Abstract
Particularly interesting new cysteine-histidine-rich protein (PINCH) is a LIM-domain-only adaptor that plays important roles in cytoskeletal organization and extracellular matrix adhesion, migration, proliferation and survival. Mammalian cells have two functional PINCH proteins, PINCH1 and PINCH2. PINCH not only binds to Nck2 and engages in the signaling of growth factor receptors, but also forms a ternary complex with ILK and parvin (IPP complex). Normally, the IPP complex locates to focal adhesions participating in the signaling of integrins and mediating the interaction of cytoskeleton and extracellular matrix (ECM). Accumulative evidence indicates that abnormalities in PINCH signaling are involved in the pathogenesis of important diseases, such as cancers, renal diseases, cardiomyopathy, and HIV. Therefore, clarifying the functions of PINCH and its interactions with key factors is important for better understanding of signaling events both in health and disease.
Keywords: PINCH, IPP complex, ILK, Nck2, adhesion, focal adhesion, protein-protein interaction, tumor
1. PINCH and binding partners
1.1 Structure and expression
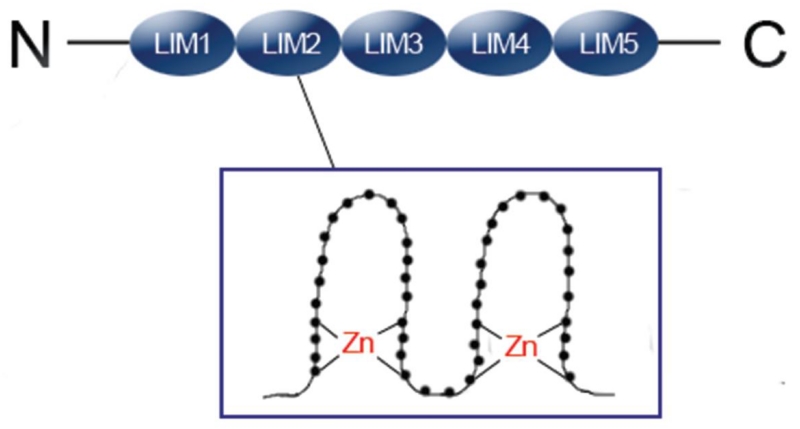
PINCH was originally found and named in 1994 during search for senescent cell antigens.1 Following the discovery of PINCH, in 2003, another member of the family, PINCH2, was discovered. PINCH was renamed to PINCH1 afterwards. There is a high sequence similarity between the two PINCH proteins, with 82% of their amino acid sequences being identical. Both proteins are composed of five LIM domains and a C-terminal putative nuclear localization/export signal (Figure 1).2, 3 LIM domain is a specialized double-zinc finger motif, through which PINCH associates with other proteins, thus serving to mediate protein-protein interactions.4 PINCH1/2 have more LIM domains than any other members in the LIM domain-containing family. Neither PINCH1 nor PINCH2 has a catalytic domain. These features make them ideal adaptor molecules to mediate the formation of multiprotein complexes. Both PINCH1 and PINCH2 are ubiquitously expressed in most mammalian tissues and organs, including the heart, lung, liver, kidney, and bladder. During mouse embryogenesis, PINCH1 expression begins at E8.5, while the expression of PINCH2 starts at E14.5. This time difference probably explains, in part, the dramatic differences between the phenotypes of their knockout mice.2
Figure 1. Schematic of PINCH showing its five LIM domain structure.
1.2 PINCH protein complexes
Although PINCH proteins have no catalytic activity, they form multiple complexes with other proteins via their five LIM domains. This largely explains how they exert their signaling function in cells. For this reason, it is of great significance to know which proteins interact with PINCH, and what the functions of the complexes are.
1.2.1 ILK-PINCH-parvin (IPP) complex
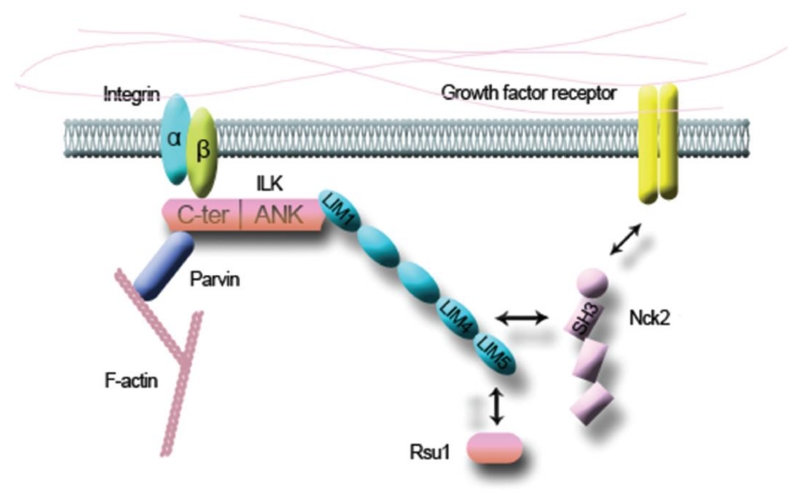
Integrins, a group of transmembrane cell adhesion receptors, play an important role in mediation of the interaction of cell and extracellular matrix (ECM). On one hand, the extracellular domain of integrin interacts with components of ECM (Figure 2). On the other hand, the cytoplasmic tail of it recruits a number of adaptors and signaling proteins, which together form a structure called focal adhesion (FA). Through FAs, integrins physically contact with the cytoskeleton and transduce signals into the cell.5 Integrin-linked kinase (ILK) was found to be able to bind the cytoplasmic tail of integrin β1 by yeast two-hybrid analysis. It was thought that ILK can not only bind to, but also phosphorylate the cytoplasmic tail of integrin β1.6 However, recent structural and genetic studies support that ILK is actually a pseudokinase that acts as an adaptor protein in FAs.7 Wu group was the first to demonstrate that PINCH can interact with ILK through its LIM1 domain (Figure 2). Besides, binding with PINCH is the prerequisite for ILK to locate to integrin-rich FAs.8, 9 Later, ILK was found to be able to bind to parvin; thus together with PINCH, they form a ternary complex known as the IPP (ILK-PINCH-parvin) (Figure 2). The formation of the IPP complex is crucial for the stability of the three proteins, and is the prerequisite for them to locate to the cell-ECM adhesion sites. Any mutations in PINCH, ILK or parvin that disrupt the formation of the complex will prevent other members from locating to FAs.10 One exception was from a recent study in that PINCH1 in ILK-deficient keratinocytes could still gather within the adhesion sites and recruited EPLIN to modulate the function of FAs.5 This suggests that some of the PINCH1 function is independent upon the presence of the IPP complex.
