RUNX1 (runt-related transcription factor 1) is a myeloid transcription factor described as recurrently mutated in de novo acute myeloid leukemia (AML; ~10%), clustering in the intermediate-risk cytogenetic group and showing prognostic adverse impact on the overall survival and disease progression.1, 2, 3 In the World Health Organization (WHO) Classification of Tumors of Hematopoietic and Lymphoid Tissues,4 AML are classified in the categories ‘AML with recurrent genetic abnormalities', ‘AML with myelodysplasia-related changes (AML-MRC)', ‘therapy-related myeloid neoplasms' and ‘AML, not otherwise specified'. AML-MRC includes cases with a myelodysplasia-related cytogenetic abnormality, a previous myeloid malignancy or showing multilineage dysplasia (MLD). MLD positive (MLD+) morphology shows dysplastic features in ⩾50% of cells in ⩾2 hematopoietic lineages.5 In 2008 ‘AML with mutated NPM1' and ‘AML with mutated CEBPA' were introduced as provisional entities and reached in 2016 a status as own entities.4, 5 Therein, the presence of MLD alone will not classify a case as AML-MRC when one of these mutations is present. Ongoing discussions now focus on RUNX1 mutations characterizing the new provisional entity ‘AML with mutated RUNX1'. However, classification of patients with MLD and RUNX1 mutation into the AML-MRC category is questionable and needs to be discussed. We therefore comprehensively analyzed 152 RUNX1-mutated AML patients by cytogenetics and molecular genetics, and especially investigated the prognostic impact of MLD. RUNX1-mutated AML showed strong associations to trisomy 13 (13/152, 9%) and mutations within genes coding for the spliceosome (88/140, 63%), and for chromatin modifiers (86/140, 61%). However, MLD did not show prognostic impact in multivariate Cox regression analyses. This supports an approach to classify RUNX1-mutated AML also as a new provisional entity irrespective of dysplastic features.
In this study, we investigated 152 AML patients at diagnosis harboring a RUNX1 mutation. The cohort comprised 49 female and 103 male, the median age was 67 years, ranging from 18 to 78 years. Ninety-nine percent of patients had a de novo AML and 1% a secondary AML. Therapy-related AML were excluded, as these are classified separately within the WHO. Forty out of 152 (26%) had allogeneic transplantation in the follow-up. All samples underwent May-Grünwald-Giemsa staining and cytochemistry. Dysplasia was assessed according to Goasguen et al.6 MLD was defined by ⩾50% dysplastic cells in ⩾2 lineages following the WHO guidelines.4, 5 In 132/152 patients all three lineages were evaluable, while in 20 cases only two hematopoietic lineages were evaluable. All patients were investigated by the standard chromosome banding analysis (cytogenetics) and a diagnostic molecular genetic approach following European Leukemia Network (ELN) guidelines.7 All patients had prognostically intermediate karyotypes according to Medical Research Council criteria (group 2).8 In addition, a next-generation sequencing-based mutational screening targeting 25 genes (Supplementary Table S2) was performed in 140/152 patients. All patients were intensively treated according to standard AML protocols.9 For further details and patients characteristics, see Supplementary Material. All patients gave written informed consent for research studies; the study design adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Board before its initiation.
Within the total cohort of 152 AML patients with RUNX1 mutations, the majority were classified as M2 and M1 according to French-American-British (FAB) criteria10 (64/152, 42% and 45/152, 30%, respectively), followed by M0 (31/152, 20%), M4 (9/152, 6%) and M6 (3/152, 2%). This confirms earlier studies that described the immature and undifferentiated morphology of RUNX1-mutated AML, reflected by the high number of M0 subtypes, as well as a comparison to a matched control cohort without RUNX1 mutation from our data base, showing only 2% of M0 cases (21/886, P<0.001; Supplementary Table S3).11 Addressing dysplasia revealed dysplastic granulopoiesis in 24% (37/152), erythropoiesis in 31% (47/152) and megakaryopoiesis in 55% (73/132) of patients. A total of 44 patients (33%) had no dysplastic features in any of the three cell lineages, 38 (29%) had unilineage dysplasia, 39 (30%) had bilineage dysplasia, whereas 11 cases (8%) had trilineage dysplasia (TLD). In four cases, a differentiation of bilineage dysplasia or TLD was not possible, as megakaryopoietic dysplasia was not evaluable. MLD was detected in 36% (54/152) of the analyzed bone marrow samples. These numbers are in line with a number of large AML studies, where MLD was found in 23–36% and TLD in 2–15% (Supplementary Table S4).
