TET2, located on chromosome 4q24, is frequently mutated (~60%) in patients with chronic myelomonocytic leukemia (CMML).1, 2 TET2 has 11 exons, and variations, especially in exon 3 have been described as a part of age-related clonal hematopoiesis.3 In a large population-based study (n=17 182), somatic variations involving DNMT3A, TET2 and ASXL1 were seen in ~11% of the population >80 years of age, and in comparison with patients without clonal hematopoiesis, were associated with an increased risk of hematological malignancies (HR- 11.1) and all-cause mortality (HR-3.7).3 In CMML, thus far, clonal TET2 mutations in the absence of clonal ASXL1 mutations (ASXL1wt/TET2mt) have been associated with favorable outcomes.1, 4 The exact mechanism behind this interaction remains to be elucidated, one potential explanation being better responses to hypomethylating agents.4 In the current larger CMML patient cohort (n=261), we describe the number and type of TET2 mutations and examine their phenotypic and prognostic effects.
Two hundred and sixty one patients with CMML were included in the study. All patients had bone marrow (BM) biopsies and cytogenetics performed at diagnosis. Targeted capture assays were carried out on BM DNA specimens obtained at diagnosis for the following genes: TET2, DNMT3A, IDH1, IDH2, ASXL1, EZH2, SUZ12, SRSF2, SF3B1, ZRSR2, U2AF1, PTPN11, Tp53, SH2B3, RUNX1, CBL, NRAS, KRAS, JAK2, CSF3R, FLT3, KIT, CALR, MPL, NPM1, CEBPA, IKZF and SETBP1, by previously described methods.1 TET2 (NM_001127208.2) coverage extended from exons 3–11, with frame shift, nonsense and missense variations considered pathogenic. Previously annotated single nucleotide polymorphisms (http//www.hapmap.org) were considered non-pathogenic. The 2008 and 2016 World Health Organization (WHO) criteria were used for CMML diagnosis and classification.5
Among the 261 study patients, 65% were males and median age was 70 years (range, 28–91). One hundred and fifty four (59%), 64 (25%) and 43 (16%) patients were classified as CMML-0, 1 and 2, respectively. At a median follow-up of 23 months, 174 (67%) deaths and 37 (14%) leukemic transformations were documented. Mutational frequencies included: ASXL1 45%, TET2 43%, SRSF2 40%, NRAS 14%, SETBP1 13%, CBL 10%, JAK2 7%, RUNX1 6%, DNMT3A 6%, U2AF1 6%, SF3B1 5%, ZRSR2 4%, Tp53 4%, IDH2 4%, KRAS 3%, c-KIT 3%, PTPN11 3% and <1% each for FLT3ITD, CALR and MPL. There were no IKZF, STAG2 or SH2B3 mutations seen.
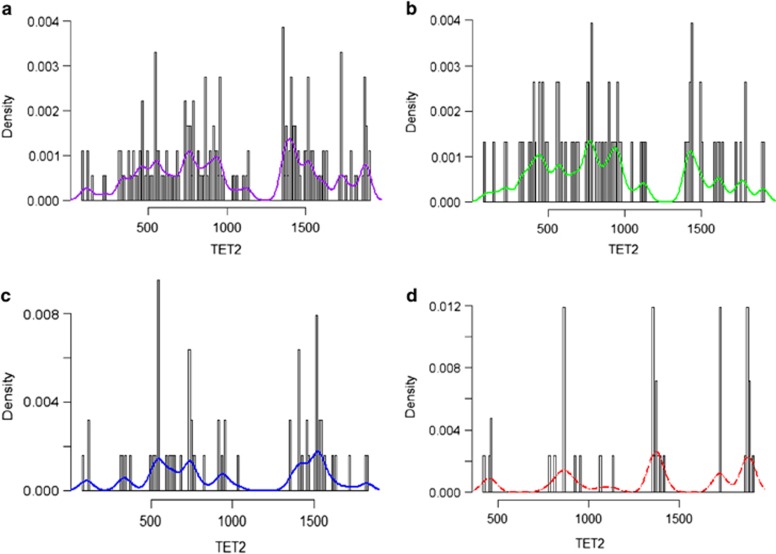
Two hundred and sixty four TET2 mutations were seen in 113 (43%) patients; these included 34 (30%) patients with frameshift, 30 (27%) with nonsense and 13 (10%) with missense mutations, whereas 36 (33%) patients had more than one type of mutation (Figure 1). Overall, 58 (52%) patients had more than 1 TET2 mutation: 55 (49%) patients had 1, 47 (41%) had 2 and 11 (10%) had ⩾3 TET2 mutations. The median variant allelic fractions (VAF) for TET2 mutations included; frameshift 43% (range, 10–92%), nonsense 47% (range, 9–100%) and missense 47% (range, 14–95%), respectively.
