Abstract
CCL18 is a chemotactic cytokine involved in the pathogenesis and progression of various disorders, including cancer. Previously, our results showed high levels of CCL18 in the serum of epithelial ovarian carcinoma patients suggesting its potential as a circulating biomarker. In this study, we determined that CCL18 expression was up‐regulated in ovarian carcinoma compared with adjacent tissue and was expressed in carcinoma cells in the tumor and not in normal ovarian epithelial cells by laser capture microdissection coupled with real‐time RT‐PCR. Moreover, correlation analysis showed that the CCL18 level was positively correlated with the metastasis of patients with ovarian cancer. Survival analysis also revealed that an increased level of CCL18 was associated with worse survival time in ovarian cancer patients. Over‐expression of CCL18 led to enhanced migration and invasion of the Skov3 ovarian cancer cell line in vitro and in vivo. Finally, proteomics analysis demonstrated that CCL18‐mediated ovarian cancer invasiveness was strongly correlated with the mTORC2 pathway. These findings suggest that the CCL18 chemokine has an important role in chemokine‐mediated tumor metastasis, and may serve as a potential predictor for poor survival outcomes for ovarian cancer. © 2015 The Authors. Molecular Carcinogenesis published by Wiley Periodicals, Inc.
Keywords: epithelial ovarian carcinoma, chemokine (C‐C motif) ligand 18, metastasis, signaling pathway
INTRODUCTION
Epithelial ovarian carcinoma (EOC) has a low incidence compared with other common cancers, however, it is the leading cause of death from gynecologic malignancies and the fifth leading cause of cancer death in women in United States 1. EOC is rarely diagnosed during its early stages, when survival rates are 90%. Most patients are diagnosed with EOC when micro‐ and macro‐metastases are already present 2. Current treatments are not effective against EOC once it reaches the late metastatic stages and the disease frequently recurs and becomes insensitive to standard therapies. Thus, management of the metastatic disease becomes a crucial problem for the treatment of EOC.
Metastatic progression of EOC is unique, as metastases that cause death spread loco‐regionally throughout the peritoneal cavity 2. Malignant cells are shed off of the primary tumor and are carried by the intraperitoneal ascitic fluid 3. Previous studies demonstrated that peritoneal ascitic fluid is full of growth factors, extracellular matrix proteins, chemokines, and other factors that support and promote peritoneal metastasis 4, 5.
Preliminary quantitative mass spectrometry and ELISA data in our laboratory suggested that the Chemokine (C‐C motif) ligand 18 (CCL18), a chemokine predominantly produced by monocyte‐derived cells with M2 phenotype 6, was highly expressed in serum of EOC patients and is a circulating EOC biomarker 7. Excessive production of CCL18 by M2 macrophages has been demonstrated in various chronic inflammations and fibrotic diseases, including Gaucher's disease and rheumatoid arthritis 8, 9. In addition, constitutive expression of CCL18 was observed in ovarian cancer, gastric cancer, and glioma 10, 11, 12, 13. However, the role of CCL18 in cancer progression is controversial. CCL18 was reported to participate in immunosuppression in ovarian cancer 13, 14 but it was associated with prolonged survival in patients with gastric cancer 11.
Understanding the role of CCL18 in tumors has been hampered by the lack of an identified receptor and by the lack of a murine orthologue. There are only three receptors have been proposed for CCL18: PITPNM3 15, GPR30 16, and CCR8 17. PITPNM3 is expressed in breast cancer cells but not on T‐cells nor B‐cells. CCL18 binding to PITPNM3 will activate Pyk2 (proline rich tyrosine kinase 2) and Src kinase PITPNM3, then play an important role in metastasis of breast cancer 18. Binding of CCL18 to GPR30 don't induce chemotaxis, but blocks both activation of GPR30 and reduce the ability of CXCL12‐dependant activation of acute lymphocytic leukemia B cells 16. CCR8 is the most recently discovered receptor for CCL18, and the effects of CCR8‐CCL18 interactions appear to induce chemotaxis of Th2 cells 17. Therefore, the signaling pathway of CCL18 and mechanism of tumor regulation need to further explore.
Our previous studies suggested that CCL18 over‐expression in the epithelial ovarian cancer cell line, Skov3, could lead to enhanced invasion, migration, and adhesion in vitro 19. However, the role of CCL18 in ovarian tumor progression and its downstream‐signaling pathways remains elusive. In this study, we aimed to investigate whether CCL18 was expressed by epithelial ovarian cancer cells resident in human specimens of EOC, whether its expression may have any functional significance in the progression of this disease and to identify signaling pathways that mediate CCL18 effects.
MATERIALS AND METHODS
Patients and Tissue Samples
Primary carcinomas of the ovaries were obtained from 59 female patients (median age 55.4 yr, range 30–82 yr) at the Affiliated Tumor Hospital of Guangxi Medical University during the period of 2005–2007. Additionally, benign ovarian tissue samples were collected from 34 cases of serous cyst adenoma and 30 cases of mucinous cyst adenoma. All samples were collected with informed consent according to the internal review and the ethics boards of the Affiliated Tumor Hospital of Guangxi Medical University.
Cell Culture and Treatment
We used five ovarian cancer cell lines (Skov3, A2780, HO8910, Ovcar3, and PEO‐1). Cell culture was performed according to standard protocols. Stock cell cultures were grown on 75‐cm2 tissue culture flasks using Dulbecco's modified eagle medium (DMEM) enriched with 10% fetal bovine serum (FBS, Gibco) and 1% antibiotic mixture (penicillin and streptomycin, Sigma) at 37°C in 5% CO2. Cells were allowed to reach 85–90% confluence before the experiments were conducted.
Laser Capture Microdissection
The microdissection method was according to Liu 20. Eight micrometer sections were first fixed with acetone for 5 min, stained with hematoxylin–eosin (HE), dehydrated in graded alcohol and xylene, and then air‐dried for 5 min. Small tumor areas (about 50 cells) on the section were selected and captured using a PixCell laser capture microscope (Arcturus Engineering Inc.). The Laser capture microdissection (LCM) parameters were as follows: a laser power of 70 mW, laser pulse duration of 1.2–3.5 ms, and laser spot size of 7.5–15 μm diameter. The tissue section was overlaid with a thermoplastic polymer membrane mounted on an optically transparent cap (CapSure, Arcturus Engineering Inc., Mountain View, CA); then, the infrared laser was pulsed over the selected cells. Those cells were then captured by focal melting of the membrane through laser activation.
