Abstract
Recent investigations strongly suggest that Raman spectroscopy (RS) can be used as a clinical tool in cancer diagnosis to improve diagnostic accuracy. In this study, we evaluated the efficiency of Raman imaging microscopy to discriminate between healthy and neoplastic thyroid tissue, by analyzing main variants of Papillary Thyroid Carcinoma (PTC), the most common type of thyroid cancer. We performed Raman imaging of large tissue areas (from 100 × 100 μm2 up to 1 × 1 mm2), collecting 38 maps containing about 9000 Raman spectra. Multivariate statistical methods, including Linear Discriminant Analysis (LDA), were applied to translate Raman spectra differences between healthy and PTC tissues into diagnostically useful information for a reliable tissue classification. Our study is the first demonstration of specific biochemical features of the PTC profile, characterized by significant presence of carotenoids with respect to the healthy tissue. Moreover, this is the first evidence of Raman spectra differentiation between classical and follicular variant of PTC, discriminated by LDA with high efficiency. The combined histological and Raman microscopy analyses allow clear-cut integration of morphological and biochemical observations, with dramatic improvement of efficiency and reliability in the differential diagnosis of neoplastic thyroid nodules, paving the way to integrative findings for tumorigenesis and novel therapeutic strategies.
Raman spectroscopy (RS) is a technique, which utilizes inelastic light scattering. It is capable of probing fundamental vibrations of biomolecules, representing a label-free optical technology tool. It is increasingly used in a rapidly expanding research area dedicated to biological tissue studies1,2,3. Presently, clinical applications of RS are extremely challenging, being severely limited mainly by the time-consuming spectral measurements. However, technological advances over the last decade have created innovative RS based tools, providing morphological investigation of large tissue areas coupled with high resolution, point-by-point spectral analysis of biochemical composition. Such a technological development has prompted a burst of rapidly growing, clinically-driven RS investigations, leading to several in vivo applications of RS in biomedicine4,5,6,7,8. With this regard, the aim of the present research is to illustrate the great potential of the RS imaging technology with increased acquisition speed and performances for improved non-destructive diagnosis and imaging of thyroid tissues.
Thyroid nodules are increasing in frequency with a parallel increase of thyroid cancer. Fine needle aspiration biopsy (FNA), followed by cytological assessment, is the best procedure for their management in a large proportion of cases. Of these, about 60–70% are classified cytologically as benign nodules, while 4–10% are deemed malignant. However, the remaining nodules (10–26% of all FNAs) have indeterminate morphological findings (inconclusive, equivocal, atypical features) and represent a clinical problem9, being the “indeterminate” category associated with a 15 to 48% prevalence of malignancy, mainly papillary thyroid carcinomas (PTC)10. As a matter of fact, histopathological evaluation of thyroid nodules, though currently representing the ‘gold standard’ for the diagnosis, is still hampered by significant levels of inter- and intra-observer variations, even among experts11. In recent years, molecular analysis of FNA biopsy from thyroid nodules has been performed in order to reduce the number of cytologically indeterminate cases, with the development of several diagnostic panels12. Through these advances, molecular diagnostics has improved the care of patients with thyroid nodules and cancer13; the cost of such procedures, however, should be reduced in order to increase their cost-effectiveness in standard medical practice. Moreover, genetic molecular analysis, even if extensive, does not provide any information about the biochemical profile of these tumors.
Aim of our work was to investigate the potential of RS imaging technology in improving the assessment of thyroid tissues and in supplying the differential diagnosis of thyroid nodules. As a potential non-destructive tool to support the histopathological evaluation, in situ RS analysis of the biochemical features of thyroid tissues was applied to discriminate (a) between healthy and PTC tissue and (b) between two variants of PTC (classical and follicular). Our results, obtained by combining morphological and biochemical observations on the same tissue section, may represent an additional step towards improved standardization and increased reliability in thyroid cancer diagnostics.
Results
Raman spectroscopic study, performed using a Raman imaging microscope (RM), was carried out for nine patients, which underwent total thyroidectomy and received a diagnosis of PTC based on FNA at the Endocrinology Unit of University Campus Bio-medico of Rome (UCBM). The detailed description of sample preparation for Raman measurements is given in the Methods section. Shortly, frozen thyroid tissue sections collected on glass slides were submitted to RS investigation. Adjacent, Haematoxylin/Eosin stained tissue slides were used as a reference for the presence of healthy and neoplastic tissue areas. Multiple Raman biochemical maps for the healthy and PTC zones were taken for each tissue section. In Table 1, the experimental dataset is fully represented, showing the distribution of the tissue samples (healthy, PTC classical variant and PTC follicular variant) for each patient, numbered consecutively from 1 to 9.
Table 1. DATASET: Thyroid glands from 9 PTC patients.
| Case | Histologic diagnosis: PTC | Number of healthy tissue maps | Number of pathological tissue maps | Total Raman maps | Total Raman spectra |
|---|---|---|---|---|---|
| 1 | classical | 2 | 2 | 4 | 626 |
| 2 | 2a) follicular 2b) classical | 2 | 2 + 2 | 6 | 799 |
| 3 | classical | 2 | 2 | 4 | 994 |
| 4 | classical | 2 | 2 | 4 | 669 |
| 5 | follicular | 2 | 2 | 4 | 1950 |
| 6 | classical | 2 | 2 | 4 | 2187 |
| 7 | classical | 2 | 2 | 4 | 329 |
| 8 | follicular | 2 | 2 | 4 | 713 |
| 9 | classical | 2 | 2 | 4 | 566 |
Total number of MAPS: 38. Total number of spectra: 8833.