Figure 2. PINCH interacts with ILK, Nck2, and Rsu1.
The N-terminal ANK repeat domain of ILK binds to the LIM1 domain of PINCH. Together with parvin, they form a ternary complex referred to as the IPP complex. Through the LIM4 domain, PINCH interacts with Nck2, which may link the integrin signaling pathway with the growth factor receptor signaling pathways. Except for ILK and Nck2, PINCH also contacts with other key factors such as Rsu1. The LIM5 domain of PINCH is responsible for the interaction with Rsu1.
What are the roles of IPP complex in FAs? In mice, deleting each of the three proteins resulted in embryonic lethality.11-13 Some common features of the knockout animals include defects in cell polarity, migration, viability and cell-ECM adhesion, which are quite similar to those caused by integrin deficiency. These results further demonstrate that the IPP complex plays a critical role in integrin-mediated cell-ECM interaction network.
In contrast to PINCH1, PINCH2 knockout mice have no obvious phenotypes. This is likely due to a compensation by PINCH1 as the expression of PINCH1 is up-regulated in tissues of the PINCH2 KO mice.14 PINCH2 can bind to ILK and form the PINCH2-ILK-parvin complex just like PINCH1 does. But this does not mean that they can completely replace each other.15 Overexpression of PINCH2 in HeLa cells did prevent the down-regulation of ILK and parvin resulted from loss of PINCH1; but it failed to rescue the defects in cell spreading and cell survival signaling.16 Furthermore, overexpression of PINCH2 in 293 cells inhibited cell spreading and migration, which may be due to destruction of the PINCH1-ILK-parvin complex.15 Although a functional redundancy between the two proteins does exists as they are often co-expressed and structurally similar, PINCH1 and PINCH2 have their own unique roles in cells as well.
Apart from binding with integrins on the membrane, IPP complex can interact with the cytoskeleton as well as many signaling proteins inside the cell. Up to now, dozens of IPP complex-related proteins have been identified. One of these proteins is parvin that binds to F-actin, thus connecting ECM with cytoskeleton.17 In the following, we will discuss PINCH-interacting proteins and their roles.
1.2.2 PINCH-Nck2 complex
Cytoplasmic protein Nck2 is one of the first proteins that was discovered to be related to PINCH. It consists of three N-terminal SH3 domains and one C-terminal SH2 domain (Figure 2). The two proteins bind to each other through the fourth LIM domain of PINCH1 and the third SH3 domain of Nck2 (Figure 2). Apart from associating with PINCH1, Nck2 can also interact with components of the growth factor receptor signaling pathways (Figure 2), such as epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor β (PDGFR-β), and insulin receptor substrate 1 (IRS-1). So through its interaction with Nck2, PINCH1 links the integrin signaling pathways with the growth factor receptor signaling pathways (Figure 2).18 Besides, Nck2 binds to and activates N-WASP, a molecule related to actin polymerization. Thus, the interaction of PINCH1 and Nck2 drives cytoskeleton assembly at cell-ECM adhesions.19
Studies using NMR spectroscopy revealed that the binding between PINCH1 and Nck2 was highly specific, but weak, which changes rapidly. The association is so weak that it cannot be detected by GST pull-down and Co-IP assays. Nevertheless, the interaction of PINCH1 and Nck2 is important for the formation of FAs. A mutation in LIM4 domain that disrupts PINCH1 binding to Nck2 prevented PINCH1 from locating to FAs.20 Furthermore, knocking down Nck2 in HeLa cells led to spreading defect. Overexpression of mutant Nck2 that is defective in PINCH1 binding failed to rescue cell morphology and motility defects. These results suggest that the interaction between PINCH1 and Nck2 plays a pivotal role in control of the progress of cytoskeleton assembly and cell-ECM interaction. 21 It is noted that Nck2 does not seem to regulate the PINCH1-mediated cell survival signaling.22
1.2.3 PINCH-Rsu1 complex
