Abstract
Nucleic acid-based aptamers are emerging as therapeutic antagonists of disease-associated proteins such as receptor tyrosine kinases. They are selected by an in vitro combinatorial chemistry approach, named Systematic Evolution of Ligands by Exponential enrichment (SELEX), and thanks to their small size and unique chemical characteristics, they possess several advantages over antibodies as diagnostics and therapeutics. In addition, aptamers that rapidly internalize into target cells hold as well great potential for their in vivo use as delivery tools of secondary therapeutic agents. Here, we describe a nuclease resistant RNA aptamer, named GL56, which specifically recognizes the insulin receptor (IR). Isolated by a cell-based SELEX method that allows enrichment for internalizing aptamers, GL56 rapidly internalizes into target cells and is able to discriminate IR from the highly homologous insulin-like growth factor receptor 1. Notably, when applied to IR expressing cancer cells, the aptamer inhibits IR dependent signaling. Given the growing interest in the insulin receptor as target for cancer treatment, GL56 reveals a novel molecule with great translational potential as inhibitor and delivery tool for IR-dependent cancers.
Keywords: aptamer, cancer therapy, IGF-1R, IR, SELEX
Introduction
Aptamers are short structured single-stranded nucleic acids that are selected by an in vitro combinatorial chemistry approach named Systematic Evolution of Ligands by Exponential enrichment (SELEX)1,2 to bind with high affinity to their target molecules. They are a promising new class of pharmaceuticals with a great potential as diagnostic and therapeutic tools.3,4 Indeed, aptamers show binding affinities and specificities similar to therapeutic antibodies, but, compared with protein-based therapeutic reagents, have many advantages, including simple and cost effective production and modification with adequate stability and no immunogenicity.5,6,7 Furthermore, aptamers that bind to cell surface can be rapidly internalized into target cells and, thus, can be developed to direct secondary reagents (small molecule drugs, radioisotopes, toxins or mi/siRNAs) to specific cells or tissues.8,9,10 This strategy is emerging as an effective mean to increase efficacy and reduce potential unwanted side effects of therapies.
In recent years, among the most promising new targets for cancer treatment, there is a growing interest in the insulin receptor (IR). IR belongs to a family of receptor tyrosine kinases (RTKs) that also includes the insulin-like growth factor 1 receptor (IGF-1R). Both, IR and IGF-1R, are tetrameric proteins sharing ~60% of amino acid homology. They are constituted of two extracellular α-chains and two β-chains that contain the transmembrane and the tyrosine kinase domains. In the receptor, α- and β-chains are linked together by disulfide bonds. The interaction of IR and IGF-1R with their ligands, insulin and insulin-like growth factors 1 and 2 (IGF-1 and IGF-2), stimulates the receptors to autophosphorylate and transphosphorylate intracellular adaptor molecules, including IR substrate proteins (IRS1–4). This leads to activate multiple downstream signaling pathways, such as the mitogen-activated protein kinase (MAPK)/Extracellular signal-regulated kinase (ERK) and the phosphatidylinositol 3-kinase (PI3-K)/AKT pathways.11,12,13,14 Activation of both receptors plays a key role in normal tissues physiology and has been implicated in cancer development and progression.15,16 While different inhibiting strategies for IGF-1R have already been developed as anticancer therapeutics (including monoclonal antibodies and small molecules),17 the importance of IR pathway in cancer development has been addressed more recently.
The mature human IR has two isoforms of alternative splicing, isoform A (IR-A) and isoform B (IR-B),18 which play different biological functions. Under physiological conditions, IR-B mediates the major metabolic effects of the receptor, whereas IR-A regulates growth and apoptosis during the embryonic development. Deregulated expression of the IR in its embryonic isoform A and overactivation of the receptor have been demonstrated in several kinds of cancer.19,20 In addition, it has been reported that IR may have an important role in the resistance to various anticancer therapies21,22 and to anti-IGF-1 receptor drugs,23 strongly indicating the potential of IR targeting.24 So far, available drugs, generated to inhibit IGF-1R, have generally no inhibiting activity on IR and anticancer strategies specifically targeting the IR are still lacking, thus representing an important challenge in oncology.25,26
In this work, we address the characterization of an aptamer, named GL56 that binds the human IR, but not the IGF-1R. The GL56 aptamer was generated by adopting a variant of the cell-based SELEX method that allows enrichment for cell-specific internalizing RNA aptamers. Our results show that the treatment of IR expressing glioma cancer cells with GL56 results in the inhibition of IR dependent signaling and in the reduction of cell viability. Further, upon binding the IR, GL56 rapidly internalize into target cells.
These data indicate that GL56 is a promising RNA-based molecule that can be further developed as novel inhibitory drug candidate and tool for delivery.
Results
Internalizing RNA aptamer enrichment strategy
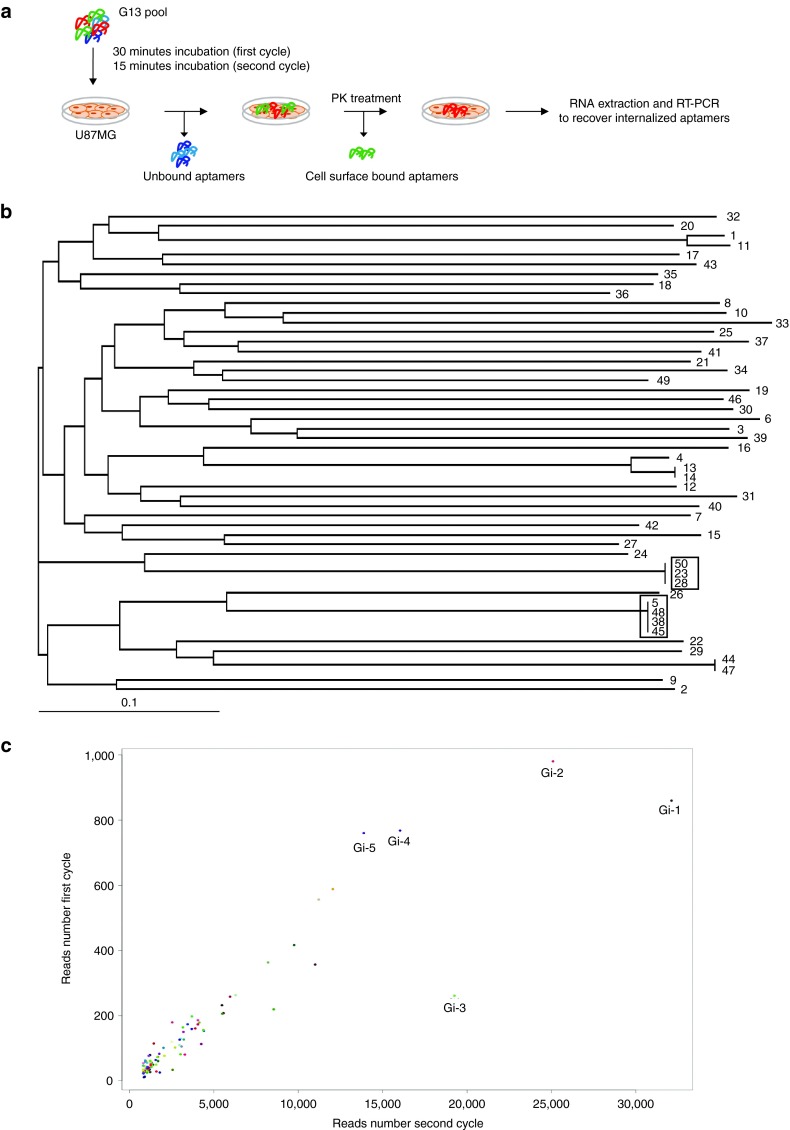
We first adopted a cell-SELEX variant to enrich for aptamers that once bound to glioma cell surface target are rapidly internalized. To this end, we chose to use as starting pool a 2′Fluoro-Pyrimidines-RNA (2′F-Py RNA) aptamer library previously subjected to 13 rounds of differential cell-SELEX onto U87MG cells (G13)27 with the final aim to preserve the glioma specificity achieved at the 13th rounds and isolate among the glioma-specific sequences, a pool of RNAs able to rapidly internalize into target cells. We performed two more rounds (intG14 and intG15) adopting the cell-internalization SELEX protocol as schematized in Figure 1a. Briefly, the G13 pool was incubated with target cells; unbound aptamers were removed and cells were treated with proteinase K (PK) to remove surface-bound aptamers; allowing to recover internalized aptamers.