Biochemical profile study
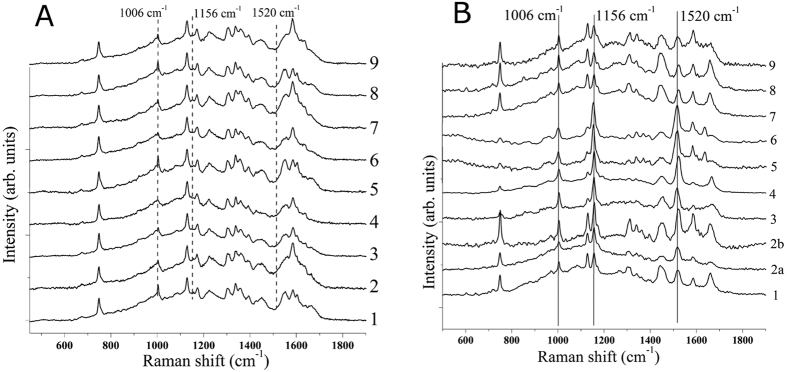
The spectra obtained by averaging the Raman biochemical maps have been classified according to the tissue type (healthy and PTC), resulting in 9 average PTC and 9 average healthy spectra, corresponding to thyroid samples from 9 patients. In Fig. 1, both sequences show the fingerprint (FP) region of spectra. In Fig. 1(A), the sequence of average Raman spectra collected upon healthy thyroid tissues is shown, whereas in Fig. 1(B), the sequence of average Raman spectra collected upon PTC tissues is presented. It should be noted that histological diagnosis evidenced the presence of zones corresponding to follicular 2(a) and to classical 2(b) PTC variants in thyroid sample of patient 2. Spectrum numbers correspond to the thyroid case numbers given in Table 1.
Figure 1.
Fingerprint region: (A) sequence of average Raman spectra collected upon healthy thyroid tissues, (B) sequence of average Raman spectra collected upon PTC tissues (2(a)- follicular variant PTC and 2(b)–classical variant PTC in thyroid sample of patient 2). Spectrum numbers correspond to the thyroid case/patient number given in Table 1.
It should be noticed that the spectra corresponding to the same type of thyroid tissue (healthy or PTC), but belonging to different patients, are very similar to each other, demonstrating good correlation in the single case and among different cases, whereas comparison between healthy and PTC groups of spectra reveals significant differences.
An accurate assignment of the major thyroid Raman bands registered in our spectra and comparison with the literature data is given in Table 2. Available Raman literature studies regarding thyroid tissue are scarce14,15,16; in this study, the applied RS technique allowed us to detect for the first time some peculiar features, characteristic for thyroid tissue. The most remarkable difference between the corresponding spectra of healthy and PCT tissues consists in the presence of three intense bands at 1006, 1156 and 1520 cm−1 in the pathological tissue, attributable to carotenoids14,17,18,19. Indeed, the comparison between Fig. 1(A) healthy and Fig. 1(B) PTC sequences of spectra provides clear evidence that PTC tissue hosts a significant presence of carotenoids, which are otherwise just trace-like in healthy tissue. The less intense Raman band at 956 cm−1 (4th carotenoid peak) was not distinguishable in our spectra, due to its low intensity (only about 10% of the 1156 cm−1 band intensity19). The 1006 cm−1 band is a mixed Raman peak, with contribution of carotenoids and phenylalanine νs (C-C) (at 1003 cm−1) (see Table 2).
Table 2. Peak positions and assignments of major Raman bands observed in thyroid healthy and PTC tissues.
| Healthy tissue peak position (cm−1) | Classical PTC tissue peak position (cm−1) | Follicular PTC tissue peak position (cm−1) | Band attribution | Reference |
|---|---|---|---|---|
| 673 | tryptophan (ring breathing) | 15 | ||
| 717 | membrane phospholipids head (C-N); adenine; lipids (CN+ (CH3)3) | 15 | ||
| 748 | 748 | 748 | DNA | 15 |
| 851 | proline&hydroxyproline (side chain vibration); tyrosine (ring breathing and Fermi doublet); glycogen | 15,16 | ||
| 919 | proline; hydroxyproline; glycogen; lactic acid | 15 | ||
| 957 | carotenoids; phosphates νs(PO43−); cholesterol; quinoid ring in-plane deformation | 15,16 | ||
| 971 | 971 | ν(C-C) wagging | 15 | |
| 994 | C-O ribose, C-C | 15,16 | ||
| 1003 | 1003 | 1003 | phenylalanine νs (C-C) | 15,17 |
| 1006 | 1006 | carotenoids | 15 | |
| 1031 | phenylalanine (δ(C-H) and C-H in-plane bending); protein (C-N stretching); carbohydrate residues of collagen | 15,16 | ||
| 1086 | 1084 | 1089 | ν(C-C) gauche; ν1(CO32−); ν3(PO43−); ν(PO2−); ν(C-C) skeletal of acyl backbone in lipid (gauche conform.) | 15,16 |
| 1128 | 1128 | 1128 | proteins (C-N stretching); carbohydrates (C-O stretching); ceramides; acyl backbone in lipid (trans conform., ν(C-C)) | 15, 16, 17 |
| 1156 | 1156 | 1156 | protein (stretching C-C and C-N) | 15 |