Another well-studied PINCH-binding protein is Ras suppressor-1 (Rsu1). LIM5 domain of PINCH1 is responsible for its interaction with Rsu1 (Figure 2). As LIM5 is the least conserved domain between PINCH1 and PINCH2, it is not surprising to find that PINCH2 cannot bind to Rsu1. This explains, at least in part, why PINCH2 does not completely compensate for the loss of PINCH1.23 Rsu1 was first discovered as a suppressor of Ras-dependent oncogenic transformation. So through the interaction of Rsu1 and PINCH1, we may have an explanation for the change of cell motility during oncogenesis.24
It was reported that Rsu1 and PINCH1 co-localized at integrin-rich adhesion sites, which increased their stability.25 This suggests that Rsu1 may be involved in important processes, such as cell adhesion and migration. Consistently, overexpression of Rsu1 in NIH3T3 cells promoted cell spreading and actin aggregation.26 In contrast, knocking down Rsu1 in 293T cells significantly inhibited cell adhesion. In the meantime, the expression of PINCH1 was down-regulated in these cells.23 Similarly, knocking down either PINCH1 or Rsu1 in MCF10A cells led to defects in cell adhesion, spreading and migration. Overexpression of PINCH1 binding-defective Rsu1 in Rsu1-deficient cells rescued the cell adhesion defect, but had no effect on defects in cell migration and formation of FAs.27 These results suggest that PINCH1 cooperates with Rsu1 in regulating cell adhesion and migration, but they have their own functions as well. PINCH1 can regulate the JNK signaling through the interaction with Rsu1.25, 26 However, the influence of Rsu1 on P38 signaling is independent of PINCH1.28
A recent study in breast cancer cells showed that ablation of Rsu1 resulted in up-regulation of PINCH1 expression, suggesting that Rsu1 acts as a negative regulator of PINCH1 to inhibit the proliferation of breast cancer cells. This result contradicts, at least in part, with the finding that Rsu1 stabilized PINCH129, further complicating the functional relationship between PINCH1 and Rsu1.
1.2.4 Pinch interactions with other factors
Thymosin beta 4 (Tβ4) is a ubiquitous protein with multiple biological functions, including protection, repair and regeneration of the mammalian heart. One important role of Tβ4 is to regulate the polymerization of actin, thus participating in the assembly of cytoskeleton. Tβ4 was shown to be able to interact with PINCH and ILK in cardiac cells. The interaction among them is important for Tβ4 to promote cardiac cell migration and survival through Akt signaling pathway.30 Treating myocardial infarction mice with Tβ4 increased the expression of ILK and PINCH1 and activated the phosphorylation of Akt.31 Thus, the beneficial effect of Tβ4 in cardiac protection may be partly attributed to its interaction with the IPP complex.
Resistance to radiotherapy is a major subject of cancer research. Akt1 signaling pathway plays an important role in this process. The expression of PINCH1 is up-regulated in many tumors. The increased PINCH1 binds to PP1α and inhibits its phosphatase activity, and, thereby, decreases Akt1 dephosphorylation by PP1α, which favors tumor cell survival under radiotherapy.32 We will discuss the relationship between PINCH1 and cancer below.
Wilms Tumor 1 (WT1) is a transcription factor that controls the expression of podocyte-specific genes. WT1 is crucial for the development of kidney during embryonic stage. As we know, TGFβ1 is a typical profibrotic factor. It was reported that treating podocytes with TGFβ1 resulted in nuclear translocation of PINCH1. In the nucleus, PINCH1 bound to WT1 and inhibited its transcriptional activation capacity.3 This study broadens our knowledge of PINCH, which not only binds to proteins in the cytoplasm, but also translocate to the nucleus where it regulates the activity of key transcription factors.
Moreover, some other proteins, including EPLIN that was recently identified and UNC-95 and UNC-98 in C. elegans can associate with PINCH.5, 7 Due to the space limitation, we will not discuss them in the review. As we have discussed above, PINCH, as one of the major components of FAs, plays an extremely important role in regulation of fundamental biological processes such as cell adhesion, migration, proliferation, and survival.
