Uematsu et al. show that ASB7 ubiquitinates DDA3, which facilitates Kif2a-mediated depolymerization of microtubules (MTs) for proteasomal degradation. The presence of MTs prevents the ASB7–DDA3 interaction, suggesting a feedback loop to appropriately regulate MT polymerization and spindle dynamics.
Abstract
Proper dynamic regulation of the spindle is essential for successful cell division. However, the molecular mechanisms that regulate spindle dynamics in mitosis are not fully understood. In this study, we show that Cullin 5–interacting suppressor of cytokine signaling box protein ASB7 ubiquitinates DDA3, a regulator of spindle dynamics, thereby targeting it for proteasomal degradation. The presence of microtubules (MTs) prevented the ASB7–DDA3 interaction, thus stabilizing DDA3. Knockdown of ASB7 decreased MT polymerization and increased the proportion of cells with unaligned chromosomes, and this phenotype was rescued by deletion of DDA3. Collectively, these data indicate that ASB7 plays a crucial role in regulating spindle dynamics and genome integrity by controlling the expression of DDA3.
Introduction
Regulation of cell division is important for normal proliferation and prevention of tumor development. The distribution of replicated chromosomes into the two daughter cells is a key event of the cell cycle and is essential for normal cell division. This step is tightly regulated by multiple molecules and signaling pathways (Kline-Smith and Walczak, 2004; Musacchio, 2015; Meunier and Vernos, 2016). Normal bipolar attachment of mitotic spindles to sister kinetochores and congression of chromosomes to the metaphase plate are achieved by dynamic turnover of microtubules (MTs; Musacchio and Hardwick, 2002). When all kinetochores are attached to the spindle and the structure is under tension, the spindle checkpoint is deactivated, and MT depolymerization provides the driving force for chromosome segregation at anaphase (Musacchio and Hardwick, 2002).
MTs switch between growing and shrinking phases and are thus dynamically unstable (Kline-Smith and Walczak, 2004). Spindle dynamics are regulated by MT nucleators (e.g., γ-tubulin), MT-associated proteins, and MT depolymerases (e.g., kinesin-13 and related proteins; Kline-Smith and Walczak, 2004). Three members of the kinesin-13 family, Kif2a, Kif2b, and mitotic centromere-associated kinesin/Kif2c, execute diverse functions in mitosis (Ems-McClung and Walczak, 2010; Walczak et al., 2013). These proteins do not walk along MTs like conventional kinesins; instead, they undergo 1D diffusion along the MT lattice with no bias toward the plus or minus end (Helenius et al., 2006). Kif2a, which localizes at spindle poles, is essential for spindle bipolarity; consequently, knockdown of Kif2a in human cells results in formation of monopolar spindles (Ganem and Compton, 2004). In contrast, in Xenopus laevis animal caps, monopolar spindles are a minor phenotype of Kif2a depletion that occurs at stage 10. The predominant phenotype, which arises at stage 10.5–11, is formation of multipolar spindles (Eagleson et al., 2015). Furthermore, Kif2a depletion increases the proportion of cells with three or more centrosomes (Eagleson et al., 2015). Thus, multipolar spindles caused by Kif2a depletion are likely to arise because of the presence of multiple centrosomes. Kif2a also contributes significantly to efficient pole coalescence, although it is not strictly required for this process (Eagleson et al., 2015).
Kif2a interacts with an MT-associated protein, DDA3 (also known as PSRC1), which functions on the mitotic spindles to control chromosome congression and segregation by regulating the dynamics of the mitotic spindle. This interaction increases the efficiency of Kif2a targeting to spindle poles (Jang et al., 2008). Knockdown of DDA3 increases the frequency of unaligned chromosomes, substantially reduces tension across sister kinetochores at metaphase, and decreases the velocity of chromosome segregation during anaphase (Jang et al., 2008). Down-regulation of DDA3 stabilizes spindle MTs, which phenocopies the effect of Kif2a knockdown. In summary, DDA3 acts as an MT-destabilizing protein in collaboration with Kif2a to regulate spindle dynamics and mitotic progression (Jang et al., 2008).
The ubiquitin–proteasome system regulates various cellular processes, including cell cycle progression, transcription, and signal transduction (Liu et al., 2015). Covalent attachment of ubiquitin to the substrate is achieved through sequential reactions by a ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). E3s are thought to be primarily responsible for substrate recognition (Skaar et al., 2014). The ECS (Elongin B/C–Cullin 5–SOCS [suppressor of cytokine signaling] box protein) family belongs to the largest RING finger E3 superfamily, the Cullin–RING ligases (Okumura et al., 2012). Cullin 5 (Cul5) is a scaffold protein that assembles multiple proteins into complexes that include a small RING finger protein (Rbx2), an adapter protein (Elongin B or C), and a substrate-targeting protein (SOCS box protein; Kile et al., 2002; Kamura et al., 2004). SOCS box proteins are divided into four major classes: the SOCS family (whose members contain an SH2 domain and SOCS box), WSB family (WD40 repeats and a SOCS box), SSB family (SPRY domain and SOCS box), and ASB family (ankyrin repeats and SOCS box; Hilton et al., 1998).
The ASB family, the largest family of SOCS box proteins, has 18 members (ASB1–18). These proteins all contain two functional domains: a C-terminal SOCS box and a variable number of N-terminal ankyrin repeats (Kile et al., 2002). The SOCS box itself contains two subdomains, the BC and Cul5 boxes, which are required in order to interact with Elongin B/C and Cul5–Rbx2 to form E3 complexes (Kamura et al., 2004; Mahrour et al., 2008).
In this study, we showed that DDA3 is targeted by ASB7 for proteasomal degradation. ASB7 polyubiquitinated DDA3 both in vivo and in vitro. Importantly, knockdown of ASB7 prevented MT polymerization and increased the proportion of cells with unaligned chromosomes. Depletion of DDA3 in ASB7 knockdown cells reverted these phenotypes. We propose that ASB7 modulates spindle dynamics and genome integrity by regulating the level of DDA3.
Results
Ubiquitination and degradation of DDA3 by ASB7
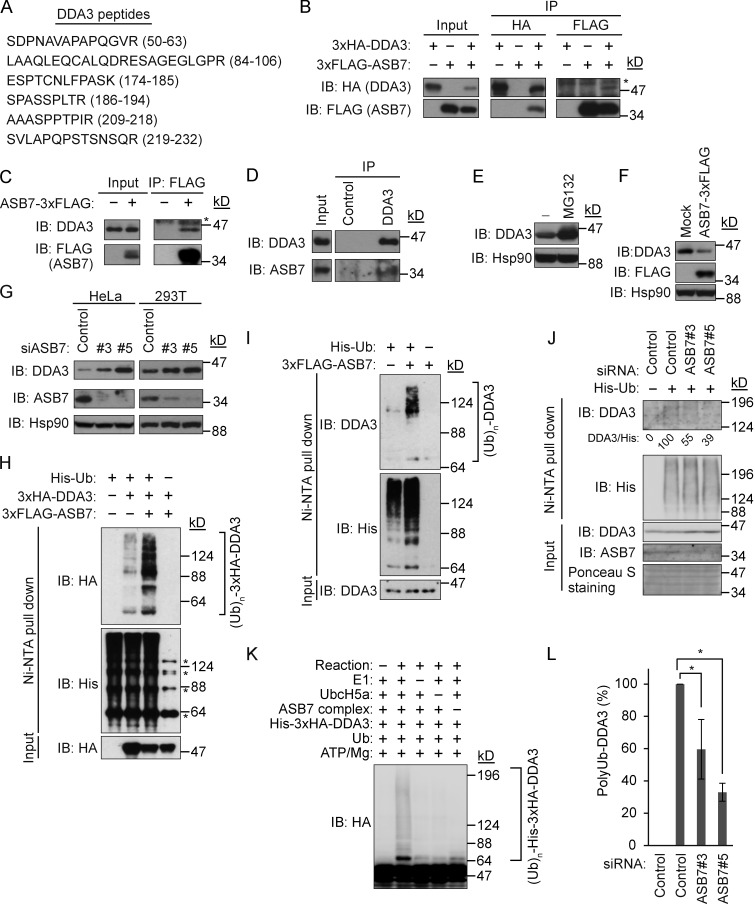
To investigate the biological function of ASB7, we subjected HEK293T cell lysates expressing FLAG-tagged ASB7 to immunoprecipitation (IP) with anti-FLAG antibodies and then analyzed the resultant immunoprecipitates by mass spectrometry. This screen identified several peptides belonging to DDA3 (Fig. 1 A). To confirm that ASB7 binds DDA3, 3×FLAG-tagged ASB7 and 3×HA-tagged DDA3 were coexpressed in HEK293T cells, and the interaction was validated by IP and immunoblot (IB) analysis (Fig. 1 B). In addition, overexpressed ASB7 bound to endogenous DDA3 (Fig. 1 C), and ultimately we were able to demonstrate association between endogenous ASB7 and endogenous DDA3 (Fig. 1 D).