Figure 1.
Characterization of TET2 mutations. Each plot is generated using all mutations from their respective categories. The relative proportion of the mutation subtype is shown on the y axis, across the length of the TET2 gene, from 0 to 2002 amino acids. The colored bar represents the density of the mutations along the gene. (a) All mutation types, (b) frameshift mutations, (c) nonsense mutations and (d) missense mutations.
Among the 113 TET2 mutated patients, 65% were males, and median age was 71 years with no significant difference in age and gender distribution between mutated and un-mutated cases, or type of TET2 mutations; however, older patients were more likely to carry multiple TET2 mutations (P=0.01) (Table 1). The frequency distribution of TET2 mutations with age was: <50 years n=15 (6%), 50–59 years n=25 (10%), 60–69 years n=83 (31%), 70–79 years n=104 (39%) and ⩾ 80 years n=37 (14%), respectively. The cytosine-to-thymidine (C:G> T:A) base pair change (transition) is often considered a somatic mutational signature of ageing.6, 7 In this cohort, C>T base pair changes proportionally comprised, 0% <50 years, 18% 50–60 years, 44% 60–69 years, 41% 70–79 years and 50% ⩾ 80 years. In addition, 73% of patients with TET2 C>T base pair changes had more than one TET2 mutation. DNMT3A mutations significantly clustered with TET2 C>T base pair changes (P=0.03), with 5 of 6 (83%) DNMT3A-mutated patients having concomitant TET2 C>T base pair changes. Incidentally, only 2 of 6 (33%) DNMT3A mutations themselves were as a result of C>T base pair changes.
Table 1. Clinical and laboratory features and subsequent events in 261 patients with World Health Organization defined chronic myelomonocytic leukemia (CMML), stratified by the presence or absence of TET2 mutations.
| Variable | All patients with CMML (n=261) | CMML patients with TET2 mutations (n=109) | CMML patients without TET2 mutations (n=152) | P-value |
|---|---|---|---|---|
| Age in years; median (range) | 70 (20–91) | 64.5 (20–87) | 70 (27–91) | 0.067 |
| Males; n (%) | 168 (64) | 9 (56) | 159 (65) | 0.48 |
| Hemoglobin g/dl; median (range) | 10.6 (6.4–16.9) | 9.6 (6.8–13.2) | 10.7 (6.4–16.9) | 0.093 |
| MCV femtoliter; median (range) | 91 (59–119) | 91 (75–112) | 91 (59–119) | 0.5 |
| WBC × 109/l; median (range) | 12.1 (1.5–264) | 12.6 (2.9–71.5) | 12 (1.5–264) | 0.83 |
| ANC × 109/l; median (range) | 5.8 (0–151) | 6.7 (1–39.2) | 5.7 (0–151) | 0.74 |
| AMC × 109/l; median (range) | 2.3 (1.0–40) | 1.7 (1.0–20) | 2.4 (1.0–40) | 0.756 |
| ALC × 109/l; median (range) | 1.7 (0–22) | 1.9 (0.4–5.6) | 1.7 (0–22) | 0.82 |
| Platelets × 109 /l; median (range) | 97 (10–840) | 112 (11–840) | 96 (10–726) | 0.45 |
| Presence of circulating immature myeloid cells; n (%) | 142 (54) | 9 (60) | 133 (55) | 0.7 |
| PB blast % median (range) | 0 (0–19) | 0 (0–19) | 0 (0–7) | 0.3 |
| BM blast % ; median (range) | 3 (0–19) | 3 (0–13) | 3 (0–19) | 0.9 |
| BM cellularity % | 80 (40–100) | |||
| Lactate dehydrogenase levels IU/ml; n (range) | 225 (84–1296) | 223 (109–294) | 225 (84–1296) | 0.48 |