| 1156 | 1156 | 1156 | carotenoids | 15 |
| 1172 | 1172 | 1172 | δ(C-H), tyrosine | 15 |
| 1205 | ν(C–C6H5); tryptophan; phenylalanine; adenine and tyrosine (ring breathing); amide III | 16 | ||
| 1225 | amide III (β sheet structure) | 15 | ||
| 1234 | a concerted ring mode | 15 | ||
| 1239 | amide III | 15, 16 | ||
| 1264 | 1264 | lipids | 15 | |
| 1307 | 1307 | 1307 | lipid and collagen (twisting, bending, wagging) | 15 |
| 1337 | 1337 | 1337 | C-H deformation (protein); amide III; glycine and proline side chain (CH2 wagging vibrations); adenine and guanine (ring breathing modes) | 15 |
| 1360 | 1360 | 1360 | tryptophan | 15 |
| 1393 | 1393 | 1393 | CH rocking | 15 |
| 1424 | lipid (CH2 scissoring); deoxyribose (B, Z-marker) | 15 | ||
| 1440/1442 | 1440/1442 | CH, CH2 and CH3 deformation; cholesterol; triglycerides (fatty acids); lipids (CH2 scissoring and CH3 bending); collagen | 15 | |
| 1445 | collagen (δ(CH2), δ(CH3) and CH2CH3 bending); phospholipids (δ(CH2), δ(CH3) and CH2CH3 bending); methylene (bending) | 15 | ||
| 1450 | 1448/1452/1464 | 1448 | CH2CH3, CH2 and CH deformation; ν(C-H); δ(CH2); methyl groups bending; methylene deformation; proteins (δ(CH) and δ(CH2)) | 15 |
| 1498 | 1498 | (C-C) stretching in benzenoid ring | 15 | |
| 1516/1518 | 1516/1518 | β-carotene ν(C-C); carotenoid (C-C and conjugated C = C stretching); porphyrin ν(C = C) | 14 | |
| 1520 | 1520 | (-C = C-) carotenoids | 15 | |
| 1545 | C6-H deformation; tryptophan | 15 | ||
| 1552 | tryptophan ν(C = C); porphyrin ν(C = C) | 15 | ||
| 1557 | 1557 | tryptophan; porphyrin ν(C = C); amide II (ν(CN) and δ(NH)); COO− (tyrosine, amide II) | 15 | |
| 1584 | 1584 | 1584 | phenylalanine δ(C-C); (C-C) olefinic stretching; hydroxyproline; acetoacetate; riboflavin; lipids | 15, 16, 17 |
| 1602 | 1602 | phenylalanine δ(C-C) | 15 | |
| 1638 | 1640 | water (intermolecular bending and very weak and broad ν2) | 15 | |
| 1660 | 1660 | 1660 | ν(C-C) cis; (C-C) groups in unsaturated fatty acids; fatty acids; lipids; ceramide backbone; amide I | 15 |
| 2852 | 2852 | νs(CH2); lipids; fatty acids | 15 | |
| 2879 | lipids and proteins (CH2 and CH) | 15 | ||
| 2888 | 2888 | lipids and proteins (CH2 asymmetric stretching) | 15 | |
| 2931 | 2931 | 2931 | CH2 asymmetric stretching | 15 |
| 2936 | 2936 | chain end CH3 symmetric band | 15 | |
| 2960 | 2960 | out-of-plane chain end asymmetric CH3 stretching | 15 | |
| 3010 | 3010 | unsaturated = CH stretching | 15 |
In addition, the high wave number (HWN) region of spectra depicts a broad band centered at 2900 cm−1 (see Fig. 2 (full range spectra)), generally assigned to proteins, lipids and fatty acids vibrational modes. The ratio between this band in PTC cases and in healthy cases is approximately 2.5, therefore, indicating that another feature of PTC is a much more intense HWN band at 2900 cm−1.
Figure 2.
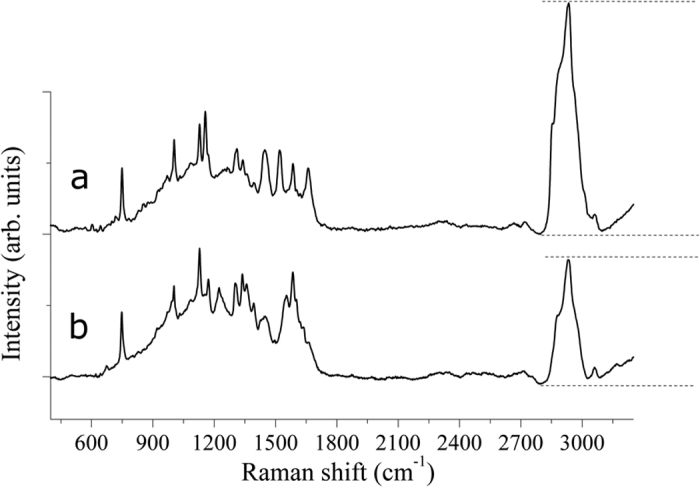
Full range Raman spectra collected upon (a) PTC and (b) healthy tissues. The shown spectra are average from the complete corresponding dataset.
RS imaging
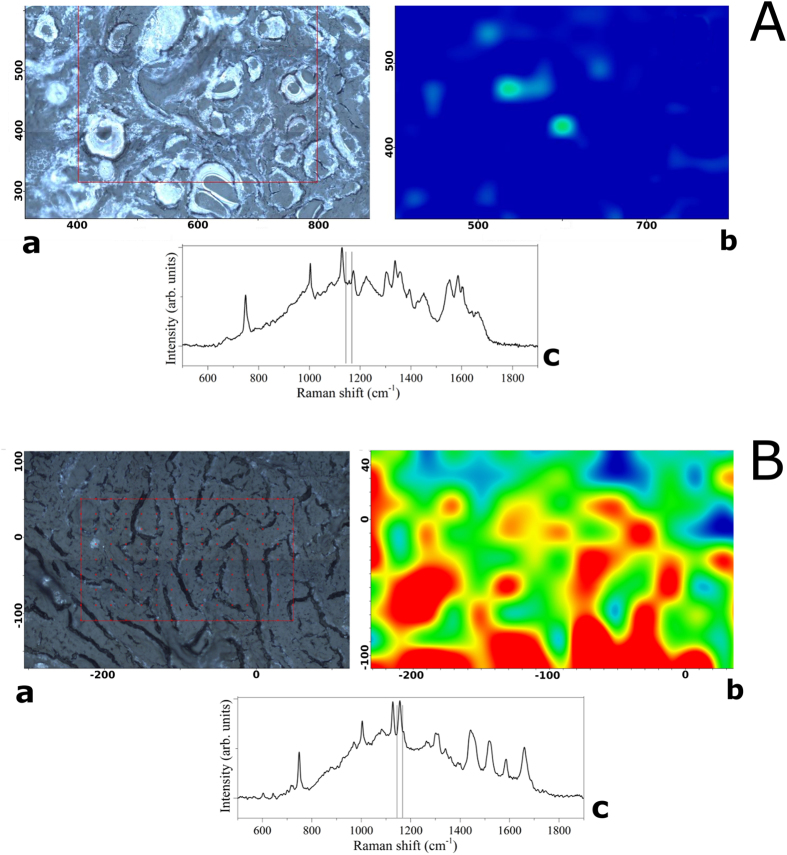
The Raman spectra collected upon a selected area provide intrinsic biochemical information that can be used for diagnosis. By selecting specific wavelengths, Raman imaging allows one to obtain different graphical results for the maps of the two tissue typologies. An example of such maps (20 μm step size and 400 × 300 μm2 area) in false colors referred to the band at 1156 cm−1 is shown in Fig. 3. In Fig. 3(A), the results obtained for healthy tissue ((a)–dark field optical image, (b)–Raman map, (c)–average reference Raman spectrum) are presented. The healthy tissue is in blue, while minimal amounts of carotenoids are represented by the yellow-green colors. In Fig. 3(B), the PTC tissue is shown ((a)–dark field optical image, (b)–Raman map, (c)–average reference Raman spectrum). In this case, significant amounts of carotenoids in PTC tissue were detected (false colors in green-yellow-red represent the corresponding increasing intensity of carotenoids).
