Abstract
Defective interfering (DI) particles are viruses that carry one or more large, internal deletions in the viral genome. These deletions occur commonly in RNA viruses due to polymerase error and yield incomplete genomes that typically lack essential coding regions. The presence of DI particles in a virus population can have a major impact on the efficiency of viral growth and is an important variable to consider in interpreting experimental results. Herein, we sought to develop a robust methodology for the quantification of DI particles within influenza A virus stocks. We took advantage of reverse transcription followed by droplet digital PCR (RT ddPCR), a highly sensitive and precise technology for determination of template concentrations without the use of a standard curve. Results were compared to those generated using standard RT qPCR. Both assays relied on the use of primers binding to terminal regions conserved in DI gene segments described to date, and internal primers targeting regions typically missing from DI particles. As has been reported previously, we observed a lower coefficient of variation among technical replicates for ddPCR compared to qPCR. Results furthermore established RT ddPCR as a sensitive and quantitative method for detecting DI gene segments within influenza A virus stocks.
Keywords: influenza virus, defective interfering particles, droplet digital PCR, incomplete viral genomes
1. Introduction
Defective interfering (DI) particles were first detected over 60 years ago and have recently been identified in natural influenza A virus (IAV) populations in human and chicken hosts (2, 7, 11, 14, 22, 24, 25). These particles carry large internal deletions in one or more gene segments but can be amplified in the context of co-infection with a second influenza A virus (2, 18). Under these circumstances, propagation of defective segment(s) further interferes with the production of intact viral progeny, reducing infectious yields and increasing particle:PFU ratio (1, 6, 12, 13, 16, 19). As a consequence, the impact of DI particles on experimental outcomes and natural IAV infections may be substantial. Xue et al. recently reported a rigorous method for preparation of influenza A virus stocks designed to limit the prevalence of DI particles (28). Despite this important advance, a robust assay for the quantification of DI segments is lacking. To address this need, we developed a method based on reverse transcription droplet digital PCR (RT ddPCR).
Droplet digital PCR allows precise determination of template copies/μl in a sample (9). We employed the Bio-Rad QX200 ddPCR platform, which uses microfluidics to partition a twenty microliter PCR mixture containing cDNA and EvaGreen dye into 20,000 nanoliter-sized water-in-oil droplets. Standard thermal cycling then leads to a strong fluorescent signal only in droplets that contain one or more templates. Subsequent scanning in a microfluidics device allows enumeration of positive and negative droplets. Template copies/μl present in the initial sample is then calculated using Poisson statistics. Key advantages of ddPCR over qPCR include improved precision and the ability to perform absolute quantification of targets in the absence of a standard curve (9, 10, 27). Specifically, absolute quantification of DNA copies is possible with ddPCR because droplets are interpreted as either positive or negative. The intensity of signal observed for positive droplets, which varies with primer/template combination, is not considered. In contrast, Ct values obtained with standard qPCR are dependent on the efficiency of amplification associated with a particular primer/template combination.
To apply droplet digital PCR technology for the measurement of DI genomes, we designed primers targeting i) nucleotides 50-150 of each gene segment, a region typically present in both DI and standard segments, and ii) an internal site lacking in DI segments described to date (Figure 1, Table 1). The ratio of internal to terminal copies/μl for a given segment indicates what proportion of the total copies is full-length. All segments were analyzed for three stocks of influenza A/Panama/2007/99 (H3N2) [Pan/99] virus of differing passage histories. Evidence of DI particles was seen in each stock, but their abundance differed substantially and as expected based on passage history. In sum, we report a sensitive and quantitative means of detecting DI particles in IAV populations.
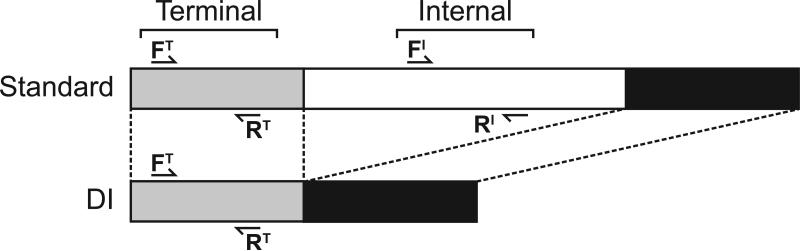
Figure 1. Strategy for detection of DI and standard gene segments by reverse transcription followed by ddPCR and qPCR.
Primers targeting the 3’ terminal region of each vRNA segment (approximately nucleotides 50-150) will bind both standard and DI cDNA templates. Primers targeting an internal region of each vRNA segment that is typically deleted in DI RNAs will bind mainly full length cDNA templates. Comparison of template copy numbers observed with terminal and internal primer sets indicates what proportion of a given vRNA is DI vs. full length.