RNA Extraction
The ovarian cancer cells captured on the cap were microcentrifuged into an Eppendorf tube. Total RNA was then extracted by using an Arcturus® Paradise® Plus RNA Extraction and Isolation Kit (Applied Biosystems) following the supplier's protocol. The RNA from cell cultures and xenografts were isolated using RNeasy Mini Reagent (Qiagen) according to the manufacturer's protocol.
QRT‐PCR Assay
Total RNA (500 ng) was reverse transcribed to cDNA using the SuperScript® III RT Kit according to the manufacturer's instructions (Applied Biosystems). Relative transcript levels of CCL18 and reference genes were quantified by real‐time PCR using HotStart‐II SYBR Green qPCR‐Master Mix (Takara, Japan) in a real‐time PCR machine (ABI 7500, Applied Biosystems). The reactions were performed in triplicate and normalized to GAPDH. Real‐time PCR data were analyzed by ΔCp (crossing point) method as R = 2[Cp sample −Cp control] 21 to generate the relative expression ratio (R) of Target gene relative to GAPDH.
Overexpression of CCL18
Sense (5′‐CTGCCCAGCATCATGAAGG‐3′) and antisense (5′‐CCTCAGGCATI'CAGCTFCAG‐3′) oligonucleotides were designed to amplify the CCL18 cDNA. CCL18 cDNA was PCR amplified from RNA extracted from ovarian cancer tissue. The ds‐cDNA with blunt ends was inserted into the PmeI site of lentivirus expression vector pWPI (pWPI was a gift from Didier Trono, Addgene plasmid # 12254). Integrity of the CCL18 construct (Lenti CCL18) was verified by sequence analysis and comparison to the human CCL18 sequence (NCBI Genbank Accession number NM_002988). Lenti CCL18 directs the expression of both CCL18 and eGFP from the EF1‐α promoter. Recombinant lentiviruses were produced in 293T cells following co‐transfection of pWPI or Lenti CCL18 with the packaging plasmids PCMM8.74 and PMD2.0G. To determine the effect of CCL18 overexpression in ovarian cancer cells, SKOV3 cells transduced with Lenti CCL18 were compared to cells transduced with the empty vector pWPI.
Proliferation Assay
To assay proliferation rates, 2.5 × 104 cells were seeded in a 96‐well plate. The cells were allowed to settle for 24 h. The proliferation rate was evaluated using the MTT assay at 1–7 d 22.
Transwell Migration and Invasion Assay
A Boyden chamber invasion assay with matrigel‐coated transwell inserts was performed with CCL18_Skov3, GFP_Skov3, and Skov3 cells over 24 h, as described previously 23. Cells were applied to the assay using media without serum and media with 10% FBS in the lower chamber was used as the chemoattractant. Relative numbers of invading cells were evaluated by MTT assay 22. Migration assays were similarly performed in the absence of matrigel. All experiments were performed in triplicate.
Tumor Xenografts
Female BalB/C nude mice (20 d) were bred and maintained under defined conditions at the Animal Experiment Center of Guangxi Medical University, and all procedures were approved by the Animal Care and Use Committee of Guangxi Medical University and conformed to the legal mandates and national guidelines for the care and maintenance of laboratory animals. Ovarian cancer cell line, Skov3, (1 × 107) transduced with lentiviruses carrying CCL18 (Lenti CCL18 also encodes for GFP) or GFP alone (WPI) were inoculated into the mammary fat pads of the mice (n = 5/group). When the xenografts were palpable (around 0.5 cm in diameter), they were collected and transplanted below the capsule of the left ovary of five mice. Tumor growth and metastasis were evaluated by monitoring GFP expression using the small animal image system (LB983, Berthold, Germany) every 3 d for 5 wk. The animals were sacrificed and tumor xenografts, intestines, kidneys and livers of the mice were harvested for further evaluation. Cryosections (4 µm) of the harvested organs were stained by H&E for histological assessment, and total RNA was extracted for qRT‐PCR analysis of human CCL18 mRNA expression.
Proteomic Sample Preparation and iTRAQ Labeling Analysis
Using Subcellular Proteome Extraction Kit (Novagen), cytoplasmic and nuclear protein were collected from xenografts. Transmenbrane protein were extracted using the Transmembrane Protein Extraction Kit (Novagen), and protein concentrations were determined by using BCA assay (Pierce). Tryptic digestion of 100 µg of protein for each condition and differential labeling of peptides with iTRAQ reagents (Applied Biosystems) was performed as described previously 24. GFP_Skov3 control samples were labeled with the 118 isobaric tag. Sample preparation and two dimensional LC MS/MS analysis using a QStar Pulsar Quadrupole TOF mass spectrometer (Applied Biosystems/Sciex) were as described previously 25. ProteinPilot (Version 2.0.1) (Applied Biosystems) was used for protein identification and quantification after searching against the human IPI version 3.28 database. A confidence cutoff for protein identification of 95% was applied, and biological modifications were included in the identification. The intensity of the 114 atomic mass unit signature mass tags generated upon MS/MS fragmentation from the iTRAQ‐labeled tryptic peptides were used to quantify the relative levels of peptides and hence proteins in each sample. Subsequently, the meaningful cutoff for up‐regulation (≥1.2) and down‐regulation (≤0.83) of proteins was finalized by the use of biological replicate method proposed by Gan et al 24.
Bioinformatics Analysis
To better appreciate the data set generated, the list of significantly altered proteins was uploaded into Ingenuity Pathway Analysis (IPA) software server (http://www.ingenuity.com) and analyzed using the Core Analysis module to rank the proteins into top biological functions including disease and disorders as well as molecular and cellular functions. The reference set and parameters for Ingenuity Pathway Analysis (IPA) on significantly altered protein list was as follows: (i) Reference set, Ingenuity Knowledge Base (genes only); (ii) Relationship to include, direct and indirect; (iii) Filter summary, consider only molecules and/or relationships where (species = human) and (cell lines = all cancer cell lines in ingenuity database).
Western Blot Analysis
Xenografted tumors were homogenized and then lysed using ice‐cold RIPA buffer (150 mM NaCl, 50 mM Tris‐HCl; pH 7.5, 1% deoxycholate, 0.1% SDS, 1% Triton X‐100, 1 mM PMSF, 1 mM Na3PO4, protease inhibitor cocktail (1:1000; Roche). Equal amounts (30 µg protein) of xenograft extracts were separated by 8–12% SDS–PAGE and transferred to PVDF membranes. The transferred samples were incubated with primary antibodies from Abcam (anti‐CCL18, anti‐EIF2S2, anti‐EIF5a, anti‐EIF5b, anti‐EIF4A1, and CDKN2A) or Cell Signaling Technology (anti‐Kras, anti‐Rictor, anti‐Raptor, anti‐Phospho‐Akt (Ser473), anti‐Akt, anti‐Phospho‐NDRG1(Thr364), NDRG1, anti‐mTOR, anti‐Phospho‐mTOR (Ser2448), anti‐Rheb, anti‐Rhoa, anti‐Rac1, anti‐Rac2, anti‐actin, anti‐Tubulin, and anti‐GAPDH) and then incubated with the corresponding IRDye 680RD secondary conjugated immunoglobulin G(Li‐CoR). The proteins were then visualized with Odyssey imaging system (Li‐CoR).
