Abstract
Eusocial insects organize themselves into behavioral castes whose regulation has been proposed to involve epigenetic processes, including histone modification. In the carpenter ant Camponotus floridanus, morphologically distinct worker castes called minors and majors exhibit pronounced differences in foraging and scouting behaviors. We found that these behaviors are regulated by histone acetylation likely catalyzed by the conserved acetyltransferase CBP. Transcriptome and chromatin analysis in brains of scouting minors fed pharmacological inhibitors of CBP and histone deacetylases (HDACs) revealed hundreds of genes linked to hyperacetylated regions targeted by CBP. Majors rarely forage, but injection of a HDAC inhibitor or small interfering RNAs against the HDAC Rpd3 into young major brains induced and sustained foraging in a CBP-dependent manner. Our results suggest that behavioral plasticity in animals may be regulated in an epigenetic manner via histone modification.
Colonies of eusocial insects organize themselves into castes comprising individuals that exhibit specific behaviors over extended periods of time. This colonial division of labor is a key adaptation responsible for the ecological and evolutionary success of eusocial insects (1–4). Different eusocial species have evolved unique strategies for regulating the expression of behavioral castes on the basis of age, morphology, and social context. The most fundamental examples of division of labor involve the differentiation of individuals into sterile (worker) and reproductive (queen) castes. In addition, workers often express a variety of specialized behaviors depending on age [e.g., the honey bee Apis mellifera (2)], body size [e.g., the fire ant Solenopsis invicta (5)], or both [e.g., formicid ants (1–4, 6)].
The principles underlying the social control of behavior and the corresponding molecular mechanisms that regulate individual behavioral plasticity have been studied primarily in solitary species, such as the fly Drosophila (7). Recently, obligately social insects, including the eusocial honey bee A. mellifera and carpenter ant Camponotus floridanus, have emerged as models of more complex behavior (8–10). Findings in these species suggest that epigenetic processes, including DNA methylation (11–15) and histone posttranslational modifications (hPTMs) (16), may play key roles in regulating caste-based behavioral plasticity.
C. floridanus workers exhibit caste-specific behaviors
To investigate the role of hPTMs in regulating ant behavioral castes, we studied C. floridanus (12), which expresses two distinct female worker caste morphologies, called minors and majors (Fig. 1A, right). These morphs are distinguished by head width and length of scape (basal antennal segment; a proxy for body size) (Fig. S1, A and B) and are produced in a 2:1 ratio in mature colonies (Fig. S1C). Although genetic factors may contribute to the quantitative variation in worker morphology (Fig. S1D), the production of minor and major castes per se is likely not caused by allelic variation. Rather, workers are genetically related supersisters (r = 0.75) resulting from a single diploid mother mating with a single haploid father (17). Further, treatment of undifferentiated larvae with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-dC) increases head width and scape length in the resulting adults (15).
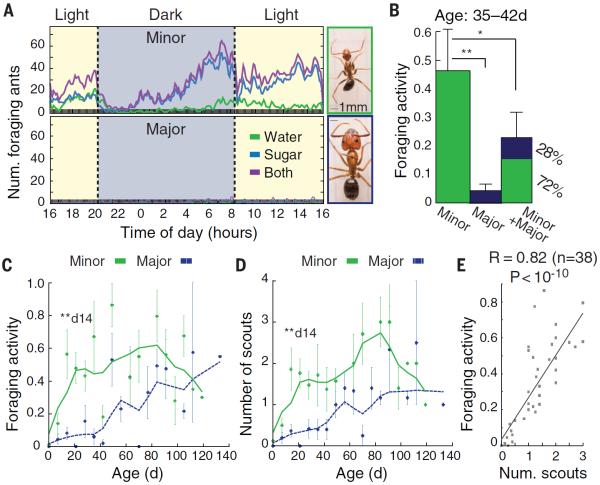
Fig. 1. Foraging and scouting behaviors depend on worker caste and age.
(A) Circadian foraging activity for minor (top) and major (bottom) workers in a single monogamous colony. Photographs show representative minor and major workers (Fig. S1, A and B). (B) Average foraging activity (defined in Fig. S2A) ± SE for 35- to 42-day-old minors and majors isolated and sugar-starved for 24 hours; rightmost column shows foraging activity for mixed cohorts of 10 majors and 10 minors of the same age. (C and D) Foraging activity (C) and number of scouts (D) for minors and majors isolated and sugar-starved for 24 hours, as a function of adult age after eclosion. Error bars denote SE over at least five independent replicates from six colonies. The earliest age of significant caste-differential behavior (day 14) is noted. Asterisks in (B) to (D) denote significance by Mann-Whitney U test: *P < 0.05, **P < 0.01. (E) Number of scouts versus foraging activity for data in (C) and (D). Pearson correlation coefficient is shown.
A survey of hPTMs in C. floridanus indicated that several hPTMs, especially the acetylation of Lys27 on histone H3 (H3K27ac), have distinct genome-wide patterns in the bodies and brains of minors and majors (16). These differences can be attributed to differential localization of the conserved acetyltransferase and transcriptional coactivator CBP [cyclic adenosine monophosphate response element–binding protein (CREB) binding protein] in each caste, and they correspond to differences in gene expression (16). In addition, a functional histone deacetylase inhibitor (HDACi), the fatty acid 10-HDA, is a major component of royal jelly, an environmental regulator of queen production in honey bees (18). Taken together, these findings suggest that hPTMs influence the generation of distinct castes in eusocial insects and that histone acetylation might regulate caste-based behavioral plasticity.
To examine caste-based behavioral plasticity in C. floridanus, we fitted the nest of a 7-year-old queen-right (i.e., containing a queen) colony with a foraging arena and counted the number and caste of each ant that foraged out to feed on water or 20% sugar water over 24-hour periods (see Materials and Methods) (19). Minors performed the vast majority of foraging, with a distinct circadian pattern and preference for sugar water (Fig. 1A). To control for effects of social interactions, we isolated 1-day-old ants and monitored their foraging behavior either in isolation for 10 days (Fig. S1, E and F) or when mixed with older, mature workers for 4 months (Fig. S1, G and H). In both cases, minors were the predominant foragers.
To determine whether caste-based foraging may be a generic feature of Camponotus ants, we also assayed the sympatric species C. tortuganus. In both species, minors foraged but majors did not (Fig. S1I), indicating that minor and major Camponotus workers display natural differences in foraging behavior (20–22).
Age correlates with behavioral plasticity in eusocial insects, including other Camponotus species (22). We therefore marked 1-day-old callows on a weekly basis in several queen-right colonies. We analyzed equal-sized cohorts of workers with identical colony background, caste morphology, and age (±48 hours) in an assay where either minors or majors were isolated from their natal nest and were water-starved (i.e., by withholding sugar) for 24 hours before foraging. Under these stringent conditions, minors showed significantly greater foraging activity than age-matched majors, although majors did forage at a low rate (Fig. 1B and Fig. S2A). Moreover, mixed cohorts of age-matched minors and majors displayed lower foraging activity than minors alone, yet only 28% of foraging was attributed to majors (Fig. 1B).
Additionally, we analyzed foraging behavior as a function of starvation time, because majors are physically larger and may have twice the food storage capacity of minors. Majors required more than 9 days of starvation to match the foraging activity of minors starved for only 24 hours (Mann-Whitney U test, P < 0.01; Fig. S2B). Thus, minors appear to be the predominant foragers in queen-right colonies (Fig. 1A) as well as in young (Fig. S1, F and H) and mature (Fig. 1B) worker cohorts.