Table 1.
Primers used for ddPCR and qPCR of Pan/99wt-His virus templates
| Gene Segment | Location | Forward Primer | Reverse Primer |
|---|---|---|---|
| PB2 | Terminal1 | 50 | 150 |
| PB2 | Internal2 | 834 | 932 |
| PB1 | Terminal | 50 | 150 |
| PB1 | Internal | 749 | 845 |
| PA | Terminal | 50 | 150 |
| PA | Internal | 857 | 935 |
| HA | Terminal | 51 | 150 |
| HA | Internal | 745 | 901 |
| NP | Terminal | 49 | 149 |
| NP | Internal | 668 | 761 |
| NA | Terminal | 50 | 152 |
| NA | Internal | 683 | 846 |
| M | Terminal | 50 | 151 |
| M | Internal | 392 | 502 |
| NS | Terminal | 69 | 194 |
| NS | Internal | 359 | 497 |
Terminal primers are located within the first (3’) 150 nt and designed to detect both defective interfering particles and standard segments.
Internal primers are beyond 359 nt and designed detect only standard segments.
2. Material and methods
2.1. PCR Primers
For DI particle detection, “terminal” and “internal” primer sets were designed to anneal specifically to a single gene segment of interest. “Terminal” primers are located within the first (3’) 150 nt of the vRNA and will detect both standard segments and defective interfering segments (4, 17, 18, 20, 22). “Internal” primers are beyond 359 nt but within the first 1000 nt and detect only standard segments (4, 17, 18, 20, 22). The primers used for Pan/99 virus are listed in Table 1. Forward and reverse primers were mixed to a final concentration of 666 nM or 2 μM for ddPCR and qPCR, respectively.
2.2. Viruses
Three virus stocks of Pan/99wt-His virus were characterized. This recombinant form of the influenza A/Panama/2007/1999 (H3N2) virus strain has been described previously (15). We selected this strain for method development due to the availability of Pan/99wt-His MDCK P3 virus, a well characterized virus stock rich in defective interfering particles (6). Briefly, Pan/99wt-His is a reverse genetics derived variant of Pan/99 virus that carries a His epitope tag and GGGS linker at the N-terminus of the HA protein. The virus stocks examined were Pan/99wt-His EP2 (egg passage 2), Pan/99wt-His 3xPP (triple plaque-purified), and Pan/99wt-His MDCK P3 (passaged in MDCK cells three times at high multiplicity of infection). The generation of Pan/99wt-His EP2 followed standard methods and comprised i) transfer of 293T cells transfected with reverse genetics plasmids to 10 day old embryonated hens’ eggs, followed by harvest of allantoic fluid containing EP1 virus; and ii) amplification of 100 PFU per egg of EP1 virus in eggs to yield the EP2 virus. Pan/99wt-His 3xPP was generated by triple plaque purification of the EP2 virus in MDCK cells, followed by a single round of amplification in eggs inoculated directly with the third plaque pick material. Preparation of Pan/99wt-His MDCK P3 virus was described previously (6). Previous characterization of this stock indicated that a large proportion of virus particles are defective and interfere with infectious virion production (6).
2.3. Sample preparation
Viral RNA was extracted from a 280 μl volume of each of the three virus stocks using the QIAamp Viral RNA Mini kit (QIAGEN). Deviations from the manufacturer's protocol included omission of carrier RNA from buffer AVL and elution in 60 μl of molecular biology grade water. Carrier RNA was omitted because we have previously found that it does not improve RNA yield, while its presence interferes with quantification of RNA product by spectrophotometry. Water was used rather than buffer AVL to ensure suitability of the sample for reverse transcription. The viral RNA was used as a template in reverse transcription reactions to produce cDNA as follows. A master mix containing reverse transcriptase buffer (1x), 0.5 μM dNTPs, 150 nM each of Uni12/Inf-1 (5’-GGGGGGAGCAAAAGCAGG) and Uni12/Inf-3 (5’-GGGGGGAGCGAAAGCAGG) primers (30), and 2 units/μl of Ribolock RNase inhibitor (ThermoScientific) were aliquoted into an 8-well PCR tube strip. Purified RNA (12 μl/reaction) and 10 units/μl Maxima reverse transcriptase (ThermoScientific) (or water for no RT reactions) were then added to the appropriate samples. Reactions were carefully capped, mixed well by vortexing, and centrifuged briefly prior to placing in a T100 thermocycler (Bio-Rad). Reactions lacking reverse transcriptase were used as negative controls in ddPCR experiments to exclude DNA contamination and gauge background fluorescence. Reverse transcription reactions were incubated at 55°C for 30 minutes, followed by heat inactivation of the reverse transcriptase at 85°C for 10 minutes. cDNA was then diluted serially in water to make working stocks for both qPCR (1:100) and ddPCR (1:500). cDNA concentrations were optimized as follows for both assays. A range of 10-fold serial dilutions of cDNA were analyzed in ddPCR and qPCR assays to ensure that starting template concentrations were within the linear range. For ddPCR, cDNA diluted 1:100 was selected for comprehensive analysis. This dilution of the cDNA yielded values of approximately 700 – 5000 copies/ul for terminal primers, depending on the target. Internal primers yielded values as low as 10 copies/ul for the MDCK P3 virus, which is still within the broad linear range of ddPCR. For qPCR, cDNA was diluted 1:500 and yielded Ct values in the range of 22 – 28, with the highest Ct values observed for internal primers on MDCK P3 templates.