Statistics
All statistical analyses were done using SPSS for Windows version 13.0 (SPSS, Chicago). Student's t test and one‐way ANOVA were applied to analyze the relationship between CCL18 expression and clinic pathological status. Kaplan–Meier survival curves were plotted, and log rank tests were done. The significance of various variables for survival was analyzed by the Cox proportional hazards model in a multivariate analysis. All experiments for cell cultures were performed independently for at least five times in triplicate. A P value < 0.05 in all cases was considered statistically significant and greater than twofold changes in transcript expression were taken as biologically significant.
RESULTS
CCL18 Expression in Ovarian Cancer Cells Correlates With Clinical Parameters
The mRNA expression levels of the CCL18 in paired cancer cells and adjacent cells isolated from 59 patients with ovarian carcinomas were determined. The ovarian cancer cells captured by LCM coupled with real‐time RT‐PCR showed that the expression of CCL18 mRNA was about 25 ± 17‐fold higher than that in the adjacent cells (Figure 1A; P < 0.001). CCL18 mRNA expression was enriched 1.9 fold in laser dissected tumor cells when compared to RNA from whole ovarian carcinoma samples. (Figure 1A; P < 0.001). Expression of CCL18 mRNA from cancer cells was also significantly increased when compared to benign ovarian and normal epithelial cells. Immunohistochemistry for CCL18 expression in ovarian cancer tissues revealed that CCL18‐positive cells were scattered in the tumor carcinomas of 59 cases of ovarian carcinomas (Figure 1D, 1, 2, 3, 4). However, no brown staining was observed in the cystic cells and the adjacent non‐neoplastic epithelia. CCL18+ cells were absent in all benign ovarian tissues (Figure 1D, 5, 6). Additionally, CCL18 protein levels in the serum of patients with ovarian cancer (150.41 ± 61.72 ng/ml) were also significantly higher than those in patients with benign disease (35.83 ± 8.40 ng/ml) or normal women (30.50 ± 16.04 ng/ml) (Figure 1C; P = 0.001).
Figure 1.
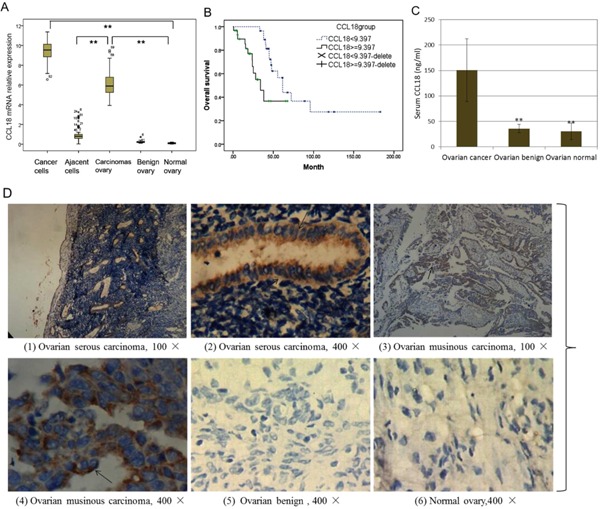
CCL18 Expression in ovarian cancer tissues and serum. (A) Expression of CCL18 mRNA from the LCM of ovarian carcinomas samples, **P < 0.001; (B) Results of overall survival (OS) analysis in patients stratified according to the CCL18 mRNA level (Kaplan–Meier test), P < 0.001; Delete: death cases. (C) CCL18 level of serum of patients with ovarian cancer and benign ovarian tissue, **P < 0.001; (D) Expression of CCL18 protein was observed by immunohistochemistry. Hemalum colors nuclei of cells were blue. CCL18 immunoreactivity in tumor cells was confined to the intercellular with brown, no staining in the nuclei of cells. Non‐epithelial ovarian carcinomas, absence of CCL18 immunostaining.
We next correlated CCL18 mRNA expression in patient ovarian carcinomas with their clinic pathological status (Table 1). The expression of CCL18 increased with higher tumor burden defined by tumor size (P = 0.024) and pelvic metastasis (P = 0.049). A Kaplan–Meier survival curve with a median follow‐up period of 47.9 months demonstrated that patients with low CCL18 (≤9.397) survive significantly longer than those with high CCL18 (>9.397) (Figure 1B; P < 0.05). In a multivariate Cox regression analysis, residual tumor and platinum resistance were associated with poor survival prognosis of patients with ovarian cancer (P = 0.038; P = 0.035), independent of other clinical covariates. However, high expression level of CCL18 (CCL18 > 9.397) in ovarian cancer has no statistical correlation with prognosis (P = 0.293), indicating that CCL18 is not an independent prognostic factor for ovarian cancer.
Table 1.
The Relationship Between the Expressions of CCL18 and Clinic Pathological Factors in Ovarian Cancer
| Factors | Classification | Cases | CCL18 mRNA | P‐value |
|---|---|---|---|---|
| FIGO stage (2012)* | I–II stage | 10 | 9.311 ± 1.010 | 0.079 |
| III–IV stage | 49 | 9.959 ± 0.914 | ||
| Pathological grade | G2 | 13 | 9.092 ± 1.164 | 0.269 |
| G3 | 46 | 9.958 ± 0.974 | ||
| Lymph node metastasis | Non‐metastasis | 27 | 9.312 ± 1.021 | 0.173 |
| Metastasis | 32 | 9.994 ± 0.894 | ||
| Liver metastasis | Non‐metastasis | 33 | 9.251 ± 0.975 | 0.089 |
| Metastasis | 26 | 9.905 ± 0.981 | ||
| Bowel and pelvic metastasis | Non‐metastasis | 12 | 9.509 ± 2.393 | 0.049* |
| Metastasis | 47 | 10.821 ± 0.995 | ||
| Residual lesions | ≤ 2 cm | 52 | 9.289 ± 0.979 | 0.024* |
| > 2 cm | 7 | 10.200 ± 0.964 |
FIGO, Federation International of Gynecology and Obstetrics.