Figure 3.
Typical example of Raman chemigram maps (1156 cm−1 band reference) for thyroid tissues: (A) healthy tissue ((a)–dark field optical image, (b)–Raman map, (c)–average reference Raman spectrum), (B) PTC tissue ((a)–dark field optical image, (b)–Raman map, (c)–average reference Raman spectrum). The red square on the right side (a) corresponds to the investigated tissue area shown on the left (b). The scale bars are expressed in μm.
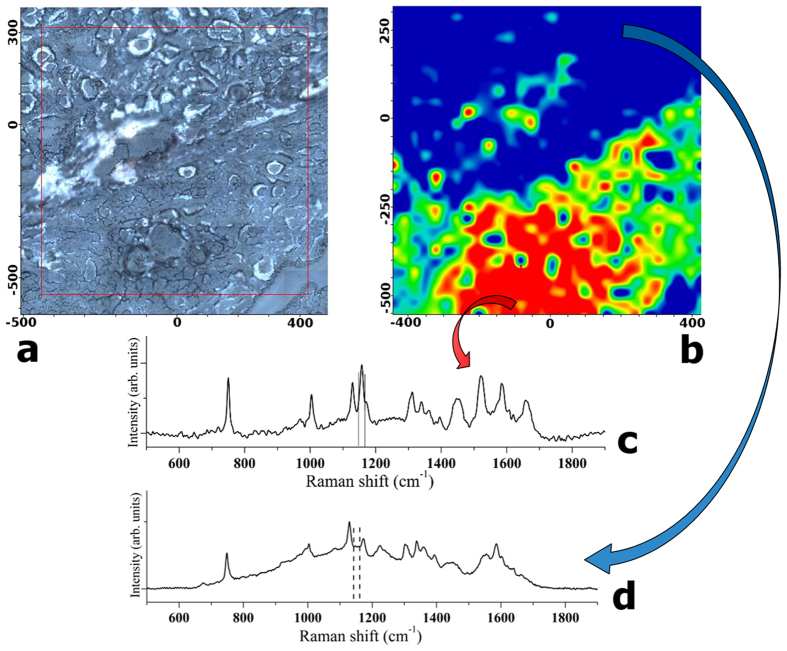
Figure 4 depicts an area in which healthy and PTC tissues are almost intermingled; however, the applied RS technique allows one to distinguish between healthy and pathologic tissue, at least in the studied cases. This should be of capital importance when dealing with precise identification of tumor margins during excision surgery for tumors with extra-capsular extension. More data are currently being collected to address exhaustively the latter issue (using, for instance, a decreased pixel step size), and the results will be reported in the near future.
Figure 4.
Typical example of Raman chemigram map (1156 cm−1 band reference) of a mixed zone of thyroid tissue (blue-healthy; red-yellow-green–PTC): ((a)–dark field optical image, (b)–Raman map, (c)–average reference Raman spectrum corresponding to healthy tissue, (d)–average reference Raman spectrum corresponding to PTC tissue). The red square on the right side (a) corresponds to the investigated tissue area shown on the left (b). The scale bars are expressed in μm.
Statistical analysis
Statistical analysis was performed on average spectra of each map, corresponding to 18 healthy tissue average spectra and 20 PTC average spectra. The FP and HWN range of Raman spectra, roughly 600 ÷ 1800 cm−1 and 2800 ÷ 3100 cm−1, were considered for statistical data treatment.
Discrimination between healthy and PTC tissue
The Principal Component Analysis (PCA) was performed on the matrix 38 × 1314 of the 38 average spectra in the range of 653 ÷ 1723 cm−1 and 2828 ÷ 3023 cm−1. It is reasonable to affirm that all the information about the hypothesized differences in composition is contained within the first 23 PCs, contributing to the total variability of the dataset with a larger % than the one represented by the 23rd PC (the 23rd PC corresponds to about 0.01%).
In order to observe whether any of the PCs have diagnostic capability (that is, the scores of the samples for that PC tend to form distinguished groups for the two tissue typologies), a two sample t-test was applied on the scores for each PC, after having controlled each time the validity of the assumption of equality between the variances of the two groups by Fisher’s test. The results of the t-test confirm that for 4 PCs (PC1, PC2, PC5, PC12) a certain separation of the mean of the healthy samples scores with respect to the one of the pathological group can be found (p-values of 0.028, 0.002, 0.002 and 0.050, respectively). Even if the reported p-values suggest a chance to use successfully this set of PCs, the check of 2D and 3D plots achievable by different combinations of such PCs shows that it is not possible. Among these combinations, for PC1–PC5, PC2–PC5 and PC5–PC12, the separation edge is simply not clear enough for diagnostic purposes. For the other cases (PC1–PC2, PC1–PC12, PC2–PC12), the test is affected by the presence of some outliers invalidating the results, when the number of observations per group is not appropriate to the degree of diversity between the distributions underlying the samples. This leads to the hypothesis that differences between tissue typologies do exist but can’t be fully revealed only by maximizing their respective variability, and truly none of the PCs by itself has diagnostic capability.