1.3 PINCH and diseases
1.3.1 PINCH and cancers
The earliest study on PINCH and tumor was demonstrated by Ann Rearden and her colleagues. They found that the expression of PINCH1 was up-regulated in tumor-associated stromal cells in patients with breast, prostate, colon, and lung carcinomas. The increase was most prominent at the invasive edges of tumors, which suggests that PINCH1, as an adhesion molecule, may play a role in tumor invasion and metastasis.33 This finding has been verified by studies from several other investigators. PINCH1 was also expressed at an elevated level in the stroma at the edge of oral squamous cell carcinoma. Patients with nodal metastasis had a much higher frequency of strong PINCH1 immunostaining compared to non-metastasis cases, suggesting a positive correlation between the expression levels of PINCH1 and tumor metastasis.34 A similar correlation of PINCH1 and nodal metastasis was observed in esophageal squamous cell carcinoma. It is possible that the expression of PINCH1 in tumor-associated stomal cells could be used to predict the ability of invasion and metastasis of esophageal squamous cell carcinoma.35 In regard to colorectal cancer, the expression of PINCH1 at invasive edges was considered as an independent prognostic indicator for patients.36, 37 A recent study on a specific subtype of colorectal cancer, mucinous adenocarcinoma, came to the same conclusion that strong expression of PINCH1 is related to poor survival.38
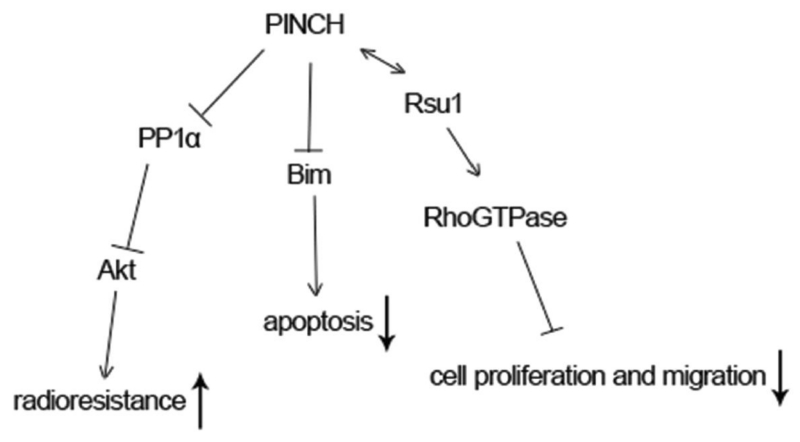
Resistance to apoptosis is an important feature of many tumor cells. One study showed that loss of PINCH1 promoted transcription of Bim and prevented it from being phosphorylated through modulating the ERK1/2 signaling. As a proapoptotic protein, Bim then entered the mitochondria to activate the apoptosis pathway (Figure 3). Thus, PINCH1 inhibits apoptosis of many tumor cells. Interestingly, the effect of PINCH1 on apoptosis does not rely on its aggregation in FAs.39
Figure 3. The roles of PINCH in tumors.
PINCH binds to PP1α and inhibits its activity, leading to increased Akt1 phosphorylation and enhanced radioresistance. PINCH prevents tumor cells from apoptosis through inhibiting Bim transcription and increasing its degradation. In hepatocytes, PINCH interacts with Rsu1, leading to decreased cell proliferation and migration.
Recent Studies showed that PINCH1 is involved in tumor resistance to radiotherapy and chemotherapy.32, 40 As mentioned above, PINCH1 activates Akt signaling through interaction with PP1α to promote survival of tumor cells under radiation and chemotherapy (Figure 3).32 It should be noted that PINCH1 regulates radiosensitivity of tumor cells under adhesion or suspension conditions.41 This suggests that there are mechanisms other than FAs for PINCH1 to mediate tumor radiosensitivity. As to clinical significance, the expression of PINCH1 could be used to predict survival of rectal cancer patients receiving radiotherapy.42
In contrast to PINCH1, the expression of PINCH2 was down-regulated in gastric cancer, which was partly due to hypermethylation in its gene. Deletion of PINCH2 in cell lines of gastric cancer increased cell migration without affecting cell proliferation. This suggests that epigenetic silencing of PINCH2 may promote invasion and metastasis of gastric cancer.43 A recent study showed that, in normal colon tissue adjacent to cancer, there was copy number amplification in the gene of PINCH2 in the recurrence-free group compared to the systemic relapse group. As to the mechanism, deletion of PINCH2 in colon cancer cells increased the levels of MMP-9 and MMP-11, which promoted tumor invasion and metastasis by autocrine and paracrine.44
Deleting PINCH1 in hepatocyte on the basis of knocking out PINCH2 globally resulted in spontaneous hepatocellular carcinomas in about 30% of the mice within one year. This is likely related to alterations in interaction between PINCH1 and Rsu1. Loss of PINCH1 diminished binding of Rsu1 to PINCH1 and decreased its expression, which led to increased hepatocyte proliferation (Figure 3).45 This seems to be contradicted with the role of PINCH1 as discussed above. It should be noted that both PINCH1 and PINCH2 were deleted in hepatocyte in this study. As we know, the two proteins have quite different roles in tumors. This may explain why the mice developed spontaneous hepatocyte carcinomas.
1.3.2 PINCH and renal diseases
PINCH also plays an important role in the kidney. Disrupting the IPP complex in podocytes reduced the foot process formation and cell-ECM adhesion, and increased the apoptosis of podocytes.46 Treating podocytes with TGF-β inhibited the formation of the IPP complex and promoted apoptosis through p38 signaling.47 In this case, PINCH1 translocated to the nucleus and interacted with WT1 to reduce the expression of Podocalyxin, a podocyte-specific protein.3 We have discussed this above. What is interesting is that TGF-β1 served to promote, rather than inhibit, the formation of the IPP complex in mesangial cells.47 Disrupting the IPP complex in mesangial cells resulted in reduced cell proliferation and matrix production.48 Besides, PINCH1 can regulate the function of the renal tubules. Treating tubular epithelial cells with TGF-β1 significantly increased the expression of PINCH1. The elevated PINCH1 then inhibited the expression of epithelium-specific proteins such as E-cadherin and ZO-1 and induced the production of fibronectin, which eventually promoted the epithelial-to-mesenchymal transition of the tubules, leading to renal fibrosis. This process is dependent on the interaction between PINCH1 and ILK. An in vivo study showed that the expression of PINCH1 was up-regulated in a time-dependent manner in the ureteral obstruction mouse model.49 In contrast to PINCH1, overexpression of PINCH2 in podocytes prevented the processes from spreading. This is probably due to a reduction of the PINCH1-ILK-parvin complex under such condition.50
So we can see from these studies, PINCH1 not only promotes the survival of podocytes through the IPP complex, but also inhibits the expression of podocyte-specific proteins by interacting with the transcription factor (i.e., WT1). On the one hand, PINCH1 protects podocytes from apoptosis; on the other hand, it promotes tubular epithelial-to-mesenchymal transition (EMT) and proliferation of the mesangial cells. It remains to be determined how PINCH1 acts so differently in different cells or under different conditions. Nonetheless, PINCH1 plays an important role in the development of renal fibrosis or even renal failure.
