Abstract
Aim:
The accumulation of disease-causing proteins is a common hallmark of many neurodegenerative disorders. Measuring the degradation of such proteins using high-throughput-compatible assays is highly desired for the identification of genetic and chemical modulators of degradation. For example, Huntington's disease (HD) is an incurable hereditary neurodegenerative disorder caused by the cytotoxicity of mutant huntingtin protein (mHTT). The high-throughput measurement of mHTT degradation is important in HD drug discovery and research. Existing methods for such purposes have limitations due to their dependence on protein tags or pan protein synthesis inhibitors. Here, we report a high-throughput-compatible pulse-chase method (CH-chase) for the measurement of endogenous tag-free huntingtin protein (HTT) degradation based on Click chemistry and Homogeneous Time Resolved Fluorescence (HTRF) technologies.
Methods:
The pulsed-labeled proteins were conjugated with biotin using the click reaction strain-promoted alkyne-azide cycloaddition (SPAAC), and the chase signals were calculated by measuring the reduction percentage of the HTT HTRF signals after pull-down with streptavidin beads.
Results:
We validated that the signals were within the linear detection range and were HTT-specific. We successfully measured the degradation of endogenous HTT in a high-throughput-compatible format using 96-well plates. The predicted changes of HTT degradation by known modifiers were observed, which confirmed that the assay is suitable for the identification of HTT degradation modifiers.
Conclusion:
We have established the first high-throughput-compatible assay capable of measuring endogenous, tag-free HTT degradation, providing a valuable tool for HD research and drug discovery. The method could be applied to other proteins and can facilitate research on other neurodegenerative disorders and proteinopathies.
Keywords: click chemistry, SPAAC, high-throughput, pulse-chase, protein degradation, Huntington's disease, PolyQ
Introduction
Protein degradation provides the ultimate regulation of protein functions. In neurodegenerative disorders, accumulation of disease-causing proteins is often the pathogenic cause of the disease, and targeting the degradation of such proteins is of both therapeutic and research interest. As an example, Huntington's disease (HD) is caused mainly by the cytotoxicity of mutant huntingtin protein (mHTT)1, and chemical or genetic modifiers of mHTT degradation may provide valuable entry points for HD therapeutic intervention2,3,4. Thus, high-throughput-compatible assays for measuring mHTT degradation are highly desired.
Existing methods for measuring the degradation of huntingtin protein (HTT) or similar proteins have limitations. The traditional 35S-based pulse-chase method5 requires the use of a radioactive isotope, which is of restricted access and is unfavorable in many laboratories. In addition, this method is dependent on the pull-down efficiency of the target protein by its antibody, which may have variations in pull-down efficiency. Most importantly, pulse-chase requires electrophoresis and is thus not compatible for high-throughput. Recently, degradation of HTT N-terminal fragments was measured using a protein tag-based imaging analysis6. The throughput of this assay is low, although it could potentially be developed into high-throughput-compatible assays. The major limitation of this assay is its dependence on the protein fusion tag, which often requires exogenous expression and may interfere with HTT degradation. Utilizing other detectable protein fusion tags will not solve these issues. Another potential method is the utilization of cycloheximide (CHX), which inhibits protein synthesis7. Under this condition, HTT synthesis is blocked, and thus the level of HTT at different time points reflects degradation. Meanwhile, the synthesis of all other proteins is also stopped, and thus CHX treatment may generate artifacts and cytotoxicity by placing the cells in an extreme condition, under which certain modulators of HTT degradation may be altered or prohibited. Prolonged treatment with CHX also leads to severe cell death due to the pan inhibition of protein synthesis. Thus, the development of high-throughput-compatible method capable of measuring endogenous, tag-free HTT degradation is needed.
We have recently applied Click chemistry technology8 to measure HTT degradation and successfully observed modulation of this process by the genetic modifier NUB1 and by proteasome or autophagy inhibitors, which are known to inhibit HTT degradation2. The detection method is still dependent on electrophoresis and Western blots and is therefore not high-throughput compatible. Here, we report the efforts to establish a high-throughput-compatible assay for measuring endogenous HTT degradation using Click chemistry and Homogeneous Time Resolved Fluorescence (HTRF) technologies.
Materials and methods
Cell culture
Mouse striatal cell lines (STHdh) were cultured in DMEM (Life Technologies, Cat No 11965) with 10% (vol/vol) FBS (Life Technologies, Cat No 10099-141) and were maintained in a 33 °C incubator with 5% CO2. Human fibroblast cells were cultured in DMEM (Life Technologies, Cat No 11965) with 15% (vol/vol) FBS (Life Technologies, Cat No 10099-141) and were maintained in a 37 °C incubator with 5% CO2. Both cell lines were obtained from Coriell Cell Repositories.