We also examined how caste and age affect the lead foragers, called scouts, which have been reported to constitute a distinct behavioral caste in a few eusocial species (1, 20, 21, 23) (see below). Scouts are the first ants to leave a nest, discover a food source, and return to the nest with this food, before recruiting additional ants to forage (movie S1 and table S1). Analysis of the number of scouts and foraging activity over 4 months revealed both caste- and age-based differences (Fig. 1, C and D). Minors exhibited significantly more foraging and scouting activity than did majors by 14 days of age (Fig. 1, C and D). Also, the foraging speed of 60-day-old scouts was greater than that of 7-day-old scouts by as much as a factor of 6 (Fig. S2C). We exclusively assayed ants that were naïve to a foraging arena, so these effects are not due to learning. Finally, the number of scouts in a cohort correlated strongly with the cohort's foraging activity (Fig. 1E); this finding suggests that scouts, which are predominantly minors, determine a cohort's overall foraging activity (24).
These results indicate that foraging and scouting behaviors in C. floridanus depend on caste morphology, age, and social context, consistent with observations in other ant species (1–3, 20–23). They also support the view that eusocial species that express polymorphic worker castes have the potential for greater colony-level behavioral complexity than species with a monomorphic worker caste, as caste morphology apparently provides an additional dimension of behavioral variation that can be controlled by colonies to optimize division of labor (1–4, 22).
Histone deacetylases inhibit foraging and scouting behaviors
Minors always foraged and scouted more than majors, regardless of age and social context; this observation is consistent with the idea that minors and majors harbor innate, molecularly determined differences that influence behavior. Given results suggesting a functional role for histone acetylation in determining caste-specific traits (16), we examined whether foraging behavior might be regulated by histone acetylation dynamics controlling gene transcription.
We first measured mRNA abundance in brains of young, age-matched minors and majors from the same colony for the five class I and II HDACs encoded in the C. floridanus genome. Orthologs of these genes have established roles in neuronal and behavioral plasticity in other insects and mammals (25–27). Both Rpd3/Hdac1a (a putative H3K27ac deacetylase) and Hdac6 showed caste-specific expression patterns (Student's t test, P < 0.003; Fig. S3).
We confirmed that both Rpd3 and Hdac6 transcripts also increased with age in minors and majors, using 25 brains evenly sampled from five colonies for each caste and age point (n = 100 brains total; Fig. 2A). These observations are consistent with specific HDACs influencing foraging behavior, either via age-dependent decreases in histone acetylation or via increased histone acetylation dynamics, as has been observed for Rpd3 in Drosophila (25).
Fig. 2. HDAC inhibition stimulates foraging and scouting behaviors.
(A) RT-qPCR analysis of mRNA abundance for Rpd3 (Hdac1a) and Hdac6 in young (1 day old) and mature (>30 days old) major and minor brains. Each bar indicates the mean ± SE over five colonies, where each measurement represents a pooled sample of five individual brains (n = 100 brains total). mRNA abundance was normalized against actin mRNA. P values were computed by Mann-Whitney U test. Primers are reported in table S2. (B) Schematic of foraging assay for VPA-treated ants. Cohorts of 28 minors and majors, age 25 to 30 days, were fed 20% sugar water with or without 10 mM VPA for 30 days (seven ants in four treatment groups). Ants were then pooled, sugar-starved for 24 hours, and assayed for foraging after attaching a foraging arena. Blue triangle denotes sugar water food source. (C) Average percent of all foraging events performed by untreated or VPA-treated minors or majors when pooled as in (B). (D) Average percent of trials in which the first scout that foraged and fed on sugar water had been provided sugar water alone or with VPA. Error bars in (C) and (D) denote SE over 46 trials from 12 colonies. (E) Schematic of foraging assay for TSA-treated ants. Cohorts of 15 minors, age 25 to 30 days, were fed 20% sugar water containing DMSO with or without 50 μM or 100 μM TSA for 42 days. Unlike in (B) to (D), treatment groups were assayed for foraging separately, without pooling, after 24 hours of sugar starvation. (F and G) Average foraging activity (F) and number of scouts (G) for DMSO- or TSA-treated minor cohorts in foraging assay shown in (E). Cohorts were tested weekly over 42 days. Error bars denote SE over six measurements per cohort for 14 (DMSO) or 18 (TSA) independent cohorts from six colonies. P values were computed by Mann-Whitney U test.
We then examined foraging and scouting behaviors after inhibiting the activity of class I and II HDACs by feeding workers a small-molecule HDACi, valproic acid (VPA) (28). As expected (28, 29), VPA treatments increased global levels of H3K27ac when fed to larvae and minors (Student's t test, P < 0.02; Fig. S4A). We fed non-lethal concentrations of 10 mM VPA (28, 29) to mixed cohorts of 25- to 30-day-old minors and majors for 30 days. Foraging was assessed after pooling a treated cohort with a corresponding untreated cohort of colony- and age-matched minors and majors (Fig. 2B). Both VPA-treated minors and majors exhibited significantly more foraging than did controls (Fig. 2C). VPA-treated minors were also the first to act as scouts by finding food in 73% of trials (Fig. 2D and Fig. S4B), whereas majors never scouted (Fig. 2D). These results indicate that increasing histone acetylation shifts the behavior of both minors and majors toward increased foraging and feeding, although minors remain the predominant foragers and scouts.
To confirm that increases in foraging activity elicited by VPA were due to HDAC inhibition, we used a second HDACi, trichostatin A (TSA), which has greater potency (i.e., inhibitory concentration, IC50) than VPA, as well as greater specificity toward histone substrates for inhibiting the removal of acetyl groups (28, 29). As with VPA treatments, we assayed foraging using 25- to 30-day-old workers. However, we repeatedly assayed foraging over 42 days (rather than once after 30 days), and we assayed treated and untreated cohorts of minors separately (rather than mixed) to avoid any potential confounding effects due to social interactions (Fig. 2E).
In this assay, relative to controls, minors fed TSA displayed significantly greater foraging activity in a dose-dependent manner (Fig. 2F). TSA also increased the number of scouts (Fig. 2G). Because only a minority of ants fed during a foraging trial, TSA does not appear to cause general locomotor hyperactivity. Furthermore, pharmacological treatment did not affect the correlation between foraging activity and the number of scouts (R=0.80; Fig. S4C) relative to controls (R = 0.82; Fig. 1E), nor did it affect the duration of a scout's return to the nest after feeding (expedition time) [Mann-Whitney U test, P<0.9 for TSA versus dimethyl sulfoxide (DMSO); Fig. S4D]. Also, TSA-treated minors showed low mortality over the 42-day treatment course (Fig. S4E), which suggests that sickness and/or hunger are unlikely explanations for increased foraging and feeding. Hence, we conclude that HDAC activity normally inhibits foraging behavior in workers.
CBP-dependent histone acetylation regulates foraging and scouting behaviors
Because of the reported correspondence between genomic regions of caste-specific hPTMs and CBP binding (16), we tested whether the stimulation of foraging behavior by HDAC inhibition depends on stabilization or enhancement of hPTMs acetylated by CBP, such as H3K27ac. We thus used the small molecule C646, a specific inhibitor of CBP's histone acetyltransferase (HAT) activity (HATi) in mammals and insects (30, 31). CBP's mammalian paralog p300 is also targeted by C646, but it is not encoded in the C. floridanus genome.