Chromosome banding analysis revealed cytogenetic aberrations in 59/152 (39%) patients. Thereof, 42 patients showed trisomies as sole abnormality, whereas only 17 showed other aberrations. In detail, 17 cases showed trisomy 8 (+8), 13 cases +13 and 4 patients each +11 and +14. Further four non-recurrent trisomies were observed. Only two cases had three aberrations, classifying for AML-MRC with complex karyotypes (⩾3 unrelated abnormalities). Although +8 is one of the most frequent recurrent cytogenetic aberrations in AML (10% of AML cases),8 +13 is a very rare event (~1% of AML);12 however, interestingly both show high incidences of RUNX1 mutations.1, 13
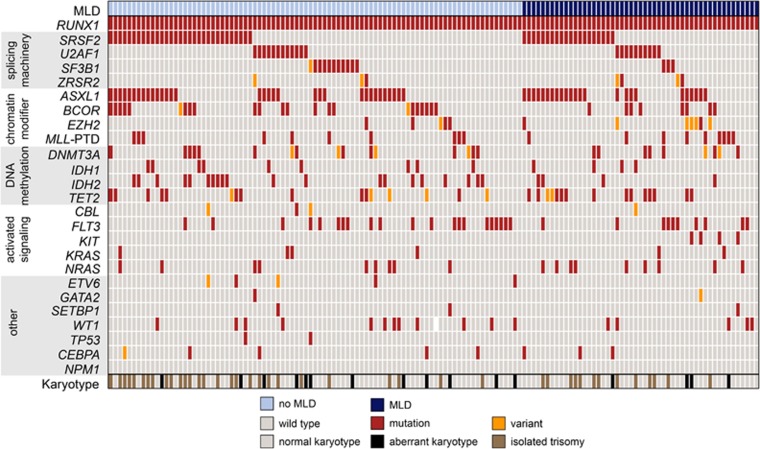
The highest mutation frequency besides RUNX1 mutations was observed for ASXL1 (41%), followed by SRSF2 (36%), FLT3 (22% p.Asp835 and internal tandem duplication), BCOR (21%), TET2 (18%), IDH2 (17%) and U2AF1 (16%). Mono-allelic CEBPA mutations were rarely detected (5%), double-mutated CEBPA was not identified, clearly differentiating these AML entities. NPM1-mutated cases (n=3) were excluded, as these cases qualify already for an own entity. Overall, 461 additional mutations were identified in 140 analyzed patients, resulting in a median number of three additional mutations (range: 0–6). Thus, in 98% of patients at least one additional mutation besides the RUNX1 mutation was observed (Figure 1). Grouping the gene mutations to cellular pathways resulted in a high number of patients, harboring at least one mutation within the splicing machinery (63%), chromatin modification (61%), followed by DNA methylation (48%) and activated signaling (40%). The high incidence of mutations within the splicing machinery as well as chromatin modification is in line with very recently published data,3 as well as the high occurrence of trisomy 13 within this RUNX1-mutated cohort, where a high incidence of SRSF2 and ASXL1 mutations have also been described previously.14 However, these molecular genetic patterns occurred in our cohort also within cases with normal karyotype, indicating that rather RUNX1 than +13 might be the trigger. In a very comprehensive study on 200 AML patients by whole-genome and whole-exome sequencing the respective pathways—splicing machinery and chromatin modification—were found to be mutated less frequently with 14% and 30%, respectively, indicating a specific genotype in RUNX1-mutated AML compared with overall AML.15 Assessing the classification according to Lindsley et al.2 would characterize these gene mutations as secondary type AML specific, whereas a RUNX1 mutation itself is classified as de novo/pan AML alteration.