SPAAC reaction
Strain-promoted alkyne-azide cycloaddition (SPAAC) is a type of click reaction. A click reaction describes a class of chemical reactions involving triazole formation from an azide and an alkyne by addition reaction. It can use bio-orthogonal or biologically unique moieties to label and detect a molecule of interest. In this study, Click-IT® Biotin DIBO Alkyne (Life Technologies, Cat No C10412) was used to form the triazole with the azide groups of the pulsed proteins. Cell lysates (in 1× TBS+1% Triton X-100+1× complete protease inhibitor, Cat No 04693116001) were mixed with 10% (vol/vol) 50 μmol/L Click-IT® Biotin DIBO Alkyne, the reaction mixture was then incubated at 37 °C in the dark for 1 h to complete the SPAAC reaction.
HTRF assay
HTRF assays were performed similarly to those previously described2,3,9. The antibody mixtures were prepared by adding the donor and acceptor antibodies into the HTRF assay buffer containing 50 mmol/L NaH2PO4, 400 mmol/L KF, 0.1% bovine serum albumin (BSA), and 0.05% Tween 20. For all the antibody pairs used in this study, the donor antibody concentration was 0.023 μg/mL, and the acceptor antibody concentration was 1.4 μg/mL in HTRF buffer. 6 μL of the lysate and 4 μL of the antibody pair mixture were then added to each well of 384-well plates (Proxiplate, PerkinElmer, Cat No 6008238) and incubated at 4 °C overnight. Plates were warmed to room temperature before they were read. Different protein concentrations were tested to ensure that the signals were in the linear range. Background corrections were performed by subtracting the background signals from blank samples.
Compound treatment
Bafilomycin A (Sigma, Cat No B1793) was diluted in Opti-MEM to a 10× concentration and was added to the cell culture medium immediately after the pulse step (see “pulse-chase” section) to reach a final concentration of 100 nmol/L. Rapamycin (Selleck, Cat No S1039) was diluted in Opti-MEM to a 50× concentration, and the final concentrations used were 0.1 μmol/L and 1 μmol/L.
cDNA and siRNA transfections
siRNAs were reversely transfected into human fibroblast cells or STHdh cells with Lipofectamine 2000 (Life Technologies, Cat No 11668) according to the manufacturer's protocol. The HTT siRNA target sequence was CAGGTTTATGAACTGACGTTA.
Cell lysis and Western blots
Cell pellets were collected and lysed on ice for 30 min in TBS+1% Triton X-100+1× complete protease inhibitor (Roche Diagnostics, Indianapolis, IN, USA. Cat No 04693116001). The samples were sonicated for 10 s and then spun at >20 000 ×g at 4 °C for 10 min. The supernatants were then extracted and loaded onto denaturing gels, which were then transferred to nitrocellulose membranes for Western blot analysis. Streptavidin-HRP (1:2500) was used to detect the biotin-conjugated proteins.
Pulse-chase
As described in the main text, this assay had the following steps, and all liquid handling was performed with electronic multi-channel pipettes or plate washer to improve the throughput: 1) Plating of cells: cells were cultured in seven 96-well plates at 104 cells per well. Three to six wells were allocated as no L-azidohomoalanine (L-AHA) control wells (control group) and the rest were used for testing (test group). If needed, siRNAs or cDNAs were transfected 48 h before the next step. 2) Starvation: cells in 96-well plates were washed twice with warm PBS. Methionine-free medium containing DMEM (Life Technologies, Cat No 21013-024), 15% FBS, and 1× GlutaMAX (Life Technologies, Cat No 35050-061) was added to the cells, and the cultures were incubated at 37 °C for 60 min to deplete the remaining free methionine. 3) Pulse: Twenty-five microliters of 300 μmol/L L-AHA were added to the test group containing 125 μL of culture medium per well (final concentration of L-AHA: 50 μmol/L). Twenty-five microliters of 300 μmol/L L-methionine (Sigma, Cat No M5308-25G) were added to the control group containing 125 μL of culture medium per well (final concentration of L-methionine: 50 μmol/L). The plates were then mixed gently and incubated at 37 °C, 5% CO2 for 12 h. 4) Chase: The cells were washed three times with PBS, and complete medium was added to the cells. The first test and the control plates were immediately collected at the same time (time zero). The remaining plates were collected at different time points (ie, every 12 h). 5) Cell lysis: The culture medium was aspirated, and the cells were lysed directly in the 96-well plates with 45 μL lysis buffer (1× TBS+1% Triton X-100+1× complete protease inhibitor). 6) Click reaction: Ten percent (vol/vol) of 50 μmol/L Click-IT® Biotin DIBO Alkyne was added to the cell lysis solutions, and the solutions were incubated at 37 °C in the dark for 1 h. 7) Immunoprecipitation: The suspension of streptavidin-conjugated magnetic beads (Thermo Scientific, Cat No 88817) was dispensed into new 96-well plates at 10 μL per well and washed with 1× TBS using a BioTek plate washer. Twenty-two μL of the samples from step 6 were transferred for immunoprecipitation. The samples were incubated at room temperature for 4 h with mixing. 8) The supernatant from step 7 (post-IP) and the remaining samples from step 6 (input) were then transferred to the 384-well plates for the HTRF measurements in order to calculate the chase signals.