Feeding 25 μM C646 to larvae significantly decreased H3K27ac throughout the genome after global between-sample normalization (Wilcoxon signed-rank test, P < 10−20; Fig. S5A). In particular, C646 treatment significantly altered H3K27ac levels for more than 800 genes [χ2 test, false discovery rate (FDR) < 0.05; Fig. S5B]. Genes affected most by C646 showed significant loss of H3K27ac (Mann-Whitney U test, P< 4× 10−4; Fig. S5C). These genes are enriched for a variety of functional terms pertaining to the structure, development, differentiation, and communication of neurons (Fisher's exact test, FDR < 0.05; Fig. S5D). This suggests that C646 treatment, despite interfering with a pleiotropic pathway of gene regulation, preferentially inhibits genes with neuronal functions.
To test whether the increased foraging activity of HDACi-treated workers depends on the HAT activity of CBP, we fed 25- to 30-day-old minors a cocktail of 50 μM TSA together with either 50 μM or 100 μM C646 and assayed foraging behavior as above with TSA alone (Fig. 2E). Treatment with 50 μM C646 did not significantly affect foraging or scouting, either alone or in combination with TSA (Fig. 3, A and B, and Fig. S4F, red and blue versus green). In contrast, treatment with 100 μM C646 essentially blocked all foraging and scouting, despite the presence of TSA at a dosage that stimulated foraging alone (Fig. 3, A and B, and Fig. S4F, blue versus purple). Indeed, delivering 100 μM C646 alone was sufficient to inhibit scouting within 14 days of treatment (Mann-Whitney U test, P < 0.05; Fig. S4G). Moreover, C646 had minimal, nonsignificant effects on scout expedition time (Fig. S4D) and mortality (Fig. S4E), which suggests that it did not affect locomotion or induce severe toxicity. These results imply that HDACi-induced foraging is mediated by histone residues that are acetylated by CBP, such as H3K27ac (32, 33).
Fig. 3. CBP-dependent acetylation regulates foraging via production of scouts.
(A and B) Average foraging activity ± SE (A) and number of scouts ± SE (B) for cohorts of 15 minors isolated at age 25 to 30 days and continuously fed DMSO, 50 μM TSA, 50 μM C646, 50 μM TSA + 50 μM C646,or 50 μM TSA+100 μM C646 ad libitum in 20% sugar water (Fig. 2E). (C) Proportion of minors that scouted for 1 to 10 consecutive foraging trials over a span of 49 days; an ant that scouted once acted as a scout for a median of 20 days (four trials, indicated by dashed line). (D) Distribution of the number of trials after isolation until an individual first scouted; dashed line indicates 15 days after isolation (three trials). Error bars in (C) and (D) denote SE over seven replicate cohorts of 15 minors, age 30 to 35 days, from four colonies. P value in (D) is from a χ2 test. (E) Average foraging activity ± SE for individual trials per treatment group over 42 days. Orange bars represent trials in which C646 was provided starting 14 days after isolation and first administration of TSA; for blue bars, C646 was provided starting immediately upon isolation in parallel with TSA. Cohorts were tested weekly over 42 days. Error bars in (A) and (E) denote SE over six measurements per cohort and 14 (green), 6 (red), 18 (purple), 11 (blue), or 8 (orange) independent cohorts sampled from six colonies. P values in (A), (B), and (E) were estimated by Mann-Whitney U test.
Because foraging activity correlates strongly with the number of scouts, and because scouting is inhibited by C646, we reasoned that CBP may be required to induce workers to become scouts. To examine this, we assayed individually marked 25- to 30-day-old minors in isolated cohorts. We found that the median duration of individual scouting behavior was 20 days, with some ants consistently scouting for the 49-day duration of the assay (Fig. 3C). Moreover, most of the workers that scouted (66%) emerged in the first 15 days (i.e., were 40 to 45 days old), rather than uniformly over time (Fig. 3D). Therefore, scouts likely represent a persistent behavioral caste expressed by select individuals in C. floridanus.
We tested whether the addition of C646 after 14 days of TSA treatment, after most scouts have already emerged (Fig. 3D), affected TSA-dependent increases in scouting and foraging. Indeed, addition of 100 μM C646 after 2 weeks of 50 μM TSA treatment by feeding failed to reverse increases in foraging and scouting (Fig. 3E, orange versus purple), in contrast to parallel treatment of 50 μM TSA together with 100 μM C646 (Fig. 3E, orange versus blue). These results suggest that CBP, via its histone acetylation activity, may serve as a licensing factor to enable the transition of a worker into the scout behavioral caste. Moreover, the fact that the timing of C646 delivery appears to be critical to elicit the behavioral response suggests that loss of foraging is not due to the toxicity of C646 or its nonspecific effects on general locomotion (Fig. S4, D and E).
We then examined how the observed changes in behavior after treatment with TSA and C646 are associated with molecular changes. We analyzed genome-wide mRNA expression in individual brains dissected from 21 minors sampled while feeding on sugar water during a scouting run (Fig. 4A and Figs. S6 and S7) (19). Hierarchical clustering analysis of the 250 genes exhibiting greatest differential expression among treatments yielded two clusters of brain samples, a TSA-treated group and a DMSO-treated or TSA + C646–treated group, consistent with our behavioral analyses (Fig. 4B). In contrast, random gene subsampling and analysis of internal spike-in control transcripts failed to partition samples by treatment group, as expected (Fig. S8).
Fig. 4. HAT and HDAC inhibitors alter gene expression and histone acetylation in scout brains.
(A) Schematic for genomics analysis. Minor scouts were sampled at the time of feeding in a foraging arena, and their central brains were dissected, homogenized, and analyzed by next-generation sequencing, with replication as indicated. (B) mRNA expression in brains of ~70-day-old minor scouts for the 250 genes with the greatest alteration between TSA treatment and DMSO or TSA + C646 treatment (n = 21 samples). FPKM denotes fragments per kilobase of transcript per million mapped reads. Left: Average (±SE) mRNA expression of these 250 genes. Right: Hierarchical clustering of samples for the 250 genes, using Mahalanobis distance and Ward's agglomeration criterion. P values were computed by bootstrap (19); BP, bootstrap probability. (C) Normalized ChIP-seq enrichment plots for H3K9ac and H3K27ac over significant dmROIs (±500 base pairs) that gain (top) or lose (bottom) enrichment with TSA treatment [P < 0.001 compared to an empirical null distribution (19)]. (D) P values for H3K27ac and H3K9ac dmROIs reflect proximity to gene transcription start sites (TSSs) ranked by absolute differential expression compared to randomly selected genes; P values were computed by Mann-Whitney U test (19). (E) Absolute change in a gene's TSS-proximal H3K27ac dmROI versus absolute change in mRNA level between TSA and DMSO samples, binned by magnitude of differential expression (top 50, 150, 300, and 600 genes). Lines denote linear regression trendlines for each group. Pearson correlation coefficients are shown; P values were computed by Student's t test. (F) Distributions of minimum distance from a dmROI to an H3K27ac-positive CBP binding site for H3K27ac and H3K9ac dmROIs. P values were computed by Mann-Whitney U test with randomly sampled ROI coordinates (n = 250 trials).
Most of the top 250 differentially expressed genes (72%) were up-regulated with TSA (Fig. 4B, left), consistent with an overall increase in histone acetylation due to TSA and overall decrease in CBP-mediated acetylation due to C646. Gene ontology analysis of the 101 genes with significant differential expression (Bonferroni P < 0.05; table S3) (19) revealed enrichment for hormone signaling, dendrite morphogenesis, synaptic transmission, and sphingolipid biosynthesis (Fisher's exact test, P < 0.01; Fig. S7C) as processes responsive to chromatin regulation (25, 33, 34). These results argue that inhibition of HATs and HDACs by chronic feeding of small-molecule inhibitors may affect the expression of specific genes in the ant central brain.
