A diffusion barrier impeding membrane molecule motion between the axon and the somatodendritic compartment develops as neurons mature and the axon initial segment (AIS) is enriched in specific molecules. Albrecht et al. analyze the mobility of lipid-anchored molecules in the AIS using single-particle tracking time course experiments and propose a new mechanistic model for the AIS diffusion barrier.
Abstract
The axon initial segment (AIS) is enriched in specific adaptor, cytoskeletal, and transmembrane molecules. During AIS establishment, a membrane diffusion barrier is formed between the axonal and somatodendritic domains. Recently, an axonal periodic pattern of actin, spectrin, and ankyrin forming 190-nm-spaced, ring-like structures has been discovered. However, whether this structure is related to the diffusion barrier function is unclear. Here, we performed single-particle tracking time-course experiments on hippocampal neurons during AIS development. We analyzed the mobility of lipid-anchored molecules by high-speed single-particle tracking and correlated positions of membrane molecules with the nanoscopic organization of the AIS cytoskeleton. We observe a strong reduction in mobility early in AIS development. Membrane protein motion in the AIS plasma membrane is confined to a repetitive pattern of ∼190-nm-spaced segments along the AIS axis as early as day in vitro 4, and this pattern alternates with actin rings. Mathematical modeling shows that diffusion barriers between the segments significantly reduce lateral diffusion along the axon.
Introduction
Neurons are highly polarized cells that bear a complex dendritic arbor and usually only a single, elongated axon emanating from the cell body. The somatodendritic domain receives input from thousands of synapses, and if a threshold is met, an action potential is generated by a large density of ion channels in the first 50–150 µm of the axon, the so-called axon initial segment (AIS; Rasband, 2010). The AIS contains a specific complement of molecules with ankyrin G (AnkG), βIV-spectrin, neurofascin, and voltage-gated ion channels that is assembled during neuronal development (Galiano et al., 2012). The motion of membrane molecules in the AIS decreases in correlation with the accumulation of AnkG, an AIS-specific cytoskeletal adaptor molecule, during neuronal development until a diffusion barrier that impedes motion of axonal membrane molecules into the somatodendritic domain and vice versa is established (Winckler et al., 1999; Nakada et al., 2003; Boiko et al., 2007). This barrier is believed to be a result of the anchorage of transmembrane molecules to the submembrane cytoskeleton meshwork (Nakada et al., 2003). These tethered proteins then act as obstacles to the lateral motion of membrane molecules, which as a result may be transiently confined within the compartments of the meshwork, a general concept known as the picket fence model (Fujiwara et al., 2002).
Recent investigations have found a periodic cytoskeletal meshwork in axons, including the AIS (Xu et al., 2012; Zhong et al., 2014; D’Este et al., 2015; Leterrier et al., 2015), that is ubiquitously found in many types of neurons (D’Este et al., 2016; He et al., 2016) and is conserved from Caenorhabditis elegans to Homo sapiens (He et al., 2016). On the other hand, electron microscopy studies have demonstrated that a dense coat of ion channels and adhesion molecules is assembled in the AIS membrane during development (Jones et al., 2014). However, whether specifically the structure of the periodic cytoskeletal meshwork as in the picket fence model or more generally the molecular crowding by a high density of membrane molecules along the AIS provides the basis for the membrane diffusion barrier remains unclear (Rasband, 2013).
Here, we perform high-density single-particle tracking (SPT) of glycosylphosphatidylinositol (GPI)-anchored molecules in the AIS over the time course of neuronal development and find that as early as day in vitro 5 (DIV5), membrane protein motion is reduced. Using high-speed SPT, we show that the motion of GPI-anchored molecules is confined to a repetitive array of small areas spaced at ∼190 nm. Correlative superresolution microscopy of the cytoskeleton demonstrated that the confinement areas and the equidistant 190-nm-spaced actin rings reported by Xu et al. (2012) are mutually exclusive. A Markov model demonstrated that the periodic confinement significantly increases transition time for individual molecules along the AIS.
Our results suggest a new mechanism for the establishment of a membrane diffusion barrier in the AIS and demonstrate that the motion of GPI-anchored molecules in the plasma membrane can be controlled by the organization of the submembrane cytoskeleton.
Results and discussion
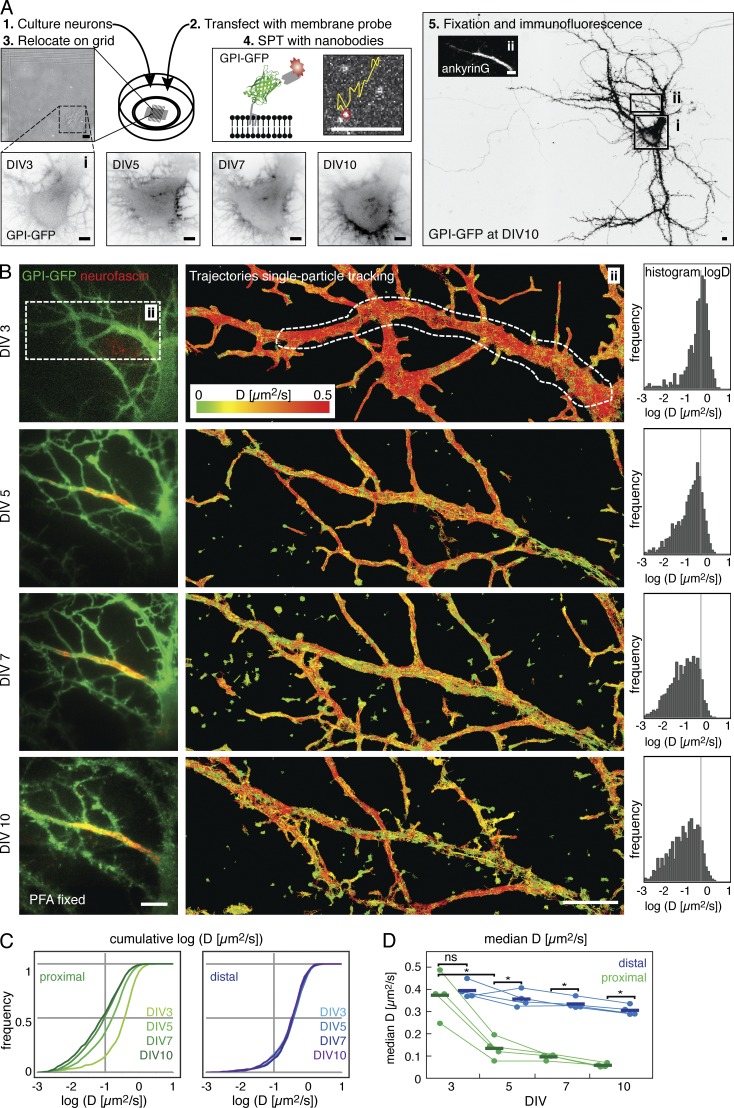
To determine at what time point during neuronal development the AIS diffusion barrier is established, we performed time-course experiments of high-density SPT on primary hippocampal neurons. To do so, we transfected neurons at DIV2 with GPI-GFP and then performed SPT via Atto647N-labeled anti-GFP nanobodies by the uPAINT (universal point accumulation for imaging in nanoscale topography) method (Giannone et al., 2010) on the presumed AIS and a segment of the distal axon at DIV3 (Fig. 1 A). Because we plated the neurons on gridded coverslips and performed experiments in the presence of Cy3B-labeled antineurofascin antibodies, we could identify the same neuron and AIS in subsequent SPT experiments at DIV5, 7, and 10. After the last experiment, we fixed the cells and performed immunofluorescence staining against AnkG to confirm the location of the AIS. When we analyzed the hundreds of obtained trajectories of single anti-GFP nanobodies according to the diffusion coefficient D, we found a decrease in the median instantaneous diffusion coefficient of GPI-GFP between DIV3 and DIV10 in the AIS (Fig. 1 B). However, we did not observe a decrease in the median diffusion coefficient in the distal axon (Fig. 1, C and D) of the same neuron. Strikingly, we observed a significant reduction in the median diffusion coefficient of GPI-GFP between DIV3 and DIV5 (Fig. 1 D) in the AIS of all tested neurons (n = 4 neurons, n = 3,390 trajectories for DIV3, n = 11,290 for DIV5, P = 0.003, paired t test of median D). This reduction was spatially correlated with the appearance of neurofascin staining (Fig. 1 B and Fig. S1).
