM. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov Today 2013; 18:43-9; PMID:22885522; http://dx.doi.org/ 10.1016/j.drudis.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev 2011; 25:440-4; PMID:21363962; http://dx.doi.org/ 10.1101/gad.2009411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 2007; 3:570-5; PMID:17643112; http://dx.doi.org/ 10.1038/nchembio.2007.16 [DOI] [PubMed] [Google Scholar]
- 35.Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 2012; 11:847-59; PMID:23123942; http://dx.doi.org/ 10.1038/nrd3823 [DOI] [PubMed] [Google Scholar]
- 36.Chothia C. Proteins. One thousand families for the molecular biologist. Nature 1992; 357:543-4; PMID:1608464; http://dx.doi.org/ 10.1038/357543a0 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A 2005; 102:1029-34; PMID:15653774; http://dx.doi.org/ 10.1073/pnas.0407152101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golas MM, Sander B, Bessonov S, Grote M, Wolf E, Kastner B, Stark H, Lührmann R. 3D cryo-EM structure of an active step I spliceosome and localization of its catalytic core. Mol Cell 2010; 40:927-38; PMID:21172658; http://dx.doi.org/ 10.1016/j.molcel.2010.11.023 [DOI] [PubMed] [Google Scholar]
- 39.Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Lührmann R. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 2010; 16:2384-403; PMID:20980672; http://dx.doi.org/ 10.1261/rna.2456210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al.. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J 2007; 26:1737-48; PMID:17332742; http://dx.doi.org/ 10.1038/sj.emboj.7601631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Azubel M, Wolf SG, Sperling J, Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol Cell 2004; 15:833-9; PMID:15350226; http://dx.doi.org/ 10.1016/j.molcel.2004.07.022 [DOI] [PubMed] [Google Scholar]
- 42.Boehringer D, Makarov EM, Sander B, Makarova OV, Kastner B, Luhrmann R, Stark H. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat Struct Mol Biol 2004; 11:463-8; PMID:15098019; http://dx.doi.org/ 10.1038/nsmb761 [DOI] [PubMed] [Google Scholar]
- 43.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 2009; 36:593-608; PMID:19941820; http://dx.doi.org/ 10.1016/j.molcel.2009.09.040 [DOI] [PubMed] [Google Scholar]
- 44.Jurica MS, Sousa D, Moore MJ, Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat Struct Mol Biol 2004; 11:265-9; PMID:14981503; http://dx.doi.org/ 10.1038/nsmb728 [DOI] [PubMed] [Google Scholar]
- 45.Ohi MD, Ren L, Wall JS, Gould KL, Walz T. Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc Natl Acad Sci U S A 2007; 104:3195-200; PMID:17360628; http://dx.doi.org/ 10.1073/pnas.0611591104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Luhrmann R, Stark H. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol Cell 2006; 24:267-78; PMID:17052460; http://dx.doi.org/ 10.1016/j.molcel.2006.08.021 [DOI] [PubMed] [Google Scholar]
- 47.Wolf E, Kastner B, Deckert J, Merz C, Stark H, Luhrmann R. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J 2009; 28:2283-92; PMID:19536130; http://dx.doi.org/ 10.1038/emboj.2009.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Lührmann R. Molecular architecture of the human Prp19/CDC5L complex. Mol Cell Biol 2010; 30:2105-19; PMID:20176811; http://dx.doi.org/ 10.1128/MCB.01505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol 2006; 26:5528-43; PMID:16809785; http://dx.doi.org/ 10.1128/MCB.00582-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 2002; 8:426-39; PMID:11991638; http://dx.doi.org/ 10.1017/S1355838202021088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z, Sim J, Griffith J, Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc Natl Acad Sci U S A 2002; 99:12203-7; PMID:12215496; http://dx.doi.org/ 10.1073/pnas.182427099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMullan G, Faruqi AR, Henderson R, Guerrini N, Turchetta R, Jacobs A, van Hoften G. Experimental observation of the improvement in MTF from backthinning a CMOS direct electron detector. Ultramicroscopy 2009; 109:1144-7; PMID:19541421; http://dx.doi.org/ 10.1016/j.ultramic.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol 2012; 415:406-18; PMID:22100448; http://dx.doi.org/ 10.1016/j.jmb.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 2013; 10:584-90; PMID:23644547; http://dx.doi.org/ 10.1038/nmeth.2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 2015; 523:47-52; PMID:26106855; http://dx.doi.org/ 10.1038/nature14548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science 2016; 351:466-75; PMID:26743623; http://dx.doi.org/ 10.1126/science.aad6466 [DOI] [PubMed] [Google Scholar]