Figure 1.
Cell-internalizing SELEX. (a) Scheme of the cell-internalizing protocol. We performed two cycles in which the G13 pool was incubated with U87MG target cells. Unbound aptamers were discarded and cells were treated with proteinase K for 30 minutes to remove cell surface-bound aptamers. Internalized aptamers were recovered by RNA extraction and RT-PCR. (b) Dendrogram by MAFFT analysis of the 50 individual sequences cloned after the two rounds of internalization. The most represented sequences are boxed. (c) Scattering plot of the 100 most abundant aptamers obtained by Illumina deep sequence analyses of the two cycles of internalization. The top five aptamers are indicated. SELEX, Systematic Evolution of Ligands by Exponential enrichment; RT-PCR, reverse transcription polymerase chain reaction.
The enriched final pool was, then, cloned and ~50 individual clones were sequenced. We identified four families of related aptamers (Figure 1b) with the most enriched being seq5 (identical to seq38-45–48, boxed) and seq23 (identical to seq28-50, boxed) that cover ~8 and 6% of the clones analyzed, respectively. In addition, the two pools intG14 and intG15 were also subjected to high throughput sequencing (HTS), using Illumina sequencing technology, in order to have a broader and accurate analysis. The obtained reads were filtered based on aptamers constant region. Over the hundred most abundant sequences (see Supplementary Figure S1), the top five aptamers (named Gi-1–5) covered ~30% of all individual sequences (Table 1) and were consistently enriched during the two internalization rounds (Figure 1c). Notably, three of the most represented sequences in HTS analysis were also found by cloning (Table 2). Among the obtained sequences, seq12 was identical to GL21, an anti-Axl aptamer previously identified from the original glioma enriched pool,27,28 whereas seq13 was identical to Gint4, that has been recently published as inhibitor of PDGFRβ.29 Within the most enriched PK-resistant sequences obtained both from cloning and HTS, seq5/Gi-2 (hereafter indicated as GL56), already found in the final glioma selection pool (Kd versus U87MG target cells of 63 nmol/l),27 was chosen for further characterization.
Table 1. Enrichment of the top five aptamers over the hundred most abundant sequences from HTS.
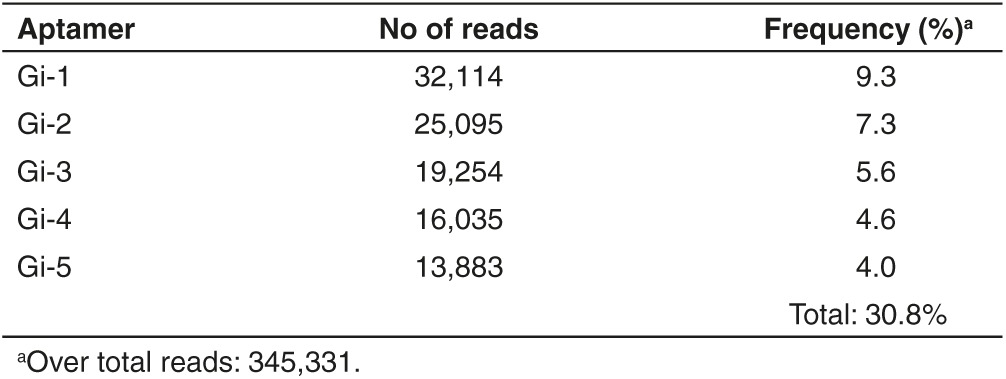
Table 2. Correspondence of the three most represented sequences from both illumina sequencing and cloning.

Validation of GL56 aptamer target
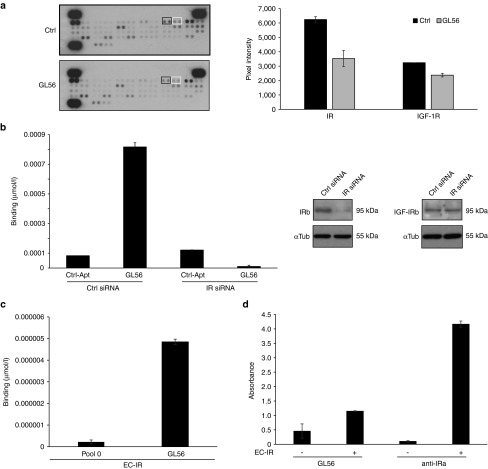
Since the selection was performed without prior knowledge of the aptamer target molecules, as a first attempt, we addressed the identification of GL56 functional targets. Aptamers selected by cell-SELEX strategies often recognize cell surface receptor and are endowed of inhibitory activity.28,29,30 Thus, by using an antiphospho-RTK antibody array, we first determined whether GL56 might inhibit some of the RTKs present on the array. To this purpose, U87MG cells were serum starved, pretreated with 200 nmol/l of GL56 and stimulated by serum for 15 minutes. Cell lysates were then analyzed for RTKs inhibition. As shown (Figure 2a), among the receptors whose serum-dependent phosphorylation was significantly reduced in the presence of GL56, we found that the aptamer treatment inhibits the phosphorylation of the IR, and at a lesser extent that of the cognate IGF-1 receptor, indicating IR as candidate target for GL56.
Figure 2.