CCL18 Enhances Proliferation of Ovarian Cancer Cell Lines In Vitro
We found that five ovarian cancer cell lines showed very low levels of CCL18 expression in vitro compared to levels in ovarian carcinoma tissues by QRT‐PCR (Figure 2A, P = 0.000). The Skov3 cell line was transduced with Lenti CCL18 to induce over‐expression of CCL18 (Figure 2B, C). We performed in vitro assays to examine the effects of CCL18 on the growth behavior of Skov3 cells. CCL18_Skov3 cells showed significant (P < 0.05) increase in growth when compared to the control cells (Figure 2F). To determine the role of CCL18 on in vivo tumor growth, CCL18‐transduced and control Skov3 cells were injected subcutaneously in nude mice and monitored for tumor growth. CCL18 overexpression did not obviously change the primary tumor size (Figure 2E).
Figure 2.
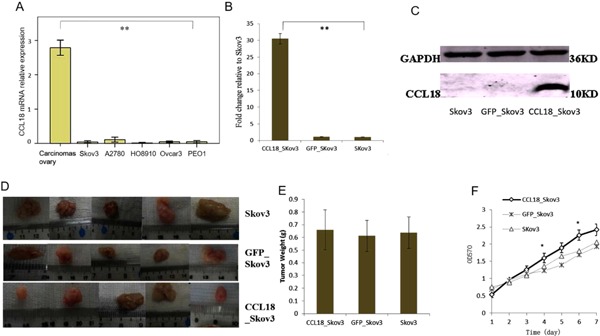
Effects of CCL18 overexpression on the proliferation of Skov3 cells. (A) Expression of CCL18 mRNA in different ovarian cancer cell lines. (B) Transfections of CCL18 was detected by real time fluorescence quantitative PCR. (C) Transfections of CCL18 was detected by western blot. (D) Effects of over‐expression CCL18 on subcutaneous tumor formation, five repeats. (E) Comparison of tumor weight in over‐expression CCL18 after xenograft growth 30 d, five repeats. (F) The effect of CCL18 on growth of Skov3 cell line in vitro by using MTT assay. *P < 0.05; **P < 0.001.
CCL18 Enhances Invasion and Migration of Ovarian Cancer Cells
First, we examined the effects of over expression of CCL18 on the migration of Skov3 cells as determined by the transwell migration assay. The effect of CCL18 on invasiveness of Skov3 cells was evaluated using the Matrigel invasion assay system. As shown in Figure 3A, migration of Skov3_CCL18 cells was increased. In line with the results from the migration assays, CCL18 also enhanced the invasion of Skov3 cells (Figure 3B).
Figure 3.
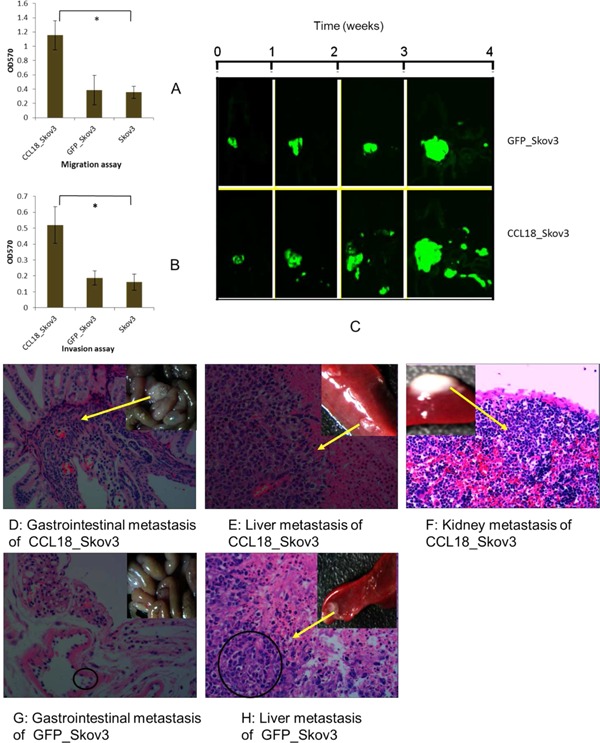
Influence of CCL18 on ovarian cancers invasion and metastasis in vitro and in vivo by orthotopic transplantation model of human ovarian cancer in nude mice. (A) The effects of CCL18 on the migration of Skov3 cells line in vitro. (B) The effects of CCL18 on the invasion of Skov3 cells line in vitro. (C) The effects of CCL18 on metastasis of ovarian tumors in vivo using a fluorescence stereomicroscope. (D–H) Hematoxylin and eosin staining (H&E) in metastasic organs, 200×. Hemalum colors nuclei of cells was blue, cytoplasm was eosin dyed red. Malignant tumor cells contained multiple or irregular nuclei. Over‐expressed CCL18 led to massive metastasis in the gastrointestinal tract, livers, and kidneys of the mice bearing CCL18_Skov3 xenografts (Figure 3D–F), Skov3_GFP cells as compared with Skov3_GFP cells (Figure 3G,H).
To examine the effect of CCL18 on ovarian cancer invasion and metastasis in vivo, CCL18_Skov3, GFP_ Skov3, and Skov3 cells were inoculated subcutaneously in nude mice, and the generated tumor tissues were collected and transplanted below the capsule of the left ovary of five mice in each group. The growth of the tumors was observed in vivo using a fluorescence stereomicroscope. Although overexpressed CCL18_Skov3 did not obviously change the tumor size when compared to GFP_Skov3 control cells, as shown in Figure 3C, CCL18‐Skov3 cells showed enhanced migration away from the primary tumors. Hematoxylin and eosin staining (H&E) also showed that overexpressed CCL18 led to more massive metastasis in the gastrointestinal tract, livers and kidneys of the mice bearing CCL18_Skov3 xenografts (Figures 3D–F) as compared with Skov3_GFP cells (Figures 3G, H). These results suggest that CCL18 promotes invasion and metastasis of ovarian cancers in vivo.
iTRAQ Proteomics Profiling of CCL18_Skov3 and Skov3 Cell Lines
To understand the global proteomic alteration mediated by CCL18 in the CCL18_Skov3 tumors, protein extracts from the xenografts with CCL18_Skov3 cells and GFP_Skov3 cells were analyzed using the iTRAQ approach. Two biological replicates for each cell line were compared for data analysis as well as to measure the variation caused by random biological effects. Applying these cutoff thresholds (≥1.2 or ≤0.8) to the two sets of biological replicates, we identified a total of 264 proteins between CCL18_Skov3 and GFP_Skov3 cells that were significantly altered in both replicates. Out of these, 168 (64%) were up‐regulated and 96 (36%) were down‐regulated in CCL18_Skov3 xenografts compared with GFP_Skov3. The corresponding ratios of these significantly altered proteins (136 proteins, P < 0.05) were plotted to calculate the correlation between two replicates (R 2 = 0.904; Figure 4A). This indicates a measurement of the confidence of true change in protein expression from corresponding peptide level alteration.