Figure 1.
The diffusion of membrane molecules is restricted by DIV5 in hippocampal neurons. (A) Experimental design for the developmental time course in cultured primary hippocampal neurons. (1) Neurons were maintained in gridded glass-bottom Petri dishes and (2) transfected with the membrane-probe GPI-GFP. (3) Individual neurons were relocated on DIV3, 5, 7, and 10, respectively, based on their position on the grid. (4) Single-particle tracking (SPT) experiments were conducted by sparsely labeling transfected neurons with anti-GFP nanobodies conjugated to fluorescent organic dyes. (5) After SPT, neurons were fixed and immunostained for cytoskeletal and or AIS marker. The soma (i) and the proximal axon with the AIS (ii), identified by AnkG staining (inset micrograph), are outlined. (B) Results from the developmental SPT time course on a typical neuron. The AIS was identified by live immunolabeling of neurofascin. (left box) The proximal axon was neurofascin negative on DIV3 and neurofascin positive from DIV5 onwards. (middle box) Plot of trajectories of mobile particles tracked in SPT experiments color-coded for instantaneous diffusion coefficients D. (right box) Histogram of D on the proximal axon where the AIS assembles (white dashed line). Number of trajectories: DIV3, n = 787; DIV5, n = 1,815; DIV7, n = 2005; and DIV10, n = 1,067. (C) Plots of the cumulative D in the AIS (left, shades of green) and a portion of the distal axon (right, shades of blue) of the neuron shown in B. (D) Graph of the median D for all neurons (n = 4) between DIV3 and DIV10. Statistical analysis of median D by paired t test; *, P < 0.01; ns, not significant. Bars, 5 µm.
Electron microscopy of the AIS of developing neurons does not show a significant accumulation of electron-dense material such as ion channels at the AIS at this early time point (Jones et al., 2014). On the other hand, the periodic submembrane spectrin–actin meshwork becomes detectable at this stage in neuronal development at the AIS (Zhong et al., 2014; D’Este et al., 2015) and individual ion channels start to become anchored by AnkG (Brachet et al., 2010). Our results thus suggested that the periodic submembrane spectrin–actin meshwork may be responsible for the observed reduction in diffusivity of GPI-GFP between DIV3 and DIV5 and not general crowding of the plasma membrane.
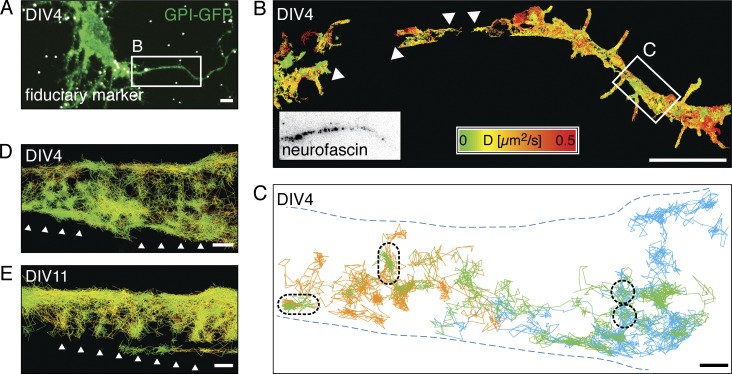
We decided to investigate the diffusivity of membrane molecules around DIV4 more closely, as this seemed to be a transition point. We reasoned that if the periodic submembrane spectrin–actin meshwork was the cause of the decrease in diffusivity, a higher spatiotemporal resolution would be required to detect such a ∼190-nm-spaced periodic pattern in the diffusion of GPI-GFP. We estimated that motion blurring of molecules with high diffusivity such as GPI-GFP made it impossible to detect sharp compartment borders with our single-molecule detection-based method (Frost et al., 2012) and that SPT with millisecond temporal and nanometer spatial resolution was required to move forward. We thus decided to use streptavidin-coated quantum dots (QDs), which are very bright and extremely photostable, and to couple them via biotinylated anti-GFP nanobodies to GPI-GFP for high-speed SPT. We reduced motion blurring by pulsing the excitation laser for 2 ms at 5- to 10-ms frame rates on our EMCCD camera and used fiduciary markers to correct for drift (Fig. 2 A). When we then performed SPT measurements with a high density of membrane-bound quantum dots to achieve high sampling of the AIS membrane and recorded tens of thousands of frames over several minutes, we generated trajectories of up to 2,200 steps with a median localization accuracy of 9 nm (Fig. 2 B).
Figure 2.
