ABSTRACT
Skin and soft tissue infections (SSTI) caused by methicillin resistant Staphylococcus aureus (MRSA) are difficult to treat. Bacteriophage (phage) represent a potential alternate treatment for antibiotic resistant bacterial infections. In this study, 7 novel phage with broad lytic activity for S. aureus were isolated and identified. Screening of a diverse collection of 170 clinical isolates by efficiency of plating (EOP) assays shows that the novel phage are virulent and effectively prevent growth of 70–91% of MRSA and methicillin sensitive S. aureus (MSSA) isolates. Phage K, which was previously identified as having lytic activity on S. aureus was tested on the S. aureus collection and shown to prevent growth of 82% of the isolates. These novel phage group were examined by electron microscopy, the results of which indicate that the phage belong to the Myoviridae family of viruses. The novel phage group requires β-N-acetyl glucosamine (GlcNac) moieties on cell wall teichoic acids for infection. The phage were distinct from, but closely related to, phage K as characterized by restriction endonuclease analysis. Furthermore, growth rate analysis via OmniLog® microplate assay indicates that a combination of phage K, with phage SA0420ᶲ1, SA0456ᶲ1 or SA0482ᶲ1 have a synergistic phage-mediated lytic effect on MRSA and suppress formation of phage resistance. These results indicate that a broad spectrum lytic phage mixture can suppress the emergence of resistant bacterial populations and hence have great potential for combating S. aureus wound infections.
KEYWORDS: bacteriophage, bacteriophage K, multi-drug resistance, Staphylococcus aureus, teichoic acid
Introduction
The rise in number of multi-drug resistant (MDR) microorganisms is a major threat to human health. Currently, it is estimated that at least 2 million infections are caused by MDR organisms every year in the United States leading to approximately 23,000 deaths.1,2,3 Methicillin resistant Staphylococcus aureus is an MDR organism of great concern in the clinical setting. MRSA is responsible for over 80,000 invasive infections and 11,285 related deaths and is the primary cause of hospital acquired infections.2 This organism has been identified in several congressional reports as threat to public health and safety.1 Additionally, the World Health Organization (WHO) has identified MRSA as organisms of international concern.4 In response to this threat, the US government devised a national strategy to combat the spread of multi-drug resistant organisms and promote the development of alternate therapies.1,2
S. aureus are gram positive cocci that can cause skin and soft tissue infections (SSTI), pneumonia, necrotizing fasciitis, and blood stream infections.5-11 S. aureus has developed a mechanism to evade the antimicrobial activity of β-lactam antibiotics by expressing an alternate form of penicillin binding protein (PBP2a) encoded by mecA.12,13 PBP2a has a reduced affinity for β-lactam antibiotics, permitting cell wall synthesis.14 On the basis of susceptibility to β-lactam antibiotics, S. aureus strains are either methicillin sensitive S. aureus (MSSA) or methicillin resistant S. aureus (MRSA). MRSA infections are divided into either health care associated (HA-MRSA) with infection originating within 4 days of hospitalization, or community acquired (CA-MRSA) infection that begins within the first day of admission.15 The USA300 pulse field type is associated with CA-MRSA and is responsible for up to 70% of S. aureus SSTIs in the US, this makes USA300 a prime target for broad spectrum phages.16,17
A potential treatment for MDR infections involves the use of phage. Lytic phage are viruses that infect bacteria, multiply and subsequently lyse the host cell.18,19 The successful therapeutic use of phage depends on the capacity to infect a broad range of pathogenic strains and a low frequency of resistance. Like bacterial resistance to antibiotic treatment, bacterial resistance to phage can develop. However, this can be reduced by utilizing a mixture of virulent phages that target diverse cell surface receptors.20,21 A candidate for combating S. aureus MDR infections is the polyvalent bacteriophage K, a well characterized member of the Myoviridae virus family.22-25 Phage K requires cell wall teichoic acid (WTA) moieties for efficient absorption into the host cell.26 WTAs are glycopolymers involved in cell wall formation and resistance to cationic antimicrobial agents.27-31 In S. aureus, most strains WTAs contain polyribitol phosphate (RboP) substituted by D-alanine and N-acetylglucosamine (GlcNAc) linked via α and β glycosidic bonds.32-34 In S. aureus, biosinthesis of WTAs commences by linking a GlcNAc moiety to undecaprenol lipids a reaction mediated by the N-acetyl glucosamine transferase, tarO. The nascent glucopolymer is modified by addition of α and β linked GlcNAc moieties a reaction catalized by tarM and tarS respectively.29,35 Upon completion, WTAs are exported to the extracellular environment where it is linked to peptidoglycan (PG).
In search for alternate treatments for MRSA infections, 7 lytic phage were isolated from sewage samples. These phage are polyvalent in nature individually affecting 70% to 91% of 170 S. aureus clinical isolates. Additionally, we demonstrate that phage K was virulent in 82% of the isolates. The combination of phage K and the novel phage have a synergistic effect on bacterial growth inhibition. These results indicate that a phage mixture can enhance growth inhibition effects of phage in S. aureus and reduce the formation of phage-resistance. Electron microscopy studies show that all novel phage are members of the Myoviridae virus family. Also, the β-N-acetyl glucosamine covalent modification of teichoic acids was identified as required for lytic activity of phage SA0414ᶲ1, SA0414ᶲ2, SA0420ᶲ1, SA0456ᶲ1, SA0470ᶲ1, SA0482ᶲ1 and SA11987ᶲ1, suggesting teichoic acids are the main receptor.
Materials and methods
Bacterial strains and growth methods
The S. aureus clinical isolate collection was available at NMRC-Silver Spring. MRSA strains were isolated from hospital staff, outpatients, and inpatients over a 5-year period. Strains with the prefix NSC or NSI were collected at the Naval Medical Research Unit-6 (NAMRU-6) in Lima and Iquitos, Peru, respectively. Community acquired S. aureus isolates were collected from the community hospital at Fort Benning, GA. Strains were grown at 37°C in Tryptic soy broth (TSB). Solid media contained 1.5% (wt/vol) bacteriological agar (BD).