Figure 4.
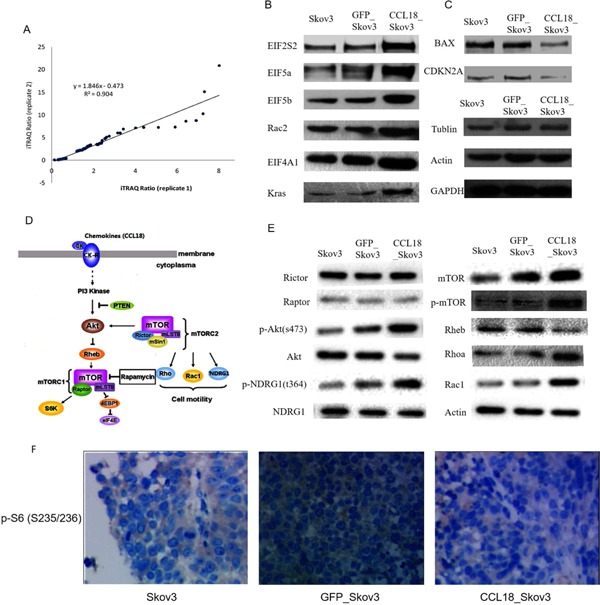
Proteomics analysis of xenograftion with CCL18_Skov3 and Skov3 cell lines using iTRAQ approach. (A) Plotting the iTRAQ ratios of the final 136 altered proteins from both biological replicates identified the correlation coefficient (R 2) at 0.904, which interpreted acceptable variation of iTRAQ ratios of the significantly regulated proteins in both biological replicates. (B) Western blot studies to validate differential expression pattern of 8 candidate proteins. Up‐regulation of EIF2S2, EIF5a, EIF5b, EIF4A1, Rac2, Kras. (C) Down‐regulation of BAX and CDKN2A in xenografts were observed by Western blot studies. (D) The schema of CCL18‐stimulated mTOR signaling. (E) Western blot analysis of mTOR signaling on mTOR biomarkers. (F) Immunohistochemical images of p‐S6 (S235/236) staining in tumor xenografts, 200×.
Cellular Pathways Involved in CCL18 Signaling
The significantly altered proteins from the iTRAQ experiment were analyzed by Ingenuity Pathway Analysis software (IPA). Two Upstream Regulators of CCL18_Skov3 cells were selected by IPA, namely PDGFRA (P‐value of overlap: 1.95E‐02) and KRAS (P‐value of overlap: 3.87E‐02). Table 2 shows significantly altered proteins related to EIF2, mTOR, EIF4, and p70S6K and p53 signaling in CCL18_Skov3 cells (cutoff thresholds, ≥2 or ≤0.5).
Table 2.
Functional Classification of the Significantly Altered Proteins Related to Signaling in CCL18_Skov3 Cells
| Unused protein score | % Sequence coverage | Accession number | Name | Species | iTRAQ ratio* replicate 1 | Replicate 2 | Expression pattern** |
|---|---|---|---|---|---|---|---|
| EIF2 signaling | |||||||
| 8.27 | 40.24 | P20042 | Eif2s2 | 9606 | 6.194 | 10.28016 | ↑ |
| 10.79 | 77.27 | P63241 | eif5a | 9606 | 4.285 | 1.294196 | ↑ |
| 2.16 | 11.56 | O60841 | Eif5b | 9606 | 2.399 | 2.964831 | ↑ |
| 1.83 | 8.547 | Q9BY44 | Eif2a | 9606 | 2.722 | 7.726806 | ↑ |
| 8 | 25.21 | O00303 | Eif3f | 9606 | 2.644 | 9.120109 | ↑ |
| 15.89 | 55.87 | P05198 | EIF2S1 | 9606 | 2.644 | 3.404082 | ↑ |
| mTOR signaling | |||||||
| 13.71 | 81.25 | P15153 | Rac2 | 9606 | 4.742 | 4.018 | ↑ |
| 3.85 | 12.5 | Q15382 | Rheb | 9606 | 0.597 | 0.283139 | ↓ |
| 6.62 | 23.2 | Q96AY3 | Fkbp10 | 9606 | 2.236 | 2.419 | ↑ |
| 10.08 | 59.04 | P62753 | RPS6 | 9606 | 2.159 | 2.582 | ↑ |
| 2 | 13.67 | Q96B36 | AKT1 | 9606 | 0.431 | 0.350 | ↓ |
| 2.25 | 26.56 | A4D2P1 | rac1 | 9606 | 2.432 | 2.818 | ↑ |
| 8.05 | 59.59 | P61586 | Rhoa | 9606 | 2.582 | 3.436 | ↑ |
| Regulation of EIF4 and p70S6K signaling | |||||||
| 20.22 | 64.04 | P60842 | EIF4A1 | 9606 | 2.208 | 2.117 | ↑ |
| 3.64 | 27.49 | P38919 | Eif4a3 | 9606 | 2.368 | 1.959 | ↑ |
| 2 | 8.756 | P06730 | eif4e | 9606 | 2.271 | 2.655 | ↑ |
| p53 signaling | |||||||
| 2 | 13.54 | Q07812 | BAX | 9606 | 0.471 | 0.291 | ↓ |
| 4 | 20.21 | O00425 | igf2bp3 | 9606 | 0.373 | 0.273 | ↓ |
| 2.37 | 30.13 | P42771 | CDKN2A | 9606 | 0.231 | 0.174 | ↓ |
| 6.28 | 43.43 | P06493 | Cdk1 | 9606 | 3.908 | 1.600 | ↑ |
iTRAQ ratio, iTRAQ ratio between CCL18_Skov3 cells and GFP_Skov3 cells were demonstrated two biological replicates, respectively.
Expression pattern, iTRAQ ratio ≥ 1.2 of proteins were defined up‐regulation and iTRAQ ratio ≤ 0.83 were down‐regulation in CCL18_Skov3 cells. Finalized by the use of method.