GL56 aptamer targets IR. (a) Lysates from U87MG left untreated (Ctrl) or treated with GL56 and, then, stimulated with 20% FBS were subjected to Human Phospho-RTK arrays. Left, RTK arrays. Boxes indicate phospho-IR (black boxes) and phospho-IGF-1R (white boxes) signals. Right, Quantization performed with ImageJ (v1.46r) of phospho-IR and phospho-IGF-1R signals. (b) U87MG cells were transfected with a siRNA specific for IR (IR siRNA) or a control siRNA (Ctrl siRNA). Left, GL56 or a control unrelated aptamer (Ctrl-Apt) binding ability was measured by RT-qPCR at 200 nmol/l concentration. Right, IR and IGF-1R expression levels were analyzed by immunoblotting with IR beta (IRβ) and IGF-1R beta (IGF-1Rβ) specific antibodies. Anti-αTubulin (αTub) antibody was used to confirm equal loading. (c) GL56 or G0 starting pool (used as control) binding to the IR purified extracellular domain (EC-IR) was measured by filter binding coupled to RT-qPCR. (d) The interaction of biotinylated GL56 with EC-IR was analyzed by ELASA assay. An antibody specific for IR-α (anti-IRα) was used as positive control. In a-d error bars depict standard deviation values on analytical replicates. IGF-1R, insulin-like growth factor receptor 1; IR, insulin receptor; FBS, fetal bovine serum; RT-qPCR, reverse transcription quantitative polymerase chain reaction; ELASA, enzyme-linked apta-sorbent assay.
To validate IR as target of GL56, we first determined whether its binding ability could be hampered by downregulating its expression. As shown in Figure 2b, the binding to U87MG cells, as assessed by reverse transcription quantitative polymerase chain reaction (RT-qPCR, left panel), was abolished upon transfection with a siRNA specific for IR (right panel).
Next, we analyzed aptamer ability to recognize the soluble extracellular domain of the receptor (EC-IR) by filter binding followed by RT-qPCR (Figure 2c), and by an aptamer-based enzyme-linked apta-sorbent assay (ELASA) (Figure 2d). As shown, GL56 showed a good binding on EC-IR.
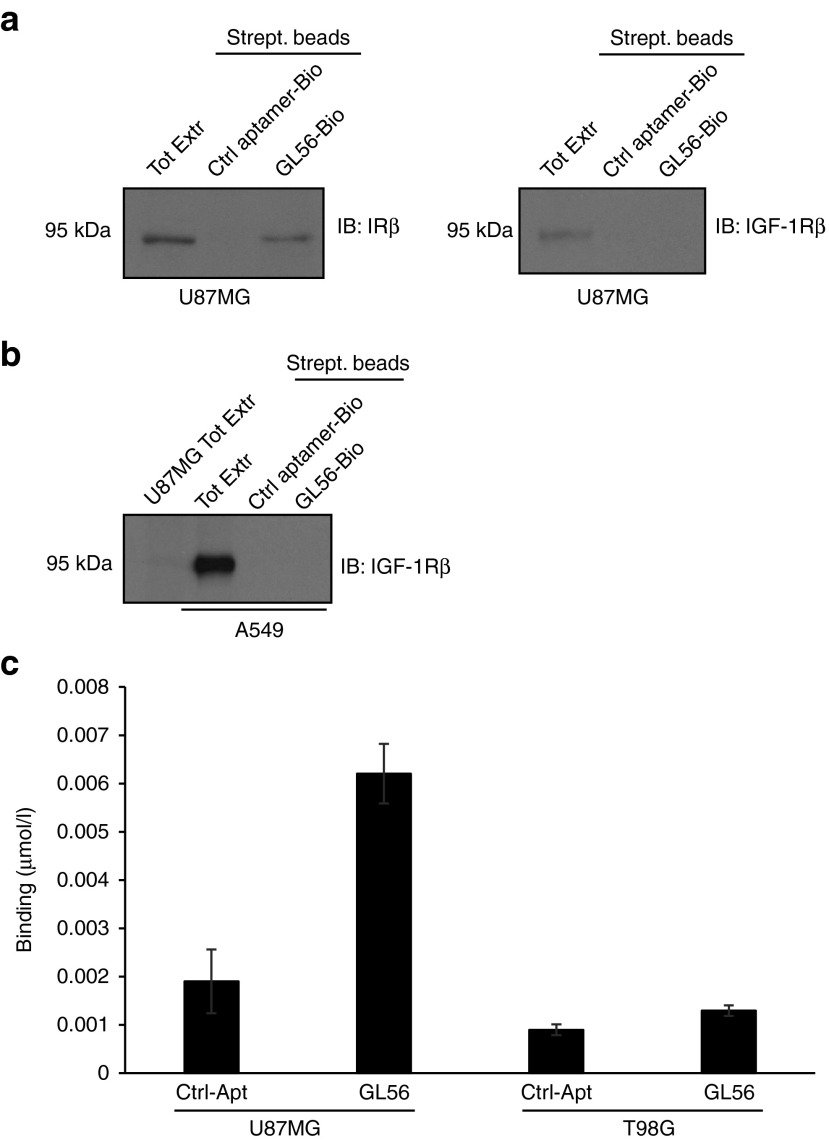
To establish the specific interaction of GL56 with IR, we then applied an aptamer-mediated affinity pull down strategy based on biotin-streptavidin purification. U87MG cells were treated with biotin-labeled GL56 and total cell extracts were purified on streptavidin-coated beads followed by immunoblotting with anti-IR or anti-IGF-1R antibodies. As shown, GL56 is able to interact with IR whereas no binding was obtained with an unrelated 2′F-Py RNA, used as a negative control (Figure 3a, left panel). Moreover, in the same cells the GL56 aptamer failed to pull down the IGF-1R (Figure 3a, right panel), indicating that it poorly interacts with this receptor. In order to exclude that the poor efficacy of binding to IGF-1R merely reflects the low levels of the receptor in U87MG cells, we repeated the pull down experiment using the nonsmall cell lung cancer (NSCLC) A549 cell line. We found that no interaction with IGF-1R was detected even in A549 cells that express significantly higher levels of IGF-1R than U87MG (Figure 3b and Supplementary Figure S2). To further support IR as selective target of GL56, we also determined its binding potential to the glioma-derived T98G cells that express low levels of IR (see Supplementary Figure S2). As assessed by RT-qPCR (Figure 3c), in comparison with an unrelated aptamer used as control, GL56 aptamer binding correlates with IR expression. Indeed the aptamer binds IR positive U87MG whereas does not bind the IR negative/very low expressing T98G cells even in presence of IGF-1R (see Supplementary Figure S2), further sustaining its ability to specifically recognize the IR.
Figure 3.
GL56 binging specificity. (a) and (b) Aptamer-mediated pull-down. Indicated cell lines were incubated with the biotinylated unrelated aptamer or GL56, cell lysates were purified on streptavidin beads and immunoblotted with anti-IRβ or anti- IGF-1Rβ antibodies. 20 µg of total cell extracts (Tot Extr) were loaded as references. (c) Binding by RT-qPCR of control unrelated aptamer (Ctrl-Apt) or GL56 aptamer on U87MG and T98G cells. Error bars depict standard deviation values on analytical replicates. IGF-1R, insulin-like growth factor receptor 1; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
Taken together, these results indicate bona fide that the GL56 aptamer recognizes target cells through the binding to the extracellular domain of IR and strongly support the ability of GL56 to preferentially bind this receptor with respect to the highly homologous IGF-1R.