Figure 1.
Molecular, cytogenetic and morphological characterization of AML patients with RUNX1 mutation. Illustration of all 140 analyzed cases, each column represents one patient. All 25 analyzed genes, the occurrence of trisomies as sole aberration or other cytogenetic aberrations, as well as the presence of MLD (multilineage dysplasia) are given for each patient. CEBPA was single mutated in all mutated cases. Light blue: cases without MLD, dark blue: cases with MLD, light gray: wild-type gene and normal karyotype, red: mutation, orange: variant, brown: aberrant karyotype with isolated trisomy, black: other aberrant karyotype and white: no data available.
MLD− patients showed a higher blast count than MLD+ cases (62 vs 46%, P<0.001), a higher incidence of +13 (12 vs 2% in MLD+, P=0.032), IDH2 mutation (23 vs 8% in MLD+, P=0.035), but no KIT mutation (0 vs 8% in MLD+, P=0.016). All other clinical parameters, chromosomal alterations and additional gene mutations did not differ significantly between the MLD− and MLD+ patients (Supplementary Table S1). Furthermore, there was no difference in RUNX1 mutation localization and mutation type between MLD+ and MLD− patients (Supplementary Figure S1).
In univariate analyses, the overall survival was adversely influenced by MLD+ (20 vs 31 months (mo), P=0.039), ⩾3 mutations in addition to RUNX1 (20 vs 39 mo, P=0.003), mutations within the spliceosome (23 vs 43 mo, P=0.036), DNMT3A (20 vs 36 mo, P=0.032), NRAS (12 vs 31 mo, P=0.026) and U2AF1 (21 vs 33 mo, P=0.039; Supplementary Figure S2). In multivariate Cox regression, only ⩾3 mutations retained the independent adverse prognostic influence (Table 1).
Table 1. Univariate and multivariate Cox regression analyses for the overall survival considering the covariates MLD and ⩾3 mutations, MLD and spliceosome mutations and MLD and mutations in the genes DNMT3A, NRAS and U2AF1.
|
Cox regression | ||||||||
|---|---|---|---|---|---|---|---|---|
|
Univariate |
Multivariate |
|||||||
| P-value | HR |
95% CI |
P-value | HR |
95% CI |
|||
| Lower | Upper | Lower | Upper | |||||
| Overall survival | ||||||||
| MLD | 0.041 | 1.656 | 1.022 | 2.685 | 0.118 | 1.494 | 0.904 | 2.470 |
| ⩾3 mutations | 0.003 | 2.176 | 1.297 | 3.649 | 0.005 | 2.108 | 1.255 | 3.539 |
| MLD | 0.041 | 1.656 | 1.022 | 2.685 | 0.099 | 1.528 | 0.923 | 2.529 |
| Spliceosome mutations | 0.038 | 1.763 | 1.031 | 3.015 | 0.050 | 1.714 | 1.001 | 2.935 |
| MLD | 0.041 | 1.656 | 1.022 | 2.685 | 0.066 | 1.634 | 0.969 | 2.755 |
| DNMT3A | 0.036 | 2.044 | 1.048 | 3.985 | 0.056 | 1.924 | 0.982 | 3.768 |
| NRAS | 0.031 | 2.389 | 1.085 | 5.258 | 0.058 | 2.246 | 0.972 | 5.190 |
| U2AF1 | 0.042 | 1.898 | 1.023 | 3.521 | 0.203 | 1.541 | 0.792 | 3.001 |
Abbreviations: CI, confidence interval; HR, hazard ratio; MLD, multilineage dysplasia.
In conclusion, MLD- and MLD+ RUNX1-mutated AML differ in some associations to genetic markers, such as +13 or IDH2 mutation status without prognostic impact in multivariate analysis. However, in RUNX1-mutated AML, the overall pattern shows a specific landscape with high incidences of trisomies (such as +8 and +13), and mutations in the spliceosome and in chromatin modifiers, characterizing a unique secondary type AML phenotype.2 RUNX1-mutated AML shows shorter event-free survival,1 and we found ⩾3 mutations as independent prognostic marker influencing prognosis. However, the detection of MLD has no independent influence in multivariate analysis. We therefore strongly support the classification of RUNX1-mutated AML as a provisional entity in the new WHO classification, but without further consideration of dysplastic features such as MLD.