Validation of iTRAQ Data on Selected Candidates
Eight candidate proteins of relative signaling were chosen to validate using Western blot studies. EIF2S2, EIF5a, EIF5b, EIF4A1, Rac2, and Kras were found to be up‐regulated (Figure 4B) whereas BAX and CDKN2A were down‐regulated in the CCL18_Skov3 cell line (Figure 4C). These results verified the differential expression patterns derived from the iTRAQ experiments.
mTOR Signaling in the CCL18_ Skov3 Cell Line
Our study identified seven proteins belonging to the mTOR pathway to be significantly up‐regulated in CCL18_Skov3 cells (Table 2). Xenografts with over‐expressed CCL18 increased RhoA, Rac1, phosphorylation of Akt S473, and NDRG1 T346 (Figure 4E). Phosphorylation of Akt S473 is the best‐characterized mTORC2 activity 26. mTORC2 also activates SGK1. Phosphorylation of the SGK1‐specific substrate NDRG1 on T346 has appeared as a reliable biomarker for mTORC2 signaling 27 However, phospho‐S6 (S235/236), a marker of mTORC1 activity, was not detected (Figure 4F). This data indicates that CCL18 expression was associated with elevated mTORC2 signaling in the Skov3 xenograft model. Increased mTORC2 signaling was not associated with elevated expression of Rictor.
DISCUSSION
Cancer‐related inflammation is known as the “seventh hallmark of cancer” 28. Chemokines cause neoplastic transformation and are abundantly expressed in chronic inflammatory conditions in the tumor microenvironment 29. Chemokine receptors and their corresponding chemokine ligands have been demonstrated to play a pivotal roles in cancer metastasis to vital organs as well as regional lymph nodes, the most frequent site of cancer metastasis 30. Cytokines such as CCR7 and CXCR4 were expressed more often in breast cancer tumors that had metastasized to bone when compared to visceral metastases 31. Here, we showed that CCL18, a characteristic C‐C chemokine, was abundantly expressed in primary ovarian carcinomas cells and correlates with the bowel and pelvic metastasis of patients with ovarian cancer (Table 1).
CCL18, also known as dendritic cell chemokine, pulmonary and activation‐regulated chemokine, and macrophage inflammatory protein 4 32, is a chemotactic cytokine that is expressed by a broad range of lymphocytes. The primary targets of CCL18 include helper T lymphocytes (CD4+), cytotoxic T lymphocytes (CD8+), naive T lymphocytes (CD45RA+), and B lymphocytes. Previous reports showed that CCL18 was involved in the pathogenesis and progression of various disorders, including cancer. In patients with systemic sclerosis, for instance, levels of CCL18 have been associated with the complication of interstitial lung disease 33, and idiopathic pulmonary fibrosis 34. However, the role that CCL18 has in cancer patients seems to depend on tumor type. Leung and coauthors 11 demonstrated that CCL18 expression was associated with a better survival in gastric patients. On the contrary, Chen et al. 15 reported that CCL18 was thought to promote the invasiveness of cancer cells by triggering integrin clustering and enhancing their adherence to the extracellular matrix, suggested a negative predictor for the survival of patients with breast cancer. Similar controversies could also be seen in ovarian cancer, bladder cancer, pancreatic cancer, and skin cancer 35, 36, 37, 38.
Previously, we found that CCL18 were significantly increased in patients of ovarian cancer and could be used as a detection biomarker, moreover, CCL18 over‐expression in ovarian cancer Skov3 cells could lead to enhance invasion, migration, and adhesion in vitro 7, 19. To investigate whether CCL18 predicts poor prognosis of ovarian cancer, we examined carcinoma tissue of patients with EOC for expression of CCL18. Previous studies suggested that CCL18 might be released by a subset of macrophages 15, 33, however our results indicated that CCL18 mRNA was up‐regulated in ovarian carcinoma cells compared with tumor adjacent cells, with no detectable expression in normal ovarian epithelial cells (Figure 1A). It implied that CCL18 was produced by carcinoma cells in the tumor. Moreover, immunohistochemistry also suggested that high expression of CCL18 in EOC patients was from the tumor cells themselves. Correlation analysis showed that the CCL18 level was positively correlated with bowel and pelvic invasion and tumor size (Table 1). Survival analysis also revealed that an increased level of CCL18 was associated with worse OS in EOC patients (Figure 1B).
Interestingly, we found that CCL18 was quite low in ovarian cancer cell lines relative to tumor tissues (Figure 2A). We have also found that CCL18 was induced when tumor cell lines were co‐culture with stromal fibroblasts and mesothelial cells (not shown in this paper). To determine the biological significance of up‐regulation of CCL18 expression in ovarian cancer cells, we overexpressed CCL18 in the Skov3 cell line. Continuous expression of CCL18 proteins enhanced cellular proliferation over 7 d in vitro. Overexpression of CCL18 enhanced migration and invasion of the ovarian cancer cell line, Skov3 (Figure 3A–B). In addition, CCL18 increased peritoneal and gastrointestinal metastases of mice by using orthotopic transplanted model (Figure 3C–H). According to these properties observed for Skov3 cells, CCL18 might enhance tumor progression, metastasis and invasion that may translate into the poorer overall survival in patients as shown in Figure 1B.
Although the functions of CCL18 have been reported in many cancers, downstream‐signaling pathways of CCL18 have been difficult to characterize because there is no murine analog of CCL18. Although three possible receptors for CCL18 have been recently identified 15, 16, 18, only PITPNM3‐CCL18 binding induced Pyk2 and Src mediated signaling, and enhanced metastasis of breast cancer 18. Further characterization of downstream‐signaling pathways would lead to better understanding of the physiological role of CCL18 in ovarian cancer.
The current advances in the fields of proteomics and bioinformatics provide the opportunity to study changes in global proteome level of cells at any given time point. Particularly, labeling approaches such as iTRAQ (isobaric tags for relative and absolute quantitation) allows multiplexing of up to eight samples to identify the relative abundance of proteins in different samples within a single liquid chromatography mass spectrometry (LC‐MS) based proteomics experiment 39, 40. An 8‐plex iTRAQ workflow involving 2D LC (two‐dimensional liquid chromatography) MS/MS (tandem mass spectrometry) was adopted for the identification of differentially expressed proteins between the CCL18 _Skov3 cell lines and control cells from an EOC orthotopic xenograft model. We identified a total of 264 proteins whose expression was significantly altered Skov3 cells expressing CCL18; 168 (64%) proteins were up‐regulated and 96 (36%) proteins were down‐regulated. The significantly altered proteins from the iTRAQ experiment were functionally analyzed and classified to interpret the molecular events relevant to the pathophysiology of EOC.
Two proteins upregulated in the CCL18_Skov3 cells were the upstream regulators PDGFRA and KRAS. PDGFRA was an isoform of the PDGFR family of tyrosine kinase receptors increased in patients with gastrointestinal stromal tumors or myeloid malignancies associated with hypereosinophilia 41. Studies elucidating PDGFRα signaling in processes ranging from profibrotic signaling, angiogenesis, and oxidative stress to epithelial‐to‐mesenchymal transition point toward PDGFRα as a potential therapeutic target in hepatic fibrosis and liver cancer 42. The protein product of the normal KRAS gene performs an essential function in normal tissue signaling, and the mutation of a KRAS gene is an essential step in the development of many cancers 43. One potential consequence of CCL18 expression is the upregulation of these two regulators which may contribute to the pathogenicity of the disease.