2.4. DI particle detection by droplet digital PCR (ddPCR)
A master mix was made for each 1:500 diluted cDNA (4.4 μl/reaction) and 2x QX200 ddPCR EvaGreen Supermix (Bio-Rad) (11 μl/reaction). 15.4 μl of master mix was aliquoted into each well of an 8-well PCR tube strip then 6.6 μl of the appropriate primer set (“terminal” or “internal” for a particular gene segment) was added. Reactions were carefully capped, mixed well by vortexing, and centrifuged briefly to remove any liquid from the cap. A multichannel pipet was used to transfer 20 μl of PCR reaction to the sample wells of a DG8 cartridge for droplet generation (Bio-Rad). 70 μl of Droplet Generation Oil for EvaGreen (Bio-Rad) was then added to the oil wells of the DG8 cartridge. A gasket was then fitted onto the cartridge holder and placed in the QX200 droplet generator (Bio-Rad). Following droplet generation, the 40 μl volume of droplets was carefully transferred using a multichannel pipet to a Twin-Tech 96-well PCR plate (Eppendorf). Eight samples can be processed simultaneously in the droplet generator. For larger samples sets, additional droplets were generated as described above until all samples were processed. Once all droplets were transferred to the 96-well plate, the plate was heat-sealed with pierceable foil. Without mixing or spinning, the plate was placed into a C1000 Touch deep well thermocycler (Bio-Rad). An optimized protocol was adapted from that provided by the manufacturer: 95°C for 5 minutes, [95°C for 30 seconds, 57.5°C for 1 minute] ×40, 4°C for 5 minutes, 90°C for 5 minutes. The ramp rate for all steps was 2°C/s, as recommended by the manufacturer. The plate was then allowed to come to room temperature before reading using the QX200 Droplet Reader and QuantaSoft software (Bio-Rad). No RT control samples were included in each assay and results obtained from these negative controls were used to define positive vs. negative droplets in samples. Data was collected for three biological replicates, each representing an independent RNA extraction performed on different days. Two technical replicates were done for each biological replicate and results of technical replicates were averaged prior to calculating T:I ratio. Data was analyzed using GraphPad Prism software to determine the ratio of terminal:internal template copies/μl and Student's t-test with Welch's correction was used to assess statistical significance. P-values less than 0.05 were considered significant.
2.5. DI particle detection by quantitative PCR (qPCR)
A master mix was made for each 1:100 diluted cDNA (7.5 μl/reaction) and SsoFast EvaGreen Supermix (Bio-Rad) (12.5 μl/reaction). 20 μl of master mix was added directly to the appropriate well of a 96-well PCR plate then 5 μl of the appropriate primer set (“terminal” or “internal” for a particular gene segment) was added to yield a final reaction volume of 25 ul. The 96-well plate was then carefully sealed with adhesive plastic and centrifuged in a Sorvall ST 16 tabletop centrifuge (ThermoScientific) at 3000 RPM for 2 minutes. The following optimized qPCR protocol was used: 95°C for 30 seconds, [95°C for 5 seconds, 57.5°C for 5 seconds] ×40, 66°C for 5 seconds, 91°C in a CFX96 Real-Time System (Bio-Rad). Data was collected for three biological replicates and four technical replicates were done for each biological replicate (the same cDNA preparations were used for both ddPCR and qPCR to ensure consistency). Data was analyzed using GraphPad Prism software to determine terminal:internal ratios following conversion of Ct values to 2^(-Ct) and averaging of technical replicates. Student's t-test with Welch's correction was used to assess statistical significance and P-values less than 0.05 were considered significant. No-template controls were used to check for the presence of primer dimers and contaminants.