To assess whether TSA and C646 affect brain gene expression via alterations in chromatin structure, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for H3K27ac and H3K9ac, two hPTMs implicated in neuronal and behavioral plasticity (27, 35), using tissue from nine of the scout brains also used for RNA-seq (Fig. 4A). We normalized these data using input lysate as well as internal spike-in lysate (ChIP-Rx) from the evolutionarily divergent ant H. saltator (12, 19, 36).
We identified regions of interest (ROIs) that were differentially marked by H3K9ac or H3K27ac between DMSO and TSA treatments (dmROIs) (Fig. 4C). These dmROIs covered 0.33% (H3K9ac) and 1.5% (H3K27ac) of the genome, respectively, and predominantly occurred in noncoding regions (~90%; Fig. S9A). Notably, dmROIs exhibited both gains and losses in histone acetylation in TSA-treated samples (Fig. 4C), possibly from acclimation to chronic treatment.
We then assessed whether dmROIs occur near differentially expressed genes. Indeed, dmROIs for H3K27ac were significantly enriched among the top 500 differentially expressed genes, with the greatest significance for the top 100 genes (Fig. 4D and Fig. S9, B and C), whereas dmROIs for H3K9ac showed no significant enrichment near these genes (Fig. 4D). Furthermore, we found a mild but significant correlation between the magnitude of a gene's differential expression and the differential enrichment of the nearest H3K27ac dmROI, with correlation coefficients increasing with differential expression (0.11 < R < 0.31; Fig. 4E). These results are consistent with a chromatin-based model of gene regulation underpinning foraging behavior.
If CBP mediates the observed, drug-dependent changes in chromatin and gene expression, dmROIs should also cluster near binding sites for CBP (16, 35). Using a genome-wide annotation of CBP binding sites (16), we identified 3173 putatively active CBP sites with H3K27ac enrichment (Fig. S9D). Like dmROIs, these binding sites were also enriched near the top differentially expressed genes (Mann-Whitney U test, P < 0.001 for the top 1000 genes; Fig. S9E). Furthermore, dmROIs for H3K27ac, but not H3K9ac, were proximal to H3K27ac-positive CBP binding sites (median distance of 0.6 kb versus 8.7 kb from center of CBP site; Fig. 4F). Lastly, 475 H3K27ac dmROIs showed overlap with CBP sites by at least 50%. These data suggest that chronic drug treatment may induce both direct and indirect effects involving multiple histone residues and multiple regulatory factors (30–33). Taken together, these results are consistent with our prior observation of differential H3K27ac in the brains of minors and majors (16) and suggest that CBP may modulate foraging behavior in workers in part by regulating histone acetylation levels at specific genomic loci in the brain.
HDAC inhibition induces and sustains minor-like behavior in majors
We next sought to discern why minors and majors are differentially predisposed to forage (Fig. 1) and why majors treated with HDAC inhibitors forage and scout less than minors (Fig. 2, C and D, and Fig. S10A). Given that CBP regulates the production of scouts (an age-based caste), we reasoned that differential activity of CBP and HDACs in the brains of young minors and majors might likewise establish behavioral states tied to morphological caste.
To evaluate this model, we first identified 160 genes that exhibited significant differential expression by caste in brains of 1-day-old workers from the same colony (Bonferroni P < 0.01; Fig. 5A and table S4) (19). These caste-differential genes pertain to synaptic transmission, synapse structure, and neurotransmitter release (Fisher's exact test, FDR < 0.25; Fig. S11C). Furthermore, these genes were particularly responsive to TSA and C646 treatments in mature minor scout brains (Student's t test, R = 0.43, P < 10−7; Fig. 5B)—a trend also seen for the entire transcriptome (R = 0.10, P < 10−15) but not for random gene subsets (99.9%; Fig. 5B, inset).
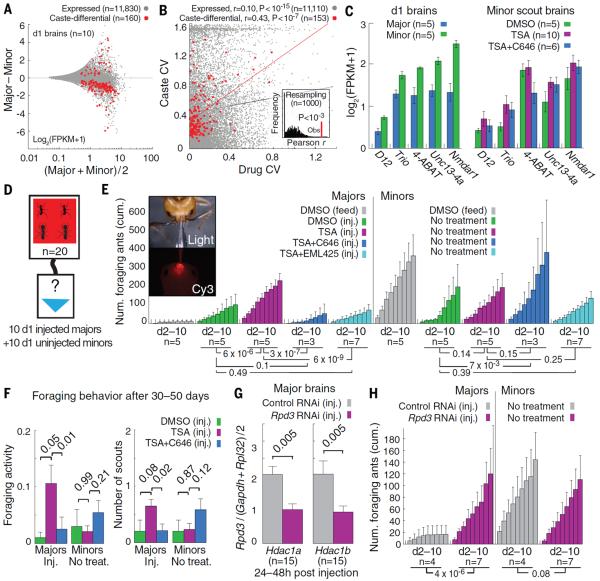
Fig. 5. RPD3 and CBP mediate epigenetic reprogramming of major workers.
(A) Differential versus average scatterplot comparing mRNA levels for brains dissected from individual 1-day-old majors and minors (n = 5 per caste) from one colony. Caste-differential genes were identified by Mann-Whitney U test (P<0.05) and gene-specific SE (Bonferroni P<0.01). (B) Scatterplot comparing coefficients of variation (CVs) in gene expression between drug treatments and caste. Pearson correlation coefficients are shown. Inset shows correlation coefficients for 1000 random sets of 153 genes sampled from 11,110 expressed genes. (C) mRNA levels for select caste-differential genes in 1-day-old worker brains (left) and minor scout brains (right). Error bars denote SE among biological replicates. 4-ABAT, 4-aminobutyrate aminotransferase; Unc13-4a, mammalian uncoordinated homology 13, 4a ortholog. (D) Schematic of piggyback foraging assay. Majors and minors received no injection (gray) or 0.5% DMSO, 50 μM TSA, 50 μM TSA + 100 μM C646, or 50 μM TSA + 100 μM EML425 and were placed in a nest box attached to a foraging arena with 20% sugar water (blue triangle) (Fig. S1E). (E) Cumulative foraging over the course of 9 days for pooled cohorts of minor and major nestmates, measured by time-lapse photography every 6 min. Uninjected cohorts were fed 20% sugar water containing 0.5% DMSO. Error bars denote SE over replicate cohorts. See table S5 for colony background and mortality information. Photographs (and movie S2) depict brain injection procedure. P values were estimated by Mann-Whitney U test using counts of daily foraging events. (F) Long-term assessment of foraging activity (left) and number of scouts (right) for the same injected majors and control minors from (E). (G) RT-qPCR analysis of mRNA abundance for Rpd3 (Hdac1a and Hdac1b) in major brains 24 to 48 hours after injection with nontargeting (control) RNAi or Rpd3 RNAi. Each bar indicates the mean ± SE over measurements from 15 brains. Abundance was normalized against Gapdh1 and Rpl32. (H) Cumulative foraging over the course of 9 days for pooled cohorts of minors and majors, as in (D). P values in (E) to (H) were computed by Mann-Whitney U test.
Among these jointly caste- and drug-responsive genes were those encoding D12, a component of the ATAC HAT complex (37); Rho guanosine triphosphate exchange factor Trio, which regulates dendrite morphogenesis (38); and N-methyl-d-aspartate receptor (NMDAR) 1, which regulates olfactory learning and memory (39) (Fig. 5C). These observations are consistent with a classic response threshold model (10) whereby caste-specific behavioral states are established with chromatin regulatory factors, which suppress behavioral plasticity by regulating genes that modulate the brain's sensitivity to environmental cues, in turn contributing to stable neuroanatomical differences between behavioral castes (40, 41).