Bacteriophage isolation
Phage K samples were purchased from the American Type Culture Collection (ATCC19685-B1) and propagated in Staphylococcus hyicus. The novel phage group were isolated from untreated sewage obtained from the municipal water treatment plant at Frederick, MD. Three 500 ml batches of influent sewage water were collected and fortified with 15 g of TS, 1 mL of each MRSA strain (NSC0414, NSC0420, NSC0456, NSC0470, NSC0482, NSC0409 and NSC0419) and incubated for 24 h at 37°C. Following incubation, 1 ml was centrifuged for 5 min at 8000 g and the the supernatant sterilized by 1 min of centrifugation at 8000 g with a 0.22 µm microfuge filter (Costar® Cat#8160). A total of 10 µl of filtrate were screened for lytic phage via plaque assays using the soft agar overlay technique.36 Plates were incubated at 37°C for 24 h before scoring for the presence of plaques. Plaques were purified 3 times by removing plugs and overnight elution in SM buffer (50 mM Tris pH 7.5, 100 mM NaCl, 8 mM MgSO4 and 0.01% gelatin) followed by amplification in the host strain. Phage titer was assessed by plating 10-fold serial dilutions and calculating the plaque forming units per ml (PFU).
Host range analysis and phage comparison
The host range of the novel phage group and phage K was examined via an efficiency of plating assay (EOP). . 3.5 µl of a 10-fold dilution series starting at 1×108 PFU/ml were spotted on TSA seeded with 800 µl of bacteria at 0.6 OD600.37 Plaque formation at any dilution is considered an indication of phage virulence. Phage species specificity was tested by spotting 10μl of 1×108 PFU/ml on bacterial lawns of S. cohnii, S. epidermitis, S. haemoliticus, S. saprophiticus, S. sciurii, S. xylosus and S. hyicus. A clear area was considered indicative of virulence.
Phage purification
The corresponding S. aureus host strain was grown to 0.1 OD600 at 37°C. Cells were infected with phage at a multiplicity of infection (MOI) of 0.5 and incubated at 37°C until the culture was clear. The lysate was cleared via centrifugation at 10,000 g for 10 min and 360 g (10% w/v) of polyethylene glycol 8000 (PEG) was added to the supernatant and precipitated overnight at 4°C. The solution was centrifuged at 5,000 g for 1 h, the supernatant decanted, and the pellet resuspended in 5 ml of SM buffer. Next, 0.75 g of Cesium chloride per ml of precipitate was added and mixed by inversion. The sample was centrifuged on a 90 Ti rotor at 58,000 g at 4°C for 24 h. The resulting band was retrieved and dialyzed by using a 10,000 Da MWC Slide-A-Lyzer® dialysis cassette (Pierce, cat#66373) in 4 L of dialysis buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl). After 24 h the dialysis buffer was exchanged and dialyzed for 4 h. Phage was collected from the dialysis cassette and titered.
Phage DNA purification
Purified phage (150 µl) was mixed with 6 µl of 0.5 M EDTA pH 8, 10 µl of Proteinase K (20 mg/ml) and 7.5 µl of 10% SDS and incubated for 1 h at 56°C. Once cooled to room temperature, 3 extractions with 150 µl of Tris-EDTA saturated phenol pH 7.0 and 3 extractions with 150 µl of Chloroform: Isoamyl alcohol 24:1. The supernatant was mixed with 10 µl of 3 M Sodium acetate and 1 ml of 100% ice cold ethanol overnight at −20°C. The DNA sample was centrifuged at 10,000 g for 5 min and the pellet washed 3 times with 750 µl of 70% isopropanol. The pellet was air dried for 5 min and resuspended in 50 µl of distilled water. The DNA concentration was determined by reading OD260 on a spectrophotometer.
Pulse-field gel electrophoresis (PFGE)
Purified phage DNA (20 µg) was digested with EcoR1 restriction endonuclease for 3 h at 37°C. The digested samples were loaded into a 1% pulse field agarose gel prepared with 0.5X TBE and resolved for 11 h at 14°C with a pulse rate of 30 V/s and a switch time of 1–6 seconds under constant buffer recirculation. Upon completion, the gel was incubated for 10 min in 0.5X TBE containing 0.1 µg/ml ethidium bromide followed by three, 10 min washes with 0.5X TBE and imaged on a BioRad® gel documentation system.
Electron microscopy
Standard methods of sample preparation were employed for transmission electron microscopy (TEM) and scanning electron microscopy (SEM). Briefly, Cesium chloride purified bacteriophages (1×105-5×107 total phage) were fixed in 4% paraformaldehyde with 1% glutaraldehyde in 0.1 M Sodium cacodylate buffer for 2 h. After fixation, a portion of each sample was spread onto carbon coated copper grids, washed with water to remove fixative, and negatively stained with 1% Uranyl acetate. The grids were imaged with an FEI Tecnai T12 TEM at 100 kV. The remaining sample was processed for SEM analysis. The fixed samples were washed 3 times with 0.1 M Sodium cacodylate buffer, post fixed for 1 h with 1% Osmium tetroxide buffer, washed, and then immersed in a 0.5% Uranyl acetate solution for 1 h. The samples were subsequently dehydrated through an ethanol series, critically point dried, and coated with gold-palladium. The coated SEM samples were imaged in an FEI Quanta 200 FEG SEM at 5 kV.
Preparation of microtiter plates for OmniLog® assay
Microtiter plates (96 well) were prepared as follows. 90μl of TS broth with 0.1% v/v tetrazolium dye was added to each well. 10μl of 1×108 PFU/ml of each phage were added to the first well and diluted 10-fold down to 10 PFU per well. 10μl of 0.4 OD600 of bacteria (4×106 cells) were added to each well for a final volume of 100μl corresponding to a multiplicity of infection (MOI) range of 2.5 to 2.5×10−5. Media and phage only controls were added. The 96 well plates were incubated in the OmniLog® system (Biolog®) at 37°C for 48 h. Phage mixtures were prepared at equal volumes of phage for a final titer of 1×108 PFU/ml.
Results
Isolation of novel S. aureus phage
In an effort to isolate broad spectrum lytic phage against MRSA strains, 7 MRSA clinical isolates were used as hosts to isolate phage from environmental sources, primarily raw sewage; a rich source of environmental phage.23 Seven plaque-forming phage were identified in their corresponding S. aureus host strains SA0414ᶲ1, SA0414ᶲ2, SA0420ᶲ1, SA0456ᶲ1, SA0470ᶲ1, SA0482ᶲ1, and SA11987ᶲ1 henceforth referred to as “novel phage group.” Phage SA0414ᶲ1 and SA0414ᶲ2 are independent phage that were isolated against strain NSC0414 (data not shown).