- 57.Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature 2016; 530:298-302; PMID:26829225; http://dx.doi.org/ 10.1038/nature16940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-Å resolution. Science 2015; 349:1182-91; PMID: 26292705; http://dx.doi.org/26803803 10.1126/science.aac8159 [DOI] [PubMed] [Google Scholar]
- 59.Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science 2015; 349:1191-8; PMID: 26292707; http://dx.doi.org/26803803 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- 60.Nguyen TH, Galej WP, Fica SM, Lin PC, Newman AJ, Nagai K. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr Opin Struct Biol 2016; 36:48-57; PMID:26803803; http://dx.doi.org/ 10.1016/j.sbi.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim DN, Nguyen CT, Bathe M. Conformational dynamics of supramolecular protein assemblies. J Struct Biol 2011; 173:261-70; PMID:20854912; http://dx.doi.org/ 10.1016/j.jsb.2010.09.015 [DOI] [PubMed] [Google Scholar]
- 62.Chacon P, Tama F, Wriggers W. Mega-Dalton biomolecular motion captured from electron microscopy reconstructions. J Mol Biol 2003; 326:485-92; PMID:12559916; http://dx.doi.org/ 10.1016/S0022-2836(02)01426-2 [DOI] [PubMed] [Google Scholar]
- 63.Coelho Ribeiro Mde L, Espinosa J, Islam S, Martinez O, Thanki JJ, Mazariegos S, Nguyen T, Larina M, Xue B, Uversky VN. Malleable ribonucleoprotein machine: protein intrinsic disorder in the Saccharomyces cerevisiae spliceosome. PeerJ 2013; 1:e2; PMID:23638354; http://dx.doi.org/ 10.7717/peerj.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Roon AM, Loening NM, Obayashi E, Yang JC, Newman AJ, Hernandez H, Nagai K, Neuhaus D. Solution structure of the U2 snRNP protein Rds3p reveals a knotted zinc-finger motif. Proc Natl Acad Sci U S A 2008; 105:9621-6; PMID:18621724; http://dx.doi.org/ 10.1073/pnas.0802494105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Q, Rymond BC. Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol Cell Biol 2003; 23:7339-49; PMID:14517302; http://dx.doi.org/ 10.1128/MCB.23.20.7339-7349.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skjaerven L, Martinez A, Reuter N. Principal component and normal mode analysis of proteins; a quantitative comparison using the GroEL subunit. Proteins 2011; 79:232-43; PMID:21058295; http://dx.doi.org/ 10.1002/prot.22875 [DOI] [PubMed] [Google Scholar]
- 67.Booth CR, Meyer AS, Cong Y, Topf M, Sali A, Ludtke SJ, Chiu W, Frydman J. Mechanism of lid closure in the eukaryotic chaperonin TRiC/CCT. Nat Struct Mol Biol 2008; 15:746-53; PMID:18536725; http://dx.doi.org/ 10.1038/nsmb.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thomä NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 2008; 135:1213-23; PMID:19109893; http://dx.doi.org/ 10.1016/j.cell.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grinthal A, Adamovic I, Weiner B, Karplus M, Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc Natl Acad Sci U S A 2010; 107:2467-72; PMID:20133745; http://dx.doi.org/ 10.1073/pnas.0914073107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zachariae U, Grubmuller H. A highly strained nuclear conformation of the exportin Cse1p revealed by molecular dynamics simulations. Structure 2006; 14:1469-78; PMID:16962977; http://dx.doi.org/ 10.1016/j.str.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 71.Kobe B, Gleichmann T, Horne J, Jennings IG, Scotney PD, Teh T. Turn up the HEAT. Structure 1999; 7:R91-7; PMID:10378263; http://dx.doi.org/ 10.1016/S0969-2126(99)80060-4 [DOI] [PubMed] [Google Scholar]
- 72.Zachariae U, Grubmuller H. Importin-β: structural and dynamic determinants of a molecular spring. Structure 2008; 16:906-15; PMID:18547523; http://dx.doi.org/ 10.1016/j.str.2008.03.007 [DOI] [PubMed] [Google Scholar]
- 73.Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev 1994; 8:1974-83; PMID:7958871; http://dx.doi.org/ 10.1101/gad.8.16.1974 [DOI] [PubMed] [Google Scholar]
- 74.Kramer A, Gruter P, Groning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol 1999; 145:1355-68; PMID:10385517; http://dx.doi.org/ 10.1083/jcb.145.7.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wells SE, Neville M, Haynes M, Wang J, Igel H, Ares M Jr. CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev 1996; 10:220-32; PMID:8566755; http://dx.doi.org/ 10.1101/gad.10.2.220 [DOI] [PubMed] [Google Scholar]