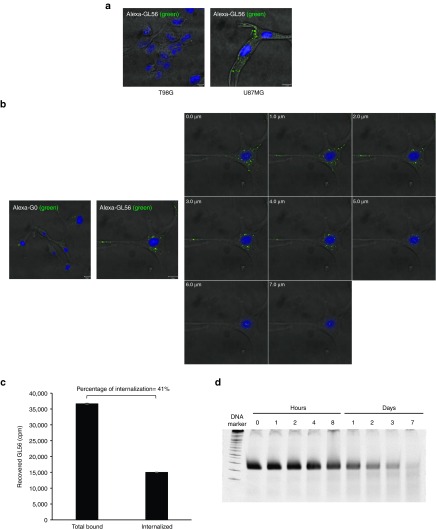
GL56 cell uptake and serum stability
Since the GL56 aptamer has been selected adopting a SELEX protocol optimized for enrichment of internalizing aptamers, we analyzed the aptamer cell uptake. Binding by confocal microscopy using Alexa-488-labeled GL56 aptamer, showed aptamer internalized into U87MG target cells following 30 minutes of incubation (Figure 4a). As expected, no signal was instead observed in T98G cells. To confirm the rapid GL56 cell uptake, we carried out Z-axis microscopy at 15 minutes of incubation. As shown in Figure 4b, labeled aptamer punctate spots were visible in each section of the Z-stacking acquisition (Figure 4b), thus indicating aptamer intracellular localization.
Figure 4.
GL56 internalization. (a, b) Confocal microscopy analysis. (a) Alexa-488 labeled-GL56 was incubated on U87MG or T98G for 30 minutes. (b) Alexa-488 labeled-GL56 or G0 pool (as control) were incubated on U87MG for 15 minutes (upper panel) and single confocal Z layers (of 1 µm each) were acquired (lower panel). In (a,b) Nuclei stained with DAPI are shown in blue. Scale bars: 10 µm. (c) Total bound or internalization of GL56 onto U87MG was measured by cell binding with radiolabeled aptamer. The background values obtained with the G0 pool were subtracted from the values obtained with the GL56. The percentage of internalized aptamer relative to total bound is indicated. Error bars depict standard deviation values on analytical replicates. (d) GL56 serum stability were measured by incubating the aptamer at 4 μmol/l in 90% human serum for indicated times. At each time point, RNA-serum samples were collected and evaluated by electrophoresis with 15% denaturing polyacrylamide gel. Gel was stained with ethidium bromide. DAPI, 4',6-Diamidino-2-Phenylindole.
Moreover by cell-binding assay, we determined that GL56 rapidly internalizes into U87MG target cells getting ~40% of total bound GL56 found intracellularly (PK resistant/total) at 15 minutes of incubation (Figure 4c).
Taken together, these results confirm that GL56 specifically binds glioma target cells and rapidly internalizes, thus representing a highly promising candidate as carrier for tissue specific internalization.
An important issue for RNA drugs is represented by serum stability that is limited by endogenous serum nucleases. GL56 aptamer contains 2′-F-Py modifications that are expected to increase endonuclease resistance.31 To evaluate the GL56 serum stability, we incubated the aptamer in 90% human serum for increasing times up to 1 week. Serum-RNA samples were then analyzed by denaturing polyacrylamide gel electrophoresis (Figure 4d). As shown, the aptamer was stable up to 8 hours and then was gradually degraded, reaching about 50% at 24 hours.
GL56 inhibits insulin-mediated signaling pathways
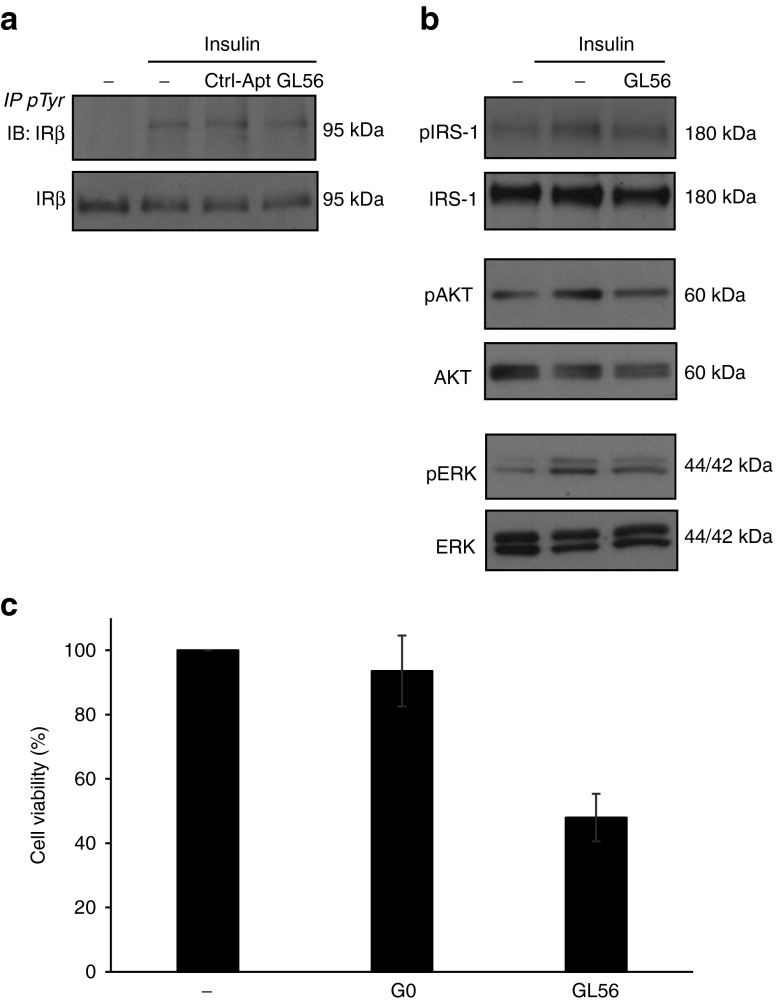
As a next step, we asked whether the GL56 aptamer could interfere with IR ligand-dependent signaling. Ligand binding to α-subunits of the receptor stimulates the intrinsic tyrosine kinase activity of the β-subunits resulting in the activation of multiple downstream signaling pathways, such as the MAPK/ERK and the PI3-K/AKT pathways.11,12,13,14 We first determined whether, upon binding, the GL56 aptamer inhibits the activity of the receptor itself. To this end, serum starved U87MG cells were stimulated for 5 minutes with insulin in the absence or in the presence of GL56 and the levels of phospho-IR were monitored by immunoprecipitation. Notably, IR phosphorylation levels were reduced in the presence of the GL56 aptamer, but not in the presence of a control unrelated aptamer (Figure 5a). We thus monitored the levels of insulin-dependent activation of the downstream effectors AKT and ERK1/2. As shown in Figure 5b, GL56 treatment was able to reduce the extent of phospho-AKT and phospho-ERK1/2 following 5 minutes of insulin stimulation and, accordingly, IR substrate protein IRS1 phosphorylation was also inhibited (Figure 5b). It has been demonstrated an important function of IR in regulating cell growth in several kind of cancers.13,24,32 Thus, we analyzed whether GL56 may suppress glioma cell viability by interfering with IR activation. As assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, the aptamer strongly reduces U87MG insulin-dependent cell viability giving ~50% of inhibition following 2 days of treatment (Figure 5c).