The Linear Discriminant Analysis (LDA) algorithm was first applied on the 38 × 23 matrix obtained considering the scores of the 38 samples for the first 23 PCs. Various combinations of all or part of the PCs were tried in order to find the discriminant function with the simplest mathematical solution for the classification: the best result (f1) was found using the PCs 1, 2, 5, 8, 10, 11, 12 and its analytical expression is:
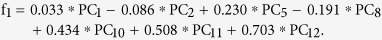 |
The scores for f1 are shown in Fig. 5. In this case, having previously verified by Fisher’s test that the variances of the sample scores on f1 within the two groups are not significantly different (F = 1.049; dfN = 19; dfD = 17), applying the two sample t-test to the same sample scores a value of t = 14.415 is obtained for 36 degrees of freedom: as a result, the two groups shown in Fig. 5 are considered significantly different at a confidence level of 0.001.
Figure 5. Projection of healthy and PTC samples along f1.
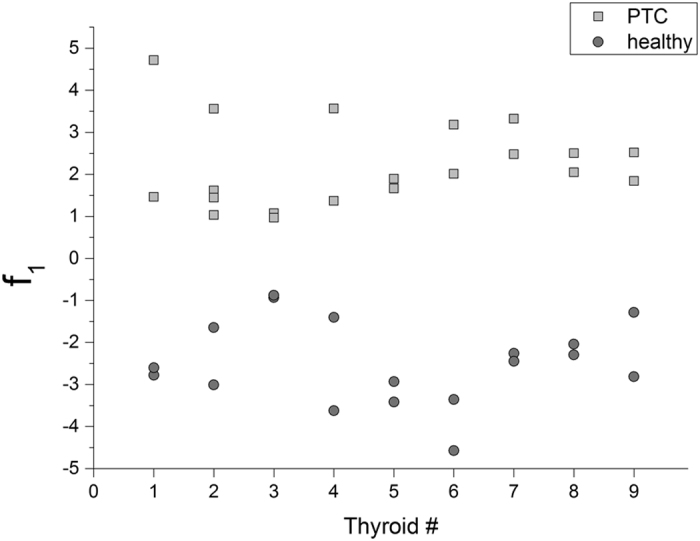
The leave-one-out cross-validation method, employed as internal tool to test the accuracy of f1, classified the 100% of the samples correctly. It should be also noticed that in Fig. 5 the groups are characterized by values ranging exclusively along the positive semiaxis of f1 (PTC tissue samples) or along the negative one (healthy tissue samples).
Discrimination between two PTC variants (classical and follicular)
The PCA was performed on the matrix 20 × 1314 of the 20 average spectra of PTC tissue samples in the 653 ÷ 1723 cm−1 and 2828 ÷ 3023 cm−1 ranges of spectra. The analysis of the variance values explained by each PC revealed that the first 17 components explain almost the totality of the variance (the 18th PC represents <0.01%).
In this case, the values for F-test indicate that variances are not significantly different among the various PCs, except for PC2, and the t-test confirms that, for none of the PCs, the means of the scores for the two groups are significantly different (confidence level of 5%).
The LDA algorithm was applied also in this case and the result is the function f2, obtained combining the PCs 1, 4, 5, 6, 7, 8, 10, 11, 12, 15. Its analytical expression is:
 |
The scores for f2 are shown in Fig. 6. Having previously verified by Fisher’s test that sample variances of the two groups are not significantly different (F = 0.816; dfN = 13; dfD = 5), applying the two sample t-test to the sample scores on f2, a value of t = −15.737 is obtained for 18 degrees of freedom: as a result, the two groups shown in Fig. 6 are significantly different at a confidence level of 0.001. The leave-one-out cross-validation method classified the 95.0% of the samples correctly: all the follicular variant PTC samples were attributed to the right group and, among the classical variant PTC samples, only one has been misclassified. Nevertheless, it is possible to classify the 100% of samples correctly, considering as a criterion the fact that the values of the two PTC variants are distributed along the opposite semiaxes of f2 (positive for follicular variant and negative for classical variant PTC).
Figure 6. Projection of two PTC variants along f2.
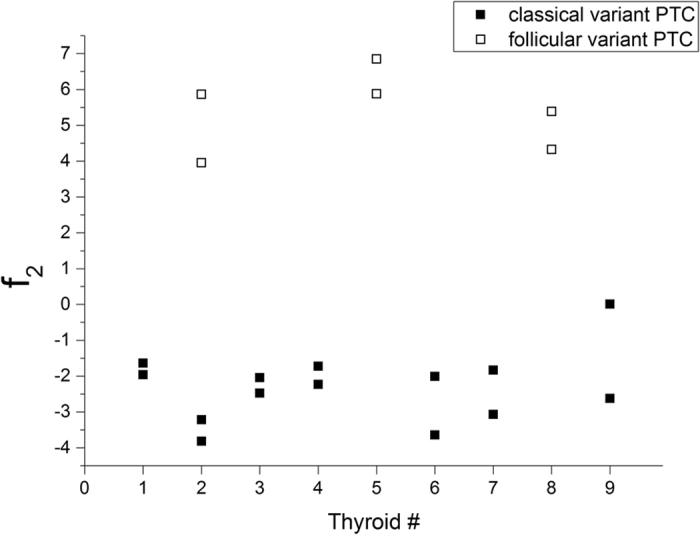
Summarizing the results of this section, we can affirm that Raman spectroscopy is able to discriminate between healthy and PTC tissues of thyroid with 100% of sensitivity, specificity and accuracy and to discriminate between classical and follicular variants of PTC with 93% of sensitivity, 100% of specificity and 95% of accuracy, by means of the leave-one-out cross-validated LDA.