Further analysis of CCL18 dysregulated proteins by IPA revealed an enrichment of significantly altered proteins that defined four signaling pathways: EIF2, mTOR, EIF4/p70S6K, and p53 signaling. One interesting pathway related to tumor metastasis, mTOR signaling, was further examined.
The mammalian target of rapamycin (mTOR) signaling pathway was proposed to be an attractive target for cancer therapeutics because the mTOR pathway is up‐regulated in various cancer types and is associated with multiple cellular responses 44. mTOR is a serine/threonine kinase, which exists in two complexes: mTORC1 (containing mTOR, Raptor, etc.) and mTORC2 (containing mTOR, Rictor, etc.) 26. mTORC1 is partially sensitive to rapamycin treatment, while mTORC2 is known to be rapamycin‐insensitive 26, 27. mTORC1 links PI3K signaling to promote cell growth and proliferation, activates hypoxia‐inducible factor‐1‐dependent glycolysis (HIF1) and stimulates angiogenesis in many types of cancer 45, 46. mTORC2 is required for the development of PTEN loss‐induced prostate cancer in mice, suggesting a central role in mediating PI3K‐dependent carcinogenesis 47, 48. Inhibition of mTORC2 but not mTORC1 up‐regulates E‐cadherin expression and inhibits cell motility in human renal cell carcinoma 49. Here, we demonstrated that CCL18 promoted Akt S473 and NDRG1 T346 phosphorylation (Figure 4E) while S6 phosphorylation was not changed (Figure 4F). Akt is commonly thought to be the most important mTORC2 effector and a primary mediator of chemotherapy resistance 50. We showed that overexpression of CCL18 led to upregulation of the mTORC2 pathway, including activation of AKT (Figure 4E).
Finally, our study suggested that the CCL18 chemokine expression in human ovarian may play an important role in the pathogenicity of EOC through the induction of regulators and the activation of signaling pathways including mTORC2 signaling pathways. These results also highlight the potential importance of CCL18 as a cancer target, and provide new insights into its role in mediating metastasis of EOC which will hopefully lead to new treatment strategies.
ACKNOWLEDGMENTS
The authors thank Prof. Hong Jin (Fudan University, China) for her Proteome technical assistance and also thank Dr. Kenneth Garson (Ottawa Hospital Research Institute, Ottawa, Canada) for his critical review of the manuscript. This work was supported by the grants from the National Natural Science Foundation of China (No. 30560157) and the Guangxi Natural Science Foundation of China (No. 0832230, 2013GXNSFAA019248, 2011GXNSFC018020, and 2012GXNSFAA053163).
[This article was corrected in January 2016 after original online publication because the wrong author was marked as the corresponding author. This article was also updated in October 2016 to correct the copyright line.]
REFERENCES
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin 2013; 63:11–30. [DOI] [PubMed] [Google Scholar]
- 2. Cannistra SA. Cancer of the ovary. N Engl J Med 2004; 351:2519–2529. [DOI] [PubMed] [Google Scholar]
- 3. Lengyel E. Ovarian cancer development and metastasis. Am J Pathol 2010; 177:1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barbolina MV, Kim M, Liu Y, et al. Microenvironmental regulation of chemokine (C‐X‐C‐motif) receptor 4 in ovarian carcinoma. Mol Cancer Res 2010; 8:653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim M, Rooper L, Xie J, Kajdacsy‐Balla AA, Barbolina MV. Fractalkine receptor CX(3)CR1 is expressed in epithelial ovarian carcinoma cells and required for motility and adhesion to peritoneal mesothelial cells. Mol Cancer Res. 2012; 10:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kodelja V, Muller C, Politz O, Hakij N, Orfanos CE, Goerdt S. Alternative macrophage activation associated CC‐chemokine‐1, a novel structural homologue of macrophage inflammatory protein‐1 alpha with a Th2‐associated expression pattern. J Immunol 1998; 160:1411–1418. [PubMed] [Google Scholar]
- 7. Wang Q, Li D, Zhang W, Tang B, Li QQ, Li L. Evaluation of proteomics‐identified CCL18 and CXCL1 as circulating tumor markers for differential diagnosis between ovarian carcinomas and benign pelvic masses. Int J Biol Markers 2011; 26:262–273. [DOI] [PubMed] [Google Scholar]
- 8. Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol 2009; 27:451–483. [DOI] [PubMed] [Google Scholar]
- 9. Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol 2005; 78:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang CY, Lee YH, Leu SJ, et al. CC chemokine ligand 18/pulmonary activation‐regulated chemokine expression in the CNS with special reference to traumatic brain injuries and neoplastic disorders. Neuroscience 2010; 165:1233–1243. [DOI] [PubMed] [Google Scholar]
- 11. Leung SY, Yuen ST, Chu KM, et al. Expression profiling identifies chemokine (C‐C motif) ligand 18 as an independent prognostic indicator in gastric cancer. Gastroenterology 2004; 127:457–469. [DOI] [PubMed] [Google Scholar]
- 12. Schutyser E, Struyf S, Proost P, et al. Identification of biologically active chemokine isoforms fromascitic fluid and elevated levels of CCL18/pulmonary and activation‐regulated chemokine inovarian carcinoma. J Biol Chem 2002; 277:24584–24593. [DOI] [PubMed] [Google Scholar]
- 13. Zohny SF, Fayed ST. Clinical utility of circulating matrix metalloproteinase‐7 (MMP‐7), CC chemokine ligand 18 (CCL18) and CC chemokine ligand 11 (CCL11) as markers for diagnosis of epithelial ovarian cancer. Med Oncol 2010; 27:1246–1253. [DOI] [PubMed] [Google Scholar]
- 14. DeNardo DG, Barreto JB, Andreu P, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009; 16:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Yao Y, Gong C, et al. CCL18 From tumor associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 2011; 19:541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Catusse J, Wollner S, Leick M, Schröttner P, Schraufstätter I, Burger M. Attenuation of CXCR4 responses by CCL18 in acute lymphocytic leukemia B cells. J Cell Physiol 2010; 225:792–800. [DOI] [PubMed] [Google Scholar]
- 17. Islam SA, Ling MF, Leung J, Shreffler WG, Luster AD. Identification of human CCR8 as a CCL18 receptor. J Exp Med 2013; 210:1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li HY, Cui XY, Wu W, et al. Pyk2 and Src mediate signaling to CCL18‐induced breast cancer metastasis. J Cell Biochem. 2014; 115:596–603. [DOI] [PubMed] [Google Scholar]
- 19. Zhang W, Yang YZ, Li L, Wang Q. Study of growth, invasion and metastasis on ovarian epithelial cancer cell line with CCL18 over‐expression by mediated in vitro. Zhonghua Fu Chan Ke Za Zhi 2013; 48:528–531. [PubMed] [Google Scholar]
- 20. Angen L. Laser capture microdissection in the tissue biorepository. J Biomol Tech 2010; 21:120–125. [PMC free article] [PubMed] [Google Scholar]
- 21. Butler GS, Dean RA, Morrison CJ, Overall CM. Methods in molecular biology. In: Clarke IM, editor. New York: Springer‐Verlag Inc; 2010. pp. 451–470. [DOI] [PubMed] [Google Scholar]
- 22. Twentyman PR, Luscombe M. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br J Cancer 1987; 56:279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larson Y, Liu J, Stevens PD, et al. Tuberous sclerosis complex 2 (TSC2) regulates cell migration and polarity through activation of CDC42 and RAC1. J Biol Chem 2010; 285:24987–24998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Livak K. ABI Prism 7700 Sequence Detection System User Bulletin #2 Relative quantification of gene expression. ABI company publication. 1997& 2001.