High-speed SPT shows that diffusing GPI-GFP molecules revisit small areas in the AIS. (A) Merged fluorescence micrograph of a DIV4 hippocampal neuron expressing GPI-GFP (green) after PFA fixation with fiduciary markers (white) for drift correction and image correlation. (B) Plot of SPT trajectories of anti-GFP nanobody-coupled quantum dots (n = 3,375) on GPI-GFP. Trajectories are color-coded according to the instantaneous diffusion coefficient. The AIS was identified by live immunolabeling of neurofascin (box). Not all areas were covered homogeneously by trajectories (arrowheads). (C) Plot of selected long trajectories (>500 steps, orange, green, and blue) along a segment with reduced lateral mobility (white box in B). The trajectories cover the axon inhomogeneously and show local zones, which are revisited by individual QDs (black dashed lines). (D) Same plot of area as in C, with all trajectories >20 steps plotted (n = 231) and color-coded according to the instantaneous diffusion coefficient as in B. Arrowheads emphasize a pattern emerging from the distribution of the trajectories. (E) Plot of trajectories of anti-GFP nanobody-coupled QDs on the AIS of a DIV11 neuron expressing GPI-GFP (n = 501). Trajectories are color coded as in B. Arrowheads emphasize an emergent pattern similar to that in D. Bars: (A and B) 5 µm; (C–E) 200 nm.
We then analyzed subsets of the longest trajectories and found that specific areas were often revisited by individual QDs, suggesting that they preferred certain domains in the plasma membrane (Fig. 2 C). Indeed, when we plotted all measured trajectories, they seemed to form a striped pattern perpendicular to the direction of propagation of the axon both early in the developmental time course (DIV4; Fig. 2 D) and at later stages (DIV11; Fig. 2 E).
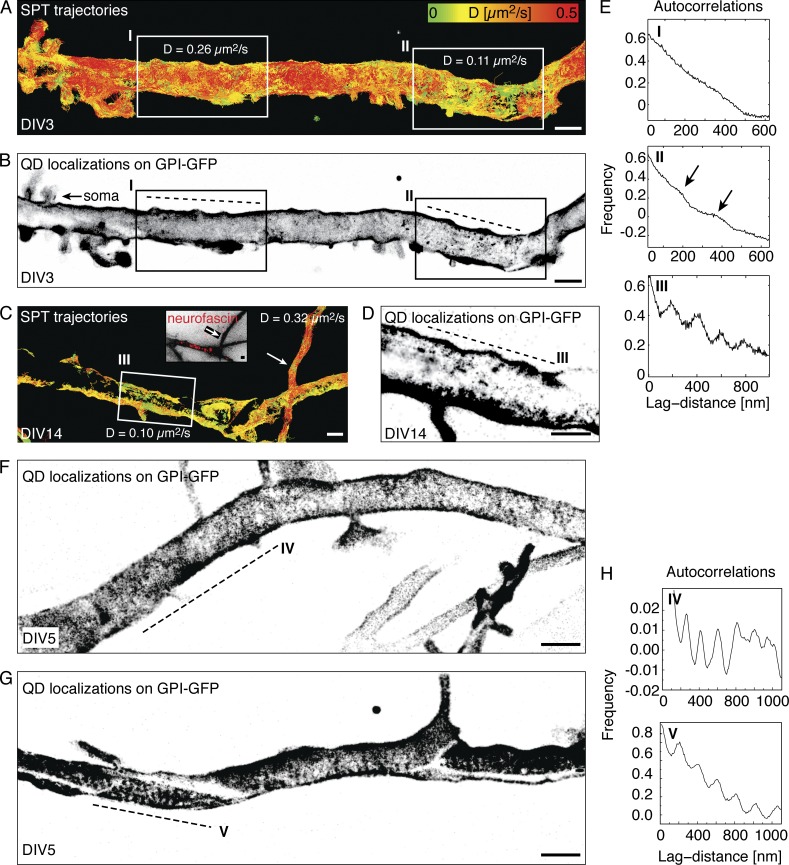
To analyze this pattern, we next reconstructed images from the cumulative localizations of all detected QDs in all captured frames and found an alternating pattern perpendicular to the axon of zones with a lower and higher density of localizations (Fig. 3). The periodic submembrane spectrin–actin meshwork becomes established in the proximal axon as early as DIV2, before AIS-specific markers accumulate (Zhong et al., 2014; D’Este et al., 2015). Likewise, the earliest time point at which a periodic pattern of GPI-GFP localizations emerged in an area showing low instantaneous diffusion coefficient trajectories was DIV3 (Fig. 3, A and B, inset II). We performed many measurements during the developmental time course and on later time points and consistently found periodic patterns on the proximal axon where the AIS is located (Fig. 3, C and D, inset III), albeit often only along segments that reached a sufficiently high localization density (Fig. 3 D, Fig. 4, and Fig. S2). When we calculated the spatial autocorrelation along the direction of propagation of the axon, we found a clear periodicity of ∼190 nm, consistent with the spacing of the AIS cytoskeleton (Fig. 3, E and H). These stripes were stable in their localization and could be found in the same place when measured again after 10 min or more (unpublished data). We detected the same periodicity for a model transmembrane protein with one fluorescent protein each on the cytosolic and exoplasmic side of a single transmembrane domain (Fig. S3, A and B; Keller et al., 2001; Albrecht et al., 2015). However, we could not detect a periodic pattern in SPT localizations in the few portions of the distal axon we imaged (Fig. S3, C and D).
Figure 3.
Accumulated positions of GPI-GFP molecules from SPT show a periodic pattern of ∼190 nm along the AIS that emerges as early as DIV3 during development. (A) Plot of SPT trajectories on the proximal axon of a DIV3 hippocampal neuron expressing GPI-GFP color-coded according to the instantaneous diffusion coefficient (D). Two areas are emphasized: one with a high (inset I) and one with a lower mean D (inset II). (B) Reconstructed image from all localizations acquired during SPT of QDs corresponding to the trajectories in A. (C) Trajectories color-coded for D on the proximal axon of a DIV 14 neuron. In a neurofascin-positive area (inset III), the lateral mobility is reduced compared with the distal axon that loops back to the neuron (arrow). The periodic pattern was observed directly on the AIS in DIV14 neurons. (D) Reconstructed image of all localizations from the SPT experiment in C. (E) Autocorrelations of localizations along the axon (dashed lines) reveal a periodicity of ∼190 nm emerging in areas of low D trajectories in insets II and III, but not in the area of high D in inset I. (F and G) A periodic pattern was observed along the entire proximal axon where the AIS is located but was usually more prominent in some regions (dashed line lines in IV and V). (H) Autocorrelations along segments IV and V show periodically arranged stripes of localizations. Bars, 1 µm.