Figure 5.
GL56 inhibitory activity. (a) U87MG cells were serum starved overnight, left untreated or pretreated for 3 hours with 400 nmol/l GL56 aptamer or control aptamer (Ctrl-Apt) and, then, stimulated for 5 minutes with 100 nmol/l insulin alone or in presence of 400 nmol/l each aptamer. Cell lysates were immunoprecipitated with anti-(phospho)-tyrosine (pTyr) antibody and immunoblotted with anti-IRβ or were immunoblotted with anti-IRβ antibody. (b) Serum-starved U87MG were left untreated or pretreated for 3 hours with 400 nmol/l GL56 aptamer and stimulated for 5 minutes with 100 nmol/l insulin alone or in presence of 400 nmol/l GL56. Phopsho-AKT (pAKT), phospho-ERK1/2 (pERK), and phopsho-IRS1 levels were monitored by immunoblotting. Filters were probed with AKT, extracellular signal-regulated kinase (ERK) or IRS1 antibodies to confirm equal loading. (c) Serum starved U87MG were grown in Dulbecco's modified essential medium (DMEM) containing 0.5% fetal bovine serum (FBS) and 100 nmol/l insulin either in absence or in presence of 400 nmol/l GL56 or G0 pool, used as negative control. Treatments were renewed each 24 hours. Cell viability was measured following 48 hours.
The above results indicate that GL56, because of its binding to IR, blocks the receptor activation hampering the insulin-dependent signaling, thus representing a promising inhibitor candidate.
Discussion
In this study, we have developed a cell-based SELEX approach to enrich for internalizing RNA aptamers and by such an approach, we have identified a novel anti-IR aptamer. Starting from a pool coming from the round 13 (G13) of selection on U87MG glioblastoma cells, we performed two further PK-based rounds. Obtained sequences were analyzed both by cloning and HTS (Figure 1, Tables 1 and 2). Among the most represented sequences, we further characterized an aptamer named GL56 by demonstrating its ability to specifically bind to IR (Figures 2, 3, and 4). The binding culminated in inhibiting insulin-dependent signaling (Figure 5).
This study provides two important findings. First, it describes a simple method to favor the isolation of RNAs that are internalized into the target cells. Internalizing aptamers have been successfully used as targeting moieties for the in vivo selective delivery of secondary reagents to cells proving to be an effective tool to improve the therapeutic efficacy.9 Even if several variants of the original SELEX protocol have led to raise aptamers that bind at high affinity and specificity to membrane receptors, this methodology does not guarantee the selection of internalizing aptamers. Our approach allow to rapidly achieve this goal, avoiding large prescreening and may be used alternatively or in combination to the already proposed cell-internalization SELEX.33,34
In our selection, we aimed at preserving the glioma specificity of aptamers and at isolating among them, those sequences able to rapidly internalize into target cells. Therefore, we used as starting pool an highly enriched library previously selected for molecules that target proteins expressed on the highly tumorigenic U87MG cell surface (from 13 rounds of SELEX). We subjected that library to two additional internalization rounds, based on a PK-treatment. Notably the presence of sequences (i.e., the anti-Axl aptamer GL21 aptamer and GL56), previously identified from the original glioma selection27 within the final internalizing pool, indicates the maintenance of glioma targeting.
The second and most important finding, herein described, is the characterization of GL56 anti-IR aptamer. As recently emerged, IR, in addition to regulate important aspects of cellular physiology,35,36,37 is overexpressed and overactivated in several cancers19,20 and interfering with its expression (by shRNA) results in the inhibition of tumor growth and metastasis.13 These data, together with the involvement of the receptor in drug resistance,21,22 strongly indicate that therapeutic strategies directed against the receptor may represent an important challenge in oncology. To date, several inhibitory molecules, including monoclonal antibodies and small tyrosine kinase inhibitors, have been developed against the cognate receptor IGF-1R, but they have shown a limited clinical success.38 The overexpression and overactivation of IR-A has been reported as one of the most important reason for the failure of IGF-1R targeted drugs39 that has generally no inhibiting activity on IR. These data suggest that the cotargeting of IR and IGF-1R may increase therapy efficacy and underline the need of anti-IR strategies that are still lacking.
Here, we develop an aptamer-based molecule as promising IR-targeting drug for a specific and selective tumor therapy. By using a phospho-RTK array, we identified IR as target candidate of the GL56 aptamer. As assessed by different biochemical approaches, we proved that GL56 specifically recognizes the human IR and acts as a neutralizing ligand for the receptor by inhibiting its downstream signaling and reducing in vitro cell viability. In accordance with the high specificity of aptamers, GL56 discriminate IR from the cognate receptor IGF-1R in our experimental conditions. Notably, the aptamer contains 2′-F-Pyrimidines that protect it against nuclease degradation giving persistent serum stability, thus improving its in vivo applicability.
Furthermore, the ability of the aptamer not only to bind IR and to inhibit its activity, but also to be rapidly and specifically internalized within the target cells, strengths its therapeutic potential and opens the concrete possibility to develop this molecule as carrier for cell-selective delivering. Indeed, the use of aptamer-based therapeutic conjugates is growing rapidly and represents an effective strategy to increase efficacy and reduce toxicity effects of therapies.40,41
Recently, DNA-aptamers against IR have been reported42,43 that provide either imaging or agonistic tools. Although their potential use in metabolic diseases, they do not have any potentiality as cancer therapeutics and have no sequence similarity with GL56.
In conclusion, GL56 aptamer provide the first example of an anti-IR inhibitory agent. Taken together our results represent an initial characterization of this molecule and may serve as a platform for the future development of novel aptamer-based targeted therapies for cancer.
Materials and methods
Cell culture. Cell lines were purchased from the ATCC (LG Standards, Milan, Italy). Human Glioblastoma U87MG and T98G were grown in Dulbecco's modified Eagle's medium (DMEM), whereas human NSCLC A549 cells were grown in Roswell Park Memorial Institute medium (RPMI). Media were supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 100 U/ml penicillin/streptomycin. All cell culture reagents were purchased from Sigma (St Louis, MO).