The S. aureus phage are polyvalent in nature
To determine the spectrum of the novel phage group, a set of 170 S. aureus clinical isolates was obtained from different geographical regions to enrich strain diversity. This collection includes MSSA and MRSA from both, community and health care settings. The S. aureus isolates were screened for lytic activity by efficiency of plating (EOP) assays. The EOP assay determines the relative virulence of each phage against a S. aureus strain, thereby allowing side-by-side comparisons of phage virulence.37 In addition, spotting phages on strain-specific S. aureus lawns creates individualized phage typing profiles. The novel phage were virulent against 70% to 91% of the strains tested (Fig. 1A and 1B). Individually, phage SA0414ᶲ1 and SA0414ᶲ2 were virulent against 80% and 74% of the isolates, respectively. This establishes a distinction between these independent isolates of strain NSC0414. Phage SA0470ᶲ1 affected 85%, phage SA0420ᶲ1 affected 91%, phage SA0456ᶲ1, SA0482ᶲ1 and SA11987ᶲ1 each affected 78%, 84% and 82% of isolates (Fig. 1A and 1B). For HA-MRSA isolates, virulence ranged from 78% to 89% whereas for CA-MRSA strains, virulence range is 43% to 93% of the isolates (Fig. 1A and 1B). In the case of methicillin sensitive (MSSA) strains; the phage were proven virulent against 85% to 93% of the strains tested.
Figure 1.
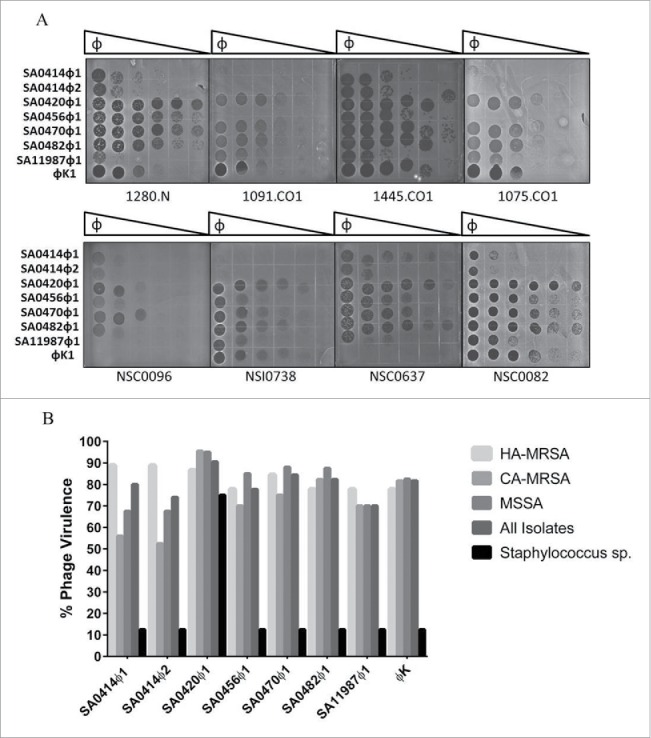
Efficiency of platting (EOP) assay for S. aureus clinical isolates. (A) 10-fold serial dilution series of phage SA0414ᶲ1, SA0414ᶲ2, SA0420ᶲ1, SA0456ᶲ1, SA0470ᶲ1, SA0482ᶲ1, SA11987ᶲ1 and phage K were spotted on several clinical isolates. A total of 1×105 phage particles were loaded in the first row. Plates were incubated overnight at 37°C and scored for the ability to inhibit bacterial growth. (B) Virulence spectra of all S. aureus phage as a percentage of strains infected. The Staphylococcus species used in this study include S. cohnii, S. epidermidis, S. hyicus, S. haemoliticus, S. saprophyticus, S. sciuri, and S. xylosus.
To assess species specificity, the S. aureus phage were tested against S. cohnii, S. epidermitis, S. haemoliticus, S. saprophiticus, S. sciurii, S. xylosus and S. hyicus. The results demonstrate that phage SA0414ᶲ1 and SA0414ᶲ2, SA0456ᶲ1, SA0470ᶲ1, SA0482ᶲ1, and SA11987ᶲ1 can only form plaques on S. aureus and S. hyicus. In contrast, SA0420ᶲ1 was capable of infecting all Staphylococcus species tested with the exception of S. xylosus (Fig. 1B). In addition, the phage did not infect gram negative bacterial species to include Escherichia coli, Enterococcus faecium, Acinetobacter baumannii, Klebsiella pneumoniae and Pseudomonas aeruginosa (data not shown). The results of these experiments indicate that the novel phage are polyvalent phage and specific to S. aureus and S. hyicus.
Previous studies demonstrated that phage K is a polyvalent S. aureus phage. The virulence of phage K was tested in this collection of clinical isolates and results show phage K is virulent against 82% of the isolates. For HA-MRSA strains, virulence was 78% compared to 87.5% of the CA-MRSA and 82.5% MSSA strains (Fig. 1A and 1B). It is noteworthy that phage K was not virulent on strains NSC0637 and NSC0096; however the novel phage infected these strains (Fig. 1A and B). This establishes a phenotypic distinction between the novel phage and phage K. In addition, phage K is virulent on a geographically independent set of strains underscoring the polyvalent nature of phage K.
Electron microscopy analysis of S. aureus phage
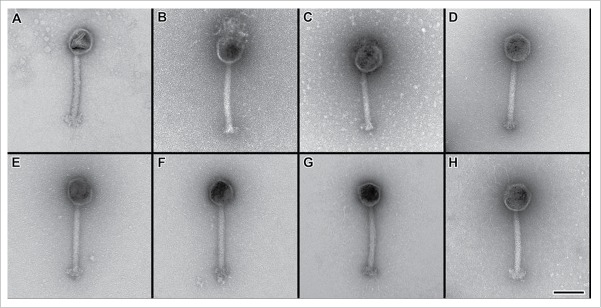
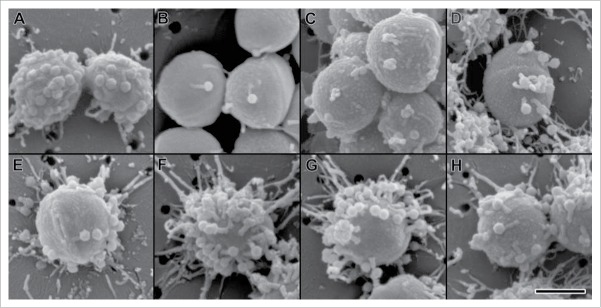
S. aureus phage are members of the Myoviridae, Syphoviridae and Podoviridae family of viruses.38 Due to the similarities in virulence spectrum of the novel phage and phage K, structural similarities among the phage are possible. To assess the morphology of the S. aureus phage, electron microscopy studies were performed on the novel phage group and contrasted to phage K. Consistent with the structural features of phage K, the novel phage group has a polyhedral shaped capsids and long contractile tails (Fig. 2A-H). Furthermore, the phage genomic DNA size is greater than 140 Kb (data not shown). These results classify the novel phage group as members of the Myoviridae family. In addition, we identified the novel phage in both the relaxed and contracted conformations (Fig. S1). The binding of the phage to S. aureus strain ATCC11987 was monitored by electron microscopy. The results indicate that multiple phage particles effectively recognize the S. aureus cell surface (Fig. 3A-H).