Statistical analysis
Statistical analysis between two groups were conducted by two-tailed unpaired Student t-test. For comparison of the time-dependent protein degradation between two groups, two-way ANOVA tests (difference between two groups of points) followed by Bonferroni post hoc tests (difference between the two points at each given time point). Significance was established at P<0.05. In all graphs, error bars indicate SEM, and the biological replicate numbers are indicated as the n numbers in the legends.
Results
Biotin conjugation by Strain-promoted Alkyne-Azide Cycloadditions (SPAAC)
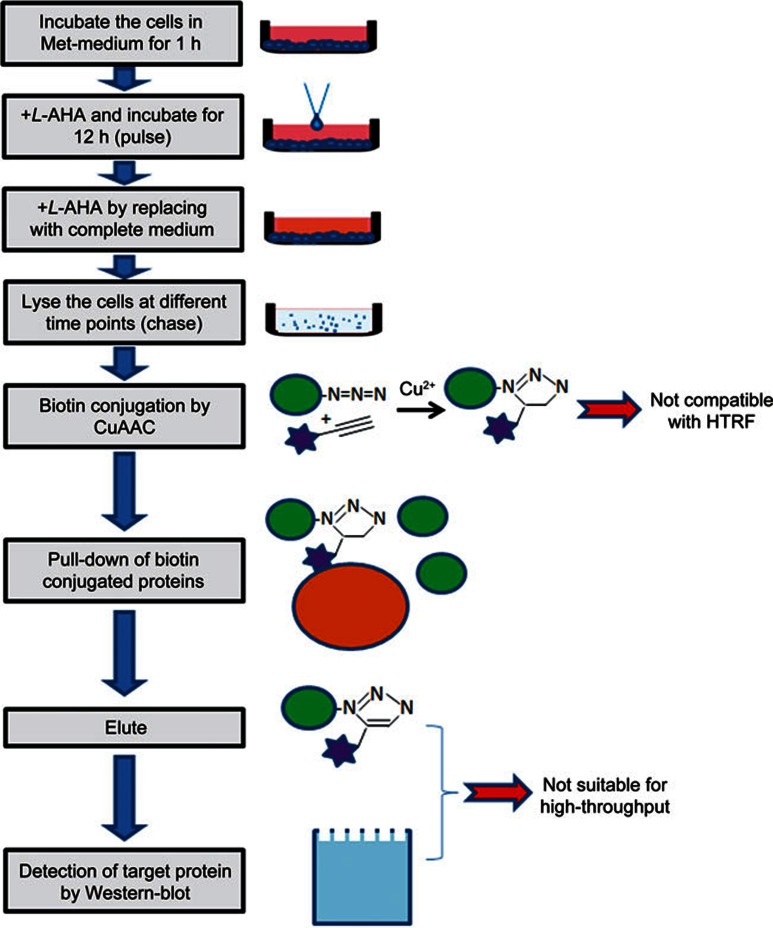
In the previous low-throughput assays utilizing Click chemistry, HTT degradation was tracked by pulse-chase2. During the pulse phase, the L-methionine is replaced by its non-toxic analog L-AHA10, which labels newly synthesized proteins with azide groups. L-AHA is then removed during the chase phase, and the azide-labeled proteins are then conjugated with biotin using Copper-Catalyzed Azide-Alkyne Cycloaddition (CuAAC). The biotin-conjugated proteins are then pulled down using streptavidin beads and eluted for Western blot detection (Figure 1).
Figure 1.
Schematic of pulse-chase experiments using Copper-Catalyzed Azide-Alkyne Cycloaddition (CuAAC) and its throughput limitations. The stars represent biotin.
We first tried to directly replace the Western blot detection with the well-established HTRF assays to achieve a higher throughput. HTRF detects HTT proteins by measuring the time-resolved fluorescent resonance energy transfer (TR-FRET) between two HTT antibodies labeled with either fluorescent donor or acceptor molecules9. We have used the 2B7/MW1 antibody pair to detect the mHTT in the HD patient fibroblast cells11, the 2B7/2166 antibody pair to detect the wild-type HTT (wtHTT) in the control patient fibroblasts12, or both the wtHTT and mHTT in the mouse striatal cells STHdhQ7/Q111 9. The effort of replacing the Western blot directly with the high-throughput-compatible HTRF assay failed because of two major reasons (Figure 1). First, the copper ion in the CuAAC reaction strongly influences the HTRF signals (Supplementary Figure S1A). It generates a non-specific signal while abolishing the HTRF signals from the target protein (Supplementary Figure S1A), possibly due to copper induced structural changes of the proteins. Removing copper after the CuAAC reaction failed to rescue the signal (data not shown). Second, all the elution conditions that we tested were not compatible with the HTRF assay (Supplementary Figure S1B). Of note, the washing and elution steps are known to generate variations due to bead loss and numerous pipetting; thus, these steps are not suitable for high-throughput assays.