Figure 1.
ASB7 interacts with DDA3 and promotes ubiquitination of DDA3. (A) Identification of DDA3 as an ASB7-interacting protein. 3×FLAG-ASB7 expressed in HEK293T cells was purified using anti-FLAG antibody and resolved by SDS-PAGE. ASB7-interacting proteins were analyzed by mass spectrometry. Identified peptides of DDA3 are shown. (B) Interaction between 3×HA-DDA3 and 3×FLAG-ASB7. HEK293T cells expressing 3×HA-DDA3 and 3×FLAG-ASB7 (as indicated) were cultured in the presence of the proteasome inhibitor MG132 (10 µM for 6 h), immunoprecipitated (IP) with anti-HA or anti-FLAG antibody, and immunoblotted (IB) with anti-HA or anti-FLAG antibody. (C) Interaction between endogenous DDA3 and 3×FLAG-ASB7. HeLa cells stably expressing 3×FLAG-ASB7 were cultured in the presence of MG132 (10 µM for 6 h), immunoprecipitated with anti-FLAG antibody, and immunoblotted with anti-DDA3 or anti-FLAG antibody. (D) Interaction between endogenous DDA3 and ASB7. HeLa cells were cultured in the presence of MG132 (10 µM for 6 h), immunoprecipitated with anti-DDA3 antibody, and immunoblotted with anti-DDA3 or anti-ASB7 antibody. (E) Accumulation of DDA3 upon exposure to MG132. HeLa cells were cultured in the presence or absence of 10 µM MG132 for 6 h and then Western blotted with antibodies against DDA3. (F) Down-regulation of DDA3 by overexpression of ASB7. HeLa cells stably expressing 3×FLAG-ASB7 were lysed and immunoblotted with anti-DDA3, anti-FLAG, or anti-Hsp90 antibody. (G) Accumulation of endogenous DDA3 by knockdown of ASB7. Two independent siRNAs (#3 and #5) targeting ASB7 were transfected into HeLa or HEK293T cells, which were cultured for 2 d and then Western blotted with anti-DDA3, anti-FLAG, or anti-Hsp90 antibody. (E–G) Hsp90 is shown as a loading control. (H) ASB7-dependent polyubiquitination of exogenous DDA3 in vivo. HEK293T cells were transfected with plasmids encoding 3×FLAG-ASB7, 3×HA-DDA3, and/or His6-tagged ubiquitin. MG132 (10 µM for 6 h) was used to detect polyubiquitination. Cell lysates were subjected to nickel–nitrilotriacetic acid (Ni-NTA) pulldown to purify proteins modified by His6-ubiquitin, followed by IB analysis with anti-HA, FLAG, or His6 antibody. Asterisks denote nonspecific bands. (I) ASB7-dependent polyubiquitination of endogenous DDA3 in vivo. HeLa cells stably expressing 3×FLAG-ASB7 were examined as in H. (J) Reduced polyubiquitination of endogenous DDA3 by knockdown of ASB7. siRNAs (#3 and #5) targeting ASB7 were transfected into HEK293T cells and examined as in I. (K) ASB7-dependent polyubiquitination of DDA3 in vitro. Recombinant ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (UbcH5a), ASB7 complex (ASB7, Cul5, Elongin B, Elongin C, and Rbx2), His6-ubiquitin, and immunopurified 3×HA-DDA3 were mixed in vitro in various combinations in the presence of ATP, incubated at 28°C for 1 h, and subjected to Western blotting with anti-HA antibody. (L) Quantification of polyubiquitinated DDA3. The signals of polyubiquitinated DDA3 shown in J were quantified. Control sample with or without His6-ubiquitin expression was set as 100 and 0, respectively. *, P < 0.02. Data represent the mean ± SD of three independent experiments.
We next investigated whether ASB7 regulates the expression of DDA3 through the ubiquitin–proteasome degradation pathway. Consistent with this idea, DDA3 accumulated in cells treated with the proteasome inhibitor MG132 (Fig. 1 E). Furthermore, DDA3 was down-regulated by stable overexpression of 3×FLAG-ASB7 in HeLa cells (Fig. 1 F). Inversely, knockdown of ASB7 dramatically up-regulated endogenous DDA3 in both HeLa and HEK293T cells (Fig. 1 G). We then sought to determine whether ASB7 promotes ubiquitination of DDA3 in vivo and in vitro. In the in vivo experiments, His6-tagged ubiquitin was expressed with or without 3×HA-DDA3 and 3×FLAG-ASB7 in HEK293T cells. Cell lysates were prepared in 8 M urea containing lysis buffer, and His6-tagged ubiquitin was pulled down with nickel–agarose beads. Polyubiquitination of both 3×HA-DDA3 (Fig. 1 H) and endogenous DDA3 (Fig. 1 I) was increased by coexpression of 3×FLAG-ASB7. In contrast, knockdown of ASB7 reduced polyubiquitination of endogenous DDA3 (Fig. 1, J and K). In vitro ubiquitination assays with purified proteins confirmed the promotion of DDA3 polyubiquitination by ASB7 (Fig. 1 L). These data clearly indicate that ASB7 is a ubiquitin ligase that activates the ubiquitination and degradation of DDA3.
ASB7 is a Cul5-interacting protein
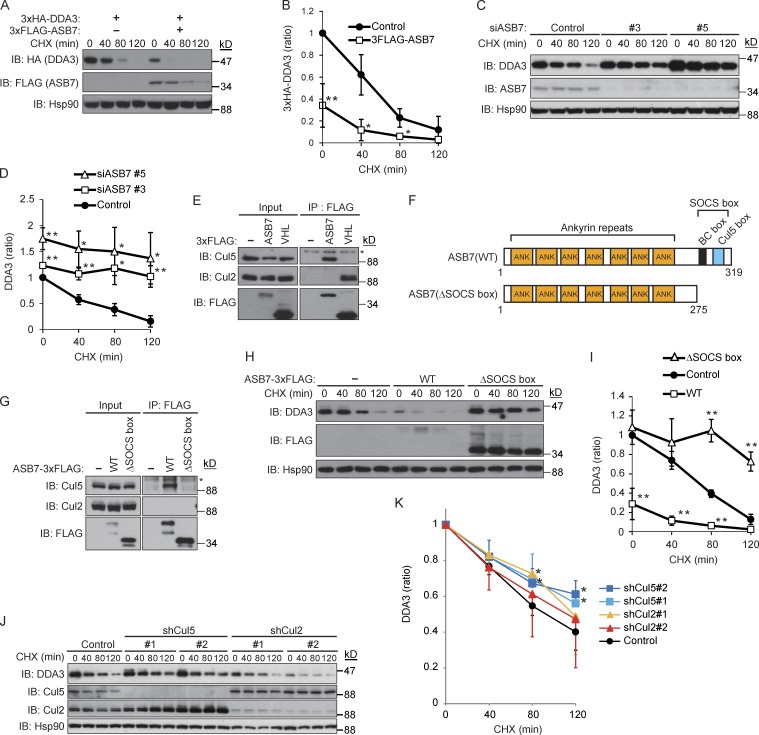
We next performed cycloheximide (CHX) chase experiments to compare the stability of 3×HA-DDA3 in the presence or absence of 3×FLAG-ASB7. As expected, 3×HA-DDA3 was destabilized by overexpression of ASB7 (Fig. 2, A and B), and knockdown of endogenous ASB7 dramatically stabilized endogenous DDA3 (Fig. 2, C and D). In IP experiments using cell lysates of HEK293T cells expressing 3×FLAG-ASB7 (Fig. 2 E), we observed an interaction between ASB7 and endogenous Cul5, but not Cul2. In contrast, the Cul2-type ubiquitin ligase pVHL interacted with endogenous Cul2, but not Cul5, under the same conditions, indicating that the interaction between ASB7 and Cul5 was selective (Fig. 2 E). Mutant ASB7 lacking the SOCS box, which is required for Cul5 binding, failed to interact with Cul5 (Fig. 2, F and G) and did not affect the rate of DDA3 degradation (Fig. 2, H and I). Moreover, knockdown of Cul5, but not Cul2, stabilized DDA3 (Fig. 2, J and K). These results demonstrate that ASB7 is a Cul5-interacting protein that regulates the turnover of DDA3.