3. Results
3.1. Assay Design
Primers were designed such that the ratio of terminal (T) to internal (I) reads would indicate the presence of DI particles (Figure 1). Since the terminal sequences are retained in DI gene segments, primers annealing in this region will detect both standard and DI segments (4, 17, 18, 20, 22). In contrast, primers binding internally will amplify cDNA derived only from standard segments (4, 17, 18, 20, 22). Thus, the presence of DI segments will increase in the ratio of terminal:internal copies/μl (T>I). Standard segments on the other hand will exhibit ~1:1 ratio of terminal to internal reads indicating no deletion (T=I). The digital approach of ddPCR allows direct comparison between results obtained with different primer sets, since variability in the efficiency of PCR amplification is not a concern. Thus, for ddPCR, a ratio of T:I reads >1.0 for any segment will indicate the presence of defective interfering particles. In contrast, for standard qPCR, results obtained with different primer sets cannot be compared directly and thus relative Ct values obtained with T and I primers are less informative. For the qPCR analysis performed herein, we first linearized qPCR data by calculating 2(-Ct), and then evaluated whether the three viruses stocks differed significantly in the ratios of 2(-CtT): 2(-CtI) observed for each segment. While this approach does not reveal levels of DI particles in the virus stocks, it allows comparison among virus stocks. Processing qPCR data in this way furthermore allowed comparison herein of the utility of ddPCR and qPCR assays for DI detection.
To ensure that comparison between ddPCR and qPCR assays would be informative, we used the same cDNA templates and primers for both asays and employed PCR supermixes from Bio-Rad that were optimized for ddPCR and qPCR, respectively. We also carried out both assays in a 96 well format with similar reaction volumes and following the manufacturers’ instructions. Finally, analysis of a wide range of serially diluted cDNAs was performed by both methods to ensure that results were within the linear range. One important difference between our ddPCR and qPCR strategies was the number of technical replicates included. For ddPCR we set up two technical replicates in each assay, while for qPCR we used four technical replicates. This choice was made based on the relative cost of the two assays. While the difference in technical replicates may bias the comparison in favor of qPCR, we nonetheless observed a lower coefficient of variation for ddPCR (see below).
3.2. Detection of DI segments by ddPCR
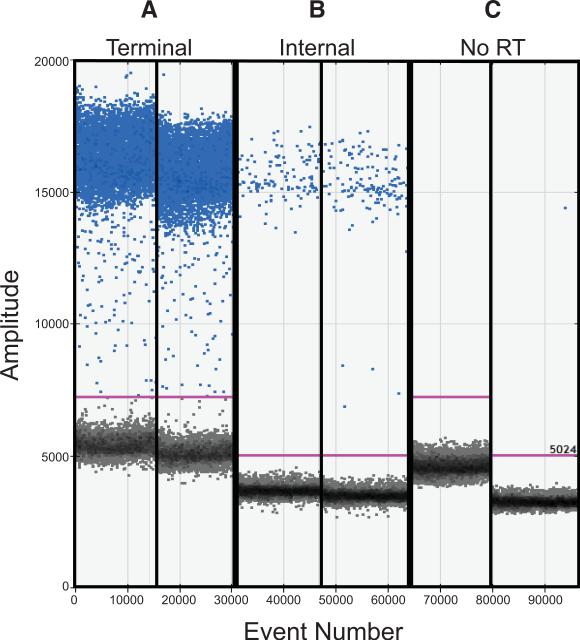
After RNA extraction and reverse transcription, ddPCR assays targeting the cDNAs of all eight viral gene segments were applied with the aim of quantifying DI gene segments in three Pan/99 virus populations of differing passage histories. The first was a standard stock grown from low dose in embryonated chicken eggs; the second was produced in eggs following triple plaque purification to limit the abundance of DI particles (12, 28); and the third was generated by serial passage in cell culture under high multiplicity conditions, which leads to enrichment of DI particles (6, 12, 28). An example of the raw data obtained by ddPCR is shown in Figure 2, with panel A displaying results acquired with terminal PB2 primers and panel B showing results obtained with internal PB2 primers, both combined with cDNA templates derived from the Pan/99wtHis-MDCK P3 virus. Panel C shows a no-RT negative control. As expected, this virus population carries many more terminal than internal PB2 sequences, yielding higher numbers of positive droplets in Figure 2A compared to Figure 2B. Data were collected in this way for all segments of the three virus stocks, using three independent RNA extracts from each stock. T:I ratios were then calculated for each biological replicate individually. The mean and standard deviation of these results are presented in Figure 3.
Figure 2. Droplet digital PCR separates droplets into template-positive and template-negative populations.