1.3.3 PINCH and HIV
Central nervous system damage is common in HIV-infected patients. The expression of PINCH was significantly higher in the brain and cerebrospinal fluid (CSF) in HIV patients compared to HIV-negative controls. Studies of HIV encephalitic patients revealed that PINCH was up-regulated primarily in neuron and ECM, especially at the inflammatory sites.51 This suggests that PINCH may be related to the development of central nervous system diseases occurred in HIV-infection patients.
TNF-α is one of the inflammatory factors that are responsible for neuron damage in HIV patients. Treating mouse cortical neurons with TNF-α not only up-regulated the expression of PINCH, but also changed its subcellular localization. The redistribution of PINCH under TNF-α treatment was essential for extension of neurites.52 This suggests that PINCH may serve as a repair signal, which protects neurons against HIV-induced damage.
Studies showed that TNF-α not only increased the expression of PINCH, but also resulted in the hyperphosphorylation of Tau (hpTau). Aggregation of hpTau had been reported in HIV-infected patients. Since PINCH can bind with Tau, it is possible that PINCH is involved in the phosphorylation or degradation of Tau.53 Unexpectedly, the amount of PINCH in CSF did not display a positive correlation with the CSF virus burden, but was related to the immune responses in patients.54 Further studies are required to reveal the exact role and significance of PINCH in central nervous system diseases induced by HIV infection.
1.3.4 PINCH and cardiovascular diseases
As discussed above, treating mice with coronary artery ligation with Tβ4 promoted the survival of cardiomyocytes and protected the function of the heart, which was due to its interaction with PINCH and ILK.30 Ablation of either PINCH1 or PINCH2 in cardiomyocytes in mice had no obvious defects in the heart structure and function at the basal level. However, loss of either of the two proteins deteriorated myocardial infarction induced by coronary artery ligation. Deletion of both PINCH1 and PINCH2 in the heart in mice resulted in severe dilated cardiomyopathy and death due to heart failure.55 These results suggest that PINCH is protective for the heart in such process as myocardial ischemia.
2. Conclusion and prospect
The structure of PINCH makes it an ideal adaptor to mediate the formation of multiprotein complexes. Except for the well-known IPP complex, PINCH interacts with other key proteins, such as Nck2 and Rsu1. Thus, PINCH serves to link one signaling pathway to another. Furthermore, PINCH translocates to the nucleus to regulate the transcription of genes. PINCH is widely expressed in cells throughout the body; therefore, it is not surprising that it is related to many diseases. On the one hand, PINCH protects cardiomyocytes and podocytes; on the other hand, it promotes tumor invasion and metastasis. Among the diseases associated with PINCH, cancer has gained the highest attention. There are a number of questions unsolved. How does the elevated PINCH in tumor-associated stromal cells at the invasive edge promotes tumor metastasis? Could the prognostic significance of PINCH in colorectal cancer be applied to other tumors? What is the value of it to guide the treatment of tumors? Besides, there are few studies of PINCH in diseases other than cancer. Most existing evidence is mostly restricted to in vitro cell experiments and a few mouse models. Obviously, more studies of PINCH in human diseases are required in the future. A better understanding of PINCH proteins in human diseases will help on providing new ideas for treating diseases.
Acknowledgments
The authors of this review were supported, in part, by the National Institutes of Health Grants (AR068950 and AR064874), by the National Natural Science Foundation of China Grants ((81630066, 81472049, and 81500160), and by the Shenzhen Municipal Science and Technology Innovation Council Grant (JCYJ20150331101823686).