Figure 2.
Morphological analysis of novel S. aureus phage. Transmission electron micrographs of negative stained bacteriophages (A) Bacteriophage K, (B) Bacteriophage SA0414ᶲ1, (C) Bacteriophage SA0414ᶲ2, (D) Bacteriophage SA0420ᶲ1, (E) Bacteriophage SA0456ᶲ1, (F) Bacteriophage SA0470ᶲ1, (G) Bacteriophage SA0482ᶲ1 (H) Bacteriophage SA11987ᶲ1. Scale bar equals 100 nm.
Figure 3.
Scanning electron micrographs of phage bound to cells of Staphylococcus aureus. S. aureus strain ATCC11987 cells bound by the novel phage on membrane filters with 100 nm pores. (A) Bacteriophage K, (B) Bacteriophage SA0414ᶲ1 (C) Bacteriophage SA0414ᶲ2 (D) Bacteriophage SA0420ᶲ1, (E) Bacteriophage SA0420ᶲ1 (F) Bacteriophage SA0456ᶲ1, (G) Bacteriophage SA0470ᶲ1, (H) Bacteriophage SA0482ᶲ1, (I) Bacteriophage SA11987ᶲ1. Scale bar equals 500 nm.
The S. aureus novel phage group and phage K are distinct from each other
Due to consistency in morphologies and lytic spectra between phage K and the novel phage, it was considered possible that these could be independent environmental isolates of phage K (Fig. 1A, 1B, and 2A). To assess this, EcoR1 DNA restriction profiles of the novel phage were compared with phage K via pulse field gel electrophoresis (PFGE). The results show that phage K generates a pattern distinct from phage SA0420ᶲ1, SA0456ᶲ1, SA0470ᶲ1, SA0482ᶲ1, and SA11987ᶲ1 (Fig. 4). Additionally, comparison of the novel phage digestion patterns shows no difference in EcoRI digestion patterns, suggesting they are closely related (Fig. 4). Collectively, these results strongly suggest that the S. aureus phage are novel in nature and are not environmental isolates of phage K.
Figure 4.
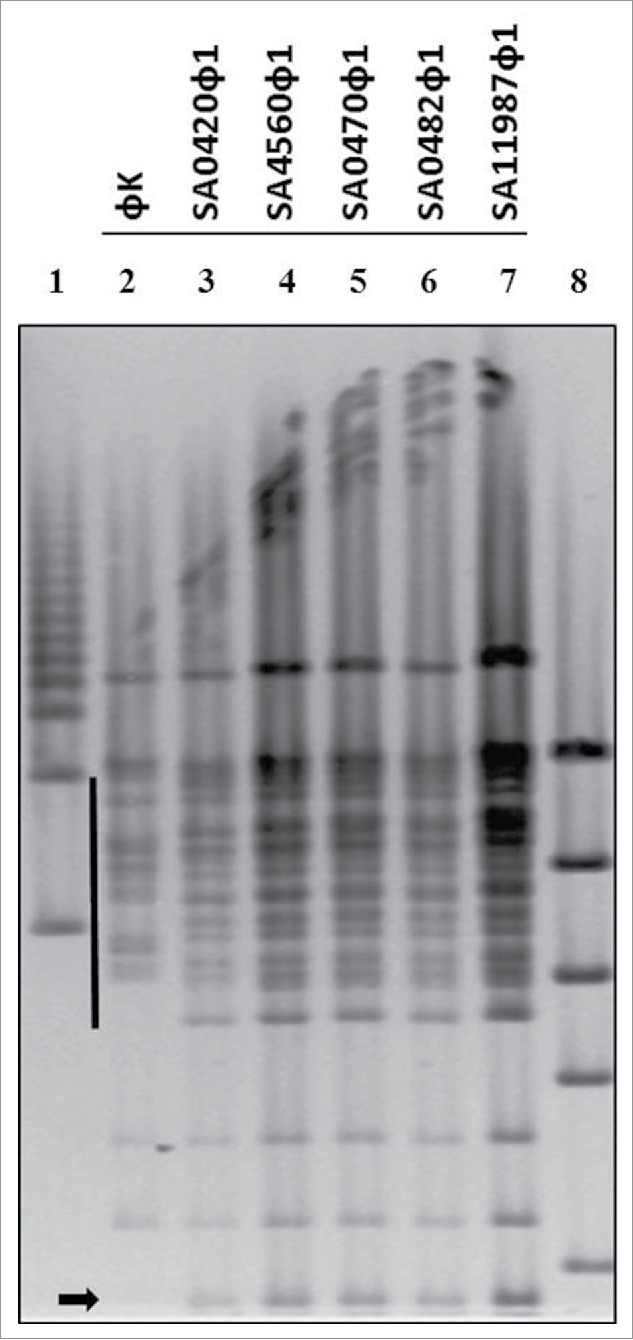
Analysis of S. aureus phages by Pulse field DNA analysis. Pulse field gel electrophoresis (PFGE) of purified S. aureus bacteriophage DNA digested with EcoR1. The solid line and arrow indicate regions where band patterns are distinct from phage K in comparison members in the novel phage group. Lanes 1 and 8 are molecular weight markers.
In vitro inhibition of S. aureus growth by the novel phage group and phage K mixtures are synergistic
Studies indicate S. aureus can develop resistance to the lytic effects of phage infection.39 To determine if resistance to the novel phage group and phage K is observed among the clinical isolates, a growth curve analysis was performed utilizing the OmniLog® automated system. Our laboratory has modified the system to monitor and characterize phage infection at continuous intervals providing insight into phage resistance and effectiveness.40 Phage K incubation with strain NSI0016 resulted in bacterial growth inhibition for 8 hours, indicating that resistance to Phage K arises in this strain (Fig. 5A). Phage SA0420ᶲ1, SA0456ᶲ1 and SA0482ᶲ1 inhibit bacterial growth for 10, 12, and 15 h, respectively. However, phage resistance is also evident after these incubation periods. Media-only or phage-only controls show baseline oxidation levels of reporter dye (Fig. 5A). These results suggest that all phage are virulent against strain NSI0016, but insufficient to control growth for time spans longer than 8–15 h.
Figure 5.