Because caste-specific foraging appears to become more pronounced with age (and correspondingly with social experience) (Fig. 5E, gray bars, and Fig. S1, F and H), we further hypothesized that majors may be most sensitive to TSA immediately after eclosion, prior to the establishment of molecular barriers that restrict behavioral plasticity. Unfortunately, 1-day-old workers did not scout after starvation (Fig. S10B) and hence could not receive TSA by feeding. We therefore developed a technique to deliver controlled treatment doses directly to ant brains by micro-injection (Fig. 5E, inset, table S5, and movie S2). Remarkably, injecting 1 μl of 50 μM TSA onto newly eclosed major brains robustly stimulated foraging activity despite the presence of untreated age-, colony-, and group size–matched minor nestmates (Fig. 5, D and E, and Fig. S12).
Over 10 days, majors injected with TSA upon eclosion consistently foraged more than did majors injected with DMSO (Mann-Whitney U test, P < 6 × 10−6)—and at levels similar to untreated minor nestmates (P < 0.13; Fig. 5E). These results suggest that inhibiting HDAC activity in young majors is sufficient to mitigate chromatin-based barriers that restrict caste-based behavioral plasticity.
In contrast, majors injected with 50 μM TSA and 100 μM C646 largely failed to forage (Fig. 5E, blue), consistent with the observed effects of these drugs when fed to mature minors (Fig. 3A, blue). To confirm that suppression of foraging by C646 was not due to off-target effects, we applied a second HAT inhibitor of CBP, EML425, which functions by a different mechanism than C646 (42); again, majors injected with 50 μM TSA and 100 μM EML425 exhibited little foraging (Fig. 5E, light blue).
We also examined the long-term effects of these brain injections within our starvation-based foraging assay (Fig. 2E). Remarkably, 30 to 50 days after the single-injection treatments, TSA-injected majors still displayed significantly increased foraging activity, and with more scouts, than did age- and colony-matched majors injected with DMSO (Mann-Whitney U test, P < 0.05) or TSA + C646 (P < 0.01; Fig. 5F). In contrast, untreated minor nestmates showed no significant differences in foraging among treatment groups (Fig. 5F). Taken together, these findings indicate that CBP is a likely epigenetic factor involved in establishing and maintaining morphological and age-dependent behavioral castes.
Finally, we developed and applied a transient RNA interference (RNAi) technique (19) to examine whether minor-like foraging may be induced in majors by specifically inhibiting Rpd3/Hdac1 mRNA. Rpd3 was selected because it encodes a class I HDAC putatively targeted by TSA and because its mRNA abundance increased with age in C. floridanus (Fig. 2A). RPD3 also deacetylates residues acetylated by CBP, such as H3K27ac (32). Injection of 27-nucleotide double-stranded small interfering RNA (siRNA) oligos against the Rpd3 and Rpd3-like transcripts into 1-day-old major brains led to a factor of 2 reduction in their mRNA levels in the central brain after 24 to 48 hours, relative to injection of nontargeting control double-stranded siRNAs (Fig. 5G). Moreover, Rpd3 RNAi induced minor-like foraging in injected majors (Fig. 5H). This result suggests that RPD3 normally acts to repress foraging in C. floridanus workers by removing acetyl groups from histones catalyzed by CBP.
Discussion
Our examination of foraging behavior as a caste-specific trait in the ant C. floridanus sheds light on a fundamental question in sociobiology regarding molecular determinants of division of labor in eusocial insects (1). Whereas recent studies have brought increasing attention to the role of genetic variation in caste specification (2, 8, 9, 43), our findings implicate the histone-modifying enzymes RPD3/HDAC1 and CBP as licensing and reprogramming factors underlying morphology and age-dependent social behaviors, hence revealing a key role for chromatin-based regulation of behavioral castes. C. floridanus colonies express two distinct morphological worker castes, minors and majors, and our results show that minors perform the majority of foraging and scouting for a colony (Fig. 1). HDACi-induced changes in CBP-dependent histone acetylation in the brains of mature minors reinforce and accentuate foraging and scouting (Figs. 2 and 3). In scouting ants, these behavioral changes correspond to altered transcript abundance of select neuronal genes with specific roles in synaptic transmission and olfactory learning, and to changes in histone acetylation that occur near CBP binding sites proximal to these genes (Fig. 4).
Remarkably, the delivery of a single dose of HDACi or RNAi against Rpd3 in major brains immediately after pupal eclosion is sufficient to overcome the intrinsic molecular barriers that normally inhibit major foraging and scouting (Fig. 5). Hence, regulation of caste-specific social behaviors involves an epigenomic landscape that remains plastic for a period of time after eclosion. This malleability appears to permit drug-and RNAi-mediated up-regulation of genes that are inherently expressed at lower levels in majors, resulting in “transdifferentiation” between behavioral castes.
Epigenetic control over behavioral castes, and thus division of labor, may allow colonies to adapt dynamically to drastic ecological changes within their lifetime—for example, in response to prolonged famine or colony predation, which can alter caste ratios (13). Hence, our findings may be broadly relevant to other eusocial insects that display age-based or caste-based division of labor. Further, our results suggest that CBP and HDACs might help to establish complex social interactions for other invertebrate, vertebrate, and mammalian species, in which these conserved enzymes are known to play critical roles in the regulation of behavioral plasticity as well as in learning and memory (27–29, 44, 45). Finally, our ability to alter a canonical altruistic behavior in a truly social organism by experimental perturbation of a single gene suggests that the application of increasingly versatile reverse genetic approaches in eusocial insects will allow us to expose the general organizational principles underlying complex social systems (10).
Materials and methods
Ant colonies and husbandry
Mature, queen-right colonies of C. floridanus and C. tortuganus were used in this study, collected as foundresses from Long Key and Sugar Loaf Key in the Florida Keys, USA, in 2007 and 2011. Colonies were maintained in a sealed environmental growth chamber at constant temperature (25°C) and humidity (50%) under a 12:12 light: dark cycle. Colonies were fed twice weekly with excess supplies of water, 20% sugar water (sucrose cane sugar), and Bhatkar-Whitcomb diet (46).
Caste and age identification
Adult morphological caste was identified by body size and head width. For calibration, head width and scape (basal antennal segment) length (a proxy for body size) were measured using a digital micrometer (Fisher Scientific FB70252) and stereo-microscope (Nikon SMZ-1500) for 377 workers. A worker was considered a major if her head width exceeded 2.25 mm and a minor if under 1.75 mm. Age was determined by marking gasters of 1-day-old callows with enamel paint (Testors). One-day-old ants were identified by their location among brood in the nest, their general behavior, and their light cuticle coloration (compared to a reference panel of ants aged from pupation through 30 days).
Foraging assays and analysis
Three assays were used for analysis of foraging behavior. In the whole-colony foraging assay, a foraging arena (Fig. S1G) was attached to a mature (7.5-year-old) queen-right colony containing more than 4000 workers. Petri dishes containing 15 ml each of pure water (×2) or 20% sugar water (×2) were placed in the arena. The arena was photographed every 3 min continuously for periods of 24 hours. For each photograph, the time of day and number of majors and minors feeding from water and sugar water dishes were recorded.
In the piggyback foraging assay, worker cohorts were isolated from single colonies (with specified caste, age, and number) and placed into a “piggyback” nest box (Fig. S1E). A 1.5-ml Eppendorf tube containing 20% sugar water was placed in the arena on the wall opposite the arena entrance. The arena was photographed every 6 min continuously for periods of 24 hours over the course of 10 days. The time and caste of each ant that fully entered into the foraging arena was recorded and analyzed blindly. Idle ants that spent more than 30 min in the arena without feeding on sugar or departing from the foraging arena were not counted.