SELEX method. The pool enriched from 13 rounds of differential cell-SELEX consisting of selection onto U87MG cells preceded by a counter selection on T98G,27 was incubated onto U87MG target cells at 37°C for 30 minutes in the first internalization round and for 15 minutes in the second internalization round. After five washes with DMEM serum free medium to remove unbound aptamers, cells were treated with proteinase K (Roche Diagnostics, Indianapolis, IN) for 30 minutes at 0.5 μg/μl final concentration. Cells were then washed with DMEM serum free and internalized RNA aptamers were recovered by RNA extraction and RT-PCR, as previously described.44 Cloning was performed by using TOPO-TA cloning kit (Invitrogen, Carlsbad, CA) as recommended. For HTS, sequences recovered from each internalization round were sequenced following the Illumina MySeq sequence preparation with MySeq Illumina sequencer.
GL56 aptamer. GL56 aptamer RNA sequence: 5′GGAGACAAGAAUAAACGCUCAAUGAUUUUGCAGCACUUCUUGUUAUCUUAACGAACUGUUGAUGAUUCGACAGGAGGCUCACAACAGGC 3′
Sequence regions corresponding to the primers are underlined.
RNAs were produced by in vitro transcription of the correspondent DNA, as reported,45 in the presence 2′-F-Pyrimidines (Tebu-bio Srl, Magenta, Milan, Italy). Before each treatment, aptamers were subjected to a short denaturation–renaturation step by incubating 5 minutes at 85°C, 3 minutes on ice, and 10 minutes at 37°C.
RTK antibody array. U87MG cells were serum starved overnight, pretreated with 200 nmol/l GL56 aptamer for 3 hours and then stimulated with 20% FBS in the absence or in the presence of the aptamer. Cell lysates were incubated with the phospho-RTK array (R&D Systems, Minneapolis, MN) following manufacturer's instructions.
Cell binding assay by RT-qPCR. Cells were seeded in 3.5-cm plates and treated with 200 nmol/l of GL56 for 15 minutes at 37°C in the presence of 100 µg/ml polyinosine used as a nonspecific competitor (Sigma). Following three washes with PBS to remove unbound RNA, bound RNA was recovered by TRIzol (Life Technologies) containing 0.5 pmol/ml of CL4 aptamer (CL4: 5′GCCUUAGUAACGUGCUUUGAUGUCGAUUCGACAGGAGGC3′), used as a reference control. The amount of bound RNAs was determined by performing RT-qPCR, as reported,46 with the following primers:
GL56 (Forward): 5′ TAATACGACTCACTATAGGGAGACAAGAATAAACGCTCAA 3′; GL56 (Reverse): 5′ CGACAGGAGGCTCACAACAGGC 3′; CL4 (Forward): 5′ GCCTTAGTAACGTGCTTT 3′; and CL4 (Reverse) 5′ GCCTCCTGTCGAATCG 3′.
At each experiment, cells cultured were counted and obtained data were normalized to the CL4 reference control and to cell number.
For IR silencing experiment, seeded cells (1.4 × 105 cells/3.5-cm plate) were transfected with siRNA specific for IR or control siRNA 100 nmol/l (TriFECTa RNAi Kit; IDT Integrated DNA Technologies, Coralville, IA) by using Lipofectamine 2000 and Opti-MEM I reduced serum medium (Invitrogen). After 5-hours, complete culture medium was added to the cells and incubation was prolonged up to 48 hours before binding assay.
Filter binding by RT-qPCR. Filter binding assay was performed by incubating 40 nmol/l purified IR extracellular domain (R&D Systems) with 1 nmol/l of GL56 or G0 starting pool used as control, in 100 µl PBS supplemented with bovine serum albumin (BSA) 0.01%. After 30 minutes of incubation at 37°C, samples were filtered through HAWP 0.45 µm nitrocellulose filters (Millipore, Bedford, MA) pretreated with PBS. Filter were washed three times with PBS and the retained RNAs were eluted by cutting the filters and incubating over night at 42°C in 1 ml elution buffer (20 mmol/l Tris HCl pH 7.8, 0.2% Sodium Dodecyl Sulphate and 0.3 mol/l sodium acetate) containing 0.5 pmol of CL4 aptamer used as a reference control. Then, RNAs were precipitated and the amount of bound RNAs was determined by performing RT-qPCR, as reported.46
Aptamer-based ELASA. C96 maxisorp nunc-immunoplate (Thermo Fisher Scientific, Waltham, MA) were left untreated or were coated with 3.2 pmoles of purified IR extracellular domain (R&D Systems) over night at 4°C. Wells were blocked for 2 hours at room temperature with PBS containing 3% BSA, washed two times with PBS and, then, incubated for 2 hours at room temperature in PBS 0.01% BSA with 200 nmol/l GL56.T aptamer previously biotinylated by in vitro transcription with 5′-biotin GMP (TriLinK Biotechnologies, San Diego, CA). Following one wash with PBS, samples were incubated with horseradish peroxidase (HRP)-conjugated Streptavidin (Thermo Fisher Scientific) for 1 hour at room temperature and washed two times with PBS. Signals were reveled with TMB substrate solution (Thermo Fisher Scientific) and stopped with stop solution for TMB substrate (Thermo Fisher Scientific). Absorbance at 450 nm was measured with Multiskan FC Microplate Photometer (Thermo Fischer Scientific). As positive control, wells were incubated with an antibody specific for IR alpha subunits (concentration: 1:1,000, Santa Cruz Biotechnology, Dallas, TX) and a HRP-conjugated specific secondary antibody (concentration 1:5,000, Santa Cruz Biotechnology) was used to reveal the signal. The assay was performed in duplicate.
Binding and internalization with rabio-labeled aptamers. Cells were seeded in 24-well plates (3.5 × 104 cells/well) and incubated with 5′-[32P]-labeled-GL56 or G0 starting pool, used as control (100 nmol/l of in 200 µl of DMEM serum free) for 15 minutes at 37°C in the presence of 100 µg/ml polyinosine as a nonspecific competitor (Sigma). Cells were washed five times with 500 µl DMEM and bound sequences were recovered in 300 µl of sodium dodecyl sulfate (SDS) 1%. To check internalization, before recovering, cells were treated with 0.5 μg/μl proteinase K (Roche Diagnostics, Indianapolis, IN) for 30 minutes at 37°C to remove cell surface bound aptamers. The recovered radioactivity was counted. Values were corrected for aptamer specific activity (cpm/pmol) and the background values detected with G0 were subtracted from those obtained with GL56 aptamer. Internalization was expressed as percentage of internalized aptamer relative to total bound aptamer.
Aptamer-mediated pull-down. GL56 aptamer was biotinylated at 3′-end by incubating with Terminal Transferase and Biotin-ddUTP (Roche, Basel, Switzerland) according to manufacturing instructions. Cells were incubated for 30 minutes at room temperature with 200 nmol/l biotinylated GL56 or an unrelated biotinylated aptamer, used as negative control, in serum free culture medium. After PBS washes, cells were lysed with 10 mmol/l Tris-HCl pH 7.5 containing 200 mmol/l NaCl, 5 mmol/l ethylenediaminetetraacetate (EDTA), 0.1% Triton X-100 and protease inhibitors. Cell extracts (400 µg in 0.4 ml lysis buffer) were purified by incubating with 200 µl-streptavidin beads (Thermo Fischer Scientific) for 2 hours with rotation. Beads were washed three washes with PBS, bound proteins were recovered by adding Laemmli buffer and then analyzed by immunoblotting with IR-beta or IGFR-1-beta specific antibodies (Cell Signaling Technologies, Danvers, MA).