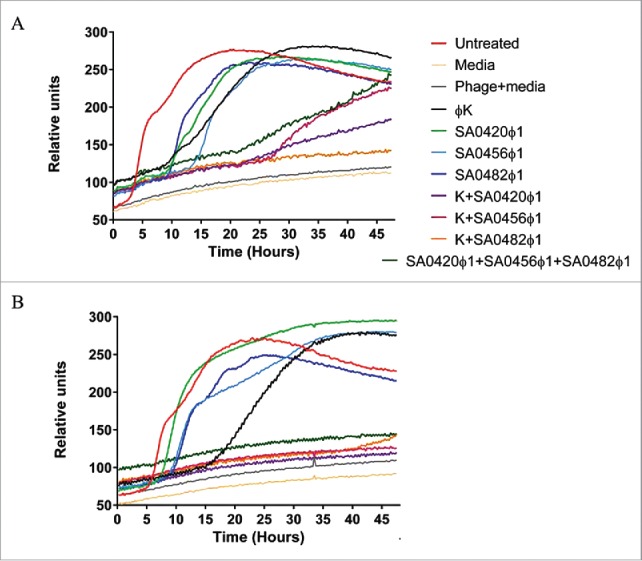
Analysis of bacteriophage cocktails on CA-MRSA and HA-MRSA Strains via the OmniLog® system. (A) Strain NSI0016 monitored for 48 h on the OmniLog® system. (B) Strain 3195.CO1 monitored for 48 h on the OmniLog® system. A total of 4×106 cells were added per well for each of the MRSA strains NSI0016 and 3195.CO1. The phage was added to a final Multiplicity of infection (MOI) of 2.5 and growth measured every 15 min for 48 h. The plots represent triplicate experiments, all combinations contain identical total phage quantities.
To determine if emergence of phage-resistance could be reduced by using phage combinations, phage SA0420ᶲ1, SA0456ᶲ1 or SA0482ᶲ1, were tested with phage K (Fig. 5B). The combination of phage K with phage SA0420ᶲ1 inhibited growth for 20 h while SA0456ᶲ1 growth was inhibited for 26 h. In the case of phage SA0482ᶲ1, the combination with phage K inhibited growth of NSI0016 for 36 h. The effects of these phage combinations are more pronounced on strain 3195.CO1, where phage SA0420ᶲ1, SA0456ᶲ1 and SA0482ᶲ1 in combination with phage K completely inhibit bacterial growth (Fig. 5B). To assess the possible additive effects of SA0420ᶲ1, SA0456ᶲ1, and SA0482ᶲ1 on S. aureus, a 3-phage mixture was prepared to a MOI identical to the single phage treatment. The results demonstrate that this phage combination can synergize to inhibit growth of NSI0016 and 3195.CO1 (Fig. 5A and 5B). These results show that combining phage K with SA0420ᶲ1, SA0482ᶲ1, SA0456ᶲ1 or a combination of the novel phage has a synergistic growth inhibition effect, suggesting the combination may reduce formation of phage resistance.
The S. aureus phage require cell wall teichoic acid molecules for infection
Studies have suggested that phage K utilizes teichoic acid molecules as cell surface receptors for infection.26 The tarO gene encodes the N-acetyl glucosamine transferase, the enzyme responsible for initiating WTA synthesis in S. aureus. The catalytic activity of the tarO gene product can be inhibited by treatment with tunicamycin.27,41 To elucidate the cell surface receptor utilized by the novel phage group, tunicamycin was used as an inhibitor of the N-Acetyl glucosamine transferase activity. Addition of 0.1 μg/ml of tunicamycin caused a 1000-fold inhibition in the ability of the novel phage group to infect bacteria (Fig. 6A). In comparison, phage K Infection was inhibited 100-fold by tunicamycin treatment (Fig. 6A). To assess the contribution of WTAs to phage infection of strains from our S. aureus collection, a sub-set of strains were screened for tunicamycin-mediated inhibition of phage infection. Exposure of strain 1028.N to tunicamycin caused a 100,000 fold inhibition of S. aureus infection by the novel phage group. This strongly suggests that teichoic acid molecules are involved in infection of the S. aureus clinical isolates in this study (Fig. 6B).
Figure 6.
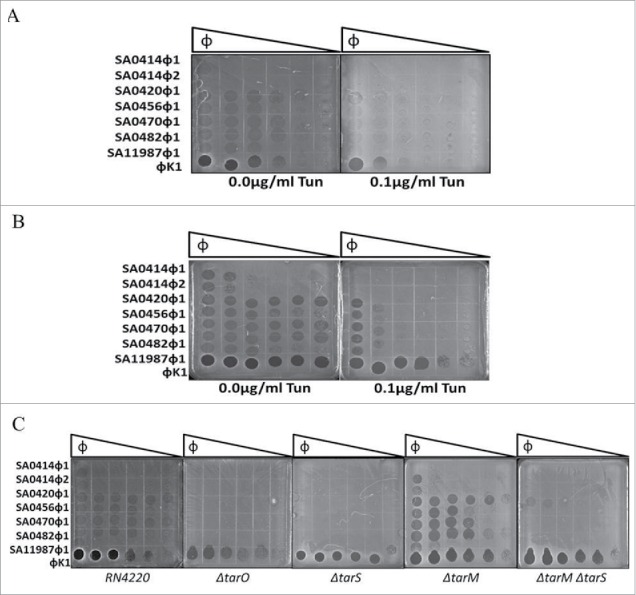
Cell wall teichoic acids are required for bacteriophage virulence. (A) Strain RN4220 infected with phage SA0414ᶲ1 SA0414ᶲ2, SA0420ᶲ1, SA0470ᶲ1, SA0482ᶲ1, SA0456ᶲ1, SA11987ᶲ1 and phage K in the absence or presence of 0.1 μg/ml tunicamycin. (B) Strain 1028.N CA-MRSA isolate treated with 0.1 μg/ml tunicamycin. (C) Strain RN4220 mutant derivatives tarO, tarS, tarM, tarM/tarS genes of the teichoic acid biosynthesis pathway treated with all phage in this study. Plates were incubated at 37°C for 12 h, a total 1×105 PFU/ml of each phage was used as the start point for a 10-fold serial dilution.
To further assess the WTA pathway involvement in the novel phage group infection of S. aureus, a series of null mutants were used. The strain RN4220 was selected as host due to the absence of a capsule, prophage, and restriction digestion modification systems known to produce immunity against phage infection.29 EOP assay show that the novel phage group infect strain RN4220, albeit at a lower efficiency than phage K. Infection by phage SA0414ᶲ1 and SA0414ᶲ2 was only evident at the highest phage titer (Fig. 6C). These results indicate that this strain background is suitable for assessing the contribution of WTA biosynthetic enzymes to infection by the novel phage group. To assess further the involvement of the WTA pathway, isogenic null mutants of tarO, tarM, tarS and the ΔtarM ΔtarS double mutant were utilized.27,29,35 The ΔtarO strain is impervious to infection by the novel phage group, however, the effects on phage K lysis inhibition are minimal on this assay (Fig. 6C). EOP on ΔtarS mutant show complete inhibition of the lytic activity of the novel phage group, implicating β-GlcNAc moieties of the WTAs in phage infection. In contrast, the deletion of ΔtarM enhances the lytic activity of the phage. The ΔtarM ΔtarS double mutant shows a lytic phenotype that resembles the ΔtarS single mutant (Fig. 6C). These results indicate that β-GlcNAc moieties on WTA are essential for the novel phage infection of S. aureus.