Figure 2.
Destabilization of DDA3 by ASB7. (A) HEK293T cells expressing 3×HA-DDA3 with or without 3×FLAG-ASB7 were exposed to 50 µg/ml CHX for 40, 80, or 120 min. The lysates were subjected to Western blotting with antibodies against HA, FLAG, or Hsp90. (B) The intensities of HA-DDA3 bands in A were normalized to those of the corresponding Hsp90 bands and plotted as a ratio of the normalized value without ASB7 expression at 0 h. (C) Stabilization of endogenous DDA3 by knockdown of ASB7. Two independent siRNAs (#3 and #5) targeting ASB7 were transfected into HeLa cells and cultured for 2 d, followed by CHX treatment as in A. The lysates were subjected to Western blotting with antibodies against DDA3, ASB7, or Hsp90. (D) The intensities of DDA3 bands in C were normalized to those of the corresponding Hsp90 bands and plotted as a ratio of the normalized value of control cells at 0 h. (E) ASB7 interacts with Cul5 but not Cul2. HEK293T cells expressing 3×FLAG-ASB7 or 3×FLAG-pVHL (Cul2-type ubiquitin ligase as a control) were cultured in the presence of MG132 (10 µM for 6 h), immunoprecipitated (IP) with anti-FLAG antibody, and immunoblotted (IB) with anti-Cul2, anti-Cul5, or anti-FLAG antibody. (F) Schematic representation of ASB7 mutants used in this study. (G) Interaction between the SOCS box of ASB7 and Cul5. HeLa cells stably expressing 3×FLAG-ASB7(WT or ΔSOCS box) were lysed, immunoprecipitated with anti-FLAG antibody, and immunoblotted with anti-Cul2, anti-Cul5, or anti-FLAG antibody. (E and G) Asterisk denotes a nonspecific band. (H) Inactivation of ASB7 by deletion of the SOCS box. HeLa cells stably expressing 3×FLAG-ASB7(WT or ΔSOCS box) were exposed to CHX as in A. The lysates were subjected to Western blotting with antibodies against DDA3, FLAG, or Hsp90. (I) The intensities of DDA3 bands in H were normalized to those of the corresponding Hsp90 bands and plotted as a ratio of the normalized value of control cells at 0 h. (J) Endogenous DDA3 was stabilized by knockdown of Cul5, but not Cul2. Two independent siRNAs (#1 and #2) targeting Cul2 or Cul5 were transfected into HeLa cells and cultured for 2 d, followed by CHX treatment as in A. The lysates were subjected to Western blotting with antibodies against DDA3, Cul2, Cul5, or Hsp90. (A, C, H, and J)_Hsp90 is shown as a loading control. (K) The intensities of DDA3 bands in J were normalized to those of the corresponding Hsp90 bands and plotted as a ratio of the normalized value of control cells at 0 h. For all graphs, *, P < 0.05; **, P < 0.01. Data represent the mean ± SD of three independent experiments (B, D, and I) and four independent experiments (K).
Cell cycle–dependent regulation of DDA3 degradation by ASB7
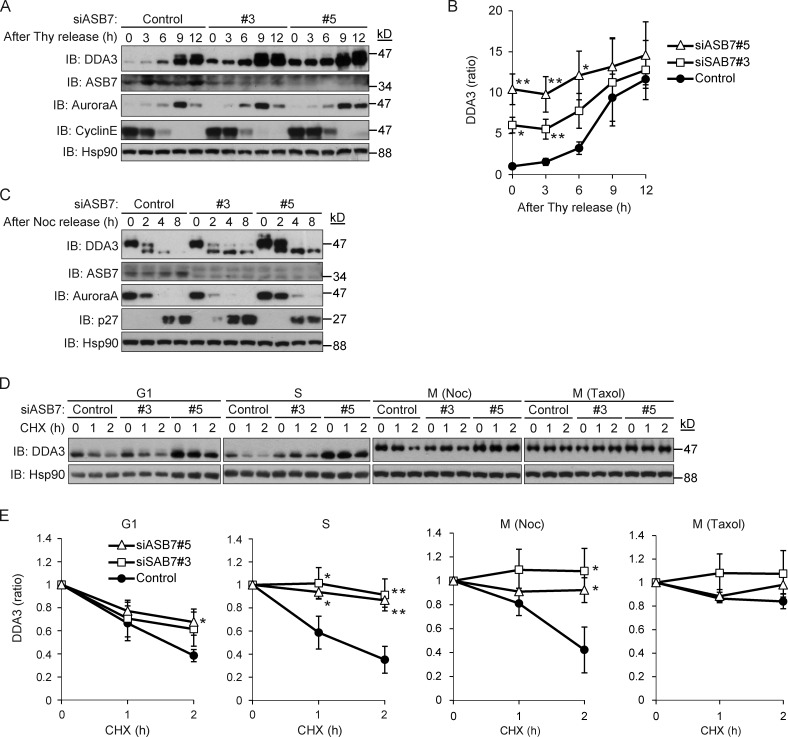
DDA3 is phosphorylated during the M phase (Jang et al., 2008, 2011), implying that the degradation of DDA3 by ASB7 is cell cycle dependent. Accordingly, we examined the expression of DDA3 throughout the cell cycle. To this end, control or ASB7 knockdown HeLa cells were synchronized in the G1/S phase by a double thymidine block and then released into fresh media (Fig. 3, A and B). As reported previously, DDA3 was up-regulated and phosphorylated in the G2/M phase (Jang et al., 2008). Knockdown of ASB7 clearly increased the level of DDA3 from the G1/S through the G2/M phases but did not affect phosphorylation of DDA3 in the G2/M phase. The expression patterns of cyclin E and Aurora A, which are markers of the S and M phases, respectively, indicated that ASB7 depletion did not affect cell cycle progression from the G1/S to G2/M phase. Next, we examined the expression of DDA3 after the G2/M phase. For these experiments, cells were synchronized in the G2/M phase by nocodazole and released into fresh media (Fig. 3 C). In control cells, both the abundance of DDA3 and the levels of phosphorylated DDA3 suddenly decreased upon progression from the G2/M to the G1 phase. In contrast, ASB7 depletion slightly increased the expression of DDA3 from the G2/M phase through the G1 phase but did not alter cell cycle progression. We further examined the stability of DDA3 in each cell cycle phase in CHX chase experiments (Fig. 3, D and E). In the G1 phase, knockdown of ASB7 slightly stabilized DDA3, whereas silencing of ASB7 dramatically stabilized DDA3 in the S phase. Moreover, phosphorylated DDA3 was degraded in nocodazole-arrested (M phase) control cells, but not in ASB7 knockdown cells. In contrast, when cells were arrested in M phase using taxol, DDA3 was stable even in control cells (Fig. 3, D and E). Together, these observations demonstrate that DDA3 is degraded by ASB7 in a cell cycle–dependent manner.
Figure 3.