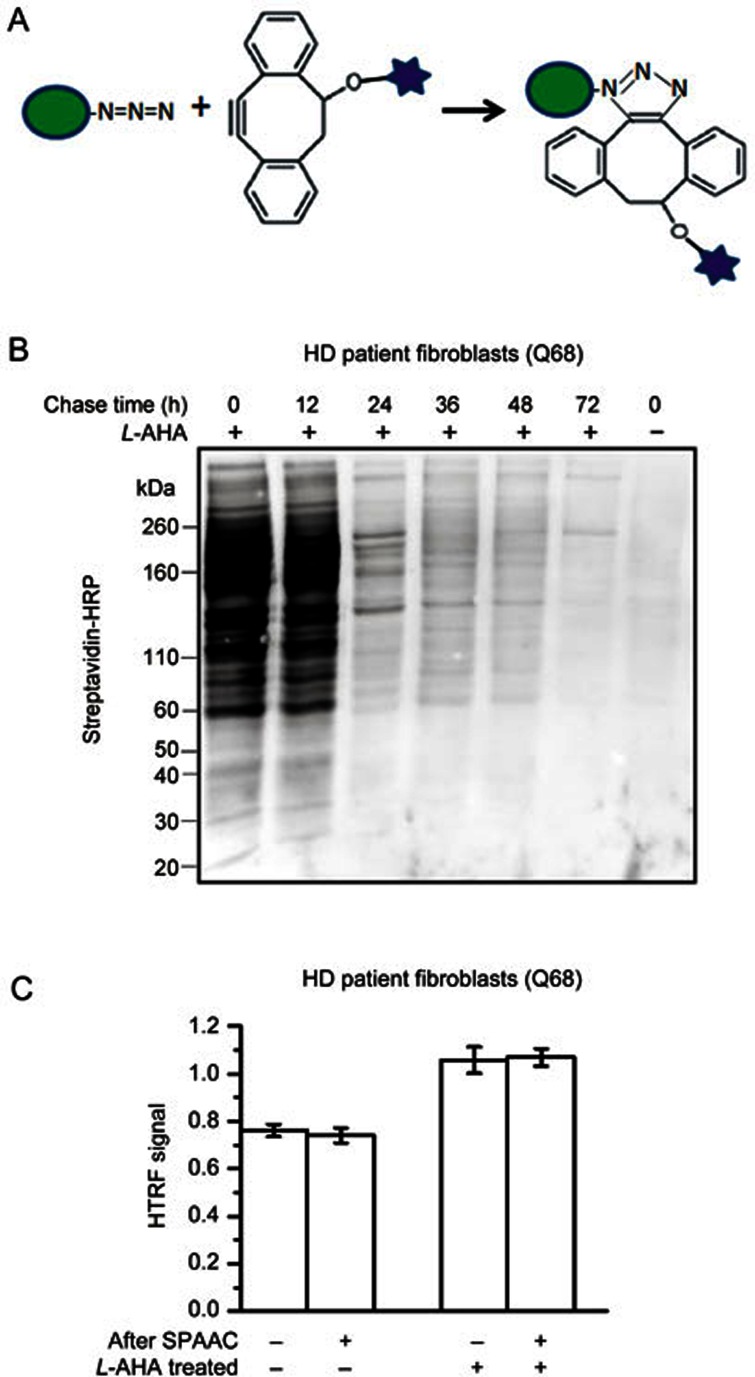
To solve the first problem, we utilized SPAAC13 instead of CuAAC to conjugate the azide labeled proteins with biotin (Figure 2A). Under our optimized conditions using biotin DIBO alkyne, SPAAC specifically conjugates biotin to the L-AHA treated samples, and the degradation of the biotin-conjugated proteins during the chase-phase is also evident by Western blot detection (Figure 2B). Importantly, SPAAC does not influence the HTRF signals (Figure 2C), confirming that SPAAC could be used for our high-throughput-compatible assays.
Figure 2.