In the starvation-induced foraging assay, worker cohorts were isolated from single colonies as above and placed into “standard” 195c nest boxes (Fig. S1G). Seven larvae (instar 2 to 4) were included to facilitate acclimation to the new nest. A 1.5-ml Eppendorf microcentrifuge tube containing 20% sugar water was provided and replaced weekly. Twenty-four hours prior to assay, the tube of sugar water was replaced with a 1.5-ml tube containing ultrapure water filtered by Milli-Q (EMD Millipore). At assay time (14:00 to 17:00 hours), a foraging arena was attached to the 195c nest box, and a half weigh boat containing 500 μl of 20% sugar water was placed within the arena at the far side relative to the tube leading to the nest. Video recordings were made for each foraging trial lasting 70 to 75 min and were analyzed blindly.
Time and caste of each ant that entered the foraging arena from the nest and fed on (i.e., contacted and consumed) sugar water for at least 10 s (one feeding event) were recorded. An ant must have returned to the nest, then reentered the foraging arena and refed to be counted again. Also, the time that the first feeding ant (a scout) returned to the nest (by descending beneath a piece of red acetate film covering a nest cavity) was recorded, as well as the number of scouts, defined as the number of individual ants feeding before the first scout returned to the nest. Scout expedition time was computed as the difference between the time the first scout returned to the nest and the time that she began to feed on sugar water. Quantitation of each foraging trial video recording was used to estimate a foraging activity statistic that summarized the foraging plus feeding rate of a worker cohort. This statistic (Fig. S2A) was computed as the number of scouts plus the number of additional feeding events occurring within the first 20 min after the first scout's return to the nest.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) sample preparation and analysis
Individual brains dissected from 1-day-old ants were rinsed in sterile phosphate-buffered saline solution (PBS) and transferred to 1.5-ml microcentrifuge tubes containing 500 μl of ice-cooled PBS. Brain tissue was dissociated by sonication using a Diagenode Bioruptor Standard device for 30 s on low power. Total RNA was purified from individual brains by phenol:chloroform extraction and purification using RNase-free Agencourt AMPure XP beads (Beckman Coulter). cDNA was produced from total RNA using SuperScript III Reverse Transcriptase (Invitrogen). Abundance of specific mRNA transcripts was estimated using Power SYBR Green PCR Master Mix (Life Technologies) on a Real Time qPCR machine (Applied Biosystems 7900HT). Relative transcript abundance was estimated using the delta Ct method (19).
Chromatin immunoprecipitation
Chromatin lysate and subsequent immunoprecipitated double-stranded DNA (dsDNA) were prepared using antibodies recognizing H3K9ac (Active Motif 39137, Lot 102), H3K27ac (Abcam ab4729, Lot GR132150-4), and total H3 (Abcam ab1791, Lot GR135488-1) by processing larvae or adult ants as described (16, 19).
Foraging assays for pharmacologically treated ants
Analysis of VPA (Calbiochem) was performed using the starvation-induced foraging assay. For a single foraging trial, four groups of seven minor or major 25- to 30-day-old ants (age-matched within 72 hours) were isolated from a queen-right colony and placed into 28c nest boxes (seven ants in each of four boxes). Ants were paint-marked to distinguish each individual. VPA was dissolved in liquid Bhatkar (or no VPA as control) to final desired concentration and replaced daily for 32 to 42 days (depending on number of starvation days). After 32 days of treatment, all 28 ants were pooled into a common 195c nest box (Fig. S1G) containing seven larvae (instar 2 to 4). The ants were starved with water for 24 hours, then assayed for foraging by attaching a 79c foraging arena (Fig. S1G) and recording the time of every foraging and feeding event for each individual ant. If a cohort failed to exhibit a single feeding event within an hour of assay, the foraging arena was detached and the ants were starved for another 24 hours, for a maximum of 10 days. In the rare case of multiple feeding events by the same individual ant, only the first feeding event was recorded.
For TSA and C646 treatments, foraging assays were conducted using cohorts of 15 workers of the same age (within 48 hours) and colony. No larvae were provided, as the isolated ants were kept in the same 195c nest box for the duration of the trial. Pharmacological small molecules TSA (Sigma-Aldrich) and C646 (Calbiochem) were dissolved in DMSO and then in 20% sugar water to the desired final concentration. Ants were permitted to drink this solution ad libitum. Fresh aliquots were prepared from stock every 48 hours. During 24-hour starvation periods, fresh aliquots were prepared by dissolving a compound into water; otherwise, aliquots were dissolved in 20% sugar water. Each cohort was assayed for foraging activity 7, 14, 21, 28, 35, and 42 days after isolation, unless indicated otherwise. Foraging activity, number of scouts, and expedition time were computed as described above.
Preparation of RNA-seq libraries
Whole brains were dissected from individual mature minor scouts fed DMSO, 50 μM TSA, or 50 μM TSA with either 50 μM or 100 μM C646 for 45 days. Scouts were identified, sampled, and frozen on dry ice while feeding on sugar water after foraging, during a starvation-induced foraging assay. Each dissected brain was rinsed in PBS and transferred to a 1.5-ml microcentrifuge tube containing 100 μl of prechilled PBS (on ice). Brain tissue was dissociated using a Diagenode Bioruptor Standard device for 30 s on low power. A 15-μl aliquot representing 15% of cells in a single brain [estimated to be 20,000 to 25,000 cells (47)] was taken for RNA-seq; this represents a sufficient sampling of cells to recapitulate whole-brain transcriptome patterns (Fig. S6) and allows remaining cells to be processed for ChIP-seq.
Total RNA was purified by phenol:chloroform extraction and purification using RNase-free Agencourt AMPure XP beads (Beckman Coulter). 1 μl of ERCC RNA Spike-In Mix (Ambion) was added to each 15-μl aliquot of total RNA to facilitate transcript-level normalization relative to cell number and to control for technical variation during library preparation. The mRNA fraction of RNA was purified by selection for polyadenylated transcripts using oligo(dT)25 Dynabeads (Invitrogen) as described (48), and RNA fragmentation was performed for 8 min at 94°C. cDNA was produced from total RNA using SuperScript III Reverse Transcriptase (Invitrogen), and second-strand synthesis was performed using deoxyuridine triphosphate (dUTP) to preserve strand specificity (48). Strand-specific RNA-seq libraries were produced for these dUTP-incorporated dsDNA libraries after one round of linear amplification (19).
RNA-seq libraries from individual 1-day-old minor and major brains were processed as above, except that whole brains were dissected from 10 1-day-old minors and majors sampled from the same colony at the same time and the same location in the nest. All 10 brains were dissected in a single session; all RNA-seq libraries were prepared from the entire brain samples as a single batch; all libraries were bar-coded and sequenced on the same flowcell.
All dsDNA libraries were prepared using the NEB NextUltra DNA Library Prep Kit for Illumina (New England BioLabs) and were amplified by PCR for 15 cycles, purified using Agencourt Ampure XP beads, and quantified by Qubit 2.0 Fluorometer (Invitrogen) and Kapa qPCR (Kapa Biosystems). Libraries were sequenced on an Illumina NextSeq 500 Desktop Sequencer.