Cell cycle–dependent regulation of DDA3 by ASB7. (A) Stabilization of DDA3 in S phase by knockdown of ASB7. Two independent siRNAs (#3 and #5) targeting ASB7 were transfected into HeLa cells and synchronized in S phase. The cells were released and harvested at the indicated times. The lysates were subjected to Western blotting with antibodies against DDA3, ASB7, Aurora A, cyclin E, or Hsp90. (B) The intensities of DDA3 bands in A were normalized to those of the corresponding Hsp90 bands and plotted as a ratio of the normalized value of control cells at 0 h. (C) Knockdown of ASB7 had no effect on the stability of DDA3 in M phase. Two independent siRNAs (#3 and #5) targeting ASB7 were transfected into HeLa cells and synchronized in M phase. The cells were released and harvested at the indicated times. The lysates were subjected to Western blotting with antibodies against DDA3, ASB7, Aurora A, p27, or Hsp90. (D) ASB7-dependent destabilization of DDA3 in S phase. Two independent siRNAs (#3 and #5) targeting ASB7 were transfected into HeLa cells and synchronized in the G1, S, or M phase. Nocodazole or taxol was used to synchronize cells in M phase. The cells were then exposed to 50 µg/ml CHX for 1 or 2 h. The lysates were subjected to Western blotting with antibodies against DDA3 or Hsp90. (A, C, and D) Hsp90 is shown as a loading control. (E) The intensities of DDA3 bands in D were normalized to those of the corresponding Hsp90 bands and plotted as a ratio of the normalized value of control cells at 0 h. For all graphs, *, P < 0.05; **, P < 0.01. Data represent the mean ± SD of three independent experiments.
MT-dependent regulation of DDA3 degradation by ASB7
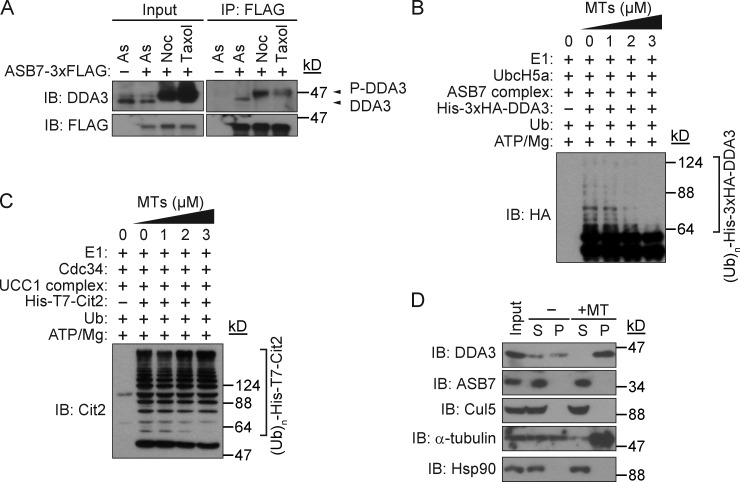
DDA3 is phosphorylated on Ser225 during mitosis (Jang et al., 2010). To investigate the effect of DDA3 phosphorylation on ASB7 binding and the stability of DDA3, we generated phospho-mimic (S225D) and nonphosphorylatable (S225A) variants of DDA3 and examined their stability and ability to interact with ASB7. Both mutants interacted with ASB7, and their degradation rates were similar to that of DDA3 (wild type [WT]; Fig. S1, A and B), indicating that the regulation of DDA3 by ASB7 is independent of Ser225 phosphorylation. MTs, the main constituents of the spindle, protect spindle assembly factors against degradation by the anaphase-promoting complex/cyclosome (APC/C; Song et al., 2014). Nocodazole is an MT-depolymerizing agent, whereas taxol is an MT-polymerizing agent; therefore, our observation that DDA3 was stable in cells treated with taxol but not with nocodazole (Fig. 3, D and E) suggested that polymerized bundled MTs might inhibit degradation of DDA3 by ASB7. To test this possibility, we compared the interaction between DDA3 and ASB7 in nocodazole- and taxol-treated conditions (Fig. 4 A). Although ASB7 interacted with both unphosphorylated and phosphorylated DDA3, the binding of ASB7 to phosphorylated DDA3 was much weaker in taxol-treated cells than in nocodazole-treated cells (Fig. 4 A).
Figure 4.
Prevention of polyubiquitination of DDA3 by MTs. (A) Interaction between ASB7 and nonphosphorylated or phosphorylated DDA3. HeLa cells stably expressing 3×FLAG-ASB7 or control cells were treated with 0.1 µg/ml nocodazole or 1 µM taxol for 16 h. Cells were further incubated in the presence of 10 µM MG132 for 4 h, and suspended cells in the culture medium were lysed, immunoprecipitated (IP) with anti-FLAG antibody, and immunoblotted (IB) with anti-DDA3 or anti-FLAG antibody. As, asynchronized. (B) Prevention of polyubiquitination of DDA3 by MTs in a dose-dependent manner. Different amounts of MTs were added to the in vitro polyubiquitination reactions. (C) MTs had no effect on polyubiquitination of Cit2. Different amounts of MTs were added to the in vitro polyubiquitination reactions. (D) ASB7 does not interact with MTs. Whole-cell lysates (input) of HeLa cells were prepared and incubated with or without taxol-stabilized MTs. Samples were centrifuged, and supernatant (S) and pellet (P) fractions were analyzed by immunoblotting with antibodies against DDA3, ASB7, Cul5, α-tubulin, or Hsp90. Hsp90 does not bind to MTs and was used as a negative control.
We then investigated whether MTs could negatively affect the polyubiquitination of DDA3 by ASB7 in vitro. Addition of MTs prevented polyubiquitination of DDA3 in a dose-dependent manner (Fig. 4 B) but had no effect on polyubiquitination of Cit2, a target of another ubiquitin ligase, UCC1 (Fig. 4 C; Nakatsukasa et al., 2015). We next examined whether ASB7 could bind to MTs by cosedimentation assay using HeLa whole-cell lysates and exogenously prepared taxol-stabilized MTs (Fig. 4 D). In the absence of exogenous MTs, DDA3 was found in both the supernatant and pellet fractions, but the amount of DDA3 in the pellet fraction was increased by the addition of MTs. In contrast, ASB7 and Cul5 were found only in the supernatant fraction under both conditions, indicating that ASB7 does not interact with MTs. These data support the idea that MT-bound DDA3 evades recognition by ASB7, resulting in its stabilization. Based on these findings, we conclude that degradation of DDA3 by ASB7 is regulated by MTs.
Regulation of MT polymerization by ASB7 and DDA3
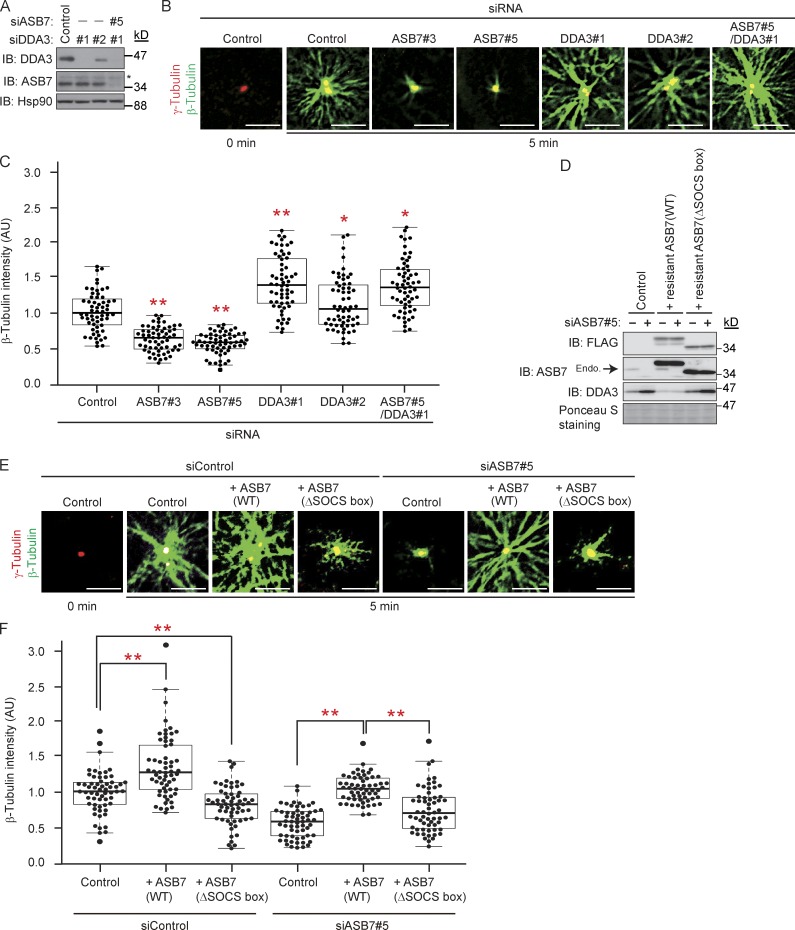
Because DDA3 facilitates Kif2a-mediated depolymerization of MTs (Jang et al., 2008), we speculated that MT polymerization might be regulated by ASB7-mediated DDA3 degradation. To test this hypothesis, we performed MT regrowth assays. On ice in the presence of nocodazole, MTs in HeLa cells were completely depolymerized, as reported previously (Sankaran et al., 2005). We then quantitated newly nucleated MTs by monitoring the fluorescence intensity of β-tubulin around centrosomes (Fig. 5, A–C; Sankaran et al., 2005). As reported previously, knockdown of DDA3 increased MT regrowth from centrosomes (Jang et al., 2008). In contrast, ASB7 knockdown clearly down-regulated MT regrowth, which reverted to the level in the DDA3 single knockdown when cells were also deprived of DDA3. We next questioned whether ASB7 complementation is able to rescue the phenotypes observed by ASB7 knockdown. We established cell lines stably expressing siRNA-resistant ASB7 (WT or ΔSOCS box; Fig. 5 D). As expected, accumulation of DDA3 by knockdown of ASB7 was rescued by complementation with ASB7(WT) but not ASB7(ΔSOCS box). Functionally, overexpression of ASB7(WT) in control cells increased MT regrowth possibly by down-regulating DDA3. In contrast, overexpression of ASB7(ΔSOCS box) reduced MT regrowth possibly by dominant-negatively stabilizing DDA3 (Fig. 2, H and I). Most importantly, reduced regrowth of MT by ASB7 knockdown (Fig. 5, B and C) was rescued by complementation with ASB7(WT) but not with ASB7(ΔSOCS box) (Fig. 5, E and F). These results indicate that ASB7 regulates MT polymerization through down-regulation of DDA3.