Biotin conjugation by Strain-promoted Alkyne-Azide Cycloadditions (SPAAC). (A) Schematic of biotin conjugation via SPAAC using DIBO-biotin. The stars represent biotin. (B) Western blot with streptavidin-HRP for the detection of biotin-conjugated proteins in the L-AHA treated (+) or control (−, L-methionine treated) samples at different chased time points. (C) HTRF results of the lysates from HD patient fibroblasts (Q68) using the 2B7/MW1 antibody pair showing that SPAAC does not influence HTRF signals. Mean±SEM; n=4.
Detection of biotin-conjugated HTT using HTRF
To solve the second issue, we needed to find methods that skip the elution step for the detection of biotin-conjugated HTT. We first attempted this by using paired HTRF antibodies targeting biotin (streptavidin-labeled donor or acceptor) and HTT (HTT antibodies labeled with donor or acceptor). This method failed to detect any HTT specific signals, likely because streptavidin was captured by a large pool of all the different proteins conjugated with biotin after SPAAC, while a single HTT antibody was not specific enough to isolate the HTT signal. In addition, in some cases, we need to distinguish mHTT from wtHTT. Antibodies targeting the polyglutamine (polyQ) tract need to be used for this purpose, and a single polyQ antibody may not be sufficient to distinguish mHTT from other polyQ proteins.
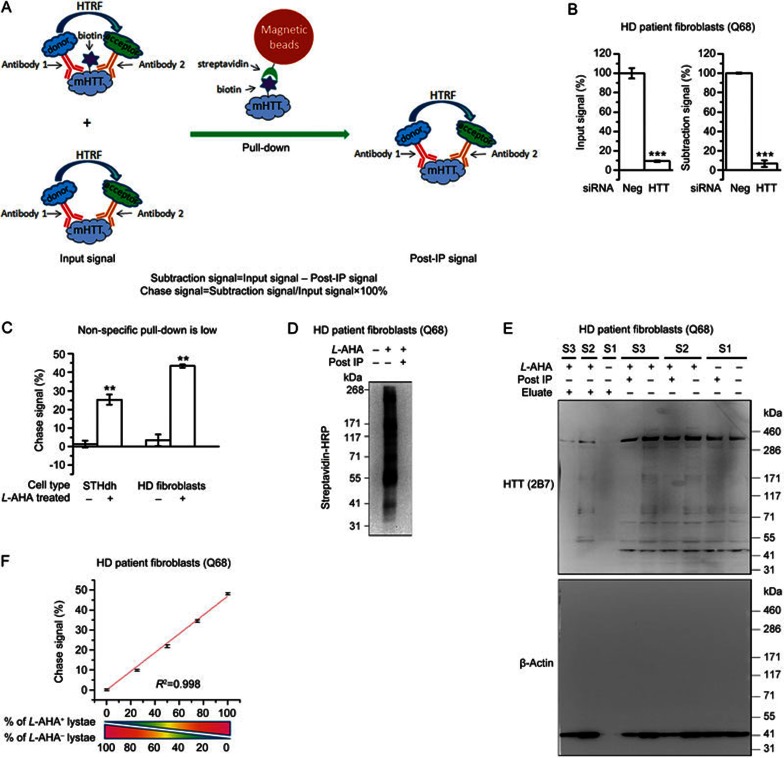
Thus, we designed the subtraction method to detect biotin-conjugated mHTT (Figure 3A). Basically, two antibodies targeting mHTT were used for HTRF detection to ensure specificity. The total HTRF signal contributed by both the biotin-conjugated and unconjugated mHTT was subsequently measured to determine the “input signal.” The samples were then incubated with bovine serum albumin (BSA) pre-treated streptavidin beads, allowing the biotin-conjugated mHTT to be pulled down by the beads. The remaining mHTT in the supernatant was then measured (referred to as the “post-IP signal”), and the reduction of the HTRF signal (referred to as the “subtraction signal”) was calculated by subtracting the post-IP signal from the input signal. To correct for possible variation in loading, the subtraction signal was normalized to the input signal, and the resulting value represents the percentage of biotin-conjugated mHTT (referred to as the “chase signal”) in total mHTT. The same principle could be applied to wtHTT and other target proteins. Utilizing this design, we successfully performed all the steps in 96-well plates in a high-throughput-compatible manner, and we performed all the subsequent experiments (except the Western blots) in this format.
Figure 3.
Detection of biotin-conjugated mHTT by HTRF. (A) Schematic picture of the subtraction method. (B) Knock-down of HTT by siRNA reduces the chase signal. Mean±SEM; n=3; **P<0.01 by t tests. (C) The chase signal from the control (L-methionine treated) compared to the L-AHA treated samples showing that the non-specific pull-down is low (%). Mean±SEM; n=6; **P<0.01 by t tests. (D) Western blot with streptavidin-HRP showing the pull-down efficiency. (E) Representative Western blots with anti-HTT antibody 2B7 and anti-β-actin antibody showing that the biotin-conjugated HTT and β-actin protein is pulled down. S1, S2 and S3 indicate three different samples: S1 is the sample from cells without L-AHA treatment and shows no difference before and after IP. S2 and S3 are biological replicates of samples from L-AHA-treated cells. (F) Linear relationship between the chase signal and the proportion of L-AHA-treated samples mixed with the control samples. The small residue from the non-specific pull-down has been corrected. Mean±SEM; n=4; Linear regression was performed, and R2 indicates the regression coefficient.