Discussion
Our results have demonstrated the feasibility and reproducibility of RS to discriminate between normal thyroid tissue and PTC, and between classical and follicular variants of PTC, on the basis of their biochemical fingerprints. This finding is of great relevance for the development of a RS optical biopsy system to investigate thyroid tissue alterations. Based on the experimental results obtained in this work, we can attest the significant carotenoids presence in the PTC tissues with respect to the healthy tissue, in which their absence or minimal and localized presence was detected (see Figs 1, 3 and 4). To the best of our knowledge, this is the first experimental evidence of carotenoids presence in the neoplastic thyroid tissue.
Papillary thyroid carcinoma has been extensively investigated with multiplatform molecular analysis and the area of unknown genomic alteration has been reduced substantially from 25% to less than 4%13. Detailed study of the genomic background, however, is not sufficient to fully investigate the mechanisms that lead to neoplastic transformations, which, in turn, would lead to the identification of more accurate diagnostic, prognostic and predictive makers. For example, BRAFV600E gene mutation, currently considered as a driver of molecular alteration in classic variant PTC, has been recognized in over 70% of benign nevus without neoplastic progression20. Spectroscopy methods applied in clinics should provide the best possible sensitivity, specificity and accuracy in order to minimize false definitions. Ideally, a new method should be performed as an in situ analysis encompassing the assessment of multiple cellular constituents and allowing paired morphological and biochemical analysis. RS is among the few available methods fulfilling the above requirements.
In our study, we considered the FP and the HWN range of thyroid Raman spectra, both presenting changes while passing from healthy to PTC areas and, therefore, both important for diagnostic utility, as confirmed also by authors6 for cervical tissue.
RS has the ability to identify specific tumor expression molecules and molecular species involved in tumorigenesis and progression. For instance, Talari et al.14 claimed the 956, 1006, 1156–1157, 1524–1528 cm−1 Raman peaks as “carotenoids absent in normal tissue”. Puppels et al.21 investigated carotenoids located in human lymphocyte subpopulations and natural killer cells, evidencing a high carotenoids concentration in the CD4 + lymphocytes, and proposed to investigate the possible mechanisms behind the protective role of carotenoids against the development of cancers. The increased intensities at 1159 and 1527 cm−1, assigned to carotenoids have been identified also in the Raman spectra of brain tumors14 and neurinomas22. Talari et al.14 suggested that carotenoids can be used as Raman biomarkers in breast cancer pathology.
Resonance Raman is probably one of the best methods to study the properties of carotenoids in complex media, such as, for example, binding sites of biological macromolecules in living organisms19. In this case, i.e. when the wavelength of the excitation laser is in the range of electronic absorption band of molecules of interest, resonance Raman intensities may be enhanced up to 6 orders of magnitude, as compared to normal Raman scattering23. In the present work, the enhancement of carotenoid Raman bands was obtained using 532 nm laser wavelength, which lies in the range of carotenoids UV/Vis absorption region24.
The coupled histopathological and Raman biochemical observations performed in this work highlighted that carotenoids are mainly present in cellular areas of PTC, so that their presence seems to be related to the neoplastic thyrocytes within the tumor tissue. Our study suggests that these characteristics could be used as Raman biomarkers in the PTC pathology. However, the mechanism underlying potential oncogenic effects of carotenoids is still unknown, since very little is known about the biochemical content of neoplastic cells, especially what regards lipids, lipoproteins and lipophilic substances that are commonly lost in routinely processed histological samples. Among human tissues, different normal cells are able to utilize carotenoids, i.e. beta-carotene is reported as a local supply of vitamin A in the skin and melanocytes25. However, physiological mechanism for carotenoids uptake in normal thyrocytes is not reported, and our results raise the hypothesis of a carotenoid-related pathway for the PTC oncogenesis. This is just a working hypothesis, which needs accurate validation, but nevertheless underlines the presence of carotenoids in neoplastic thyrocytes, as it happens in other organs15,22,23.
In conclusion, we performed the RS investigation and biochemical mapping of healthy thyroid tissue and of PTC (classical and follicular variants). The obtained results demonstrate the great potential of RS to support histopathological evaluation, increasing the reliability of cancer diagnostics. On the basis of the results of multivariate statistical model, carried out by the leave-one-out cross-validated LDA, we can affirm that RS is able to discriminate between healthy and PTC tissues of thyroid with 100% of sensitivity, specificity and accuracy and to discriminate between classical and follicular variants of PTC with 93% of sensitivity, 100% of specificity and 95% of accuracy. The achieved diagnostic sensitivity, specificity and accuracy are compatible with the clinical use, both for the PTC diagnosis and in the differential diagnosis between classical and follicular variants of PTC, the latter being a significant challenging point for thyroid nodules evaluation.
Only a few literature studies report RS investigations of thyroid tissue and neoplasia16,17. The distinctive trait of our RS analysis is that for the first time it has been performed on tissue sections, combined with microscopic assessment of the very same areas. The method is highly cost-effective, being based solely on the analysis of unstained cryostatic tissue sections. Our results are likely to represent a significant advance in the imaging of thyroid tissues, leading to subsequent clinical application by improving diagnostic accuracy and reducing inter-observer variability.
This study is the first demonstration of the presence of significant amounts of carotenoids in thyroid neoplasms, suggesting that carotenoids could be used as a Raman biomarker for the PTC pathology. In this regard, combination of the histological and Raman microscopy analysis approaches may open a new way to integrative findings with wide implications for basic pathobiology, tumor classification schemes and therapeutic strategies.
Methods
Ethics statement
The study was approved by the Ethical Committee of the UCBM (prot. 33.15 TS ComEt CBM). Before surgical procedures, the informed consent was collected. Enrolled patients are known to the pathologist and recorded in a codified file with an anonymous ID code, which was also registered in the institutional software database of the Pathology Unit of the UCBM. All personally identifiable information was recorded in a codified file. All experiments were performed in accordance with the principle of Good Clinical Practice (GCP) and the ethical principles contained in the current version of the Declaration of Helsinki.