Figure 5.
Regulation of MT polymerization by ASB7 and DDA3. (A) Knockdown of ASB7 and/or DDA3. Two independent siRNAs (#1 and #2) targeting DDA3 or siRNA targeting ASB7 were transfected into HeLa cells. The lysates were subjected to Western blotting with antibodies against DDA3, ASB7, or Hsp90. Hsp90 is shown as a loading control. Asterisk denotes a nonspecific band. (B) MT regrowth assay using ASB7 and/or DDA3 knockdown HeLa cells. After incubation for 5 min to allow MT regrowth, cells were fixed and immunostained with anti–β-tubulin or anti–γ-tubulin. (C) Box plots of β-tubulin intensities after MT regrowth. About 20 cells from five randomly selected loci were examined in each experiment. The experiment was repeated three times independently, and a total of 60 cells were examined. The mean of normalized fluorescence intensity, expressed in arbitrary units (AU) of fluorescence, was calculated. The box plot shows the normalized expression values: bottom line outside the box, 10th percentile; bottom border of the box, 25th percentile; middle line within the box, median; upper border of the box, 75th percentile; top line outside the box, 90th percentile. Each dot indicates the intensity of an individual cell. (D) Complementation of ASB7 in ASB7 knockdown cells prevents DDA3 accumulation. siRNA targeting ASB7 or control was transfected into HeLa cells with stably expressing siRNA-resistant ASB7-3×FLAG (WT or ΔSOCS box). The lysates were subjected to Western blotting with antibodies against FLAG, ASB7, or DDA3. Ponceau S staining is shown as a loading control. Endogenous ASB7 is indicated by an arrow. (E) MT regrowth assay using ASB7 knockdown or ASB7-complemented HeLa cells performed as in B. Bars, 5 µm. (F) Box plots of β-tubulin intensities after MT regrowth calculated as in C. For all graphs, *, P < 0.05; **, P < 0.01.
Regulation of genome integrity by ASB7 and DDA3
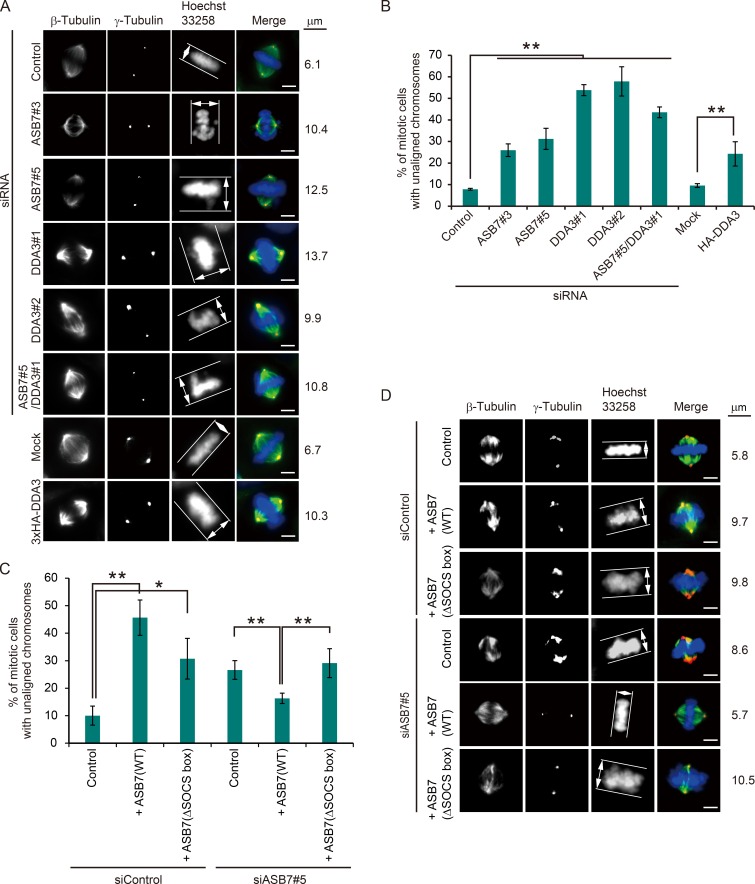
Knockdown of DDA3 also results in a high frequency of unaligned chromosomes (Jang et al., 2008). To elucidate the biological role of ASB7-mediated DDA3 degradation, we analyzed the effect of ASB7 and DDA3 on chromosome alignment at metaphase (Fig. 6, A and B). We followed the method previously established (Woo Seo et al., 2015). We determined the metaphase plate width by measuring the axis distance between the two lines at the edges of the DNA (Fig. 6 A, white lines with double-headed arrow). We considered chromosome misalignment to be when the distance was longer than 7 µm. As reported previously, when DDA3 was silenced, >50% of mitotic cells contained unaligned chromosomes. Unexpectedly, knockdown of ASB7 also significantly increased the proportion of cells with misaligned chromosomes (by 25–30%) relative to that in control cells. Importantly, DDA3 overexpression phenocopied the effect of ASB7 silencing, and, when DDA3 was cosilenced along with ASB7, the proportion of cells with misaligned chromosomes was elevated further. It also should be noted that there was a significant difference regarding chromosomal misalignment between siDDA3#1 and siASB7#5/siDDA3#1. Because particular ubiquitin ligases can target several substrates for proteasomal degradation (Zheng et al., 2016), it is possible that ASB7 targets another unidentified substrate for proteasomal degradation, and it might regulate chromosome alignment. In contrast, the increase of cells with chromosome misalignment by knockdown of ASB7 was partially rescued by complementation with ASB7(WT) but not with ASB7(ΔSOCS box) (Fig. 6, C and D). However, the complementation of ASB7(WT) did not completely rescue chromosome misalignment as in control cells, possibly because ASB7 was overexpressed and DDA3 was slightly more down-regulated than in control cells. Overexpression of ASB7 (WT or ΔSOCS box) in control cells increased cells with chromosome misalignment possibly by down-regulating and stabilizing DDA3, respectively (Fig. 2 F and Fig. 6, C and D). Collectively, these data indicate that the fine-tuned expression of DDA3 is crucial for normal chromosome alignment in the M phase.
Figure 6.