The input and subtraction signals needed to be specific to the target-protein in order to obtain correct measurements. While the HTRF antibody pairs had been previously validated, the specificity needed to be further confirmed after the Click and pull-down procedures. To confirm these signals as target-protein specific, we knocked down HTT using siRNA and tested the subtraction signals. Both the input signal and the subtraction signal of mHTT (detected by the 2B7/MW1 antibody pair) were knocked down as expected (Figure 3B).
Another factor that may influence the measurement is that the streptavidin beads may pull-down a small fraction of unconjugated proteins. In addition, the pull-down efficiency may vary among different samples, influencing the results. In fact, the classical elution method has similar issues as well. In our assay, the non-specific pull-down of mHTT are minimal (Figure 3C), whereas the pull-down efficiency is very high, and almost all the biotin-conjugated proteins were pulled-down by the streptavidin beads (Figure 3D). We further confirmed this by Western-blots for HTT and β-actin. Consistently, a portion of HTT and β-actin were pulled down and present only in the eluate for L-AHA treated samples but not in the non-L-AHA treated control samples (Figure 3E). Noticeably, the dominant HTT bands in the Q68 fibroblasts are full-length (Figure 3E, upper panel), suggesting that the degradation detected by our method is mainly contributed by the degradation of the full-length HTT protein in the Q68 fibroblasts. Similarly, the full-length protein is the dominant HTT fragment in the STHdh cells as well, based on our previous study3. To further confirm that the HTRF measurement of the chase signal is accurate, we mixed the lysates from L-AHA treated cells with the those from control (L-methionine treated) cells at different ratios and measured the chase signal after the whole procedure, including SPAAC, pull-down, and HTRF. The chase signal exhibits a clear linear relationship with the percentage of L-AHA treated samples (Figure 3F), thus confirming that the measurement is accurate.
The data above confirm that the chase signal faithfully measures the biotin-conjugated mHTT protein.
Measurement of mHTT degradation and its modulation
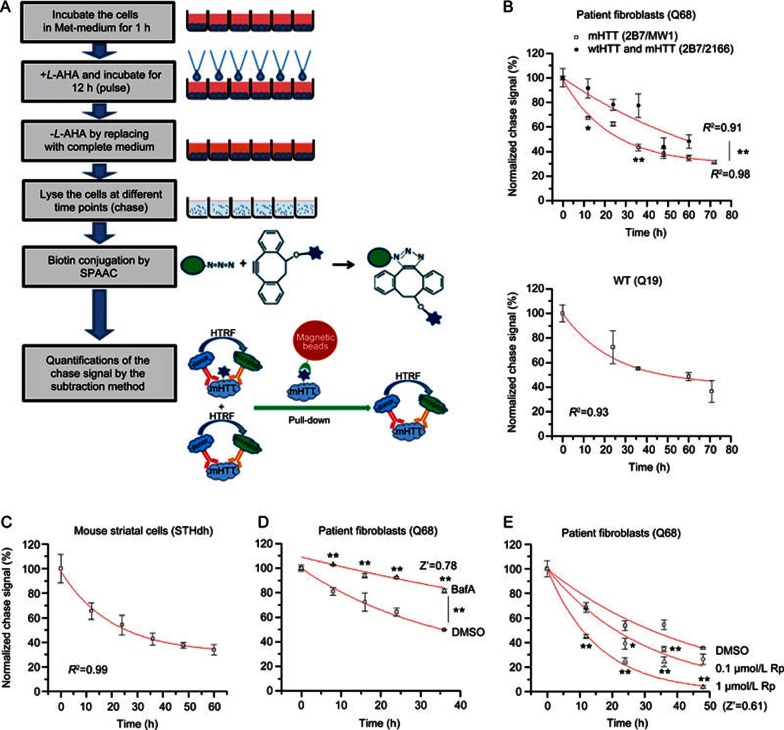
Based on the data above, we established a high-throughput-compatible assay for the measurement of endogenous mHTT degradation. The assay could also be applied to other proteins of interest. Basically, the cells are starved to deplete free L-methionine. During the pulse phase, the L-methionine is replaced by L-AHA, which is removed during the chase phase when the cells are lysed at different time points. Azide-labeled proteins in the lysates are then conjugated with biotin by SPAAC, and the remaining L-AHA labeled mHTT is measured by HTRF using the subtraction method to plot the degradation curve (Figure 4A).