Raw data obtained by ddPCR with cDNA derived from Pan/99wt-His MDCK P3 virus are shown. The fluorescence amplitude of each droplet analyzed is indicated on the Y-axis and the event number, or order in which droplets were screened, is plotted on the X-axis. Positive droplets are colored blue and negative droplets are grey. A) Duplicate samples combined with terminal primers targeting PB1 show a high number of positive droplets. B) Duplicate samples combined with internal primers targeting PB1 show relatively few positive droplets. C) Negative controls in which no-RT control samples were combined with each pair of primers are shown.
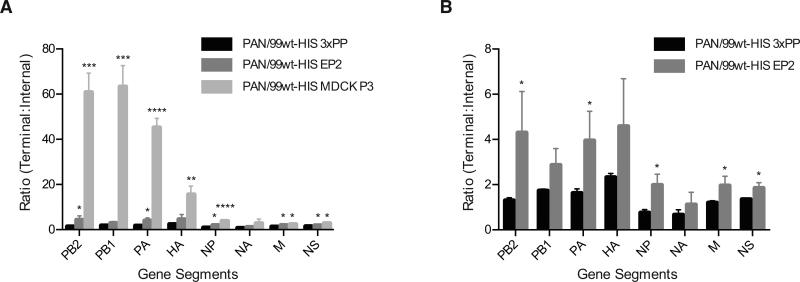
Figure 3. Defective interfering particles can be detected using droplet digital PCR.
The ratio of template copies/μl obtained with terminal and internal primers is plotted for all eight gene segments. A) Results obtained for all three virus stocks are plotted together. B) Results obtained for PAN99wt-His EP2 and triple plaque-purified (3xPP) stocks only are shown, to facilitate visualization of differences. Mean of three biological replicates is plotted and error bars indicate standard deviation. Asterisks show statistically significant differences relative to the corresponding segment of PAN99wt-His 3xPP virus (t-test). (* p<0.05; ** p<0.01; *** p<0.001; ****p<0.0001)
ddPCR results were analyzed in two ways: by one sample t-test comparing mean T:I ratios for each segment to the expected mean of 1.0, and by two sample t-test comparing results obtained with the triple plaque-purified virus stock, Pan/99wt-His 3xPP, to those collected for each of the other stocks. Results of the one sample t-tests indicated that multiple segments within all three virus stocks have T:I ratios significantly >1.0 (p<0.05). The most extreme case was the Pan/99wtHis-MDCK P3 virus, with T:I > 1.0 for all segments except NA. While the Pan/99wtHis-EP2 stock exhibited higher T:I ratios than the Pan/99wtHis-3xPP virus, the former virus showed greater variation among biological replicates; as a result, for Pan/99wtHis-EP2 virus the ratio of T:I was significantly >1.0 for only PB1 and NS segments. PB1, PA, HA and NS of the Pan/99wtHis-3xPP virus stock were all found to have T:I ratios >1.0, although the magnitude of the difference was relatively small (T:I = 2.3 for HA was the highest for this virus stock). Consistent with previous reports, the longest gene segments (PB2, PB1, and PA) were most frequently affected, yielding the highest T:I ratios (2, 3, 14, 22). It is notable, however, that this approach allowed also allowed detection of deletions in other segments (Figure 3).
Results of the two sample t-tests are indicated in Figure 3 with asterisks and facilitate comparison of data obtained by ddPCR and to that generated using qPCR.
3.3 Detection of DI segments by qPCR
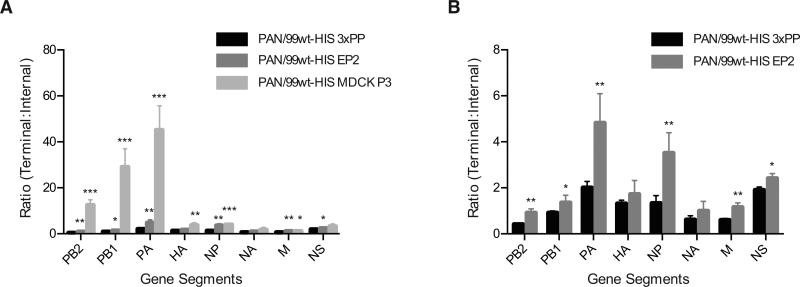
To evaluate the utility of ddPCR relative to more standard methodologies, we performed a series of qPCR experiments using the same primer sets and the same cDNAs that were used to generate the ddPCR data described above. Again, all eight segments of each virus stock were examined. Due to the lower cost of the qPCR assay relative to the ddPCR assay, samples were set up with four technical replicates for qPCR whereas two technical replicates were used for ddPCR. In contrast to ddPCR, Ct values obtained by qPCR are primer-dependent. For this reason, ratios >1.0 of 2(-CtT) : 2(-CtI) cannot be interpreted as evidence of DI segments in the virus population. These ratios can, however, be interpreted in relative terms. Thus, a significantly higher 2(-CtT): 2(-CtI) for Pan/99wt-His EP2 compared to Pan/99wt-His 3xPP indicates that the former virus stock has more DI RNAs present for a given segment than the latter virus stock. When analyzed in this way, the qPCR results obtained were comparable to those generated by ddPCR. Namely, segments in the Pan/99wt-His MDCK P3 and EP2 virus stocks that had significantly higher ddPCR T:I ratios than the corresponding segments in the Pan/99wt-His 3xPP stock also exhibited significantly higher 2(-CtT): 2(-CtI) ratios as determined by qPCR (Figure 4). Thus, both ddPCR and qPCR were sufficiently sensitive and consistent between biological replicates to reveal an increased presence of DI segments relative to the 3xPP stock. Although technical replicates showed lower variability by ddPCR compared to qPCR (see below), differences among biological replicates (that is, independent RNA extracts from each virus stock) were similar for both assays. ddPCR offers an advantage over qPCR, however, in that it allows conclusions to be drawn about the prevalence of DI segments in each virus stock, rather than only allowing comparisons between virus stocks.