Regulation of chromosome alignment by ASB7 and DDA3. (A) Maximum projections from deconvolved z stacks of representative ASB7, DDA3, and/or Kif2a knockdown HeLa cells or HeLa cells stably overexpressing 3×HA-DDA3, as well as control cells, immunostained for β- or γ-tubulin. DNA was stained with Hoechst 33258. (B) Proportion of mitotic cells with unaligned chromosomes. More than 150 mitotic HeLa cells were counted, and percentages of cells with unaligned chromosomes are shown. (C) Proportion of mitotic cells with unaligned chromosomes. More than 150 mitotic HeLa cells were counted, and percentages of cells with unaligned chromosomes are shown. *, P < 0.05; **, P < 0.01. Data represent means ± SD. (D) Maximum projections from deconvolved z stacks of representative ASB7 knockdown or ASB7-complemented HeLa cells, immunostained for β- or γ-tubulin. DNA was stained with Hoechst 33258. Bars, 5 µm.
Discussion
The ASB family is composed of 18 members, ASB1–18. Several of these proteins interact with Cul5 and Rbx2 and act as components of ubiquitin ligase complexes (Kohroki et al., 2005). In this study, we demonstrated that ASB7 polyubiquitinates DDA3, resulting in its degradation by the proteasome.
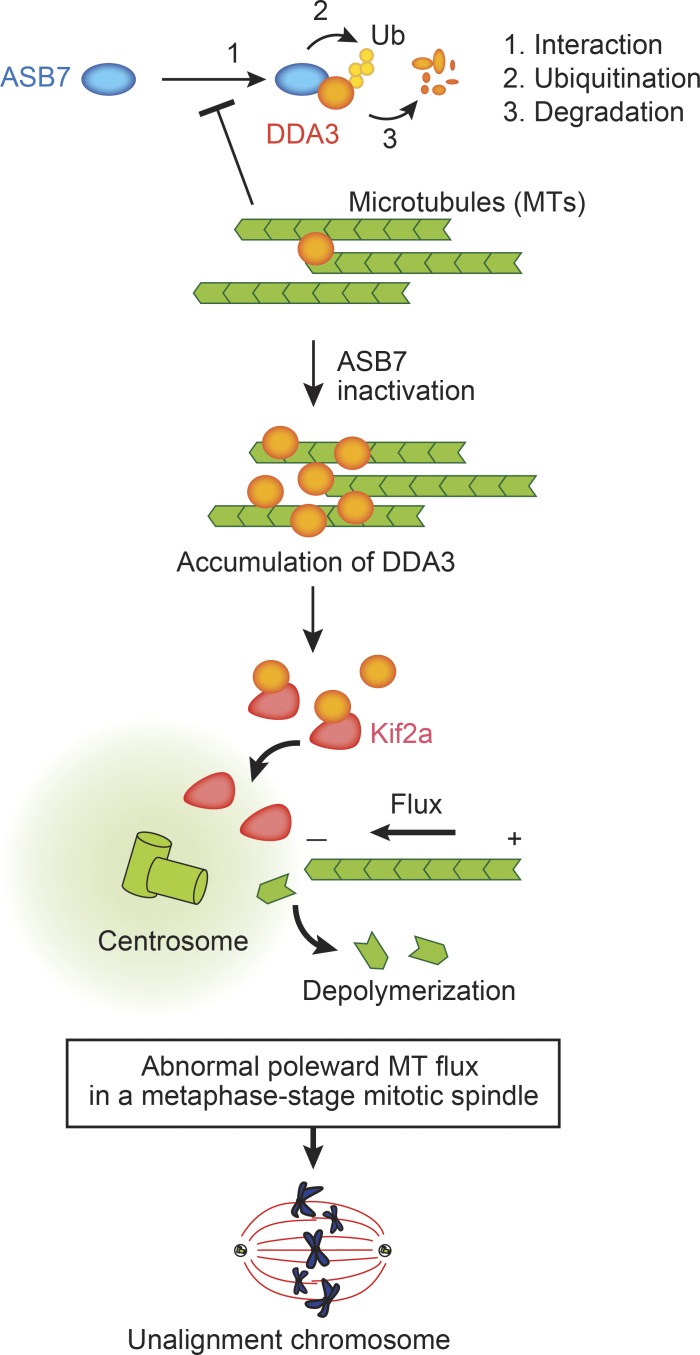
We propose that ASB7 is a novel regulator of cell division, as illustrated in Fig. 7. In this model, ASB7 interacts with and polyubiquitinates DDA3 and promotes proteasomal degradation to regulate the expression of DDA3 appropriately. In contrast, MTs prevent the interaction between ASB7 and DDA3. ASB7 destabilizes DDA3, which results in the stabilization of MTs. Then, it prevents the interaction between ASB7 and DDA3. Thus, there seems to be a feedback loop to appropriately regulate MT polymerization. Absence or inactivation of ASB7 increases DDA3 expression and depolymerization of MTs, in collaboration with Kif2a. The abnormal poleward MT flux in the M phase deranges the tension and orientation of spindle MTs, thereby preventing normal chromosome alignment.
Figure 7.
Model of regulation of cell division by ASB7. ASB7 targets DDA3 for polyubiquitination and proteasomal degradation. MTs prevent interaction between ASB7 and DDA3. Inactivation or loss of ASB7 results in accumulation of DDA3, which influences MT flux in collaboration with Kif2a. The abnormal poleward MT flux impedes normal chromosome alignment in metaphase.
DDA3 was up-regulated and stabilized in the M phase, and the presence of MTs prevented ASB7-mediated polyubiquitination of DDA3 in vitro (Figs. 3 and 4). Therefore, MTs might regulate either the ubiquitin ligase activity of ASB7 or the interaction between DDA3 and ASB7. Because the MT-stabilizing agent taxol, but not the MT-depolymerizing agent nocodazole, stabilized DDA3 and prevented the interaction between DDA3 and ASB7 (Fig. 3, D and E; and Fig. 4 A), we conclude that MTs inhibit the interaction between DDA3 and ASB7.
The APC/C induces degradation of proteins that stabilize the mitotic spindle, and loss or accumulation of such spindle assembly factors can result in aneuploidy and cancer (Peters, 2006). Some of these factors, such as HURP, NuSAP, or Tpx2, which associate with and stabilize MTs (Wittmann et al., 2000; Gruss et al., 2001; Koffa et al., 2006; Ribbeck et al., 2006; Silljé et al., 2006; Wong and Fang, 2006), are protected from APC/C-dependent degradation by the presence of MTs (Song et al., 2014). Thus, temporal and spatial regulation of spindle assembly factors by the APC/C is important for accurate cell division (Song et al., 2014). Together, these observations demonstrate that MT-dependent stabilization or degradation of cell cycle–related proteins is critical for accurate cell division. MT-dependent stabilization of DDA3 recruits Kif2a to the mitotic spindle and spindle poles, decreasing the steady-state levels of spindle MTs by increasing the rates of mitotic spindle turnover and MT depolymerization (Jang et al., 2008). Thus, temporal and spatial evasion of ASB7-dependent degradation by DDA3 is crucial for normal cell division.
Knockdown of DDA3 increased the proportion of cells with unaligned chromosomes, as reported previously (Fig. 6; Jang et al., 2008; Jang and Fang, 2011), as did either knockdown of ASB7 or overexpression of DDA3 (Fig. 6). Thus, tight regulation of the DDA3 level is essential for accurate chromosome alignment. A candidate partner of DDA3, centromere-associated protein E (CENP-E), accumulates at kinetochores in unaligned chromosomes in mitotic cells depleted of DDA3, suggesting that DDA3 modulates the function of CENP-E during chromosome alignment (Jang and Fang, 2011). CENP-E is a dimeric kinesin of the kinesin-7 family and a plus end–directed kinetochore motor; depletion of CENP-E results in chromosome misalignment with a few unattached chromosomes around the spindle pole (Putkey et al., 2002). Furthermore, CENP-E is responsible for alignment of chromosomes at the metaphase plate (Cai et al., 2009) and is also involved in the gliding of mono-oriented kinetochores alongside adjacent K fibers to their proper metaphase positions (Kapoor et al., 2006).
Our observation that ASB7 influences genome integrity through degradation of DDA3 suggests that the ASB7–DDA3 axis is dysregulated in tumors. A search of an integrated cancer microarray database provided by Oncomine revealed that DDA3 expression is significantly up-regulated in human colorectal cancers, as confirmed by two independent statistical analyses (TCGA database, https://tcga-data.nci.nih.gov/tcga/; Gaedcke et al., 2010). In contrast, ASB7 expression is down-regulated in these tumors (TCGA database; Kaiser et al., 2007). Thus, ASB7 may prevent colorectal cancer by regulating the expression of DDA3. These observations suggest that drugs preventing the activity or down-regulating the expression of DDA3 represent a novel strategy for cancer therapy.