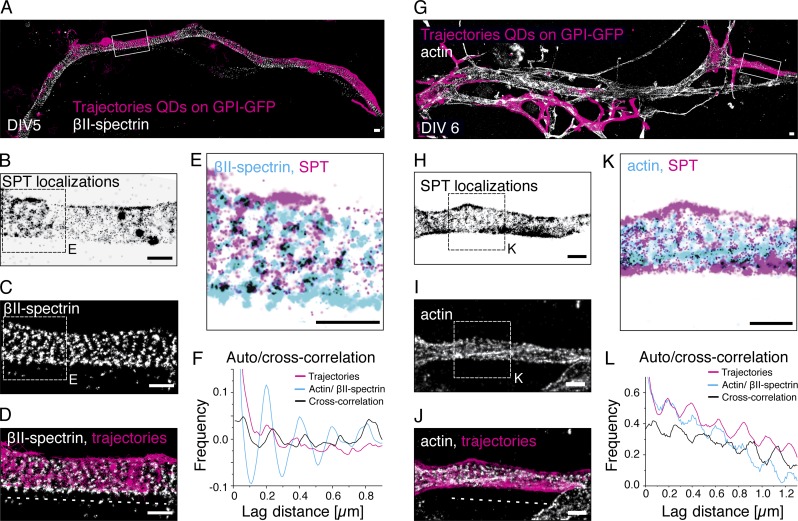
Figure 4.
The periodic confinement areas overlap with βII-spectrin immunostaining and are excluded from actin rings. (A) Proximal axon of a DIV5 hippocampal neuron with trajectories from SPT of GPI-GFP with QDs overlaid with a superresolution micrograph of βII-spectrin. (B) Reconstructed image of all localizations from SPT in the region of interest (white box, A) shows a regular pattern along the axis of the axon. (C) Superresolution micrograph of βII-spectrin confirms periodicity of the submembrane cytoskeleton in the region of interest. (D) Overlay of trajectories (magenta) and βII-spectrin (white). (E) Overlay of localizations from SPT (magenta) and cytoskeletal marker βII-spectrin (cyan). Regions where both overlap are black. (F) Auto- and cross-correlation along the axon (dashed line, D) confirms that both SPT localizations and βII-spectrin are arranged periodically at ∼190 nm. The cross-correlation places clusters of localizations from SPT on top of βII-spectrin with an offset of ∼25 nm. (G) Proximal axon of a DIV6 hippocampal neuron with trajectories from SPT of GPI-GFP with QDs overlaid with a superresolution micrograph of actin. (H) Reconstructed image of all localizations from SPT in region of interest (white box, G) shows a regular pattern along the axis of the axon. (I) Superresolution micrograph of actin confirms periodicity of the submembrane cytoskeleton in the region of interest. (J) Overlay of trajectories (magenta) and cytoskeletal marker actin (white). (K) Overlay of localizations from SPT (magenta) and cytoskeletal marker actin (cyan). Regions where both overlap are black. (L) Auto- and cross-correlation along the axon (J, dashed line) confirms that both SPT localizations and actin are arranged periodically at ∼190-nm spacing. The cross-correlation places clusters of localizations from SPT in between the actin rings with an offset of ∼20 nm. Bars, 500 nm.
Our results strongly suggested that the global reduction of diffusion in the AIS was locally correlated with the periodic submembrane spectrin–actin meshwork in the AIS. To investigate the spatial relationship between the periodic pattern of GPI-GFP localizations and the underlying cytoskeleton, we performed correlative SPT and superresolution imaging of different components of the axonal cytoskeleton. For this, we fixed and immunostained neurons after SPT experiments and then performed superresolution imaging of the cytoskeleton of the same AIS. First, we overlaid SPT measurements with direct stochastic optical reconstruction microscopy images of the C terminus of βII-spectrin, which is located at the center of the spectrin tetramer that connects the actin rings (Xu et al., 2012; Video 1). We found extensive spatial overlap between the particle trajectories and βII-spectrin immunostaining (Fig. 4, A–E). A spatial correlation analysis showed a strong cross-correlation between GPI-GFP and βII-spectrin positions (Fig. 4 F), which was also apparent in the plot of GPI-GFP versus βII-spectrin localization density (Fig. 4 E).
When instead we correlated GPI-GFP trajectories with superresolution imaging of actin via phalloidin AF647, GPI-GFP localizations and actin staining seemed mutually exclusive (Fig. 4, G–K; and Video 2). Consistently, the cross-correlation analysis between GPI-GFP and actin showed a ∼100-nm phase shift, corresponding to half a period of the cytoskeleton (Fig. 4 L), which became also apparent in the localization density (Fig. 4 K). From this, we concluded that GPI-GFP molecules are confined in their lateral motion in the neuronal plasma membrane between actin rings.
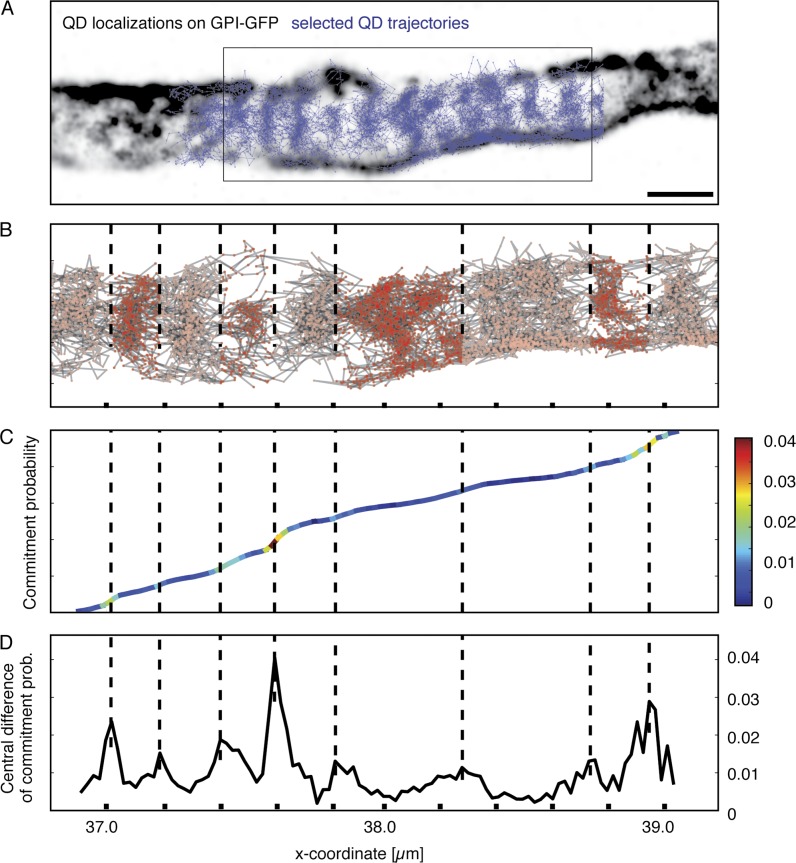
Finally, we devised a mathematical model to ask whether the periodic areas are merely areas with higher and lower density of freely diffusing molecules or whether the observed molecules indeed experienced resistance on their path down the axonal plasma membrane consistent with a diffusion barrier. We chose an area of axon with a very high density of long trajectories (Fig. 5 A). We then constructed a Markov model (Prinz et al., 2011), where the axon space between the ends of the box in Fig. 5 B is discretized in many small areas and the transition probabilities between these areas are extracted from the experimental SPT data. The Markov model can then compute kinetic properties for timescales much longer than the individual trajectory lengths. To measure global diffusivity along the axon, we computed the so-called commitment probability (Fig. 5 C), which is the probability with which a protein that is located at the given axonal position will reach the right boundary before reaching the left boundary (box in Fig. 5 A). For free diffusion, the commitment probability would be a straight line; for obstacles along the path, the model would find steps; and for a complete block, the model would fail. The commitment probability clearly shows steps (Fig. 5 C), which is a hallmark of metastable states along the axon (i.e., regions within which the dynamics is relatively fast and transitions between them are rare). To pinpoint the locations of these steps, the slope of the commitment probability is plotted in Fig. 5 D and color-coded in Fig. 5 C. The local maxima of this function correspond to the step positions of the commitment probability, and most of those occur at ∼190 nm distances. Dashed vertical lines in Fig. 5 (B–D) map these positions, showing that steps of the commitment probability correspond to low-density regions of the protein localizations and thus to localizations of actin rings. Thus, proteins move within axon stripes and between two actin rings by normal diffusion. They can also jump to adjacent axon stripes (i.e., cross the actin rings), but this event is rare, and thus, the transport of proteins along the axon is slowed down compared with the diffusivity within each segment.