Figure 4.
Measurement of mHTT degradation and its modulation. (A) Schematic picture of the high-throughput-compatible CH-chase assay. (B) The degradation curve measured by CH-chase in HD patient fibroblasts (Q68, upper panel) and the control patient fibroblasts (Q19, lower panel). Mean±SEM; n=4; R2 indicates the regression coefficient of the curve fitting using the exponential decay function (Y=Ae-X/t). Both mHTT degradation (2B7/MW1) and total HTT degradation (2B7/2166) have been measured in the Q68, and the mHTT degradation is significantly faster; (C) Similar to (B). The degradation curve measured by CH-chase in mouse striatal cells (STHdhQ7/Q111). Mean±SEM; n=3; (D) The change of mHTT degradation by treatment with autophagy inhibitor bafilomycin A (upper trace) compared to the DMSO-treated control (lower trace) in HD patient fibroblasts (Q68). Mean±SEM; n=2 (biological duplicates) for each sample at each time points because it is the usual setting for high-throughput screenings. Z′=1–3×(SD of DMSO+SD of BafA)/(average of BafA–average of DMSO). The signals at the final time point were utilized to calculate the Z′ value. (E) The change of mHTT degradation by treatment with the mTOR inhibitor rapamycin, which activates autophagy and accelerates mHTT degradation. Concentrations of 0.1 μmol/L and 1 μmol/L of rapamycin (0.1 μmol/L Rp and 1 μmol/L Rp) were tested compared to the DMSO-treated control in HD patient fibroblasts (Q68). Mean±SEM; n=4 (biological replicates) for each sample at each time points. Z′=1–3×(SD of DMSO+SD of 1 μmol/L Rp)/(average of DMSO–average of 1 μmol/L Rp). For (B), (D) and (E), *P<0.05, **P<0.01 by two-way ANOVA (difference between two groups of points) followed by Bonferroni post hoc tests (difference between the two points at each given time point). The signals from compounds treated samples are all compared with the DMSO-treated controls.
Utilizing this high-throughput-compatible assay, we detected the degradation of HTT in HD patient fibroblasts and control patient fibroblasts (Figure 4B) and the degradation of HTT (including both mHTT and wtHTT) in mouse striatal cells (STHdhQ7/Q111) (Figure 4C, half-life 25.5±1.9 h). The degradation curves are consistent with the predicted exponential decay (Figure 4B, 4C). In Q68 cells, the mHTT half-life (detected by 2B7/MW1) is 27.8±3.4 h, and the HTT half-life (including both mHTT and wtHTT) is 57.5±6.1 h. In the WT cells (Q19), the wtHTT half-life 48.1±4.6 h. These data suggest that mHTT degradation is faster than wtHTT degradation, which is consistent with previous reports6. However, our data are not sufficient to confirm the conclusion made by the other group. The detected half-lives of both mHTT and wtHTT are shorter than the half-life previously reported in neurons6, likely because fibroblasts are dividing cells and have a dilution effect of the chased protein, whereas neurons are post-mitotic. The faster degradation rate of mHTT is likely explained by the elevated chaperon-mediated degradation or other cellular processes that clear misfolded proteins. The mHTT is misfolded and expected to activate chaperon-mediated degradation, whereas the wtHTT is generally not misfolded; therefore, cells are not expected to spend extra energy to increase the rate of degradation.
To confirm that the assay is able to identify chemical or genetic modifiers of mHTT degradation, we tested the effect of known modifiers. Consistent with previous studies2,14, the autophagy inhibitor bafilomycin A slows down mHTT degradation (Figure 4D). On the other hand, the autophagy activator rapamycin15 significantly increased the mHTT degradation rate in a dose-dependent manner (Figure 4E). In both cases, the calculated Z' values are larger than 0.5 (Figure 4D, 4E), indicating excellent screening capacity16. Thus, our assay is capable of identifying modifiers of mHTT degradation.
Discussion
We have established a high-throughput-compatible assay to measure mHTT degradation. Similar assays could be developed to detect the degradation of other proteins that cause neurodegenerative disorders and could be used for drug or target screening of these diseases. The assay is based on Click chemistry and HTRF, and therefore we named it CH-chase.
The assay is high-throughput-compatible, and it is not dependent on protein fusion tags, radioactive isotope labeling, or protein synthesis inhibitors, making it advantageous over most other assays that measure protein degradation. It has not escaped our scope that another potential advantage of our assay is the detection condition. Our assay detects mHTT degradation in the cell lysates under natural conditions, and thus it may preserve the original structure of the protein. Our assay could also be modified to detect the mHTT aggregates by using the antibody pairs that selectively target the aggregated form of mHTT17.