Abbreviations
- PINCH
particularly interesting new cysteine-histidine-rich protein
- EMT
epithelial-to-mesenchymal transition
- ECM
extracellular matrix
- IPP
ILK-PINCH-parvin
- FA
focal adhesion
- ILK
integrin-linked kinase
- EGFR
epidermal growth factor receptor
- PDGFR-β
platelet-derived growth factor receptor β
- IRS-1
insulin receptor substrate 1
- Rsu1
Ras suppressor-1
- Tβ4
thymosin beta 4
- NCK2
NCK adaptor protein 2
- WT1
Wilms Tumor 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rearden A. A new lim protein containing an autoepitope homologous to “senescent cell antigen”. Biochem Biophys Res Commun. 1994;201:1124–1131. doi: 10.1006/bbrc.1994.1822. [DOI] [PubMed] [Google Scholar]
- 2.Braun A, Bordoy R, Stanchi F, Moser M, Kostka GG, Ehler E, Brandau O, Fassler R. Pinch2 is a new five lim domain protein, homologous to pinchand localized to focal adhesions. Exp Cell Res. 2003;284:239–250. doi: 10.1016/s0014-4827(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 3.Wang D, Li YJ, Wu CY, Liu YH. Pinch1 is transcriptional regulator in podocytes that interacts with wt1 and represses podocalyxin expression. Plos One. 2011:6. doi: 10.1371/journal.pone.0017048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawid IB, Breen JJ, Toyama R. Lim domains: Multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 5.Karakose E, Geiger T, Flynn K, Lorenz-Baath K, Zent R, Mann M, Fassler R. The focal adhesion protein pinch-1 associates with eplin at integrin adhesion sites. Journal of cell science. 2015;128:1023–1033. doi: 10.1242/jcs.162545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannigan GE, LeungHagesteijn C, FitzGibbon L, Coppolino MG, Radeva G, Filmus J, Bell JC, Dedhar S. Regulation of cell adhesion and anchorage-dependent growth by a new beta(1)-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 7.Qin J, Wu CY. Ilk: A pseudokinase in the center stage of cell-matrix adhesion and signaling. Curr Opin Cell Biol. 2012;24:607–613. doi: 10.1016/j.ceb.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu YZ, Li FG, Goicoechea S, Wu CY. The lim-only protein pinch directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol Cell Biol. 1999;19:2425–2434. doi: 10.1128/mcb.19.3.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li FG, Zhang YJ, Wu CY. Integrin-linked kinase is localized to cell-matrix focal adhesions but not cell-cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the pinch-binding amk repeats. Journal of cell science. 1999;112:4589–4599. doi: 10.1242/jcs.112.24.4589. [DOI] [PubMed] [Google Scholar]
- 10.Zhang YJ, Chen K, Tu YZ, Velyvis A, Yang YW, Qin J, Wu CY. Assembly of the pinch-ilk-ch-ilkbp complex precedes and is essential for localization of each component to cell-matrix adhesion sites. Journal of cell science. 2002;115:4777–4786. doi: 10.1242/jcs.00166. [DOI] [PubMed] [Google Scholar]
- 11.Li SH, Bordoy R, Stanchi F, Moser M, Braun A, Kudlacek O, Wewer UM, Yurchenco PD, Fassler R. Pinch1 regulates cell-matrix and cell-cell adhesions, cell polarity and cell survival during the peri-implantation stage. Journal of cell science. 2005;118:2913–2921. doi: 10.1242/jcs.02422. [DOI] [PubMed] [Google Scholar]
- 12.Sakai T, Li SH, Docheva D, Grashoff C, Sakai K, Kostka G, Braun A, Pfeifer A, Yurchenco PD, Fassler R. Integrin-linked kinase (ilk) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Gene Dev. 2003;17:926–940. doi: 10.1101/gad.255603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montanez E, Wickstrom SA, Altstatter J, Chu HY, Fassler R. Alpha-parvin controls vascular mural cell recruitment to vessel wall by regulating rhoa/rock signalling. Embo J. 2009;28:3132–3144. doi: 10.1038/emboj.2009.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanchi F, Bordoy R, Kudlacek O, Braun A, Pfeifer A, Moser M, Fassler R. Consequences of loss of pinch2 expression in mice. Journal of cell science. 2005;118:5899–5910. doi: 10.1242/jcs.02686. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YJ, Chen K, Guo LD, Wu CY. Characterization of pinch-2, a new focal adhesion protein that regulates the pinch-1-ilk interaction, cell spreading, and migration. J Biol Chem. 2002;277:38328–38338. doi: 10.1074/jbc.M205576200. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda T, Chen K, Shi X, Wu C. Pinch-1 is an obligate partner of integrin-linked kinase (ilk) functioning in cell shape modulation, motility, and survival. J Biol Chem. 2003;278:51324–51333. doi: 10.1074/jbc.M309122200. [DOI] [PubMed] [Google Scholar]
- 17.Olski TM, Noegel AA, Korenbaum E. Parvin, a 42 kda focal adhesion protein, related to the alpha-actinin superfamily. Journal of cell science. 2001;114:525–538. doi: 10.1242/jcs.114.3.525. [DOI] [PubMed] [Google Scholar]