Figure 5.
The periodic confinement areas are separated by diffusion barriers. (A) Overlay of a reconstructed image of SPT localizations (black) on the AIS of a DIV5 neuron with trajectories (blue). The area selected for Markov model analysis is emphasized (box), and the model describes the diffusion from the left end to the right end of the area. (B) Plot of trajectories in area used for analysis and boundaries detected by the Markov model. x axis indicates distance, and tick marks are 200 nm. From the left, a ∼200-nm spacing of detected boundaries is apparent. As the pattern becomes more complex and is not perpendicular to axon propagation, boundaries are not as easily identified by the algorithm. (C) Plot of the commitment probability from the left to the right. The value corresponds to the local probability that a molecule will move to the right boundary rather than the left boundary next. Thus, the commitment probability indicates the progress of the transport process from left to right. In the case of pure random motion, a proportional increase in commitment would be detected along the axon. The slope of the graph is color-coded to emphasize local barriers detected as sudden changes in commitment between areas of proportional increase. (D) Plot of local change of the commitment probability. Peaks indicates positions where the commitment probability makes a step change and thus positions of dynamical boundaries (color-coded in C, corresponding to low-density regions in A and B).
To quantify this slowdown effect, we compare the mean first passage time between the ends of the box in Fig. 5 A using the Markov model with the mean first passage time expected from Brownian motion with the measured diffusion constant. We found that the diffusion constant computed from the step-length distribution is equivalent in axon and stripe directions, suggesting free diffusion within domains. Overall, however, transport along the axon is between 3 and 10 times slower than expected from pure diffusion, consistent with a diffusion barrier.
Our results show that between the third and the fifth day in culture of developing hippocampal neurons, a periodic array of barriers to the motion of GPI-anchored GFP in the plasma membrane is created in the AIS that localizes to actin rings. At this time, the full density of ion channels, adhesion molecules (Jones et al., 2014), and extracellular matrix (Frischknecht et al., 2009) at the AIS is not established, ruling out a major role of these assemblies. Earlier work has observed the establishment of a diffusion barrier in the AIS at DIV7–10 (Nakada et al., 2003; Boiko et al., 2007); however, the technological possibilities at the time allowed only the observation of a few molecules that were found to be virtually immobile. In contrast, we find a macroscopic reduction in diffusion coefficients of mobile molecules in the AIS that correlates with a compartmentalization of membrane protein motion between actin rings. These results strongly suggest that the actin rings themselves or associated structures act as a physical barrier to membrane protein motion even in the outer plasma membrane leaflet. Mathematical modeling indeed confirms that periodic obstacles to membrane protein motion are located between localization rings. Our work thus provides a new mechanistic model for the AIS diffusion barrier in which a periodic array located to actin rings reduces membrane protein motion.
Because the periodic actin meshwork extends all along the axon in mature neurons but we do not detect periodic patterns of GPI-GFP localizations in the distal axon, it seems that besides the actin rings, additional AIS properties or AIS-specific factors may contribute to diffusion barrier formation. Several possible mechanisms for barriers to membrane protein motion have been proposed (Trimble and Grinstein, 2015), including interaction with submembrane organelles or lipid composition. Interestingly, a specific cisternal organelle localizes to the AIS (Bas Orth et al., 2007) that may interact with the membrane at contact sites. The AIS may also have a specific lipid composition that is influenced by the periodic actin rings, but unfortunately, although discussed (Rasband, 2010), thorough analyses of the AIS lipidome are missing.
Regarding other functions of the periodic cytoskeletal structure, our work opens several questions. It is known that the overexpression of βII-spectrin induces a periodic cytoskeletal structure in dendrites (Zhong et al., 2014) and that a fraction of dendrites stained with a fluorogenic analogue of the actin-stabilizing drug jasplakinolide exhibit periodic actin rings. It will be important to investigate whether such structures inhibit membrane protein motion as well and if the restriction of membrane protein motion contributes to AIS maturation, function, or relocation (Grubb and Burrone, 2010).
Beyond the development of the AIS, the finding that the motion of membrane molecules can be confined by the plasma membrane–associated cytoskeleton is important for all of cell biology. That the motion of a membrane molecule can be confined by actin corrals has long been proposed (Kusumi et al., 2005). Indeed, it is clear that transmembrane molecules can be caught by submembrane structures (Tomishige and Kusumi, 1999) and that whereas lipids seem to undergo Brownian motion in the plasma membrane at the macroscopic scale, at the nanoscopic scale, they exhibit subdiffusive motion (Fujiwara et al., 2002; Eggeling et al., 2009).
In recent years, the organization of the plasma membrane and its control by cellular mechanisms is being investigated with new scrutiny using several novel experimental approaches. Stimulated emission-depletion fluorescence correlation spectroscopy has demonstrated that the motion of lipids in the plasma membrane can be increased by an actin-depolymerizing drug (Andrade et al., 2015) and a combination of fluorescence correlation spectroscopy with accurate temperature control of the sample showed that actin polymerization influences the motion of GPI-anchored proteins (Saha et al., 2015). However, these studies could not spatially resolve a correlation between actin and moving molecules, and the mechanism behind these findings remains unclear.