- 25. Gan CS, Chong PK, Pham TK, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J Proteome Res 2007; 6:821–827. [DOI] [PubMed] [Google Scholar]
- 26. Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell 2007; 12:9–22. [DOI] [PubMed] [Google Scholar]
- 27. Quesada AE, Nguyen ND, Rios A, Brown RE. Morphoproteomics identifies constitutive activation of the mTORC2/Akt and NF‐κB pathways and expressions of IGF‐1R, Sirt1, COX‐2, and FASN in peripheral T‐cell lymphomas: Pathogenetic implications and therapeutic options. Int J Clin Exp Pathol 2014; 7:8732–8739. [PMC free article] [PubMed] [Google Scholar]
- 28. Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer‐related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis 2009; 30:1073–1081. [DOI] [PubMed] [Google Scholar]
- 29. Candido J, Hagemann T. Cancer‐related inflammation. J Clin Immunol 2013; 33:S79–S84. [DOI] [PubMed] [Google Scholar]
- 30. Matsuo Y, Takeyama H, Guha S. Cytokine network: New targeted therapy for pancreatic cancer. Curr Pharm Des 2012; 18:2416–2419. [DOI] [PubMed] [Google Scholar]
- 31. Cabioglu N, Sahin AA, Morandi P, et al. Chemokine receptors in advanced breast cancer: Differential expression in metastatic disease sites with diagnostic and therapeutic implications. Ann Oncol 2009; 20:1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol 2004; 78:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prasse A, Pechkovsky DV, Toews GB, et al. CCL18 as an indicator of pulmonary fibrotic activity in idiopathic interstitial pneumonias and systemic sclerosis. Arthritis Rheum 2007; 56:1685–1693. [DOI] [PubMed] [Google Scholar]
- 34. Prasse A, Probst C, Bargagli E, et al. Serum CC‐chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2009; 179:717–723. [DOI] [PubMed] [Google Scholar]
- 35. Urquidi V, Kim J, Chang M, Dai Y, Rosser CJ, Goodison S. CCL18 in a multiplex urine‐based assay for the detection of bladder cancer. PLoS ONE 2012; 7:e37797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li D, Duell EJ, Yu K, et al. Pathway analysis of genome‐wide association study data highlights pancreatic development genes as susceptibility factors for pancreatic cancer. Carcinogenesis 2012; 33:1384–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miyagaki T, Sugaya M, Suga H, et al. Increased CCL18 expression in patients with cutaneous T‐cell lymphoma: Association with disease severity and prognosis. J Eur Acad Dermatol Venereol 2013; 27:e60–e67. [DOI] [PubMed] [Google Scholar]
- 38. Pettersen JS, Fuentes‐Duculan J, Suárez‐Fariñas M, et al. Tumor‐associated macrophages in the cutaneous SCC microenvironment are heterogeneously activated. J Invest Dermatol 2011; 131:1322–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine‐reactive isobaric tagging reagents. Mol Cell Proteomics 2004; 3:1154–1169. [DOI] [PubMed] [Google Scholar]
- 40. Unwin RD, Griffiths JR, Whetton AD. Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC‐MS/MS. Nat Protoc 2010; 5:1574–1582. [DOI] [PubMed] [Google Scholar]
- 41. Velghe AI, Van Cauwenberghe S, Polyansky AA, et al. PDGFRA alterations in cancer: Characterization of a gain‐of‐function V536E transmembrane mutant as well as loss‐of‐function and passenger mutations. Oncogene 2014; 33:2568–2576. [DOI] [PubMed] [Google Scholar]
- 42. Kikuchi A, Monga SP. PDGFRα in liver pathophysiology: Emerging roles in development, regeneration, fibrosis, and cancer. Gene Expr 2015; 16:109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kranenburg O. The KRAS oncogene: Past, present, and future. Biochim Biophys Acta 2005; 1756:81–82. [DOI] [PubMed] [Google Scholar]
- 44. Efeyan A, Sabatini DM. MTOR and cancer: Many loops in one pathway. Curr Opin Cell Biol 2009; 22:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC‐mTOR pathway in human disease. Nat Genet 2005; 37:19–24. [DOI] [PubMed] [Google Scholar]
- 46. Yecies JL, Manning BD. MTOR links oncogenic signaling to tumor cell metabolism. J Mol Med 2011; 89:221–228. [DOI] [PubMed] [Google Scholar]
- 47. Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006; 22:159–168. [DOI] [PubMed] [Google Scholar]
- 48. Guertin DA, Stevens DM, Saitoh M, et al. MTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell 2009; 15:148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maru S, Ishigaki Y, Shinohara N, Takata T, Tomosugi N, Nonomura K. Inhibition of mTORC2 but not mTORC1 up‐regulates E‐Cadherin expression and inhibits cell motility by blocking HIF‐2α expression in human renal cell carcinoma. J Urol 2013; 189:1921–1929. [DOI] [PubMed] [Google Scholar]
- 50. Abedini MR, Muller EJ, Bergeron R, Gray DA, Tsang BK. Akt promotes chemoresistance in human ovarian cancer cells by modulating cisplatin‐induced, p53‐dependent ubiquitination of FLICE‐like inhibitory protein. Oncogene 2010; 29:11–25. [DOI] [PubMed] [Google Scholar]