Thyroid tissues
This prospective monocentric study has been approved by the Ethical Committee of the UCBM (prot. 33.15 TS ComEt CBM), and all patients gave the written informed consent. Nine patients that received a diagnosis of PTC based on FNA at the Endocrinology Unit of UCBM were enrolled for this study. These patients underwent total thyroidectomy at the Surgical Unit of the same Institution. At the time of surgery the removed specimens were immediately submitted unfixed to the Pathology Unit in an appropriately labeled container. After completion of the gross examination of the specimen by the pathologist, resection margins were marked with black ink. Pathological sampling was carried out in agreement with international guidelines for handling surgical specimens26. A tissue slice of about 1 × 1 × 0.3 cm3 was then obtained, including both healthy and neoplastic areas, avoiding surgical margins, and the slice was frozen on a metallic cold-plate inside the cryostat. A 5 μm cryostatic section was cut and stained with Haematoxylin/Eosin, in order to confirm the presence of healthy and neoplastic tissue zones, as well as the transition area between them. Additional sections were cut at 20 and 30 μm of thickness, collected on separate glass slides and stored unstained at −20 °C until the Raman evaluation. The surgical samples were subsequently fixed in buffered formalin and embedded in paraffin for permanent sectioning. Diagnosis, grading and staging were performed, in agreement with the 7°th edition of TNM27.
Dataset/casuistry
A number of thyroid tissue areas was histologically identified and diagnosed as healthy or pathological (Haematoxylin/Eosin staining of frozen samples) by experienced pathologist (A.C.). By means of the RS imaging, 18 maps have been obtained from healthy and 20 from pathological areas. In Table 1, the experimental dataset is fully represented, showing the distribution of the tissue samples (healthy, PTC classical variant and PTC follicular variant) for each of the nine patients.
Raman spectroscopic measurements
Raman spectra were recorded using a Thermo Fisher Scientific DXRxi Raman microscope at the following conditions: 532 nm laser source; 200–3400 cm−1 full range grating; 10× and 50× objectives; 25 μm confocal pinhole, 5 (FWHM) cm−1 spectral resolution. The RM instrument indicated above guarantees a fast change of experimental parameters, for better measurements procedure optimization, and does not require consumable reagents and staining treatments. As a first step, the collection of a number of mosaic images at low magnification (10×) using the RM has been carried out, providing the generic overview information on the tissue morphology and allowing one to individuate and evaluate regions of interest. After that, the region of interest was investigated collecting spectra at high magnification (50×).
Preliminary measurements were performed, in order to optimize the experimental parameters to provide a high signal-to-noise (S/N) ratio and to minimize tissue fluorescence. A 5th order polynomial correction was used to compensate the tissue fluorescence. A laser power of 8 mW measured at the sample has been applied as the best compromise between the signal quality and the undesired tissue burning. The exposure time was 0.8 sec, as a suitable compromise to achieve a good spectra quality and to shorten the overall acquisition time. At least 50 exposures were averaged to obtain a high S/N ratio. Laser spot size was about 700 nm (50× objective). Various Raman maps ranging from 100 × 100 μm2 up to 1 × 1 mm2, collecting several hundreds of spectra per map, were obtained. Step size employed for small maps (i.e. 100 × 100 μm) was of about 2 μm, while for larger maps with lateral dimension ranging from several hundred of μm up to 1 mm, the average step size of about 50 μm was used. Typical collection time for each map was about 4/6 hours, both for small and large maps.
The background-subtracted Raman spectra were further normalized for the area under the curve for standardization of the tissue Raman intensities. No pre-treatment was performed on tissue samples before RS examination. To assess intra-sample variability, multiple measurements were carried out at different regions within the same sample.
Statistical analysis
Statistical analysis was performed with a supervised approach on average spectra of each map, aiming to generate a model for classifying tissues. The same number of maps (two) from each typology of tissue present in each patient (healthy, PTC classical variant and PTC follicular variant) were employed, corresponding to 18 healthy tissue average spectra and 20 PTC average spectra. The FP and HWN range of Raman spectra, roughly 600 ÷ 1800 cm−1 and 2800 ÷ 3100 cm−1, respectively, were selected for statistical data analysis treatment. The remaining spectral range was not considered, as non meaningful from the point of view of the contained biochemical information. The collected Raman data were processed performing multivariate analysis, used for complex systems with high internal variability. Two principal statistical procedures were performed on the dataset: Principal Component Analysis and Linear Discriminant Analysis.
At first, PCA was carried out in order to reduce the initial high dimensionality of the dataset and to verify whether, in a subset of dimensions, the internal variability of the spectra can reveal on its own differences among tissue typologies that can be considered diagnostic.
LDA was applied on the same components observed in PCA to verify if differences among the typologies exist due to differences among the means of samples belonging to different tissue groups. Moreover, the implementation of the algorithm on principal components let to optimize the process of classification on the basis of a reduced number of input variables. The LDA was tested by using the leave-one-out cross validation method, in order to estimate the accuracy of our model in predicting unknown samples.
Additional Information
How to cite this article: Rau, J. V. et al. RAMAN spectroscopy imaging improves the diagnosis of papillary thyroid carcinoma. Sci. Rep. 6, 35117; doi: 10.1038/srep35117 (2016).
Acknowledgments
Authors are grateful to the Thermo Fisher Scientific Company for DXRxi Raman system supplied in the frames of the Thermo Raman Seed Unit Program (2016).
Footnotes
Author Contributions J.V.R. and A.C. conceived and developed the methodology; M.F. and V.G. performed Raman spectroscopy data acquisition and their statistical analysis treatment; A.C. and C.T. carried out the histopathological evaluation of tissues; M.C. and P.C. performed clinical and surgical evaluation; all the co-authors discussed the results with a major contribution of J.V.R., A.C., M.C., P.P. and A.O.M.; all the co-authors contributed to the manuscript writing with a major contribution of J.V.R., A.C. and A.O.M. Technical and instrumental support and training was provided by M.R. The overall study was supervised by J.V.R.