How a GPI-anchored molecule in the exoplasmic membrane leaflet of the plasma membrane can be confined by the submembrane cytoskeleton is subject of intensive investigation. It is known that transbilayer interactions of the alkyl chains of lipids can confine molecules in supported membrane bilayers (Spillane et al., 2014) and likely also in the plasma membrane of cells (Raghupathy et al., 2015). In the future, it will be important to investigate whether individual GPI-anchored or transmembrane molecules exhibit confined motion in protein-dense areas of the plasma membrane of cultured cells as well (Saka et al., 2014), as such areas, according to the picket fence model, should capture lipid-anchored molecules.
Our work opens several exciting questions regarding the function of the periodic axonal cytoskeleton and the global regulation of membrane protein motility by the cytoskeleton. Future work will elucidate the functional consequences of membrane protein compartmentalization in the AIS on membrane transport and neuronal polarization and a possible compartmentalization of the dendritic and distal axonal plasma membrane.
Materials and methods
Ethics statement
Humane killing for preparation of rat primary hippocampal neurons conformed to local King’s College London ethical approval under the UK Supplementary Code of Practice, The Humane Killing of Animals under Schedule 1 to the Animals (Scientific Procedures) Act 1986, or in accordance with the guidelines issued by the Swiss Federal Act on Animal Protection, respectively. All efforts were made to minimize animal suffering and to reduce the number of animals used.
Cell culture
Primary hippocampal neurons were prepared from embryonic day18 Sprague–Dawley rats (Charles River) as previously described (Kaech and Banker, 2006). Neurons were maintained in neurobasal medium with B27 supplement and GlutaMAX (all Thermo Fisher Scientific) on poly-lysine (Sigma-Aldrich)–coated 18-mm-diameter #1.0 glass coverslips (Menzel) or µGrid glass-bottom Petri dishes (ZellKontakt GmbH) at 37°C in a CO2-controlled humidified incubator. After 3 d, the cytostatic cytosine β-d-arabinofuranoside (Sigma-Aldrich) was added at a final concentration of 5 µM. Neurons were transiently transfected with protein fusion constructs between DIV2 and DIV8 using Lipofectamine 2000 (Thermo Fisher Scientific). The GPI-GFP protein fusion construct was a gift from the Helenius laboratory (ETH Zurich, Zurich, Switzerland; Keller et al., 2001). The L-YFP-GT-mHoneydew-46 construct was based on L-YFP-GT46, a gift from P. Keller (Max Planck Institute for Cell Biology and Genetics, Dresden, Germany; Keller et al., 2001), and modified by inserting mHoneydew between the transmembrane domain and the CD46 domain to increase the size of the cytosolic part of the fusion protein.
SPT
Anti-GFP nanobodies (ChromoTek) were labeled with biotin-sulfo-N-hydroxysuccinimide (Thermo Fisher Scientific) by standard N-hydroxysuccinimidyl ester chemistry according to the manufacturer’s protocol using a threefold molar excess resulting in approximately one biotin per nanobody. The labeled nanobody was purified from the excess of unreacted biotin using three 3-kD MWCO desalting columns (Zeba Spin; Thermo Fisher Scientific). Biotin–anti-GFP nanobodies were conjugated to 1 nM Qdot 705 streptavidin (Thermo Fisher Scientific) at a ratio of 3:1 on the day of the experiment. Nanobody-conjugated QDs were added before image acquisition at a concentration of ∼10 pM. A single image series of 25,000–50,000 frames, which took 2–5 min, was recorded with 2- to 10-ms exposure time and 2- to 3-ms pulsed laser illumination. The irradiance at the sample was ∼2 kW/cm2. The EMCCD camera was run in its high-speed modality using the 20-MHz readout.
100-nm red-fluorescent (580/605) beads (Thermo Fisher Scientific) were added as fiduciary markers for drift correction and image correlation.
SPT using Atto647N-coupled anti-GFP nanobodies (ChromoTek) was done using the uPAINT method. Nanobodies were added immediately before image acquisition at a concentration of ∼25 pM, and multiple image series (typically 10–20) of 500 frames were recorded with 25-ms exposure time and 5-ms laser illumination time. A lower irradiance of ∼0.5 kW/cm2 was used to reduce the extent of photobleaching.
SPT experiments were all performed in live-cell imaging buffer (145 mM NaCl, 5 mM KCl, 10 mM glucose, 10 mM Hepes, 2 mM CaCl2, 1 mM MgCl2, 0.2% [wt/vol] BSA, and 10 mM ascorbate; Ewers et al., 2014) on a heated microscope stage.
The lateral localization accuracy was 25–30 nm for the tracking of Atto647N and 10–15 nm for the QDs. Instantaneous diffusion coefficients D2–4 for individual trajectories were calculated from the mean square displacement.
Immunofluorescence staining
Before fixation, the neurons were briefly rinsed in warm PBS. For staining of βII-spectrin and AnkG, neurons were fixed for 15 min with 4% (wt/vol) PFA (Sigma-Aldrich) in PBS. For actin staining, neurons were extracted and fixed with glutaraldehyde as previously described (Xu et al., 2012). Samples were permeabilized in 0.2% (vol/vol) Triton X-100 (Sigma-Aldrich) for 30 min, blocked for 30 min with ImageIT (Thermo Fisher Scientific), and then washed twice in PBS and subsequently blocked for 1 h in 10% (vol/vol) horse serum and 1% (wt/vol) BSA in PBS (blocking buffer).
As primary antibodies, we used monoclonal mouse βII-spectrin antibody (clone 42, which targets a sequence close to the C terminus of βII-spectrin; BD), monoclonal mouse AnkG antibody (clone 65; Neuromab), and monoclonal neurofascin antibody (clone A12/18; Neuromab) conjugated in-house to Cy3 by N-hydroxysuccinimidyl ester chemistry. As secondary antibody, we used donkey anti–mouse AF647 (A31571; Thermo Fisher Scientific). Staining with antibodies was performed in blocking buffer using a 1-h incubation for primaries and secondaries, respectively. To label actin filaments, samples were labeled with Alexa Fluor 647–conjugated phalloidin (A22287; Invitrogen) overnight at 4°C or ∼1 h at room temperature. A concentration of ∼0.5 µM phalloidin in blocking buffer was used.
Microscope setups
Standard immunofluorescence microscopy was performed on a spinning-disk confocal microscope (inverted Olympus IX71; Roper Scientific). Superresolution imaging and SPT experiments were performed on a custom-built setup. In brief, a 473-nm laser (100 mW; Laserglow Technologies), a 556-nm laser (200 mW; Laserglow Technologies), and a 643-nm laser (150 mW; Toptica Photonics) were focused onto the back-focal plane of an NA 1.49, 60×, total internal reflection fluorescence objective (Olympus). A quad-edge dichroic beamsplitter (405/488/532/635 nm; Semrock) was used to separate fluorescence emission from excitation light. Emission light was filtered by a quad-band bandpass filter (446/523/600/677 nm; Semrock) and focused by a 500-mm tube lens onto a back-illuminated electron multiplying charge coupled device chip (Evolve Delta; Photometrics).