| Next-generation sequencing analysis; n (%) | ||||
| Epigenetic regulators | ||||
| DNMT3A | (45) | (50) | (45) | 0.7 |
| IDH1 | 4 (2) | 0 (0) | 4 (2) | 0.6 |
| IDH2 | 11 (4) | 0 (0) | 11 (4) | 0.38 |
| Chromatin regulation | ||||
| ASXL1 | 120 (50) | 6 (37) | 114 (51) | 0.3 |
| EZH2 | 3 (1) | 0 (0) | 3 (1) | 0.656 |
| SUZ12 | 0 | 0 (0) | 0 (0) | − |
| Transcription factors | ||||
| RUNX1 | 16 (6) | 2 (12) | 14 (6) | 0.27 |
| Spliceosome components | ||||
| SF3B1 | 13 (5) | 4 (25) | 9 (4) | 0.0001 |
| SRSF2 | 105 (40) | 1 (6) | 104 (42) | 0.0042 |
| U2AF1 | 16 (6) | 2 (12) | 14 (6) | 0.2 |
| ZRSR2 | 5 (3) | 0 (0) | 5 (2) | 0.8 |
| Cell signalling | ||||
| JAK2 V617F | 17 (7) | 1 (6) | 16 (7) | 0.9 |
| CALR | 1 (0.5) | 0 (0) | 1 (0.5) | 0.8 |
| MPL | 1 (0.4) | 0 (0) | 1 (0.5) | 0.8 |
| SH2B3 | 1 (0.5) | 0 (0) | 1 (0.5) | 0.8 |
| CBL | 25 (10) | 0 (0) | 25 (10) | 0.4 |
| KRAS | 8 (3) | 0 (0) | 8 (3) | 0.5 |
| NRAS | 37 (14) | 2 (16) | 35 (14) | 0.8 |
| PTPN11 | 6 (2) | 2 (12) | 4 (2) | 0.005 |
| CSF3R | 3 (1) | 0 (0) | 3 (1) | 0.7 |
| C-KIT | 7 (3) | 1 (6) | 6 (2) | 0.4 |
| FLT3TKD | 1 (0.5) | 0 (0) | 1 (0.5) | 0.8 |
| NPM1 | 0 | 0 (0) | 0 (0) | − |
| Tumor suppressor genes | ||||
| Tp53 | 10 (4) | 2 (12) | 9 (4) | 0.09 |
| PHF6 | 0 | 0 (0) | 0 (0) | − |
| Others | ||||
| SETBP1 | 34 (13) | 2 (12) | 32 (13) | 0.9 |
| IKZF | 0 | 0 (0) | 0 (0) | − |
| 2008 WHO morphological subtypes; n (%) | ||||
| CMML-1 | 221 (84) | 13 (81) | 208 (85) | 0.7 |
| CMML-2 | 40 (16) | 3 (19) | 37 (15) | |
| 2016 WHO morphological subtypes; n (%) | ||||
| CMML-0 | 154 (59) | 10 (62) | 144 (59) | 0.9 |
| CMML-1 | 65 (25) | 4 (25) | 61 (25) | |
| CMML-2 | 42 (16) | 2 (12) | 40 (16) | |
| aSpanish Cytogenetic risk stratification; n (%) | ||||
| Low | 180 (72) | 11 (69) | 169 (72) | 0.1 |
| Intermediate | 43 (17) | 1 (6) | 42 (18) | |
| High | 27 (11) | 4 (25) | 23 (10) | |
| aMayo-French cytogenetic risk stratification; n (%) | ||||
| Low | 180 (72) | 11 (69) | 169 (72) | 0.4 |
| Intermediate | 57 (23) | 3 (19) | 54 (23) | |
| High | 13 (5) | 2 (12) | 11 (5) | |
| Mayo prognostic model; n (%) | ||||
| Low | 89 (34) | 3 (20) | 86 (35) | 0.2 |
| Intermediate | 83 (32) | 4 (27) | 79 (32) | |
| High | 87 (34) | 8 (53) | 79 (32) | |
| Molecular Mayo model; n (%) | ||||
| Low | 26 (10) | 0 (0) | 26 (11) | 0.5 |
| Intermediate-1 | 72 (28) | 4 (25) | 68 (28) | |
| Intermediate-2 | 79 (30) | 6 (37) | 73 (30) | |
| High | 81 (31) | 6 (37) | 75 (31) | |
| GFM CMML prognostic model; n (%) | ||||
| Low | 119 (46) | 9 (56) | 110 (45) | 0.4 |
| Intermediate | 92 (36) | 6 (37) | 86 (35) | |
| High | 48 (18) | 1 (6) | 47 (19) | |
| Leukemic transformations; n (%) | 37 (14) | 4 (25) | 33 (13) | 0.2 |
| Deaths; n (%) | 174 (67) | 10 (62) | 164 (67) | 0.7 |
Abbreviations: ALC, absolute lymphocyte count; AMC, absolute monocyte count; ANC, absolute neutrophil count; BM, bone marrow; CMML, chronic myelomonocytic leukemia; FAB, French American British; GFM, Groupe Français des Myélodysplasies; MCV, mean corpuscular volume; PB, peripheral blood; WBC, white blood cell count; WHO, World Health Organization.