- 76.Tan D, Marzluff WF, Dominski Z, Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science 2013; 339:318-21; PMID:23329046; http://dx.doi.org/ 10.1126/science.1228705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westbrook J, Feng Z, Chen L, Yang H, Berman HM. The Protein Data Bank and structural genomics. Nucleic Acids Res 2003; 31:489-91; PMID:12520059; http://dx.doi.org/ 10.1093/nar/gkg068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. The Protein Data Bank and the challenge of structural genomics. Nat Struct Biol 2000; 7 Suppl:957-9; PMID:11103999; http://dx.doi.org/ 10.1038/80734 [DOI] [PubMed] [Google Scholar]
- 79.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res 2010; 38:W545-9; PMID:20457744; http://dx.doi.org/ 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu R, Zhang R, Bargassa M, Joachimiak A. Midwest Center for Structural Genomics to be published [Google Scholar]
- 81.Kimberlin CR, Bornholdt ZA, Li S, Woods VL Jr, MacRae IJ, Saphire EO. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci U S A 2010; 107:314-9; PMID:20018665; http://dx.doi.org/ 10.1073/pnas.0910547107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Q, He J, Lynn B, Rymond BC. Interactions of the yeast SF3b splicing factor. Mol Cell Biol 2005; 25:10745-54; PMID:16314500; http://dx.doi.org/ 10.1128/MCB.25.24.10745-10754.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al.. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2012; 44:47-52; http://dx.doi.org/ 10.1038/ng.1032 [DOI] [PubMed] [Google Scholar]
- 84.Berglund JA, Rosbash M, Schultz SC. Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines. RNA 2001; 7:682-91; PMID:11350032; http://dx.doi.org/ 10.1017/S1355838201002187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frilander MJ, Meng X. Proximity of the U12 snRNA with both the 5′ splice site and the branch point during early stages of spliceosome assembly. Mol Cell Biol 2005; 25:4813-25; PMID:15923601; http://dx.doi.org/ 10.1128/MCB.25.12.4813-4825.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan D, Ares M Jr. Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol 1996; 16:818-28; PMID:8622683; http://dx.doi.org/ 10.1128/MCB.16.3.818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sikand K, Shukla GC. Functionally important structural elements of U12 snRNA. Nucleic Acids Res 2011; 39:8531-43; PMID:21737423; http://dx.doi.org/ 10.1093/nar/gkr530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benecke H, Luhrmann R, Will CL. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J 2005; 24:3057-69; PMID:16096647; http://dx.doi.org/ 10.1038/sj.emboj.7600765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci 2008; 33:2-8; PMID:18054235; http://dx.doi.org/ 10.1016/j.tibs.2007.10.003 [DOI] [PubMed] [Google Scholar]
- 90.Marsh JA, Teichmann SA. Protein flexibility facilitates quaternary structure assembly and evolution. PLoS Biol 2014; 12:e1001870; PMID:24866000; http://dx.doi.org/ 10.1371/journal.pbio.1001870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs). J Mol Biol 2006; 362:1043-59; PMID:16935303; http://dx.doi.org/ 10.1016/j.jmb.2006.07.087 [DOI] [PubMed] [Google Scholar]
- 92.Hicks JM, Hsu VL. The extended left-handed helix: a simple nucleic acid-binding motif. Proteins 2004; 55:330-8; PMID:15048824; http://dx.doi.org/ 10.1002/prot.10630 [DOI] [PubMed] [Google Scholar]
- 93.Gsponer J, Babu MM. The rules of disorder or why disorder rules. Prog Biophys Mol Biol 2009; 99:94-103; PMID:19344736; http://dx.doi.org/ 10.1016/j.pbiomolbio.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 94.Weber G, Trowitzsch S, Kastner B, Luhrmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 2010; 29:4172-84; PMID:21113136; http://dx.doi.org/ 10.1038/emboj.2010.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schwalbe M, Ozenne V, Bibow S, Jaremko M, Jaremko L, Gajda M, Jensen MR, Biernat J, Becker S, Mandelkow E, et al.. Predictive atomic resolution descriptions of intrinsically disordered hTau40 and α-synuclein in solution from NMR and small angle scattering. Structure 2014; 22:238-49; PMID:24361273; http://dx.doi.org/ 10.1016/j.str.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 96.Semrad K. Proteins with RNA chaperone activity: a world of diverse proteins with a common task-impediment of RNA misfolding. Biochem Res Int 2011; 2011:532908; PMID:21234377; http://dx.doi.org/ 10.1155/2011/532908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem 1995; 270:20871-4 [DOI] [PubMed] [Google Scholar]
- 98.Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA 2004; 10:1855-60; PMID:15525706; http://dx.doi.org/ 10.1261/rna.7121704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kovacs D, Rakacs M, Agoston B, Lenkey K, Semrad K, Schroeder R, Tompa P. Janus chaperones: assistance of both RNA- and protein-folding by ribosomal proteins. FEBS Lett 2009; 583:88-92; PMID:19071121; http://dx.doi.org/ 10.1016/j.febslet.2008.11.049 [DOI] [PubMed] [Google Scholar]