Single-molecule localization microscopy
The buffer for superresolution imaging consisted of 0.1 M β-mercaptoethylamine/0.2 M Tris, pH 8.0, with 5% (wt/vol) glucose, 0.25 mg/ml glucose-oxidase, and 20 µg/ml catalase. The imaging laser intensity of the 643-nm laser line used was ∼2 kW/cm2. The intensity of the 473-nm activation laser was automatically adjusted to keep the mean number of localizations per frame constant (maximum intensity, ∼0.5 kW/cm2). We recorded a minimum of 25,000 frames with an exposure time of 20–35 ms.
Data analysis
Data analysis was performed in MATLAB (MathWorks). Positions of single particles were determined based on a maximum likelihood estimator by Gaussian fitting (Smith et al., 2010; Albrecht et al., 2015). Single-molecule localization microscopy images were corrected for drift based on image correlation, whereas for SPT experiments, the localizations were drift-corrected using fiducials. SPT data were registered with subsequent superresolution images based on fiducials. We used a minimum of three fiducials and performed a rigid transformation based on the MATLAB built-in routine cp2tform using nonreflective similarity. We determined a fiducial registration error of <25 nm. Auto- and cross-correlations were determined by converting coordinates to pixels and determining the correlation function based on the MATLAB built-in routine autocorr.
Modeling
For the Markov state model and commitment probability, analyses shown in Fig. 5 were done with the PyEMMA software package (Scherer et al., 2015). The axon stripe area (Fig. 5 A) was discretized by finding 1,000 centroid coordinates using the k-means method and then assigning all protein localizations to the nearest centroid (Voronoi partition). A reversible maximum likelihood Markov model (Prinz et al., 2011) was estimated from the discretized trajectory data using a time lag of one step (0.2 s). The committor probability (Fig. 5 C) was computed as described previously (Noé et al., 2009), where the starting and finishing states are chosen at the two ends of the selected axon section that exhibits a clear stripe pattern. The qualitative form of the committor function is robust with respect to changes of the lag time. A smoothing window of size 50 nm was applied to reduce statistical noise. The curve shown in Fig. 5 D was computed as a central difference from the smoothed committor function.
Online supplemental material
Fig. S1 shows that the reduction of lateral mobility coincides with the accumulation of neurofascin at the AIS between DIV3 and DIV5. Fig. S2 shows examples of the periodic pattern of localizations from SPT on the proximal axon over the time course of neuronal development. Fig. S3 shows SPT experiments of a transmembrane protein in the proximal and of GPI-GFP in the distal axon. Video 1 shows a βII-spectrin superresolution micrograph overlaid with trajectories of QDs on GPI-GFP in the AIS at DIV5. Video 2 shows an actin superresolution micrograph overlaid with trajectories of QDs on GPI-GFP in the AIS at DIV4.
Supplementary Material
Acknowledgments
The authors acknowledge support from Marie Curie Actions grant (FP7-PEOPLE-2013-IEF 630024, to C.M. Winterflood), Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung Sinergia (grant CRS113_141945, to D. Albrecht and H. Ewers), and the Deutsche Forschungsgemeinschaft (grant SFB958, to H. Ewers, M. Sadeghi, and F. Noé). This project was supported by Deutsche Forschungsgemeinschaft SFB1114/Project C03 (F. Noe).
The authors declare no competing financial interests.
Author contributions: H. Ewers and C.M. Winterflood designed research. D. Albrecht and C.M. Winterflood performed research. M. Sadeghi, F. Noé, T. Tschager, and C.M. Winterflood contributed new reagents or analytic tools. M. Sadeghi, F. Noé, C.M. Winterflood, D. Albrecht, and H. Ewers analyzed data. H. Ewers and D. Albrecht wrote the paper with input from all authors.
Footnotes
A preprint of this paper was posted in bioRxiv on March 31, 2016.
Abbreviations used:
- AIS
- axon initial segment
- D
- instantaneous diffusion coefficient
- DIV
- days in vitro
- GPI
- glycosylphosphatidylinositol
- QD
- quantum dot
- SPT
- single-particle tracking
References
- Albrecht D., Winterflood C.M., and Ewers H.. 2015. Dual color single particle tracking via nanobodies. Methods Appl. Fluoresc. 3:1–8. 10.1088/2050-6120/3/2/024001 [DOI] [PubMed] [Google Scholar]
- Andrade D.M., Clausen M.P., Keller J., Mueller V., Wu C., Bear J.E., Hell S.W., Lagerholm B.C., and Eggeling C.. 2015. Cortical actin networks induce spatio-temporal confinement of phospholipids in the plasma membrane: a minimally invasive investigation by STED-FCS. Sci. Rep. 5:11454 10.1038/srep11454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas Orth C., Schultz C., Müller C.M., Frotscher M., and Deller T.. 2007. Loss of the cisternal organelle in the axon initial segment of cortical neurons in synaptopodin-deficient mice. J. Comp. Neurol. 504:441–449. 10.1002/cne.21445 [DOI] [PubMed] [Google Scholar]
- Boiko T., Vakulenko M., Ewers H., Yap C.C., Norden C., and Winckler B.. 2007. Ankyrin-dependent and -independent mechanisms orchestrate axonal compartmentalization of L1 family members neurofascin and L1/neuron-glia cell adhesion molecule. J. Neurosci. 27:590–603. 10.1523/JNEUROSCI.4302-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachet A., Leterrier C., Irondelle M., Fache M.-P., Racine V., Sibarita J.-B., Choquet D., and Dargent B.. 2010. Ankyrin G restricts ion channel diffusion at the axonal initial segment before the establishment of the diffusion barrier. J. Cell Biol. 191:383–395. 10.1083/jcb.201003042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Este E., Kamin D., Göttfert F., El-Hady A., and Hell S.W.. 2015. STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Reports. 10:1246–1251. 10.1016/j.celrep.2015.02.007 [DOI] [PubMed] [Google Scholar]
- D’Este E., Kamin D., Velte C., Göttfert F., Simons M., and Hell S.W.. 2016. Subcortical cytoskeleton periodicity throughout the nervous system. Sci. Rep. 6:22741 10.1038/srep22741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C., Ringemann C., Medda R., Schwarzmann G., Sandhoff K., Polyakova S., Belov V.N., Hein B., von Middendorff C., Schönle A., and Hell S.W.. 2009. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 457:1159–1162. 10.1038/nature07596 [DOI] [PubMed] [Google Scholar]