Acknowledgments
We thank all clinicians for sending samples to our laboratory for diagnostic purposes and for providing the clinical information and follow-up data. In addition, we thank all the co-workers at the MLL Munich Leukemia Laboratory for approaching together many aspects in the field of leukemia diagnostics and research, especially Niroshan Nadarajah for excellent support with the graphic arts.
Author contributions
TH was responsible for the cytomorphologic analysis and was the principle investigator of the study. AS contributed to the cytogenetics, SE to the cytomorphology, KP to the mutation analyses and TA to the collection of clinical data. WK was involved in the statistical analyses. CH was responsible for the cytogenetics. MM investigated the molecular mutations, analyzed the data and wrote the manuscript. All authors read and contributed to the final version of the manuscript.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
TH, WK and CH declare part ownership of the MLL Munich Leukemia Laboratory. AS, SE, TA and MM are employed by the MLL Munich Leukemia Laboratory, KP by MLL2.
Supplementary Material
References
- Gaidzik VI, Bullinger L, Schlenk RF, Zimmermann AS, Rock J, Paschka P et al. RUNX1 Mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J Clin Oncol 2011; 29: 1364–1372. [DOI] [PubMed] [Google Scholar]
- Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015; 125: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidzik VI, Teleanu V, Papaemmanuil E, Weber D, Paschka P, Hahn J et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia 2016; e-pub ahead of print 3 May 2016; doi:10.1038/leu.2016.126. [DOI] [PubMed]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood 2016; 127: 2391–2405. [DOI] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. International Agency for Research on Cancer (IARC): Lyon, France, 2008. [Google Scholar]
- Goasguen JE, Matsuo T, Cox C, Bennett JM. Evaluation of the dysmyelopoiesis in 336 patients with de novo acute myeloid leukemia: Major importance of dysgranulopoiesis for remission and survival. Leukemia 1992; 6: 520–525. [PubMed] [Google Scholar]
- Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015; 373: 1136–1152. [DOI] [PubMed] [Google Scholar]
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010; 116: 354–365. [DOI] [PubMed] [Google Scholar]
- Buchner T, Berdel WE, Haferlach C, Haferlach T, Schnittger S, Muller-Tidow C et al. Age-related risk profile and chemotherapy dose response in acute myeloid leukemia: a study by the German Acute Myeloid Leukemia Cooperative Group. J Clin Oncol 2009; 27: 61–69. [DOI] [PubMed] [Google Scholar]
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med 1985; 103: 620–625. [DOI] [PubMed] [Google Scholar]
- Dicker F, Haferlach C, Kern W, Haferlach T, Schnittger S. Trisomy 13 is strongly associated with AML1/RUNX1 mutations and increased FLT3 expression in acute myeloid leukemia. Blood 2007; 110: 1308–1316. [DOI] [PubMed] [Google Scholar]
- Mesa RA, Hanson CA, Ketterling RP, Schwager S, Knudson RA, Tefferi A. Trisomy 13: prevalence and clinicopathologic correlates of another potentially lenalidomide-sensitive cytogenetic abnormality. Blood 2009; 113: 1200–1201. [DOI] [PubMed] [Google Scholar]
- Alpermann T, Haferlach C, Eder C, Nadarajah N, Meggendorfer M, Kern W et al. AML with gain of chromosome 8 as the sole chromosomal abnormality (+8sole) is associated with a specific molecular mutation pattern including ASXL1 mutations in 46.8% of the patients. Leuk Res 2015; 39: 265–272. [DOI] [PubMed] [Google Scholar]
- Herold T, Metzeler KH, Vosberg S, Hartmann L, Rollig C, Stolzel F et al. Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis 3. Blood 2014; 124: 1304–1311. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.