Materials and methods
Plasmid construction
cDNAs encoding human DDA3/PRSC1 (GenBank/EBI accession no. NM_032636), ASB7 (GenBank/EBI accession no. NM_198243), and Kif2a (GenBank/EBI accession no. NM_001098511) were amplified by RT-PCR from HEK293T cells and introduced into pcDNA3, pCI-neo, or pMX-puro.
Antibodies
Antibodies against FLAG (1 µg/ml; M2, F1804; Sigma-Aldrich), HA (1 µg/ml; 12CA5; Sigma-Aldrich), His6 (1 µg/L; MAB050; R&D Systems), DDA3 (0.1 µg/ml; goat, sc-160272; Santa Cruz Biotechnology, Inc.), DDA3 for IP (rabbit, BS7598; bioWORLD), Cul5 (1 µg/ml; mouse, sc-373822; Santa Cruz Biotechnology, Inc.), Cul2 (1 µg/ml; mouse, sc-166506; Santa Cruz Biotechnology, Inc.), Aurora A (0.4 µg/ml; mouse, 610939; BD), cyclin E (1 µg/ml; rabbit, sc-198; Santa Cruz Biotechnology, Inc.), p27 (0.5 µg/ml; mouse, 610242; BD), cyclin B1 (1 µg/ml; mouse, sc-245; Santa Cruz Biotechnology, Inc.), and Kif2a (1:15,000; rabbit, NB500-180; Novus Biologicals) were obtained from the indicated suppliers and used at the indicated dilutions. Rabbit anti-ASB7 antibody was generated using recombinant ASB7 purified from Escherichia coli on nickel–nitrilotriacetic acid agarose (Invitrogen). Anti-ASB7 antibody was further purified by ASB7 affinity column.
Reagents
CHX was purchased from Sigma-Aldrich, protein A–Sepharose was purchased from GE Healthcare, and MG132 was purchased from Peptide Institute Inc. Thymidine and taxol were purchased from Wako Pure Chemical Industries. Nocodazole was purchased from EMD Millipore. Hydroxyurea was purchased from Tokyo Chemical Industry.
Cell culture and transfection
HEK293T and HeLa cells were cultured as described previously (Okumura et al., 2007); HEK293T cells were transfected using polyethyleneimine. Retrovirus infection was performed as described previously (Okumura et al., 2007).
IP and IB analyses
IP and IB analyses were performed as described previously (Okumura et al., 2007).
Immunofluorescence staining
HeLa cells were fixed in methanol for 5 min at −20°C and permeabilized in 0.5% Triton X-100. The cells were then incubated overnight at 4°C with antibody against β-tubulin (1 µg/ml; mouse, E1C601-2; Eno Gene) or γ-tubulin (1:2,000; rabbit, T5192; Sigma-Aldrich) in PBS containing 0.1% BSA and 0.1% Triton X-100. Cells were washed three times with PBS, followed by incubation for 1 h at room temperature in the dark with Alexa Fluor 594 goat anti–rabbit and Alexa Fluor 488 goat anti–mouse antibodies (both at a 1:3,000 dilution; Invitrogen) in PBS containing 0.1% BSA and 0.1% Triton X-100. The cells were further incubated with 0.1 µg/ml Hoechst 33258 in PBS for 1 min, followed by extensive washing with PBS, and then photographed with a charge-coupled device camera (Axio Observer Z1; ZEISS).
Isolation and identification of ASB7-interacting proteins
The substrates of ASB7 ubiquitin ligase were identified as described previously (Kamura et al., 2004).
Cell culture synchronization
Synchronization of HeLa cells was performed as described previously (Uehara et al., 2013). In brief, siRNA was transfected into HeLa cells, which were cultured for 1 d. For synchronization in S phase, cells were incubated in the presence of 2.5 µM thymidine for 20 h, washed three times with DMEM, released for 8 h, and treated with 2.5 µM thymidine again for 14 h. For synchronization of siRNA-transfected cells in the M phase, cells were incubated in the presence of 2.5 µM thymidine for 20 h, washed three times with DMEM, released for 3 h, and treated with 0.1 µg/ml nocodazole or 1 µM taxol for 16 h. For synchronization of siRNA-transfected cells in the G1 phase, M phase–arrested cells were released for 4 h. For CHX chase experiments, siRNA-transfected cells were cultured for 2 d and synchronized in S phase by incubation in the presence of 1 mM hydroxyurea for 18 h. Mitotically arrested cells were shaken off the culture dishes and harvested.
In vitro ubiquitination
In vitro ubiquitination was performed as described previously with slight modifications (Kamura et al., 2004). Recombinant ASB7 complex was purified as described previously (Kamura et al., 1999). Recombinant His6-3×HA-DDA3 was immunopurified from the lysates of transiently transduced HEK293T cells using anti-HA antibody and protein A–Sepharose. In vitro ubiquitination assays in the presence of MTs were performed as reported previously (Song et al., 2014).
MT cosedimentation assay
MT cosedimentation assay was performed as described previously with modifications (Hergovich et al., 2003). Cell lysates were prepared by centrifugation for 30 min at 20,000 g at 4°C. 2 mg/ml tubulin (Sigma-Aldrich) was diluted in buffer A (40 mM Tris-HCl, 1 mM EGTA, 0.1 mM EDTA, 0.5 mM magnesium chloride, 100 µg/ml sucrose, 1 mM DTT, and 0.1 mM GTP, pH 6.8) and polymerized with 50 µM taxol in the presence of 2.5 mM GTP for 30 min at 37°C. 10 µg of repolymerized MTs per assay was pelleted by centrifugation for 20 min at 20,000 g at 25°C. The MTs were resuspended in 20 µl of precleared cell supernatant containing 50 µM taxol and incubated for 30 min at 37°C. Samples were then centrifuged for 20 min at 20,000 g at 25°C, and supernatants and pellets were analyzed by Western blotting.
MT regrowth assay
MT regrowth assay was performed as described previously (Sankaran et al., 2005). In brief, cells were treated with 25 µM nocodazole for 40 min on ice, washed with cold PBS, and incubated at 37°C for 5 min. Cells were fixed in cold methanol for immunostaining.
Knockdown
Knockdown of Cul2 and Cul5 was performed as described previously (Kamura et al., 2004). siRNAs targeting ASB7, DDA3, and Kif2a were transfected into HeLa cells using Lipofectamine RNAiMAX (Thermo Fisher Scientific). The target sequences for ASB7#3 and ASB7#5 were 5′-GAGGCTTCACGGCTCTTCACT-3′ and 5′-GAGAGAGGTCAAGCTGTGTGA-3′, respectively; those for DDA3#1 and DDA3#2 were 5′-AAGCAAGACTTCAGTAGCATT-3′ and 5′-CCACCGAAGTGACCCAAATTT-3′, respectively; and those for Kif2a#1 and Kif2a#2 were 5′-GGCAAAGAGATTGACCTGG-3′ and 5′-CCCTCCTTCAAGAGATAATTT-3′, respectively. siRNA-resistant ASB7 was constructed by introducing silent mutation in the siRNA target sequence for ASB7#5; 5′-TCA-3′ was substituted for 5′-AGC-3′.
Statistical analysis
Student’s two-tailed t test was used to determine the statistical significance of differences. All data are expressed as means ± SD. A p-value of <0.05 was considered statistically significant. The variance was similar between the groups compared.
Online supplemental material
Fig. S1 shows that phosphorylation of DDA3 has no effect on the interaction with ASB7 or the stabilization of DDA3.