Figure 4. Quantitative PCR allows comparison of DI segment abundance among virus stocks.
The ratio of 2(-Ct) obtained with terminal and internal primers is plotted for all eight gene segments. Note that, in this case, a ratio >1 does not directly indicate the presence of DI particles due to the potential for T and I primers to give differing efficiencies of amplification. A) Results obtained for all three virus stocks are plotted together. B) Results obtained for PAN99wt-His EP2 and triple plaque-purified (3xPP) stocks only are shown, to facilitate visualization of differences. Mean of three biological replicates is plotted and error bars indicate standard deviation. Asterisks show statistically significant differences relative to the corresponding segment of PAN99wt-His 3xPP virus (t-test). (* p<0.05; ** p<0.01; *** p<0.001; ****p<0.0001)
3.4 Comparison of coefficient of variation in ddPCR and qPCR datasets
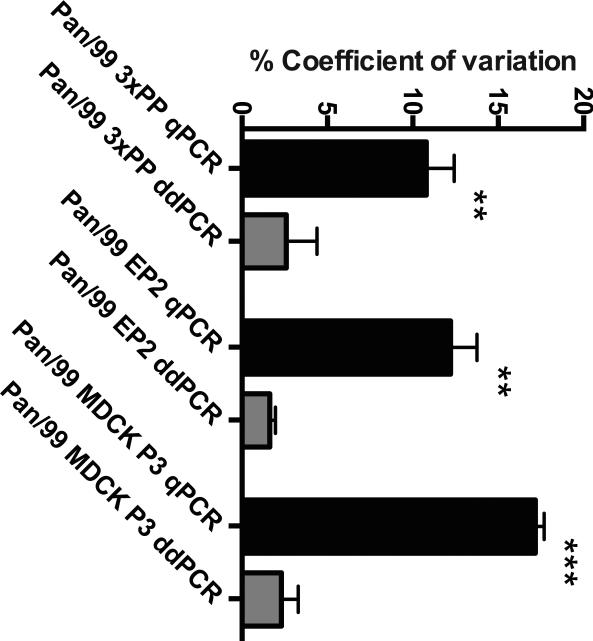
Calculation of coefficient of variation (CV) is a means of evaluating the precision of an assay. CV is equal to the standard deviation of a dataset divided by the mean. Performing this calculation is a way of normalizing standard deviation to allow comparison across datasets. We used this approach to compare ddPCR and qPCR datasets generated using the same cDNA templates and primers. For ddPCR, results of template concentration in copies/ul were evaluated. For qPCR, Ct values were first converted to a linear scale by calculating 2^(-Ct), since this conversion yields values proportional to template concentration. The %CV of technical replicates was then determined. The average %CV was determined for each biological replicate of each virus, combining all primer pairs (T & I for all eight segments). The mean and standard deviation of the three biological replicates for each virus was then determined and is plotted in Figure 5. Despite the lower number of technical replicates used for ddPCR (two, rather than four for qPCR), significantly lower coefficient of variation values were observed with ddPCR for all three virus stocks analyzed.
Figure 5. Technical replicates show a lower coefficient of variation for ddPCR compared to qPCR.