Like all the other high-throughput-compatible assays, our assay has limitations. Depending on the antibody pairs, our assay may not be able to distinguish different mHTT fragments using a single antibody pair. For the detailed changes in the degradation of specific fragments, HTRF using several different antibody pairs could be used to calculate the signals from certain fragments. This is not a major issue in the cells used in this study however, because the full-length HTT is the dominant species in these cells.
In summary, our assay provides a high-throughput-compatible method to screen for mHTT degradation modifiers, which is important for HD drug discovery. In addition, the same principle could be applied to other proteins of interest.
Author contribution
Boxun LU initiated the project and designed the experiments; Xiao-tian CUI, Shen-liang YU, and Jian-bin ZHU validated that CuAAC is not compatible with subsequent high-throughput HTRF assays; Peng WU, Xiao-li SUN, Ming-xing LU, and He-qing YANG performed the experiments to establish and validate the high-throughput pulse-chase assay.
Abbreviations
HD, Huntington's disease; mHTT, mutant Huntington protein; wtHTT, wild type Huntington protein; L-AHA, L-azidohomoalanine; TBS, Tris-based saline; CuAAC, Copper-Catalyzed Azide-Alkyne Cycloaddition; SPAAC, Strain-Promoted Alkyne-Azide Cycloaddition; TR-FRET, time-resolved fluorescent resonance energy transfer; HTRF, Homogeneous Time Resolved Fluorescence; HRP, Horseradish Peroxidase; polyQ, polyglutamine.
Acknowledgments
The study was supported by the National High Technology Research and Development Program of China (2014AA02502) and the National Natural Science Foundation of China (NSFC 31371421 and 31422024).
Footnotes
Supplementary information was available on the website of Acta Pharmacologica Sinica.
Supplementary Information
HTRF is incompatible with CuAAC and elution
References
- Rubinsztein DC, Carmichael J. Huntington's disease: molecular basis of neurodegeneration. Expert Rev Mol Med 2003; 5: 1–21. [DOI] [PubMed] [Google Scholar]
- Lu B, Al-Ramahi I, Valencia A, Wang Q, Berenshteyn F, Yang H, et al. Identification of NUB1 as a suppressor of mutant Huntingtin toxicity via enhanced protein clearance. Nat Neurosci 2013; 16: 562–70. [DOI] [PubMed] [Google Scholar]
- Yao Y, Cui X, Al-Ramahi I, Sun X, Li B, Hou J, et al. A striatal-enriched intronic GPCR modulates huntingtin levels and toxicity. Elife 2015; 4. doi:10.7554/eLife.05449. [DOI] [PMC free article] [PubMed]
- Yu S, Liang Y, Palacino J, Difiglia M, Lu B. Drugging unconventional targets: insights from Huntington's disease. Trends Pharmacol Sci 2014; 35: 53–62. [DOI] [PubMed] [Google Scholar]
- Persichetti F, Carlee L, Faber PW, McNeil SM, Ambrose CM, Srinidhi J, et al. Differential expression of normal and mutant Huntington's disease gene alleles. Neurobiol Dis 1996; 3: 183–90. [DOI] [PubMed] [Google Scholar]
- Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, Shaby BA, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol 2013; 9: 586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LLS Jr, Ward VL, Brockman RW. Inhibition of protein synthesis in vitro by cycloheximide and related glutarimide antibiotics. Biochim Biophys Acta 1965; 103: 478–85. [DOI] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl 2001; 40: 2004–21. [DOI] [PubMed] [Google Scholar]
- Liang Y, Yao Y, Lu M, Hou J, Yu S, Lu B. TR-FRET assays for endogenous Huntingtin protein level in mouse cells. J Huntingtons Dis 2014; 3: 253–9. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci U S A 2006; 103: 9482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Abramowski D, Bibel M, Bodner R, Chopra V, DiFiglia M, et al. Single-step detection of mutant huntingtin in animal and human tissues: a bioassay for Huntington's disease. Anal Biochem 2009; 395: 8–15. [DOI] [PubMed] [Google Scholar]
- Weiss A, Trager U, Wild EJ, Grueninger S, Farmer R, Landles C, et al. Mutant huntingtin fragmentation in immune cells tracks Huntington's disease progression. J Clin Invest 2012; 122: 3731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallana E, Fernandez-Megia E, Riguera R. Surpassing the use of copper in the click functionalization of polymeric nanostructures: a strain-promoted approach. J Am Chem Soc 2009; 131: 5748–50. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Rubinsztein DC. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J 2008; 275: 4263–70. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 2004; 36: 585–95. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- Baldo B, Paganetti P, Grueninger S, Marcellin D, Kaltenbach Linda S, Lo Donald C, et al. TR-FRET-based duplex immunoassay reveals an inverse correlation of soluble and aggregated mutant huntingtin in Huntington's disease. Chem Biol 2012; 19: 264–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HTRF is incompatible with CuAAC and elution