Supplementary Material
Acknowledgments
This work was supported by Japan Society for the Promotion of Science KAKENHI grants 25291023 (to F. Okumura and T. Kamura), 25860043 (to F. Okumura), 24112006 and 15K14474 (to T. Kamura), 25870312 and 15K18503 (to K. Nakatsukasa), and 13J40160 (to A. Joo-Okumura), the Uehara Memorial Foundation (to F. Okumura), and the Inamori Foundation (to F. Okumura).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- APC/C
- anaphase-promoting complex/cyclosome
- CHX
- cycloheximide
- IB
- immunoblot
- IP
- immunoprecipitation
- MT
- microtubule
- SOCS
- suppressor of cytokine signaling
- WT
- wild type
References
- Cai S., O’Connell C.B., Khodjakov A., and Walczak C.E.. 2009. Chromosome congression in the absence of kinetochore fibres. Nat. Cell Biol. 11:832–838. 10.1038/ncb1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagleson G., Pfister K., Knowlton A.L., Skoglund P., Keller R., and Stukenberg P.T.. 2015. Kif2a depletion generates chromosome segregation and pole coalescence defects in animal caps and inhibits gastrulation of the Xenopus embryo. Mol. Biol. Cell. 26:924–937. 10.1091/mbc.E13-12-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems-McClung S.C., and Walczak C.E.. 2010. Kinesin-13s in mitosis: Key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 21:276–282. 10.1016/j.semcdb.2010.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedcke J., Grade M., Jung K., Camps J., Jo P., Emons G., Gehoff A., Sax U., Schirmer M., Becker H., et al. 2010. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 49:1024–1034. 10.1002/gcc.20811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., and Compton D.A.. 2004. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 166:473–478. 10.1083/jcb.200404012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., and Mattaj I.W.. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin αon TPX2 activity. Cell. 104:83–93. 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Helenius J., Brouhard G., Kalaidzidis Y., Diez S., and Howard J.. 2006. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 441:115–119. 10.1038/nature04736 [DOI] [PubMed] [Google Scholar]
- Hergovich A., Lisztwan J., Barry R., Ballschmieter P., and Krek W.. 2003. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell Biol. 5:64–70. 10.1038/ncb899 [DOI] [PubMed] [Google Scholar]
- Hilton D.J., Richardson R.T., Alexander W.S., Viney E.M., Willson T.A., Sprigg N.S., Starr R., Nicholson S.E., Metcalf D., and Nicola N.A.. 1998. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc. Natl. Acad. Sci. USA. 95:114–119. 10.1073/pnas.95.1.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.Y., and Fang G.. 2011. DDA3 associates with MCAK and controls chromosome congression. Biochem. Biophys. Res. Commun. 407:610–614. 10.1016/j.bbrc.2011.03.081 [DOI] [PubMed] [Google Scholar]
- Jang C.Y., Wong J., Coppinger J.A., Seki A., Yates J.R. III, and Fang G.. 2008. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J. Cell Biol. 181:255–267. 10.1083/jcb.200711032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.Y., Coppinger J.A., Yates J.R. III, and Fang G.. 2010. Phospho-regulation of DDA3 function in mitosis. Biochem. Biophys. Res. Commun. 393:259–263. 10.1016/j.bbrc.2010.01.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.Y., Coppinger J.A., Yates J.R. III, and Fang G.. 2011. Mitotic kinases regulate MT-polymerizing/MT-bundling activity of DDA3. Biochem. Biophys. Res. Commun. 408:174–179. 10.1016/j.bbrc.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S., Park Y.K., Franklin J.L., Halberg R.B., Yu M., Jessen W.J., Freudenberg J., Chen X., Haigis K., Jegga A.G., et al. 2007. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8:R131 10.1186/gb-2007-8-7-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T., Koepp D.M., Conrad M.N., Skowyra D., Moreland R.J., Iliopoulos O., Lane W.S., Kaelin W.G. Jr., Elledge S.J., Conaway R.C., et al. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 284:657–661. 10.1126/science.284.5414.657 [DOI] [PubMed] [Google Scholar]
- Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R.C., Conaway J.W., and Nakayama K.I.. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055–3065. 10.1101/gad.1252404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile B.T., Schulman B.A., Alexander W.S., Nicola N.A., Martin H.M., and Hilton D.J.. 2002. The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 27:235–241. 10.1016/S0968-0004(02)02085-6 [DOI] [PubMed] [Google Scholar]
- Kline-Smith S.L., and Walczak C.E.. 2004. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 15:317–327. 10.1016/j.molcel.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Koffa M.D., Casanova C.M., Santarella R., Köcher T., Wilm M., and Mattaj I.W.. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16:743–754. 10.1016/j.cub.2006.03.056 [DOI] [PubMed] [Google Scholar]
- Kohroki J., Nishiyama T., Nakamura T., and Masuho Y.. 2005. ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett. 579:6796–6802. 10.1016/j.febslet.2005.11.016 [DOI] [PubMed] [Google Scholar]
- Liu J., Shaik S., Dai X., Wu Q., Zhou X., Wang Z., and Wei W.. 2015. Targeting the ubiquitin pathway for cancer treatment. Biochim. Biophys. Acta. 1855:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrour N., Redwine W.B., Florens L., Swanson S.K., Martin-Brown S., Bradford W.D., Staehling-Hampton K., Washburn M.P., Conaway R.C., and Conaway J.W.. 2008. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J. Biol. Chem. 283:8005–8013. 10.1074/jbc.M706987200 [DOI] [PubMed] [Google Scholar]
- Meunier S., and Vernos I.. 2016. Acentrosomal microtubule assembly in mitosis: the where, when, and how. Trends Cell Biol. 26:80–87. 10.1016/j.tcb.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Musacchio A. 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25:R1002–R1018. (published erratum appears in Curr. Biol. 2015. 25:3017) 10.1016/j.cub.2015.08.051 [DOI] [PubMed] [Google Scholar]
- Musacchio A., and Hardwick K.G.. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3:731–741. 10.1038/nrm929 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Nishimura T., Byrne S.D., Okamoto M., Takahashi-Nakaguchi A., Chibana H., Okumura F., and Kamura T.. 2015. The ubiquitin ligase SCF(Ucc1) acts as a metabolic switch for the glyoxylate cycle. Mol. Cell. 59:22–34. 10.1016/j.molcel.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Okumura F., Zou W., and Zhang D.E.. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21:255–260. 10.1101/gad.1521607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F., Matsuzaki M., Nakatsukasa K., and Kamura T.. 2012. The role of Elongin BC-containing ubiquitin ligases. Front. Oncol. 2 10.3389/fonc.2012.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656. 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Putkey F.R., Cramer T., Morphew M.K., Silk A.D., Johnson R.S., McIntosh J.R., and Cleveland D.W.. 2002. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell. 3:351–365. 10.1016/S1534-5807(02)00255-1 [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Groen A.C., Santarella R., Bohnsack M.T., Raemaekers T., Köcher T., Gentzel M., Görlich D., Wilm M., Carmeliet G., et al. 2006. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell. 17:2646–2660. 10.1091/mbc.E05-12-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran S., Starita L.M., Groen A.C., Ko M.J., and Parvin J.D.. 2005. Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol. Cell. Biol. 25:8656–8668. 10.1128/MCB.25.19.8656-8668.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé H.H.W., Nagel S., Körner R., and Nigg E.A.. 2006. HURP is a Ran-importin β-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16:731–742. 10.1016/j.cub.2006.02.070 [DOI] [PubMed] [Google Scholar]
- Skaar J.R., Pagan J.K., and Pagano M.. 2014. SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 13:889–903. 10.1038/nrd4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Craney A., and Rape M.. 2014. Microtubule-dependent regulation of mitotic protein degradation. Mol. Cell. 53:179–192. 10.1016/j.molcel.2013.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Tsukada Y., Kamasaki T., Poser I., Yoda K., Gerlich D.W., and Goshima G.. 2013. Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J. Cell Biol. 202:623–636. 10.1083/jcb.201302123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Gayek S., and Ohi R.. 2013. Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 29:417–441. 10.1146/annurev-cellbio-101512-122345 [DOI] [PubMed] [Google Scholar]
- Wittmann T., Wilm M., Karsenti E., and Vernos I.. 2000. TPX2, a novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149:1405–1418. 10.1083/jcb.149.7.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., and Fang G.. 2006. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173:879–891. 10.1083/jcb.200511132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo Seo D., Yeop You S., Chung W.J., Cho D.H., Kim J.S., and Su Oh J.. 2015. Zwint-1 is required for spindle assembly checkpoint function and kinetochore-microtubule attachment during oocyte meiosis. Sci. Rep. 5 10.1038/srep15431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Zhou Q., Wang Z., and Wei W.. 2016. Recent advances in SCF ubiquitin ligase complex: Clinical implications. Biochim. Biophys. Acta. 1866:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.