Preparation of ChIP-seq libraries
ChIP was performed on individual scout brains using 45 μl of remaining cell material (60,000 to 75,000 cells) for a subset of the same brains used for RNA-seq (above). Chromatin was prepared as described (16), except that brain tissue was cross-linked with formaldehyde for 5 min. Before immunoprecipitation, cross-linked chromatin prepared from pools of heads and thoraces from 1-day-old workers of the evolutionarily divergent ant Harpegnathos saltator was added to a final concentration of 2.5% by volume based on Qubit protein quantitation [ChIP-Rx (36)]. After mixing C. floridanus (75 μg/IP) and H. saltator (1.88 μg/IP) chromatin, immunoprecipitation was performed as described (16) using 1 μg of antibody for H3K27ac or H3K9ac. dsDNA was purified from ChIP material as described (16), and ChIP-seq libraries were prepared using the NEB NextUltra DNA Library Prep Kit for Illumina. Libraries were amplified by PCR for 15 cycles, purified using Agencourt Ampure XP beads, quantified by Qubit and Kapa qPCR, and sequenced on an Illumina NextSeq 500 Desktop Sequencer.
Analysis of RNA-seq and ChIP-seq data
Sequenced reads were aligned to a diploid version of the C. floridanus reference genome v3.0 (12) after incorporating single-nucleotide polymorphisms (SNPs) (table S9). This diploid reference was first split into two pseudo-haplotypes by randomly partitioning the two alleles at each SNP into two haploid genome sequences. Mapping was performed against each reference haplotype using Glimmr (16), with calls to Bowtie2 (49) with options (–end-to-end–very-sensitive) and allowing each read to display valid alignment for up to 10 distinct genomic locations (-k 10). Mapped reads were merged into a single alignment file by retaining the alignments for a given read that contained fewer mismatches to the particular reference haplotype. For RNA-seq, remaining unmapped reads were also aligned to a reference transcriptome containing full-length transcripts for annotated genes. Remaining unmapped reads were trimmed in an iterative manner from full-length 75-nt reads to 55-nt reads to 35-nt reads, where the 10 outer bases on either end of the read were removed at each step. Quantitation, normalization, and analysis of RNA-seq and ChIP-seq data are described in (19).
Brain injection procedure
Before eclosion, major and minor pupae were removed from their natal colony and reared in smaller nests by 15 adult minors. In 24-hour intervals, newly eclosed callows were removed from rearing nests and organized into age-matched treatment groups containing 10 majors and 10 minors. Individuals were isolated in 1.5-ml microcentrifuge tubes and cooled on ice for up to 5 min until sedated. Major workers were moved to an ice-cooled silicone platform, and their cuticles were superficially perforated at the target injection site using a sterilized steel pin (Minutae, Sphinx). Immediately after perforation, a borosilicate glass needle filled with injection material was directed to the injection site using a robotic arm (Patchman N2, Eppendorf) and the needle tip was placed just beneath the surface of the cuticle to avoid damage to underlying tissue. A Femtojet microinjector (Eppendorf) delivered 1 μl of injection material to the injection site. After injection, individuals were given 1 hour to recover before being assessed for morbidity and mortality. Successful injections were pooled in groups of 10 with age-matched minors and placed into the test arena for behavioral analysis.
RNAi-mediated mRNA knockdown and analysis
Individual major brains were injected with a 1-μl pool of 1 μM each of two different 27-nt double-stranded siRNAs targeting Rpd3 (Cflo_10463) and Rpd3-like (Cflo_10465) (Rpd3 RNAi) or a 1-μl pool of 1 μM each of two different nontargeting control double-stranded siRNAs (Control RNAi) (19). Individuals were killed 24 or 48 hours after injection. cDNA libraries were prepared from individual dissected brains as described above. RT-qPCR was performed as described above, using primers specific to Cflo_10463 and Cflo_10465 and to Gapdh1 and Rpl32 mRNAs (table S2). Quantitation was performed using the median quantity estimated based on fivefold dilution series of each primer. Rpd3 quantities were normalized to the average of Gapdh1 and Rpl32 quantities as controls. P values were calculated by Mann-Whitney U test.
Brain injection piggyback foraging assay
Cohorts of 10 majors and 10 minors, all 1 day old, were sampled from a single queen-right colony. Only majors received brain injections with a fixed delivery volume of 1 μl, containing 0.5% DMSO, 50 μM TSA, or 50 μM TSA with 100 μM C646. For RNAi, majors were injected with pools of 1 μM 27-nt duplex RNA oligos (RPD3 or nontargeting controls) (table S6). All injections were performed between 15:00 and 17:00 hours. After injection, majors were placed with their minor nestmates into a piggyback nest box with foraging arena containing a microcentrifuge tube with 1 ml of 20% sugar water. The arena was photographed every 6 min for 10 days; recorded photographs were analyzed blindly as described above.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Kronauer and I. Fetter-Pruneda for advice on brain injections, and P. Cole, D. Meyers, M. Sammons, P. Shah, and H. Yan for intellectual contributions. Supported by Howard Hughes Medical Institute Collaborative Innovation Award 2009005 (S.L.B., C.D., J.L., A.R., D.R., and L.J.Z.), NIH training grant T32HD083185 in developmental biology from the University of Pennsylvania (D.F.S.), and NIH New Innovator Award DP2MH107055 (R.B.). Sequencing data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession number GSE69553. Behavioral data are available in tables S7 and S8. C. floridanus SNPs are available in table S9, and gene annotations are available in table S10. Software used for high-throughput sequencing and gene ontology analyses is available on GitHub (https://github.com/dfsimola/glimmr).
Footnotes
SUPPLEMENTARY MATERIALS www.sciencemag.org/content/351/6268/aac6633/suppl/DC1
REFERENCES AND NOTES
- 1.Hölldobler B, Wilson EO. The Ants. Belknap/Harvard Univ. Press; Cambridge, MA: 1990. [Google Scholar]
- 2.Robinson GE. Regulation of division of labor in insect societies. Annu. Rev. Entomol. 1992;37:637–665. doi: 10.1146/annurev.en.37.010192.003225. doi: 10.1146/annurev.en.37.010192.003225; pmid: 1539941. [DOI] [PubMed] [Google Scholar]
- 3.Wilson EO. Behavioral discretization and the number of castes in an ant species. Behav. Ecol. Sociobiol. 1976;1:141–154. doi: 10.1007/BF00299195. [Google Scholar]
- 4.Fjerdingstad EJ, Crozier RH. The evolution of worker caste diversity in social insects. Am. Nat. 2006;167:390–400. doi: 10.1086/499545. doi: 10.1086/499545; pmid: 16673347. [DOI] [PubMed] [Google Scholar]
- 5.Tschinkel WR. The Fire Ants. Belknap/Harvard Univ. Press; Cambridge, MA: 2013. [Google Scholar]
- 6.Lucas C, Sokolowski MB. Molecular basis for changes in behavioral state in ant social behaviors. Proc. Natl. Acad. Sci. U.S.A. 2009;106:6351–6356. doi: 10.1073/pnas.0809463106. doi: 10.1073/pnas.0809463106; pmid: 19332792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. doi: 10.1038/nature09231; pmid: 20535202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: Social life in molecular terms. Nat. Rev. Genet. 2005;6:257–270. doi: 10.1038/nrg1575. doi: 10.1038/nrg1575; pmid: 15761469. [DOI] [PubMed] [Google Scholar]
- 9.Smith CR, Toth AL, Suarez AV, Robinson GE. Genetic and genomic analyses of the division of labour in insect societies. Nat. Rev. Genet. 2008;9:735–748. doi: 10.1038/nrg2429. doi: 10.1038/nrg2429; pmid: 18802413. [DOI] [PubMed] [Google Scholar]
- 10.Yan H, et al. Eusocial insects as emerging models for behavioural epigenetics. Nat. Rev. Genet. 2014;15:677–688. doi: 10.1038/nrg3787. doi: 10.1038/nrg3787; pmid: 25200663. [DOI] [PubMed] [Google Scholar]
- 11.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. doi: 10.1126/science.1153069; pmid: 18339900. [DOI] [PubMed] [Google Scholar]
- 12.Bonasio R, et al. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science. 2010;329:1068–1071. doi: 10.1126/science.1192428. doi: 10.1126/science.1192428; pmid: 20798317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herb BR, et al. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat. Neurosci. 2012;15:1371–1373. doi: 10.1038/nn.3218. doi: 10.1038/nn.3218; pmid: 22983211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonasio R, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. doi: 10.1016/j.cub.2012.07.042; pmid: 22885060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarado S, Rajakumar R, Abouheif E, Szyf M. Epigenetic variation in the Egfr gene generates quantitative variation in a complex trait in ants. Nat. Commun. 2015;6:6513. doi: 10.1038/ncomms7513. doi: 10.1038/ncomms7513; pmid: 25758336. [DOI] [PubMed] [Google Scholar]
- 16.Simola DF, et al. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res. 2013;23:486–496. doi: 10.1101/gr.148361.112. doi: 10.1101/gr.148361.112; pmid: 23212948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadau J, Heinze J, Hölldobler B, Schmid M. Population and colony structure of the carpenter ant Camponotus floridanus. Mol. Ecol. 1996;5:785–792. doi: 10.1111/j.1365-294x.1996.tb00374.x. doi: 10.1111/j.1365-294X.1996.tb00374.x; pmid: 8981768. [DOI] [PubMed] [Google Scholar]
- 18.Spannhoff A, et al. Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Rep. 2011;12:238–243. doi: 10.1038/embor.2011.9. doi: 10.1038/embor.2011.9; pmid: 21331099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.See supplementary materials on Science Online.