- 18.Tu YZ, Li FG, Wu CY. Nck-2, a novel src homology2/3-containing adaptor protein that interacts with the lim-only protein pinch and components of growth factor receptor kinase-signaling pathways. Molecular biology of the cell. 1998;9:3367–3382. doi: 10.1091/mbc.9.12.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CY. Pinch, n(i)ck and the ilk: Network wiring at cell-matrix adhesions. Trends Cell Biol. 2005;15:460–466. doi: 10.1016/j.tcb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Velyvis A, Vaynberg J, Yang YW, Vinogradova O, Zhang YJ, Wu CY, Qin J. Structural and functional insights into pinch lim4 domain-mediated integrin signaling. Nat Struct Biol. 2003;10:558–564. doi: 10.1038/nsb938. [DOI] [PubMed] [Google Scholar]
- 21.Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu YZ, Ng L, Wu CY, Qin J. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17:513–523. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 22.Xu Z, Fukuda T, Li Y, Zha XL, Qin J, Wu CY. Molecular dissection of pinch-1 reveals a mechanism of coupling and uncoupling of cell shape modulation and survival. J Biol Chem. 2005;280:27631–27637. doi: 10.1074/jbc.M504189200. [DOI] [PubMed] [Google Scholar]
- 23.Dougherty GW, Chopp T, Qi S, Cutler ML. The ras suppressor rsu-1 binds to the lim 5 domain of the adaptor protein pinch1 and participates in adhesion-related functions. Experimental cell research. 2005;306:168–179. doi: 10.1016/j.yexcr.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Dougherty GW, Jose C, Gimona M, Cutler ML. The rsu-1-pinch1-ilk complex is regulated by ras activation in tumor cells. Eur J Cell Biol. 2008;87:721–734. doi: 10.1016/j.ejcb.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadrmas JL, Smith MA, Clark KA, Pronovost SM, Muster N, Yates JR, Beckerle MC. The integrin effector pinch regulates jnk activity and epithelial migration in concert with ras suppressor 1. J Cell Biol. 2004;167:1019–1024. doi: 10.1083/jcb.200408090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuelli L, Cutler ML. Increased expression of the ras suppressor rsu-1 enhances erk-2 activation and inhibits jun kinase activation. Mol Cell Biol. 1996;16:5466–5476. doi: 10.1128/mcb.16.10.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Nieves R, DeSantis AI, Cutler ML. Rsu1 contributes to regulation of cell adhesion and spreading by pinch1-dependent and -independent mechanisms. J Cell Commun Signal. 2013;7:279–293. doi: 10.1007/s12079-013-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YC, Gonzalez-Nieves R, Cutler ML. Rsu1 contributes to cell adhesion and spreading in mcf10a cells via effects on p38 map kinase signaling. Cell Adhes Migr. 2015;9:227–232. doi: 10.4161/19336918.2014.972775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giotopoulou N, Valiakou V, Papanikolaou V, Dubos S, Athanassiou E, Tsezou A, Zacharia L, Gkretsi V. Ras suppressor-1 promotes apoptosis in breast cancer cells by inhibiting pinch-1 and activating p53-upregulated-modulator of apoptosis (puma); verification from metastatic breast cancer human samples. Clin Exp Metastas. 2015;32:255–265. doi: 10.1007/s10585-015-9701-x. [DOI] [PubMed] [Google Scholar]
- 30.Bock-Marquette I, Saxena A, White MD, DiMaio JM, Srivastava D. Thymosin beta 4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432:466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- 31.Sopko N, Qin YL, Finan A, Dadabayev A, Chigurupati S, Qin J, Penn MS, Gupta S. Significance of thymosin beta 4 and implication of pinch-1-ilk-alpha-parvin (pip) complex in human dilated cardiomyopathy. Plos One. 2011:6. doi: 10.1371/journal.pone.0020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eke I, Koch U, Hehlgans S, Sandfort V, Stanchi F, Zips D, Baumann M, Shevchenko A, Pilarsky C, Haase M, Baretton GB, Calleja V, Larijani B, Fassler R, Cordes N. Pinch1 regulates akt1 activation and enhances radioresistance by inhibiting pp1 alpha. J Clin Invest. 2010;120:2516–2527. doi: 10.1172/JCI41078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang-Rodriguez J, Dreilinger AD, Alsharabi GM, Rearden A. The signaling adapter protein pinch is up-regulated in the stroma of common cancers, notably at invasive edges. Cancer. 2002;95:1387–1395. doi: 10.1002/cncr.10878. [DOI] [PubMed] [Google Scholar]
- 34.Zhang JT, Li QX, Wang DW, Zhu ZL, Yang YH, Cui DS, Wang MW, Sun XF. Up-regulation of pinch in the stroma of oral squamous cell carcinoma predicts nodal metastasis. Oncol Rep. 2005;14:1519–1522. [PubMed] [Google Scholar]
- 35.Zhu ZL, Yang YH, Zhang Y, Wang ZM, Cui DS, Zhang JT, Wang MW, Sun XF. Pinch expression and its significance in esophageal squamous cell carcinoma. Dis Markers. 2008;25:75–80. doi: 10.1155/2008/473860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao JF, Arbman G, Rearden A, Sun XF. Stromal staining for pinch is an independent prognostic indicator in colorectal cancer. Neoplasia. 2004;6:796–801. doi: 10.1593/neo.04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmqvist A, Gao JF, Holmlund B, Adell G, Carstensen J, Langford D, Sun XF. Pinch is an independent prognostic factor in rectal cancer patients without preoperative radiotherapy - a study in a swedish rectal cancer trial of preoperative radiotherapy. Bmc Cancer. 2012:12. doi: 10.1186/1471-2407-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang MJ, Ping J, Li Y, Holmqvist A, Adell G, Arbman G, Zhang H, Zhou ZG, Sun XF. Prognostic significance and molecular features of colorectal mucinous adenocarcinomas: A strobe-compliant study. Medicine. 2015;94:e2350. doi: 10.1097/MD.0000000000002350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K, Tu YZ, Zhang YJ, Blair HC, Zhang L, Wu CY. Pinch-1 regulates the erk-bim pathway and contributes to apoptosis resistance in cancer cells. J Biol Chem. 2008;283:2508–2517. doi: 10.1074/jbc.M707307200. [DOI] [PubMed] [Google Scholar]