- 100.Kim H, Abeysirigunawarden SC, Chen K, Mayerle M, Ragunathan K, Luthey-Schulten Z, Ha T, Woodson SA. Protein-guided RNA dynamics during early ribosome assembly. Nature 2014; 506:334-8; PMID:24522531; http://dx.doi.org/ 10.1038/nature13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rau M, Stump WT, Hall KB. Intrinsic flexibility of snRNA hairpin loops facilitates protein binding. RNA 2012; 18:1984-95; PMID:23012481; http://dx.doi.org/ 10.1261/rna.035006.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/ 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 103.Fox-Walsh KL, Hertel KJ. Splice-site pairing is an intrinsically high fidelity process. Proc Natl Acad Sci U S A 2009; 106:1766-71; PMID:19179398; http://dx.doi.org/ 10.1073/pnas.0813128106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brock JE, Dietrich RC, Padgett RA. Mutational analysis of the U12-dependent branch site consensus sequence. RNA 2008; 14:2430-9; PMID:18824513; http://dx.doi.org/ 10.1261/rna.1189008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dietrich RC, Peris MJ, Seyboldt AS, Padgett RA. Role of the 3′ splice site in U12-dependent intron splicing. Mol Cell Biol 2001; 21:1942-52; PMID:11238930; http://dx.doi.org/ 10.1128/MCB.21.6.1942-1952.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res 2010; 38:6664-72; PMID:20529876; http://dx.doi.org/ 10.1093/nar/gkq494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev 2011; 25:445-59; PMID:21363963; http://dx.doi.org/ 10.1101/gad.2014311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res 2008; 36:2257-67; PMID:18285363; http://dx.doi.org/ 10.1093/nar/gkn073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprévote O, Séraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 2004; 23:4847-56; PMID:15565172; http://dx.doi.org/ 10.1038/sj.emboj.7600482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Lührmann R, Sattler M. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA 2006; 12:410-25; PMID:16495236; http://dx.doi.org/ 10.1261/rna.2271406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kent OA, MacMillan AM. Early organization of pre-mRNA during spliceosome assembly. Nat Struct Biol 2002; 9:576-81; PMID:12091875; http://dx.doi.org/ 10.1038/nsb822 [DOI] [PubMed] [Google Scholar]
- 112.Rino J, Carvalho T, Braga J, Desterro JM, Luhrmann R, Carmo-Fonseca M. A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput Biol 2007; 3:2019-31; PMID:17967051; http://dx.doi.org/ 10.1371/journal.pcbi.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen W, Moore MJ. The spliceosome: disorder and dynamics defined. Curr Opin Struct Biol 2014; 24:141-9; PMID:24530854; http://dx.doi.org/ 10.1016/j.sbi.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem 2008; 77:363-82; PMID:18410248; http://dx.doi.org/ 10.1146/annurev.biochem.77.062906.171838 [DOI] [PubMed] [Google Scholar]
- 115.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol 1997; 268:209-25; PMID:9149153; http://dx.doi.org/ 10.1006/jmbi.1997.0959 [DOI] [PubMed] [Google Scholar]
- 116.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction data sets. Nucleic Acids Res 2006; 34:D535-9; PMID:16381927; http://dx.doi.org/ 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pintilie G, Chiu W. Comparison of Segger and other methods for segmentation and rigid-body docking of molecular components in cryo-EM density maps. Biopolymers 2012; 97:742-60; PMID:22696409; http://dx.doi.org/ 10.1002/bip.22074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol 2010; 170:427-38; PMID:20338243; http://dx.doi.org/ 10.1016/j.jsb.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605-12; PMID:15264254; http://dx.doi.org/ 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 120.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol 2007; 157:281-7; PMID:16963278; http://dx.doi.org/ 10.1016/j.jsb.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 121.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol 1999; 125:185-95; PMID:10222274; http://dx.doi.org/ 10.1006/jsbi.1998.4080 [DOI] [PubMed] [Google Scholar]
- 122.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev 2010; 2:21-7; PMID:20174447; http://dx.doi.org/ 10.1007/s12551-009-0026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 2008; 16:673-83; PMID:18462672; http://dx.doi.org/ 10.1016/j.str.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 2009; 49:174-80; PMID:19398010; http://dx.doi.org/ 10.1016/j.ymeth.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.