References
- COST Action BM 1401 “Raman for Clinics”. Network www.raman4clinics.eu.
- Çulha M. Raman spectroscopy for cancer diagnosis: how far have we come? Bioanalysis 7(21), 2813–2824 (2015). [DOI] [PubMed] [Google Scholar]
- Kong K., Kendall C., Stone N. & Notingher I. Raman spectroscopy for medical diagnostics — from in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 89, 121–134 (2015). [DOI] [PubMed] [Google Scholar]
- Bergholt M. S. et al. Raman endoscopy for objective diagnosis of early cancer in the gastrointestinal system. J. Gastroint. Dig. Syst. S1, 008 (2013). [Google Scholar]
- Bergholt M. S. et al. Raman endoscopy for in vivo differentiation between benign and malignant ulcers in the stomach. Analyst 135, 3162–3168 (2010). [DOI] [PubMed] [Google Scholar]
- Duraipandian S. et al. Simultaneous fingerprint and high-wavenumber confocal Raman spectroscopy enhances early detection of cervical precancer in vivo. Anal. Chem. 84, 5913–5919 (2012). [DOI] [PubMed] [Google Scholar]
- Lui H., Zhao J., Mclean D. & Zeng H. Real-time Raman spectroscopy for in vivo skin cancer diagnosis. Cancer Res. 72(10), 2491–2500 (2012). [DOI] [PubMed] [Google Scholar]
- Desroches J. et al. Characterization of a Raman spectroscopy probe system for intraoperative brain tissue classification. Biomed. Opt. Express 6(7), 2380–2397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloch Z. W., Fleisher S., Li Volsi V. A. & Gupta P. K. Diagnosis of “follicular neoplasm”: a gray zone in thyroid fine-needle aspiration cytology. Diagn. Cytopathol. 26(1), 41–44 (2002). [DOI] [PubMed] [Google Scholar]
- Trimboli P. et al. Clinical characteristics as predictors of malignancy in patients with indeterminate thyroid cytology: a meta-analysis. Endocrine 46(1), 52–59 (2014). [DOI] [PubMed] [Google Scholar]
- Elsheikh T. M. et al. Interobserver and intraobserver variation among experts in the diagnosis of thyroid follicular lesions with borderline nuclear features of papillary carcinoma. Am. J. Clin. Pathol. 130(5), 736–744 (2008). [DOI] [PubMed] [Google Scholar]
- Ferris R. L. et al. American thyroid association statement on surgical application of molecular profiling for thyroid nodules: current impact on perioperative decision making. Thyroid 25(7), 760–768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N. et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159(3), 676–690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talari A. C. S., Movasaghi Z., Rehman S. & urRehman I. Raman spectroscopy of biological tissues. Appl. Spectr. Rev. 50(1), 46–111 (2015). [Google Scholar]
- Teixeira C. S. B. et al. Thyroid tissue analysis through Raman spectroscopy. Analyst 134, 2361–2370 (2009). [DOI] [PubMed] [Google Scholar]
- Li Z. et al. Surface-enhanced Raman spectroscopy for differentiation between benign and malignant thyroid tissues. Laser Phys. Lett. 11, 045602 (2014). [Google Scholar]
- Marshall C. P. et al. Carotenoid analysis of halophilicarchaea by resonance Raman spectroscopy. Astrobiology 7(4), 631–643 (2007). [DOI] [PubMed] [Google Scholar]
- Jehlicka J., Edwards H. G. & Oren A. Bacterioruberin and salinixanthin carotenoids of extremely halophilicarchaea and bacteria: a Raman spectroscopic study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 106, 99–103 (2013). [DOI] [PubMed] [Google Scholar]
- Macernis M. et al. Resonance Raman spectra of carotenoid molecules: influence of methyl substitutions. J. Phys. Chem. A 119, 56–66 (2015). [DOI] [PubMed] [Google Scholar]
- McClenahan P. et al. BRAFV600E mutation status of involuting and stable nevi in dabrafenib therapy with or without trametinib. JAMA Dermatol. 150(10), 1079–1082 (2014). [DOI] [PubMed] [Google Scholar]
- Puppels G. J., Garritsen H. S. P., Kummer J. A. & Greve J. Carotenoids located in human lymphocyte subpopulations and natural-killer-cells by Raman microspectroscopy. Cytometry 14, 251–256 (1993). [DOI] [PubMed] [Google Scholar]
- Mahadevan-Jansen A. & Richards-Kortum R. Raman spectroscopy for cancer detection: a review. Proceed. 19th Intern. Conf. IEEE/EMBS Oct 30–Nov 2 1997 Chicago, Il USA.
- Rimai L., Kilponen R. G. & Gill D. Excitation profiles of laser Raman spectra in the resonance region of two carotenoid pigments in solution. J. Am. Chem. Soc. 92, 3824–3825 (1970). [Google Scholar]
- V. V. & A. A. Carotenoids. Volume 4: Natural Functions (ed. Britton G., Liaaen-Jensen S. & Pfander H.) 60 (BirkhäuserVerlag, P.O. Box 133, CH-4010 Basel, Switzerland. ISBN 3-7643-7498-3 BirkhäuserVerlag, Basel–Boston–Berlin, 2008). [Google Scholar]
- Andersson E., Vahlquist A. & Rosdahl I. Beta-carotene uptake and bioconversion to retinol differ between human melanocytes and keratinocytes. Nutr. Cancer 39(2), 300–306 (2001). [DOI] [PubMed] [Google Scholar]
- Rosai J. Rosai and Ackerman’s Surgical Pathology (10th edition) (Mosby Elsevier, 2011). [Google Scholar]
- Edge S. B. et al. editors. AJCC Cancer Staging Manual (7th edition) (New York, NY: Springer, 2010). [Google Scholar]