- 40.Holmqvist A, Holmlund B, Ardsby M, Pathak S, Sun XF. Pinch expression in relation to radiation response in co-cultured colon cancer cells and in rectal cancer patients. Oncol Rep. 2013;30:2097–2104. doi: 10.3892/or.2013.2673. [DOI] [PubMed] [Google Scholar]
- 41.Sandfort V, Eke I, Cordes N. The role of the focal adhesion protein pinch1 for the radiosensitivity of adhesion and suspension cell cultures. Plos One. 2010:5. doi: 10.1371/journal.pone.0013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmqvist A, Holmlund B, Ardsby M, Pathak S, Sun XF. Pinch expression in relation to radiation response in co-cultured colon cancer cells and in rectal cancer patients. Oncol Rep. 2013;30:2097–2104. doi: 10.3892/or.2013.2673. [DOI] [PubMed] [Google Scholar]
- 43.Kim SK, Jang HR, Kim JH, Noh SM, Song KS, Kim MR, Kim SY, Yeom YI, Kim NS, Yoo HS, Kim YS. The epigenetic silencing of lims2 in gastric cancer and its inhibitory effect on cell migration. Biochemical and biophysical research communications. 2006;349:1032–1040. doi: 10.1016/j.bbrc.2006.08.128. [DOI] [PubMed] [Google Scholar]
- 44.Park CH, Rha SY, Ahn JB, Shin SJ, Kwon WS, Kim TS, An S, Kim NK, Yang WI, Chung HC. Pinch-2 presents functional copy number variation and suppresses migration of colon cancer cells by paracrine activity. Int J Cancer. 2015;136:2273–2283. doi: 10.1002/ijc.29273. [DOI] [PubMed] [Google Scholar]
- 45.Donthamsetty S, Bhave VS, Mars WM, Bowen WC, Orr A, Haynes MM, Wu CY, Michalopoulos GK. Role of pinch and its partner tumor suppressor rsu-1 in regulating liver size and tumorigenesis. Plos One. 2013:8. doi: 10.1371/journal.pone.0074625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang YQ, Guo LD, Blattner SM, Mundel P, Kretzler M, Wu CY. Formation and phosphorylation of the pinch-1-integrin linked kinase-alpha-parvin complex are important for regulation of renal glomerular podocyte adhesion, architecture, and survival. J Am Soc Nephrol. 2005;16:1966–1976. doi: 10.1681/ASN.2004121112. [DOI] [PubMed] [Google Scholar]
- 47.Jung KY, Chen K, Kretzler M, Wu CY. Tgf-beta 1 regulates the pinch-1-integrin-linked kinase-alpha-parvin complex in glomerular cells. J Am Soc Nephrol. 2007;18:66–73. doi: 10.1681/ASN.2006050421. [DOI] [PubMed] [Google Scholar]
- 48.Guo L, Wu CY. Regulation of fibronectin matrix deposition and cell proliferation by the pinch-ilk-ch-ilkbp complex. Faseb J. 2002;16:1298–+. doi: 10.1096/fj.02-0089fje. [DOI] [PubMed] [Google Scholar]
- 49.Li YJ, Dai CS, Wu CY, Liu YH. Pinch-1 promotes tubular epithelial-to-mesenchymal transition by interacting with integrin-linked kinase. J Am Soc Nephrol. 2007;18:2534–2543. doi: 10.1681/ASN.2007030315. [DOI] [PubMed] [Google Scholar]
- 50.Shi XH, Qu H, Kretzler M, Wu CY. Roles of pinch-2 in regulation of glomerular cell shape change and fibronectin matrix deposition. Am J Physiol-Renal. 2008;295:F253–F263. doi: 10.1152/ajprenal.00070.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rearden A, Hurford R, Luu N, Kieu E, Sandoval M, Perez-Liz G, Del Valle L, Powell H, Langford TD. Novel expression of pinch in the central nervous system and its potential as a biomarker for human immunodeficiency virus-associated neuro degeneration. J Neurosci Res. 2008;86:2535–2542. doi: 10.1002/jnr.21701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jatiani A, Pannizzo P, Gualco E, Del-Valle L, Langford D. Neuronal pinch is regulated by tnf-alpha and is required for neurite extension. J Neuroimmune Pharm. 2011;6:330–340. doi: 10.1007/s11481-010-9236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langford D, Ozdemir AY, Rom I, Kovalevich J, Yen W, Adiga R, Dave R. Pinch in the cellular stress response to tau-hyperphosphorylation. J Neuroimmune Pharm. 2013;8:423–423. doi: 10.1371/journal.pone.0058232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adiga R, Ozdemir AY, Carides A, Wasilewski M, Yen W, Chitturi P, Ellis R, Langford D. Changes in pinch levels in the csf of hiv plus individuals correlate with hptau and cd4 count. J Neurovirol. 2014;20:371–379. doi: 10.1007/s13365-014-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang XQ, Sun YF, Ye MQ, Scimia MC, Cheng HQ, Martin J, Wang G, Rearden A, Wu CY, Peterson KL, Powell HC, Evans SM, Chen J. Targeted ablation of pinch1 and pinch2 from murine myocardium results in dilated cardiomyopathy and early postnatal lethality. Circulation. 2009;120:568–576. doi: 10.1161/CIRCULATIONAHA.109.864686. [DOI] [PMC free article] [PubMed] [Google Scholar]