Discussion
Reducing antimicrobial agent resistance is crucial for development of therapies against multiple-drug resistance in organisms such as S. aureus.4 The results outlined in this article describe 7 novel polyvalent S. aureus phage virulent against 70% to 91% of the clinical isolates in our collection (Fig. 1A and 1B). Phage K has been reported as a broad spectrum phage against MRSA strains.22 Here, we have expanded on those studies by finding phage K virulent against 82% of the of S. aureus strains (Fig. 1A and 1B). Importantly, the novel phage complements the virulence of phage K. For example, in strains NSC0096 and NSC0637, phage K is ineffective yet members of the novel phage group are virulent against these same strains (Fig. 1A). This makes phage K and all members of the novel phage group suitable candidates for a phage mixture for therapeutic treatment of S. aureus infections. Either phage K or the novel phage group fail to infect gram negative bacteria and are specific to S. aureus, showing minimal staphylococcal species cross virulence by infecting S. hyicus (Fig. 1B). One exception was phage SA0420ᶲ1 who infected several Staphylococcus species including S. cohnii, S. epidermitis, S. haemoliticus, S. saprophiticus, and S. sciurii. This suggests that phage SA0420ᶲ1 targets cell surface receptors common to all Staphylococcus species tested.
Electron micrograph studies demonstrate the novel phage group are members of the Myoviridae family, characterized by a polyhedral capsid, a long contractile tail and genomes larger than 140 Kb (Fig. 2A-H).38 Indeed, our study determined that genome sizes of all the members of the novel phage group are 140 Kb or larger, consistent with members of the Myoviridae family (data not shown). The polyvalent nature of the novel phage showed a similarity with phage K, however, restriction analysis demonstrates otherwise (Fig. 4). Further distinction comes from lytic spectrum EOP analysis, where comparison of equal titers generates virulence profiles distinct for each strain-phage combination reflecting genetic differences among the novel phage (Fig. 1A and 1B). In the case of phage SA0414ᶲ1 has an average non-contracted tail size of 90.6nm and an average contracted tail size of 53.8 nm and an average capsid size 98.5 nm. In contrast, SA0414ᶲ2 has an average non-contracted tail size of 206.5 nm, an average contracted tail size of 112.5 nm and an average capsid size of 105.5 nm. Even though their lytic spectra are very similar, the difference in size establishes a morphological distinction between these phage (Fig. 1A, 1B and data not shown). Taken together, the results indicate that the novel phage are polyvalent in nature and effective against S. aureus clinical isolates.
This study shows that phage K resistance develops in strain NSI0016 after 8 h. Phage resistance is prevented or delayed by the addition of phage SA0420ᶲ1, SA0456ᶲ1 or SA0482ᶲ1. Different degrees of enhanced virulence are observed when combining phage K with SA0420ᶲ1, SA0456ᶲ1 or SA0482ᶲ1 (Fig. 5A and 5B). Additionally, the combination of phage SA0420ᶲ1, SA0456ᶲ1 and SA0482ᶲ1 effectively reduced phage-resistance on NSI0016 and 3195.CO1 (Fig. 5A and 5B). The synergistic effects observed in phage K combinations suggest that the novel phage group utilized an infection mechanism distinct from phage K. However, the additive effects observed in the SA0420ᶲ1, SA0456ᶲ1 and SA0482ᶲ1 triple combination indicate that there are differences in the mechanisms of infection among them. Perhaps, differences in novel phage affinity toward cell surface receptors may alter phage replication kinetic in a particular host resulting in distinct latent periods and bust sizes.
Previous studies have implicated cell wall teichoic acids as important components for absorption of phage in the Myoviridae virus family.26 Treatment with tunicamycin, an inhibitor of N-acetyl glucosaminidase activity, led to the inhibition of infection by the novel phage group suggesting a role of this modification in the infection mechanism (Fig. 6A). Importantly, the effect of tunicamycin is not confined the RN4220 strain for treatment of clinical isolate strain 1028.N1 shows a 10,000 fold reduction in phage infection (Fig. 6B). In other clinical isolates, a 10-fold reduction is observed (data not shown). The deletion of the N-acetyl glucosamine transferase, ΔtarO; impedes infection by members of the novel phage group, implicating cell wall teichoic acids as a key component for infection (Fig. 6C). Furthermore, the deletion of ΔtarS, the β N-acetyl glucosaminidase, rendered the cells insensitive to infection by the novel phage group. This suggests that β-N-acetyl glucosamine modification of cell wall teichoic acids act as cell surface receptors for the novel phage group (Fig. 6C). Interestingly, the deletion of tarM, the α-N-acetyl glucosaminidase, enhances the ability of the novel phage group to infect strain RN4220 (Fig. 6C). This may be due to increased β-N-acetyl glucosamine modification of WTAs enhancing binding of the novel phage group. Alternatively, the α-N-acetyl glucosaminidation may have an inhibitory effect on phage infection. Of note is the fact that the extent of WTA modification in S. aureus varies among strains. Some S. aureus strains have been reported to exclusively harbor α or β-GlcNAc modifications in WTAs while other strains consist of a mixture of both modifications.27,33,34 This level of heterogeneity could explain the difference in phage susceptibility among S. aureus isolates. Saliently, phage K infection was not affected by ΔtarS or ΔtarM and only mildly by ΔtarO, suggesting that phage K may use other receptors besides WTAs. The enhancement of growth inhibition observed in phage K combinations with SA0420ᶲ1, SA0456ᶲ1 and SA0482ᶲ1 support the notion that the novel phage group use a common cell surface receptor distinct from that of phage K.
It is noteworthy that β-N-acetyl glucosamine modifications of WTAs have been linked to potentiation of β-lactam antibiotic resistance on MRSA strains.27,29 β-N-acetyl glucosamine modifications serve as binding sites for the PBP2a, the enzyme responsible for β-lactam resistance in S. aureus, allowing for continuous cell wall synthesis. Interestingly, the WTA β-N-acetyl glucosamine modification needed for β-lactam resistance is also essential for novel phage group infection. An intriguing possibility lies in competition between the phage and PBP2a for a common site of action. Our preliminary studies suggest a phage-antibiotic synergy (PAS) effect of the novel phage group and β–lactam antibiotic (unpublished results).42-44 This suggests that the novel phage group would be suitable candidates to augment β-lactam antibiotic treatment and strengthen the notion that the novel phage group are a useful tool to combat the emergence of MDR organisms (data not shown).