Immunofluorescence analysis. For immunofluorescence analysis, GL56 or the G0 unselected pool, used as negative control, were labeled with Alexa Fluor 488 probe by using the Ulysis Nucleic Acid Labeling Kit (Invitrogen) according to instructions. Labeled RNAs were purified with a spin-column procedure by using Micro Bio-Spin P6 (Bio-Rad, Hercules, CA).
Cells were grown on glass coverslips overnight and were treated with 500 nmol/l labeled GL56 or G0 in serum free media at 37°C. Cells were washed three times with PBS, fixed with paraformaldehyde 4% in PBS for 10 minutes and coverslips were mounted on microscope slides with Prolong Gold Antifade Reagent with 4',6-Diamidino-2-Phenylindole (DAPI) (Cell Signaling Technologies). Cells were visualized by confocal microscopy using a Zeiss 510 LSM confocal microscope with a 63x oil objective. For Z-stacking acquisition, single confocal Z layers of 1 µm each were acquired.
Stability in human serum. Aptamer was incubated at 4 μmol/l concentration in 90% human serum (Type AB Human Serum provided by Euroclone, Milan, Italy) from 1 hour to 7 days. At each time point 4 μl (16 pmoles RNA) were recovered and treated for 1 hour at 37°C with 5 μl of proteinase K solution (600 mAU/ml). Following the addition of 18 μl denaturing gel loading buffer, samples were stored at −80°C. All serum-RNA samples were then analyzed on 15% denaturing electrophoresis polyacrylamide gel and visualized by staining with ethidium bromide and UV exposure.
Immunoprecipitation and immunoblot analysis. Cell extracts were prepared with buffer A (50 mmol/l Tris-HCl pH 8.0 buffer containing 150 mmol/l NaCl, 1% Nonidet P-40, 2 µg/ml aprotinin, 1 µg/ml pepstatin, 2 µg/ml leupeptin, 1 mmol/l Na3VO4) and protein concentration was determined by the Bradford assay.
For immunoprecipitation, cell extracts (250 µg) were incubated with antiphospho-tyrosine antibody (#4G10; Upstate Biotechnology, Lake Placid, MA) for 2 hours and then on protein A/G- agarose (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Following three washes with buffer A, immunoprecipitated proteins were denatured in Laemmli buffer for 10 minutes at 100°C. Cell lysates or immunoprecipitates were subjected to SDS-PAGE and electroblotted into polyvinylidene difluoride membranes (Merck Millipore, Billerica, MA). Filters were probed with primary antibodies as indicated. The primary antibodies used were: antiphospho-AKT (Ser473), antiphospho-ERK1/2, anti-AKT, antiphospho-IRS1 (Ty895), anti-IRS1, anti-IR-beta, anti-IGF-1R-beta (Cell Signaling Technologies, Danvers, MA), anti-ERK1, and anti-α tubulin (Santa Cruz Biotechnology, Dallas, TX).
Cell viability. Cells (1.6 × 103 cells/well in 96-well plates) were serum starved and then grown in DMEM containing 0.5% FBS and 100 nmol/l insulin either in absence or in presence of 400 nmol/l GL56 or G0 pool, used as negative control. Treatments were renewed each 24 hours. Cell viability was assessed by using CellTiter 96H AQueous One Solution cell Proliferation Assay (Promega, Madison, WI) measuring the absorbance at 492 nm with Multiskan FC Microplate Photometer (Thermo Fischer Scientific).
SUPPLEMENTARY MATERIAL Figure S1. Clustering of sequences from HTS. Figure S2. IR and IGF-1R expression.
Acknowledgments
M.I. designed and performed the majority of the experiments, interpreted results and assisted with manuscript preparation. A.R., R.F., S.C., M.C., and S.N. performed and/or assisted with several experiments. G.S. performed the HTS analyses. C.L.E. coordinated the research, secured the funding, guided the experimental design, and the preparation of the manuscript. V.d.F. and G.C. secured the funding, provided intellectual input and assisted the preparation of the manuscript. We thank L. Baraldi and F. Moscato for technical assistance. This work was supported by funds from: AIRC # 13345 (V.d.F.) and # 10620 (G.C.); the Italian Ministry of Economy and Finance to the CNR for the Project FaReBio di Qualità (V.d.F.); Compagnia San Paolo # 2011.1172 (V.d.F.); the Italian Ministry of Health, GR-2011-02352546 (C.L.E.). The authors declare no competing financial interests.
Supplementary Material
References
- Tuerk, C and Gold, L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Ellington, AD and Szostak, JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Zhou, G, Wilson, G, Hebbard, L, Duan, W, Liddle, C, George, J et al. (2016). Aptamers: A promising chemical antibody for cancer therapy. Oncotarget 7: 13446–13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, C, Qiu, L, Li, J, Fu, T, Zhang, X and Tan, W (2016). Cancer biomarker discovery using DNA aptamers. Analyst 141: 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, KE and Levy, M (2016). From selection hits to clinical leads: progress in aptamer discovery. Mol Ther Methods Clin Dev 5: 16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y, Liang, C, Lv, Q, Li, D, Xu, X, Liu, B et al. (2016). Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int J Mol Sci 17: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe, AD and Cload, ST (2008). SELEX with modified nucleotides. Curr Opin Chem Biol 12: 448–456. [DOI] [PubMed] [Google Scholar]
- Zhu, H, Li, J, Zhang, XB, Ye, M and Tan, W (2015). Nucleic acid aptamer-mediated drug delivery for targeted cancer therapy. Chem Med Chem 10: 39–45. [DOI] [PubMed] [Google Scholar]
- Esposito, CL, Catuogno, S and de Franciscis, V (2014). Aptamer-mediated selective delivery of short RNA therapeutics in cancer cells. J RNAi Gene Silencing 10: 500–506. [PMC free article] [PubMed] [Google Scholar]
- Kruspe, S, Mittelberger, F, Szameit, K and Hahn, U (2014). Aptamers as drug delivery vehicles. Chem Med Chem 9: 1998–2011. [DOI] [PubMed] [Google Scholar]
- Sachdev, D and Yee, D (2001). The IGF system and breast cancer. Endocr Relat Cancer 8: 197–209. [DOI] [PubMed] [Google Scholar]
- Zhang, H and Yee, D (2006). Is the type I insulin-like growth factor receptor a therapeutic target in endometrial cancer? Clin Cancer Res 12: 6323–6325. [DOI] [PubMed] [Google Scholar]
- Zhang, H, Fagan, DH, Zeng, X, Freeman, KT, Sachdev, D and Yee, D (2010). Inhibition of cancer cell proliferation and metastasis by insulin receptor downregulation. Oncogene 29: 2517–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F and Yang, Y (2014). Quercetin suppresses insulin receptor signaling through inhibition of the insulin ligand-receptor binding and therefore impairs cancer cell proliferation. Biochem Biophys Res Commun 452: 1028–1033. [DOI] [PubMed] [Google Scholar]