- 20.Calabi P, Traniello JFA. Behavioral flexibility in age castes of the ant Pheidole dentata. J. Insect Behav. 1989;2:663–677. doi: 10.1007/BF01065785. [Google Scholar]
- 21.Liang ZS, et al. Molecular determinants of scouting behavior in honey bees. Science. 2012;335:1225–1228. doi: 10.1126/science.1213962. doi: 10.1126/science.1213962; pmid: 22403390. [DOI] [PubMed] [Google Scholar]
- 22.Mersch DP, Crespi A, Keller L. Tracking individuals shows spatial fidelity is a key regulator of ant social organization. Science. 2013;340:1090–1093. doi: 10.1126/science.1234316. pmid: 23599264. [DOI] [PubMed] [Google Scholar]
- 23.Hölldobler B. Recruitment behavior in Camponotus socius (Hym. Formicidae) Z. Vgl. Physiol. 1971;75:123–142. [Google Scholar]
- 24.Stroeymeyt N, Franks NR, Giurfa M. Knowledgeable individuals lead collective decisions in ants. J. Exp. Biol. 2011;214:3046–3054. doi: 10.1242/jeb.059188. doi: 10.1242/jeb.059188; pmid: 21865517. [DOI] [PubMed] [Google Scholar]
- 25.Fitzsimons HL, Scott MJ. Genetic modulation of Rpd3 expression impairs long-term courtship memory in Drosophila. PLOS ONE. 2011;6:e29171. doi: 10.1371/journal.pone.0029171. doi: 10.1371/journal.pone.0029171; pmid: 22195015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai L-H. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. doi: 10.1038/nature05772; pmid: 17468743. [DOI] [PubMed] [Google Scholar]
- 27.Vecsey CG, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. doi: 10.1523/JNEUROSCI.0296-07.2007; pmid: 17553985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Göttlicher M, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. doi: 10.1093/emboj/20.24.6969; pmid: 11742974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho Y, Griswold A, Campbell C, Min K-T. Individual histone deacetylases in Drosophila modulate transcription of distinct genes. Genomics. 2005;86:606–617. doi: 10.1016/j.ygeno.2005.07.007. doi: 10.1016/j.ygeno.2005.07.007; pmid: 16137856. [DOI] [PubMed] [Google Scholar]
- 30.Bowers EM, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem. Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. doi: 10.1016/j.chembiol.2010.03.006; pmid: 20534345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crump NT, et al. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7814–7819. doi: 10.1073/pnas.1100099108. doi: 10.1073/pnas.1100099108; pmid: 21518915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. doi: 10.1242/dev.037127; pmid: 19700617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. doi: 10.1038/emboj.2010.318; pmid: 21131905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrader L, Simola DF, Heinze J, Oettler J. Sphingolipids, transcription factors, and conserved toolkit genes: Developmental plasticity in the ant Cardiocondyla obscurior. Mol. Biol. Evol. 2015;32:1474–1486. doi: 10.1093/molbev/msv039. doi: 10.1093/molbev/msv039; pmid: 25725431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malik AN, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat. Neurosci. 2014;17:1330–1339. doi: 10.1038/nn.3808. doi: 10.1038/nn.3808; pmid: 25195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando DA, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 2014;9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. pmid: 25437568. [DOI] [PubMed] [Google Scholar]
- 37.Suganuma T, et al. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. doi: 10.1038/nsmb.1397; pmid: 18327268. [DOI] [PubMed] [Google Scholar]
- 38.Iyer SC, et al. The RhoGEF trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLOS ONE. 2012;7:e33634. doi: 10.1371/journal.pone.0033634. doi: 10.1371/journal.pone.0033634; pmid: 22442703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia S, et al. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr. Biol. 2005;15:603–615. doi: 10.1016/j.cub.2005.02.059. doi: 10.1016/j.cub.2005.02.059; pmid: 15823532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gronenberg W, Heeren S, Hölldobler B. Age-dependent and task-related morphological changes in the brain and the mushroom bodies of the ant Camponotus floridanus. J. Exp. Biol. 1996;199:2011–2019. doi: 10.1242/jeb.199.9.2011. pmid: 9319922. [DOI] [PubMed] [Google Scholar]
- 41.Withers GS, Fahrbach SE, Robinson GE. Selective neuroanatomical plasticity and division of labour in the honeybee. Nature. 1993;364:238–240. doi: 10.1038/364238a0. doi: 10.1038/364238a0; pmid: 8321320. [DOI] [PubMed] [Google Scholar]
- 42.Milite C, et al. A novel cell-permeable, selective, and noncompetitive inhibitor of KAT3 histone acetyltransferases from a combined molecular pruning/classical isosterism approach. J. Med. Chem. 2015;58:2779–2798. doi: 10.1021/jm5019687. doi: 10.1021/jm5019687; pmid: 25730130. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, et al. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493:664–668. doi: 10.1038/nature11832. doi: 10.1038/nature11832; pmid: 23334415. [DOI] [PubMed] [Google Scholar]
- 44.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. doi: 10.1016/j.neuron.2004.06.002; pmid: 15207240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alarcón JM, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. doi: 10.1016/j.neuron.2004.05.021; pmid: 15207239. [DOI] [PubMed] [Google Scholar]
- 46.Bhatkar A, Whitcomb WH. Artificial diet for rearing various species of ants. Fla. Entomol. 1970;53:229–232. doi: 10.2307/3493193. [Google Scholar]
- 47.Ehmer B, Gronenberg W. Mushroom body volumes and visual interneurons in ants: Comparison between sexes and castes. J. Comp. Neurol. 2004;469:198–213. doi: 10.1002/cne.11014. doi: 10.1002/cne.11014; pmid: 14694534. [DOI] [PubMed] [Google Scholar]
- 48.Zhong S, Joung J-G, Zheng Y, Chen Y.-r., Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. Cold Spring Harb. Protoc. 2011. High-throughput Illumina strand-specific RNA sequencing library preparation. 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. doi: 10.1038/nmeth.1923; pmid: 22388286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.