Several other studies have identified phage with virulence against methicillin resistant S. aureus strains.22,23 However, few studies have identified phage with broad spectrum properties. In this study we have isolated 7 S. aureus polyvalent phage which possess the potential for use as treatment against antibiotic resistant S. aureus infections which may overcome the emergent phage resistance.
Supplementary Material
Abbreviations
- CA-MRSA
Community acquired methicillin resistant Staphylococcus aureus
- EOP
Efficiency of plating
- HA-MRSA
Hospital acquired methicillin resistant Staphylococcus aureus
- MDR
Multi-drug resistance
- MOI
Multiplicity of infection
- MRSA
Methicillin resistant Staphylococcus aureus
- MSSA
Methicillin sensitive Staphylococcus aureus
- PFGE
Pulse field gel electrophoresis.
- PFU
Plaque forming units
- SSTI
Skin and soft tissue infections
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the laboratory of Dr. Suzanne Walker for providing the isogenic strains with null mutations for ΔtarO, ΔtarM, ΔtarS and the ΔtarMΔtarS double mutant.
Notes on contributors
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the DHS or S&T. In no event shall DHS, NBACC, S&T or Battelle National Biodefense Institute have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. DHS does not endorse any products or commercial services mentioned in this publication.
Funding
This work was supported and funded by the Congressionally Directed Medical Research Program (Work Unit Number A1427), Naval Medical Research Center. The authors are military service members, full-time or contract employees of the US Government. This work was prepared as part of their official duties. Title 17 USC § 105 provides that “Copyright protection under this title is not available for any work of the United States Government. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the US Government. Ryan Hannah and Robert K. Pope were funded under Contract No. HSHQDC-07-C-00020 awarded by the Department of Homeland Security (DHS) Science and Technology Directorate (S&T) for the management and operation of the National Biodefense Analysis and Countermeasures Center (NBACC), a Federally Funded Research and Development Center.
References
- [1].National strategy for combating antibiotic-resistant bacteria The White House, Washington DC, USA: September 2015:1-37. [Google Scholar]
- [2].National action plan for comabating antibiotic-resistant bacteria The White House, Washington DC, USA: March 2015:1-63. [Google Scholar]
- [3].Report to the President on Combating Antibiotic Resistance Executive Office of the President President's Council of Advisors on Science and Technology, USA: September 2015:1-78. [Google Scholar]
- [4].Antimicrobial resistance: global report on surveillance World Health Organization, April 2014:1-257; http://www.who.int/drugresistance/documents/surveillancereport/en/ . [Google Scholar]
- [5].Fontanilla JM, Kirkland KB, Talbot EA, Powell KE, Schwartzman JD, Goering RV, Parsonnet J. Outbreak of skin infections in college football team members due to an unusual strain of community-acquired methicillin-susceptible Staphylococcus aureus. J Clin Microbiol 2010; 48:609-11; PMID:20007392; http://dx.doi.org/ 10.1128/JCM.02297-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chhibber S, Kaur T, Sandeep K. Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. Plos One 2013; 8:e56022; PMID:23418497; http://dx.doi.org/ 10.1371/journal.pone.0056022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cadena J, Richardson AM, Frei CR. Risk factors for methicillin-resistant Staphylococcus aureus skin and soft tissue infection in MRSA-colonized patients discharged from a Veterans Affairs hospital. Epidemiology and infection 2016;144(3):647-51. [DOI] [PubMed] [Google Scholar]
- [8].Echaniz-Aviles G, Velazquez-Meza ME, Vazquez-Larios MD, Soto-Nogueron A, Hernandez-Duenas AM. Diabetic foot infection caused by community-associated-methicillin-resistant Staphylococcus aureus (CA-MRSA) USA300. J Diabetes 2015; PMID:26119276 [DOI] [PubMed] [Google Scholar]
- [9].Kahanov L, Kim YK, Eberman L, Dannelly K, Kaur H, Ramalinga A. Staphylococcus aureus and community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) in and around therapeutic whirlpools in college athletic training rooms. J Athletic Training 2015; 50:432-7; PMID:25710853; http://dx.doi.org/ 10.4085/1062-6050-49.3.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Montazeri EA, Khosravi AD, Jolodar A, Ghaderpanah M, Azarpira S. Identification of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from burn patients by multiplex PCR. Burns 2015; 41:590-4; PMID:25441547; http://dx.doi.org/ 10.1016/j.burns.2014.08.018 [DOI] [PubMed] [Google Scholar]
- [11].Khokhlova OE, Hung WC, Wan TW, Iwao Y, Takano T, Higuchi W, Yachenko SV, Teplyakova OV, Kamshilova VV, Kotlovsky YV, et al.. Healthcare- and community-associated methicillin-resistant staphylococcus aureus (MRSA) and Fatal Pneumonia with pediatric deaths in Krasnoyarsk, Siberian Russia: Unique MRSA's multiple virulence factors, genome, and stepwise evolution. PloS One 2015; 10:e0128017; PMID:26047024; http://dx.doi.org/ 10.1371/journal.pone.0128017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol Rev 2008; 32:361-85; PMID:18248419; http://dx.doi.org/ 10.1111/j.1574-6976.2007.00095.x [DOI] [PubMed] [Google Scholar]
- [13].Ballhausen B, Kriegeskorte A, Schleimer N, Peters G, Becker K. The mecA homolog mecC confers resistance against β-lactams in Staphylococcus aureus irrespective of the genetic strain background. Antimicrobial Agents Chemother 2014; 58:3791-8; PMID:24752255; http://dx.doi.org/ 10.1128/AAC.02731-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guignard B, Entenza JM, Moreillon P. Beta-lactams against methicillin-resistant Staphylococcus aureus. Curr Opin Pharmacol 2005; 5:479-89; PMID:16095969; http://dx.doi.org/ 10.1016/j.coph.2005.06.002 [DOI] [PubMed] [Google Scholar]
- [15].Center for Disease Control and Prevention. Emerging Infections Program Network Active Bacterial Core Surveillance Report: Methicillin-Resistant Staphylococcus aureus. 2014:1-3 . [Google Scholar]