The mean and standard deviation of three biological replicates is plotted for each virus stock. Results from qPCR are shown in black and from ddPCR in grey. Significance was assessed by t-test with Welch's correction (** p<0.01; *** p<0.001)
4. Discussion
We have demonstrated that droplet digital PCR (ddPCR) can be applied to the identification of defective interfering particles in a virus preparation. Quantitative PCR (qPCR), visualization of viral RNAs or cDNAs following gel electrophoresis, and next generation sequencing have been used previously to detect DI segments in virus samples (5, 6, 14, 21, 22). However, there are caveats to each of these approaches. Quantitative PCR readouts must be interpreted in relative terms, comparing among virus stocks, since amplification efficiency is dependent on the properties of specific primer-pairs. In addition, due to reliance of qPCR quantification on serial 2-fold amplification of targets, error levels may be too high to discern small differences. These factors are important to consider when trying to identify small populations of DI segments. A limitation of both ddPCR and qPCR methods is the reliance on specific primers: unless one knows the precise sequence of a DI segment, the placement of terminal and internal primers is informed only by prior reports. As a result, terminal primers may not detect DI segments with very large deletions, while both terminal and internal primers may detect segments with small deletions. An advantage, therefore, of gel electrophoresis and next generation sequencing methods is that they are unbiased approaches not reliant on specific primers. These two methods are not, however, quantitative.
Defective interfering particles are important to monitor as they can greatly affect the phenotype of a parent virus through competitive inhibition during co-infection (1, 6, 12, 13, 16, 19). Development of a sensitive and precise assay for quantification of DI segments within a virus population will facilitate quality control of virus stocks and interpretation of data where the presence of DI particles is expected to alter outcomes. More generally, ddPCR is a versatile tool and can be optimized for many other applications, such as quantifying viral genomes (27, 29), measuring copy number variation (9), detecting rare SNPs (9, 26), and monitoring gene expression (8, 23).
Highlights.
Droplet digital PCR allows precise determination of the prevalence of defective interfering RNAs in an influenza A virus population
Relative to standard quantitative PCR, droplet digital PCR data were characterized by a lower coefficient of variation and allowed the prevalence of defective interfering RNAs to be quantified without a standard curve
Acknowledgements
This work was funded by NIH/NIAID through R01 AI099000 and Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400004C. In addition, the Georgia Research Alliance funded the purchase of droplet digital PCR instrumentation used for the work reported herein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carter MJ, Mahy BW. Incomplete avian influenza A virus displays anomalous interference. Arch Virol. 1982;74:71–6. doi: 10.1007/BF01320784. [DOI] [PubMed] [Google Scholar]
- 2.Crumpton WM, Dimmock NJ, Minor PD, Avery RJ. The RNAs of defective-interfering influenza virus. Virology. 1978;90:370–3. doi: 10.1016/0042-6822(78)90322-7. [DOI] [PubMed] [Google Scholar]
- 3.Davis AR, Nayak DP. Sequence relationships among defective interfering influenza viral RNAs. Proc Natl Acad Sci U S A. 1979;76:3092–6. doi: 10.1073/pnas.76.7.3092. PMC383769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duhaut SD, Dimmock NJ. Heterologous protection of mice from a lethal human H1N1 influenza A virus infection by H3N8 equine defective interfering virus: comparison of defective RNA sequences isolated from the DI inoculum and mouse lung. Virology. 1998;248:241–53. doi: 10.1006/viro.1998.9267. [DOI] [PubMed] [Google Scholar]
- 5.Duhaut SD, McCauley JW. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology. 1996;216:326–37. doi: 10.1006/viro.1996.0068. [DOI] [PubMed] [Google Scholar]
- 6.Fonville JM, Marshall N, Tao H, Steel J, Lowen AC. Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles. PLoS Pathog. 2015;11:e1005204. doi: 10.1371/journal.ppat.1005204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henle W, Henle G. Interference of Inactive Virus with the Propagation of Virus of Influenza. Science. 1943;98:87–9. doi: 10.1126/science.98.2534.87. [DOI] [PubMed] [Google Scholar]
- 8.Heredia NJ, Belgrader P, Wang S, Koehler R, Regan J, Cosman AM, Saxonov S, Hindson B, Tanner SC, Brown AS, Karlin-Neumann G. Droplet Digital PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods. 2013;59:S20–3. doi: 10.1016/j.ymeth.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. PMC3216358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–5. doi: 10.1038/nmeth.2633. PMC4118677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang AS, Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–7. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 12.Janda JM, Davis AR, Nayak DP, De BK. Diversity and generation of defective interfering influenza virus particles. Virology. 1979;95:48–58. doi: 10.1016/0042-6822(79)90400-8. [DOI] [PubMed] [Google Scholar]