- Ewers H., Tada T., Petersen J.D., Rácz B., Sheng M., and Choquet D.. 2014. A septin-dependent diffusion barrier at dendritic spine necks. PLoS One. 9:e113916 10.1371/journal.pone.0113916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht R., Heine M., Perrais D., Seidenbecher C.I., Choquet D., and Gundelfinger E.D.. 2009. Brain extracellular matrix affects AMPA receptor lateral mobility and short-term synaptic plasticity. Nat. Neurosci. 12:897–904. 10.1038/nn.2338 [DOI] [PubMed] [Google Scholar]
- Frost N.A., Lu H.E., and Blanpied T.A.. 2012. Optimization of cell morphology measurement via single-molecule tracking PALM. PLoS One. 7:e36751 10.1371/journal.pone.0036751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., and Kusumi A.. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157:1071–1081. 10.1083/jcb.200202050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano M.R., Jha S., Ho T.S.-Y., Zhang C., Ogawa Y., Chang K.-J., Stankewich M.C., Mohler P.J., and Rasband M.N.. 2012. A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell. 149:1125–1139. 10.1016/j.cell.2012.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G., Hosy E., Levet F., Constals A., Schulze K., Sobolevsky A.I., Rosconi M.P., Gouaux E., Tampé R., Choquet D., and Cognet L.. 2010. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J. 99:1303–1310. 10.1016/j.bpj.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb M.S., and Burrone J.. 2010. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 465:1070–1074. 10.1038/nature09160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Zhou R., Wu Z., Carrasco M.A., Kurshan P.T., Farley J.E., Simon D.J., Wang G., Han B., Hao J., et al. 2016. Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc. Natl. Acad. Sci. USA. 113:6029–6034. 10.1073/pnas.1605707113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.L., Korobova F., and Svitkina T.. 2014. Axon initial segment cytoskeleton comprises a multiprotein submembranous coat containing sparse actin filaments. J. Cell Biol. 205:67–81. 10.1083/jcb.201401045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S., and Banker G.. 2006. Culturing hippocampal neurons. Nat. Protoc. 1:2406–2415. 10.1038/nprot.2006.356 [DOI] [PubMed] [Google Scholar]
- Keller P., Toomre D., Díaz E., White J., and Simons K.. 2001. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 3:140–149. 10.1038/35055042 [DOI] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R.S., Kondo J., and Fujiwara T.. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. 10.1146/annurev.biophys.34.040204.144637 [DOI] [PubMed] [Google Scholar]
- Leterrier C., Potier J., Caillol G., Debarnot C., Rueda Boroni F., and Dargent B.. 2015. Nanoscale architecture of the axon initial segment reveals an organized and robust scaffold. Cell Reports. 13:2781–2793. 10.1016/j.celrep.2015.11.051 [DOI] [PubMed] [Google Scholar]
- Nakada C., Ritchie K., Oba Y., Nakamura M., Hotta Y., Iino R., Kasai R.S., Yamaguchi K., Fujiwara T., and Kusumi A.. 2003. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5:626–632. 10.1038/ncb1009 [DOI] [PubMed] [Google Scholar]
- Noé F., Schütte C., Vanden-Eijnden E., Reich L., and Weikl T.R.. 2009. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc. Natl. Acad. Sci. USA. 106:19011–19016. 10.1073/pnas.0905466106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz J.-H., Wu H., Sarich M., Keller B., Senne M., Held M., Chodera J.D., Schütte C., and Noé F.. 2011. Markov models of molecular kinetics: generation and validation. J. Chem. Phys. 134:174105 10.1063/1.3565032 [DOI] [PubMed] [Google Scholar]
- Raghupathy R., Anilkumar A.A., Polley A., Singh P.P., Yadav M., Johnson C., Suryawanshi S., Saikam V., Sawant S.D., Panda A., et al. 2015. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell. 161:581–594. 10.1016/j.cell.2015.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband M.N. 2010. The axon initial segment and the maintenance of neuronal polarity. Nat. Rev. Neurosci. 11:552–562. 10.1038/nrn2852 [DOI] [PubMed] [Google Scholar]
- Rasband M.N. 2013. Cytoskeleton: axons earn their stripes. Curr. Biol. 23:R197–R198. 10.1016/j.cub.2013.01.050 [DOI] [PubMed] [Google Scholar]
- Saha S., Lee I.-H., Polley A., Groves J.T., Rao M., and Mayor S.. 2015. Diffusion of GPI-anchored proteins is influenced by the activity of dynamic cortical actin. Mol. Biol. Cell. 26:4033–4045. 10.1091/mbc.E15-06-0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka S.K., Honigmann A., Eggeling C., Hell S.W., Lang T., and Rizzoli S.O.. 2014. Multi-protein assemblies underlie the mesoscale organization of the plasma membrane. Nat. Commun. 5:4509 10.1038/ncomms5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M.K., Trendelkamp-Schroer B., Paul F., Pérez-Hernández G., Hoffmann M., Plattner N., Wehmeyer C., Prinz J.-H., and Noé F.. 2015. PyEMMA 2: a software package for estimation, validation, and analysis of Markov models. J. Chem. Theory Comput. 11:5525–5542. 10.1021/acs.jctc.5b00743 [DOI] [PubMed] [Google Scholar]
- Smith C.S., Joseph N., Rieger B., and Lidke K.A.. 2010. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods. 7:373–375. 10.1038/nmeth.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane K.M., Ortega-Arroyo J., de Wit G., Eggeling C., Ewers H., Wallace M.I., and Kukura P.. 2014. High-speed single-particle tracking of GM1 in model membranes reveals anomalous diffusion due to interleaflet coupling and molecular pinning. Nano Lett. 14:5390–5397. 10.1021/nl502536u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M., and Kusumi A.. 1999. Compartmentalization of the erythrocyte membrane by the membrane skeleton: intercompartmental hop diffusion of band 3. Mol. Biol. Cell. 10:2475–2479. 10.1091/mbc.10.8.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble W.S., and Grinstein S.. 2015. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol. 208:259–271. 10.1083/jcb.201410071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler B., Forscher P., and Mellman I.. 1999. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature. 397:698–701. 10.1038/17806 [DOI] [PubMed] [Google Scholar]
- Xu K., Zhong G., and Zhuang X.. 2012. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 339:452–456. 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G., He J., Zhou R., Lorenzo D., Babcock H.P., Bennett V., and Zhuang X.. 2014. Developmental mechanism of the periodic membrane skeleton in axons. eLife. 3 10.7554/eLife.04581 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.