- [16].McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 2003; 41:5113-20; PMID:14605147; http://dx.doi.org/ 10.1128/JCM.41.11.5113-5120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McDougal LK, Fosheim GE, Nicholson A, Bulens SN, Limbago BM, Shearer JE, Summers AO, Patel JB. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrobial Agents Chemotherapy 2010; 54:3804-11; PMID:20585117; http://dx.doi.org/ 10.1128/AAC.00351-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hershey AD, Chase M. Genetic recombination and heterozygosis in bacteriophage. Cold Spring Harbor Symposia Quantitative Biol 1951; 16:471-9; PMID:14942757; http://dx.doi.org/ 10.1101/SQB.1951.016.01.034 [DOI] [PubMed] [Google Scholar]
- [19].Jensen KC, Hair BB, Wienclaw TM, Murdock MH, Hatch JB, Trent AT, White TD, Haskell KJ, Berges BK. Isolation and host range of bacteriophage with lytic activity against Methicillin-Resistant Staphylococcus aureus and potential use as a fomite decontaminant. PloS One 2015; 10:e0131714; PMID:26131892; http://dx.doi.org/ 10.1371/journal.pone.0131714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kutateladze M, Adamia R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol 2010; 28:591-5; PMID:20810181; http://dx.doi.org/ 10.1016/j.tibtech.2010.08.001 [DOI] [PubMed] [Google Scholar]
- [21].Adhya S, Merril CR, Biswas B. Therapeutic and prophylactic applications of bacteriophage components in modern medicine. Cold Spring Harb Perspect Med 2014; 4:a012518; PMID:24384811; http://dx.doi.org/ 10.1101/cshperspect.a012518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].O'Flaherty S, Ross RP, Meaney W, Fitzgerald GF, Elbreki MF, Coffey A. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Applied Environmental Microbiol 2005; 71:1836-42; PMID:15812009; http://dx.doi.org/ 10.1128/AEM.71.4.1836-1842.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, Tanji Y. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Applied Environmental Microbiol 2009; 75:4483-90; http://dx.doi.org/ 10.1128/AEM.02641-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Becker SC, Foster-Frey J, Donovan DM. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol Letters 2008; 287:185-91; PMID:18721148; http://dx.doi.org/ 10.1111/j.1574-6968.2008.01308.x [DOI] [PubMed] [Google Scholar]
- [25].O'Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J Bacteriol 2004; 186:2862-71; PMID:15090528; http://dx.doi.org/ 10.1128/JB.186.9.2862-2871.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xia G, Corrigan RM, Winstel V, Goerke C, Grundling A, Peschel A. Wall teichoic Acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol 2011; 193:4006-9; PMID:21642458; http://dx.doi.org/ 10.1128/JB.01412-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brown S, Santa Maria JP Jr, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 2013; 67:313-36; PMID:24024634; http://dx.doi.org/ 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Peschel A, Vuong C, Otto M, Gotz F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrobial Agents Chemotherapy 2000; 44:2845-7; PMID:10991869; http://dx.doi.org/ 10.1128/AAC.44.10.2845-2847.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, et al.. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci U S A 2012; 109:18909-14; PMID:23027967; http://dx.doi.org/ 10.1073/pnas.1209126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, et al.. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med 2004; 10:243-5; PMID:14758355; http://dx.doi.org/ 10.1038/nm991 [DOI] [PubMed] [Google Scholar]
- [31].Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 2008; 6:276-87; PMID:18327271; http://dx.doi.org/ 10.1038/nrmicro1861 [DOI] [PubMed] [Google Scholar]
- [32].Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol 2010; 300:148-54; PMID:19896895; http://dx.doi.org/ 10.1016/j.ijmm.2009.10.001 [DOI] [PubMed] [Google Scholar]
- [33].Jenni R, Berger-Bachi B. Teichoic acid content in different lineages of Staphylococcus aureus NCTC8325. Arch Microbiol 1998; 170:171-8; PMID:9683656; http://dx.doi.org/ 10.1007/s002030050630 [DOI] [PubMed] [Google Scholar]
- [34].Winstel V, Sanchez-Carballo P, Holst O, Xia G, Peschel A. Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. Mbio 2014; 5:e00869; PMID:24713320; http://dx.doi.org/ 10.1128/mBio.00869-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M, Holst O, Peschel A. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J Biol Chem 2010; 285:13405-15; PMID:20185825; http://dx.doi.org/ 10.1074/jbc.M109.096172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hershey AD, Dixon J, Chase M. Nucleic acid economy in bacteria infected with bacteriophage T2. I. Purine and pyrimidine composition. J General Physiol 1953; 36:777-89; PMID:13069681; http://dx.doi.org/ 10.1085/jgp.36.6.777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kutter E. Phage host range and efficiency of plating. Methods Mol Biol 2009; 501:141-9; PMID:19066818; http://dx.doi.org/ 10.1007/978-1-60327-164-6_14 [DOI] [PubMed] [Google Scholar]
- [38].Deghorain M, Van Melderen L. The Staphylococci phages family: an overview. Viruses 2012; 4:3316-35; PMID:23342361; http://dx.doi.org/ 10.3390/v4123316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rosato RR, Cameron JA. The bacteriophage receptor sites of staphylococcus aureus. Biochim Et Biophysica Acta 1964; 83:113-9; PMID:14152187 [DOI] [PubMed] [Google Scholar]
- [40].Henry M, Biswas B, Vincent L, Mokashi V, Schuch R, Bishop-Lilly KA, Sozhamannan S. Development of a high throughput assay for indirectly measuring phage growth using the OmniLog(TM) system. Bacteriophage 2012; 2:159-67; PMID:23275867; http://dx.doi.org/ 10.4161/bact.21440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pasquina LW, Santa Maria JP, Walker S. Teichoic acid biosynthesis as an antibiotic target. Curr Opin Microbiol 2013; 16:531-7; PMID:23916223; http://dx.doi.org/ 10.1016/j.mib.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kamal F, Dennis JJ. Burkholderia cepacia complex Phage-Antibiotic Synergy (PAS): antibiotics stimulate lytic phage activity. Applied Environmental Microbiol 2015; 81:1132-8; PMID:25452284; http://dx.doi.org/ 10.1128/AEM.02850-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Comeau AM, Tetart F, Trojet SN, Prere MF, Krisch HM. The discovery of a natural phenomenon, “Phage-Antibiotic Synergy.” Implications for phage therapy. Med Sci (Paris) 2008; 24:449-51; PMID:18466714; http://dx.doi.org/ 10.1051/medsci/2008245449 [DOI] [PubMed] [Google Scholar]
- [44].Comeau AM, Tetart F, Trojet SN, Prere MF, Krisch HM. Phage-Antibiotic Synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PloS One 2007; 2:e799; PMID:17726529; http://dx.doi.org/ 10.1371/journal.pone.0000799 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.