- 13.Janda JM, Nayak DP. Defective influenza viral ribonucleoproteins cause interference. J Virol. 1979;32:697–702. doi: 10.1128/jvi.32.2.697-702.1979. PMC353605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonges M, Welkers MR, Jeeninga RE, Meijer A, Schneeberger P, Fouchier RA, de Jong MD, Koopmans M. Emergence of the virulence- associated PB2 E627K substitution in a fatal human case of highly pathogenic avian influenza virus A(H7N7) infection as determined by Illumina ultra-deep sequencing. J Virol. 2014;88:1694–702. doi: 10.1128/JVI.02044-13. PMC3911586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall N, Priyamvada L, Ende Z, Steel J, Lowen AC. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog. 2013;9:e1003421. doi: 10.1371/journal.ppat.1003421. PMC3681746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLain L, Armstrong SJ, Dimmock NJ. One defective interfering particle per cell prevents influenza virus-mediated cytopathology: an efficient assay system. J Gen Virol. 1988;69(Pt 6):1415–9. doi: 10.1099/0022-1317-69-6-1415. [DOI] [PubMed] [Google Scholar]
- 17.Nayak DP, Chambers TM, Akkina RK. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr Top Microbiol Immunol. 1985;114:103–51. doi: 10.1007/978-3-642-70227-3_3. [DOI] [PubMed] [Google Scholar]
- 18.Nayak DP, Sivasubramanian N, Davis AR, Cortini R, Sung J. Complete sequence analyses show that two defective interfering influenza viral RNAs contain a single internal deletion of a polymerase gene. Proc Natl Acad Sci U S A. 1982;79:2216–20. doi: 10.1073/pnas.79.7.2216. PMC346162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayak DP, Tobita K, Janda JM, Davis AR, De BK. Homologous interference mediated by defective interfering influenza virus derived from a temperature-sensitive mutant of influenza virus. J Virol. 1978;28:375–86. doi: 10.1128/jvi.28.1.375-386.1978. PMC354277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noble S, Dimmock NJ. Characterization of putative defective interfering (DI) A/WSN RNAs isolated from the lungs of mice protected from an otherwise lethal respiratory infection with influenza virus A/WSN (H1N1): a subset of the inoculum DI RNAs. Virology. 1995;210:9–19. doi: 10.1006/viro.1995.1312. [DOI] [PubMed] [Google Scholar]
- 21.Odagiri T, Tashiro M. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J Virol. 1997;71:2138–45. doi: 10.1128/jvi.71.3.2138-2145.1997. PMC191316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saira K, Lin X, DePasse JV, Halpin R, Twaddle A, Stockwell T, Angus B, Cozzi-Lepri A, Delfino M, Dugan V, Dwyer DE, Freiberg M, Horban A, Losso M, Lynfield R, Wentworth DN, Holmes EC, Davey R, Wentworth DE, Ghedin E. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J Virol. 2013;87:8064–74. doi: 10.1128/JVI.00240-13. PMC3700204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X, Tang C, Wang W, Zhou D, Lu Z. Digital quantification of gene expression using emulsion PCR. Electrophoresis. 2010;31:528–34. doi: 10.1002/elps.200900362. [DOI] [PubMed] [Google Scholar]
- 24.Von Magnus P. Incomplete forms of influenza virus. Adv Virus Res. 1954;2:59–79. doi: 10.1016/s0065-3527(08)60529-1. [DOI] [PubMed] [Google Scholar]
- 25.Von Magnus P. Propagation of the PR8 strain of influenza A virus in chick embryos. III. Properties of the incomplete virus produced in serial passages of undiluted virus. Acta Pathol Microbiol Scand. 1951;29:157–81. doi: 10.1111/j.1699-0463.1951.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 26.Whale AS, Bushell CA, Grant PR, Cowen S, Gutierrez-Aguirre I, O'Sullivan DM, Zel J, Milavec M, Foy CA, Nastouli E, Garson JA, Huggett JF. Detection of Rare Drug Resistance Mutations by Digital PCR in a Human Influenza A Virus Model System and Clinical Samples. J Clin Microbiol. 2016;54:392–400. doi: 10.1128/JCM.02611-15. PMC4733194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White RA, 3rd, Quake SR, Curr K. Digital PCR provides absolute quantitation of viral load for an occult RNA virus. J Virol Methods. 2012;179:45–50. doi: 10.1016/j.jviromet.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Xue J, Chambers BS, Hensley SE, Lopez CB. Propagation and Characterization of Influenza Virus Stocks That Lack High Levels of Defective Viral Genomes and Hemagglutinin Mutations. Front Microbiol. 2016;7:326. doi: 10.3389/fmicb.2016.00326. PMC4803753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Y, Jia XJ, Wang HH, Fu XF, Ji JM, He PY, Chen LX, Luo JY, Chen ZW. Dynamic quantification of avian influenza H7N9(A) virus in a human infection during clinical treatment using droplet digital PCR. J Virol Methods. 2016;234:22–27. doi: 10.1016/j.jviromet.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. doi: 10.1128/JVI.01109-09. PMC2748056. [DOI] [PMC free article] [PubMed] [Google Scholar]