Cytogenetic studies were available for 250 patients with chronic myelomonocytic leukemia at diagnosis.
Compared with their un-mutated counterparts, TET2-mutated cases were less likely to have a low hemoglobin (P<0.001), include CMML-2 (P=0.007), have circulating immature myeloid cells (P=0.001), have peripheral blood (P=0.009) and BM blasts (P=0.009), and have higher-risk stratification per clinical, cytogenetic and molecularly inclusive CMML prognostic models (Table 1); these differences were not affected by the type or number of TET2 mutations. TET2 mutated cases were more likely to have a higher frequency of SRSF2 (P=0.004) and a lower frequency of ASXL1 (P=0.03), Tp53 (P=0.04) and IDH1/2 mutations (P<0.001); these associations were also not affected by the type or number of TET2 mutations.
Median survival for the entire cohort (n=261) was 24 months. In univariate analysis, survival was superior in TET2-mutated (median 33 months) versus wild-type (median 21 months) patients (P=0.03; HR 1.3 95% CI 1.12–1.86). This survival difference remained significant after adjustment for age (P=0.04), leukocyte count (P=0.017), absolute monocyte count (P=0.02), absolute lymphocyte count (P=0.02), platelet count (P=0.015), circulating immature myeloid cells (P=0.03), DNMT3A (P=0.02) and ASXL1 (P=0.045) mutations; however, significance was lost after adjustment for abnormal karyotype (P=0.32) and the Mayo Molecular Model (P=0.0003). These observations were not affected by the type or number of TET2 mutations. Finally, our previous observation regarding the survival advantage of ASXL1wt/TET2mt versus other genotypes was most apparent for patients with multiple TET2 mutations (P=0.02).1
TET2 mutations are frequent in CMML (~45%) and constitute approximately equal proportions of frameshift and nonsense mutations, while missense mutations are less frequent. Majority of TET2-mutated CMML cases harbor more than one mutant variant. Regardless, the relevance of type and number of TET2 mutations in CMML was limited to an association between older age and number of mutations, and the latter with possibly improved survival in the absence of clonal ASXL1 mutations.
Acknowledgments
The current article is supported in part by grants from the ‘The Henry J. Predolin Foundation for Research in Leukemia, Mayo Clinic, Rochester, MN, USA.' This publication was supported by CTSA Grant Number KL2 TR000136 from the National Center for Advancing Translational Science (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. We would like to acknowledge Ryan Hlady, Ph.D for his help with Figure 1.
The authors declare no conflict of interest.
References
- Patnaik MM, Lasho TL, Vijayvargiya P, Finke CM, Hanson CA, Ketterling RP et al. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J 2016; 6: e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol 2013; 31: 2428–2436. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik MM, Wassie EA, Padron E, Onida F, Itzykson R, Lasho TL et al. Chronic myelomonocytic leukemia in younger patients: molecular and cytogenetic predictors of survival and treatment outcome. Blood Cancer J 2015; 5: e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016; 127: 2391–2405. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlevede J, Droin N, Qin T, Meldi K, Yoshida K, Morabito M et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun 2016; 7: 10767. [DOI] [PMC free article] [PubMed] [Google Scholar]