- Scotlandi, K, and Belfiore, A (2012). Targeting the insulin-like growth factor (IGF) system is not as simple as just targeting the type 1 IGF receptor. Am Soc Clin Oncol Educational Book/ASCO Am Soc Clin Oncol Meeting: 599–604. [DOI] [PubMed]
- Lundby, A, Bolvig, P, Hegelund, AC, Hansen, BF, Worm, J, Lützen, A et al. (2015). Surface-expressed insulin receptors as well as IGF-I receptors both contribute to the mitogenic effects of human insulin and its analogues. J Appl Toxicol 35: 842–850. [DOI] [PubMed] [Google Scholar]
- Janssen, JA and Varewijck, AJ (2014). IGF-IR targeted therapy: past, present, and future. front endocrinol (Lausanne) 5: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca, F, Pandini, G, Scalia, P, Sciacca, L, Mineo, R, Costantino, A et al. (1999). Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol 19: 3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore, A, Frasca, F, Pandini, G, Sciacca, L and Vigneri, R (2009). Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr Rev 30: 586–623. [DOI] [PubMed] [Google Scholar]
- Aljada, A, Saleh, AM, Al-Aqeel, SM, Shamsa, HB, Al-Bawab, A, Al Dubayee, M et al. (2015). Quantification of insulin receptor mRNA splice variants as a diagnostic tumor marker in breast cancer. Cancer Biomark 15: 653–661. [DOI] [PubMed] [Google Scholar]
- Papa, V, Pezzino, V, Costantino, A, Belfiore, A, Giuffrida, D, Frittitta, L et al. (1990). Elevated insulin receptor content in human breast cancer. J Clin Invest 86: 1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore, A (2007). The role of insulin receptor isoforms and hybrid insulin/IGF-I receptors in human cancer. Curr Pharm Des 13: 671–686. [DOI] [PubMed] [Google Scholar]
- Garofalo, C, Manara, MC, Nicoletti, G, Marino, MT, Lollini, PL, Astolfi, A et al. et al. (2011). Efficacy of and resistance to anti-IGF-1R therapies in Ewing's sarcoma is dependent on insulin receptor signaling. Oncogene 30: 2730–2740. [DOI] [PubMed] [Google Scholar]
- Vincent, EE, Elder, DJ, Curwen, J, Kilgour, E, Hers, I and Tavaré, JM (2013). Targeting non-small cell lung cancer cells by dual inhibition of the insulin receptor and the insulin-like growth factor-1 receptor. PLoS One 8: e66963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P, Alex, JM and Bast, F (2014). Insulin receptor (IR) and insulin-like growth factor receptor 1 (IGF-1R) signaling systems: novel treatment strategies for cancer. Med Oncol 31: 805. [DOI] [PubMed] [Google Scholar]
- Malaguarnera, R and Belfiore, A (2011). The insulin receptor: a new target for cancer therapy. Front Endocrinol (Lausanne) 2: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchia, L, Esposito, CL, Jacobs, AH, Tavitian, B and de Franciscis, V (2009). Differential SELEX in human glioma cell lines. PLoS One 4: e7971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cerchia, L, Esposito, CL, Camorani, S, Rienzo, A, Stasio, L, Insabato, L et al. (2012). Targeting Axl with an high-affinity inhibitory aptamer. Mol Ther 20: 2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camorani, S, Esposito, CL, Rienzo, A, Catuogno, S, Iaboni, M, Condorelli, G et al. (2014). Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRβ aptamer. Mol Ther 22: 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, CL, D'Alessio, A, de Franciscis, V and Cerchia, L (2008). A cross-talk between TrkB and Ret tyrosine kinases receptors mediates neuroblastoma cells differentiation. PLoS One 3: e1643. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lok, CN, Viazovkina, E, Min, KL, Nagy, E, Wilds, CJ, Damha, MJ et al. (2002). Potent gene-specific inhibitory properties of mixed-backbone antisense oligonucleotides comprised of 2′-deoxy-2′-fluoro-D-arabinose and 2′-deoxyribose nucleotides. Biochemistry 41: 3457–3467. [DOI] [PubMed] [Google Scholar]
- Wang, CF, Zhang, G, Zhao, LJ, Qi, WJ, Li, XP, Wang, JL et al. (2013). Overexpression of the insulin receptor isoform A promotes endometrial carcinoma cell growth. PLoS One 8: e69001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Thiel, KW, Hernandez, LI, Dassie, JP, Thiel, WH, Liu, X, Stockdale, KR et al. (2012). Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res 40: 6319–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, WH, Bair, T, Peek, AS, Liu, X, Dassie, J, Stockdale, KR et al. (2012). Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS One 7: e43836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina, Y, Ellis, L, Jarnagin, K, Edery, M, Graf, L, Clauser, E et al. (1985). The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell 40: 747–758. [DOI] [PubMed] [Google Scholar]
- Ullrich, A, Bell, JR, Chen, EY, Herrera, R, Petruzzelli, LM, Dull, TJ et al. (1985). Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature 313: 756–761. [DOI] [PubMed] [Google Scholar]
- Drakas, R, Tu, X and Baserga, R (2004). Control of cell size through phosphorylation of upstream binding factor 1 by nuclear phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 101: 9272–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, HX and Sharon, E (2013). IGF-1R as an anti-cancer target–trials and tribulations. Chin J Cancer 32: 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga, R (2013). The decline and fall of the IGF-I receptor. J Cell Physiol 228: 675–679. [DOI] [PubMed] [Google Scholar]
- Thiel, KW and Giangrande, PH (2010). Intracellular delivery of RNA-based therapeutics using aptamers. Ther Deliv 1: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, F, Liu, B, Lu, J, Li, F, Li, D, Liang, C et al. (2015). Progress and challenges in developing aptamer-functionalized targeted drug delivery systems. Int J Mol Sci 16: 23784–23822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, M, Kwon, M, Kim, S, Yunn, NO, Kim, D, Ryu, SH et al. (2014). Aptamer-based single-molecule imaging of insulin receptors in living cells. J Biomed Opt 19: 051204. [DOI] [PubMed] [Google Scholar]
- Yunn, NO, Koh, A, Han, S, Lim, JH, Park, S, Lee, J et al. (2015). Agonistic aptamer to the insulin receptor leads to biased signaling and functional selectivity through allosteric modulation. Nucleic Acids Res 43: 7688–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, CL, Passaro, D, Longobardo, I, Condorelli, G, Marotta, P, Affuso, A et al. (2011). A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One 6: e24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catuogno, S, Esposito, CL and de Franciscis, V (2016). Developing aptamers by cell-based SELEX. Methods Mol Biol 1380: 33–46. [DOI] [PubMed] [Google Scholar]
- Catuogno, S, Rienzo, A, Di Vito, A, Esposito, CL and de Franciscis, V (2015). Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J Control Release 210: 147–159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.