Summary
Differences in gene sequences, many of which are single nucleotide polymorphisms, underlie some of the most important traits in plants. With humanity facing significant challenges to increase global agricultural productivity, there is an urgent need to accelerate the development of these traits in plants. oligonucleotide‐directed mutagenesis (ODM), one of the many tools of Cibus’ Rapid Trait Development System ( RTDS ™) technology, offers a rapid, precise and non‐transgenic breeding alternative for trait improvement in agriculture to address this urgent need. This review explores the application of ODM as a precision genome editing technology, with emphasis on using oligonucleotides to make targeted edits in plasmid, episomal and chromosomal DNA of bacterial, fungal, mammalian and plant systems. The process of employing ODM by way of RTDS technology has been improved in many ways by utilizing a fluorescence conversion system wherein a blue fluorescent protein (BFP) can be changed to a green fluorescent protein (GFP) by editing a single nucleotide of the BFP gene (CAC→TAC; H66 to Y66). For example, dependent on oligonucleotide length, applying oligonucleotide‐mediated technology to target the BFP transgene in Arabidopsis thaliana protoplasts resulted in up to 0.05% precisely edited GFP loci. Here, the development of traits in commercially relevant plant varieties to improve crop performance by genome editing technologies such as ODM, and by extension RTDS , is reviewed.
Keywords: oligonucleotide‐directed mutagenesis, precision gene editing, RTDS ™ , CRISPR, TALEN
Introduction
A major challenge in biology, particularly plant biology, is gene conversion. Almost a decade ago, this challenge was also known as directed gene modification or gene targeting and more recently has been termed precision gene editing. Irrespective of the name, the intent of gene conversion is to augment the genetic diversity of a specific genotype by precisely altering the sequence of a particular genome target(s) by one or more bases. These alterations may also include precise insertions or deletions in the target sequence. The oligonucleotide‐directed mutagenesis (ODM) technique for genome editing has been successfully employed in bacterial, yeast, mammalian and plant systems (Aarts et al., 2006; Gocal et al., 2015; Moerschell et al., 1988; Yoon et al., 1996). This review will specifically focus on using chemically synthesized oligonucleotides as a template for making targeted changes in these four systems.
The oligonucleotide template
Once oligonucleotide synthesis became commercially available, the automated production of short nucleic acid strands became routine. This advancement allowed not only site‐directed mutagenesis to progress beyond just editing plasmids in Escherichia coli, but to making precise changes in the nuclear genome of a variety of organisms. In the late 70s, it had been shown using site‐directed mutagenesis that one or a few nucleotides could be precisely exchanged within a plasmid template (Hutchison et al., 1978). Studies correcting the non‐sense/frameshift mutations at various positions along the cyc1 gene in Saccharomyces cerevisiae using targeted oligonucleotides extended this result to the nuclear genome (Moerschell et al., 1988; Yamamoto et al., 1992).
Less than a decade later, a technique known as chimeraplasty, a gene editing technique that utilized an RNA/DNA chimeric oligonucleotide, was developed to introduce site‐specific genomic alterations (Cole‐Strauss et al., 1996). Chimeraplasty‐mediated modification of a target sequence is accomplished using an exogenous polynucleotide, the so‐called chimeraplast, which locates its complementary sequence in the genome and harnesses the cell's inherent DNA repair system to direct the change in the gene target. The original chimeraplast ranged between 68 and 88 nucleotides in length and comprised both DNA and 2′‐O‐methyl‐modified RNA residues (see Figure 1a). These molecules were designed so that complementary bases fold to form a duplex region (homology region). The information strand (lower strand) of this molecule consists of DNA having a sequence identical to the target region except for the specific base(s) to be changed. This strand directs conversion(s) within its target. Improved conversion frequencies were achieved when modified RNA bases complementary to the target sequence were included on the top strand of the duplex (Metz, R., Frank, B., Walker, K.A., Avissar, P., Sawycky, X.L. and Beetham, P.R. unpublished data). This ‘classic’ chimeraplast design contained 5 nucleotides of DNA on the top strand between the 2′‐O‐methyl RNA bases (Figure 1a). The 2′‐O‐methyl‐modified RNA bases facilitated higher binding affinity to the target locus (Kmiec, E.B., Frank, B. and Holloman, B. unpublished data). Hundreds of chimeraplast designs have been evaluated both in vitro and in vivo for their ability to effect targeted conversions in plasmid, episomal and chromosomal targets of bacterial and eukaryotic systems (Metz, R., Frank, B., DiCola, M., Kurihara, T., Bailey, A., Walker, K.A., Avissar, P., Sawycky, X.L., and Beetham, P.R. unpublished data). Metz et al. (2002) was one of the first groups to characterize the chimeraplast and define its optimal properties. When the top strand of the chimeraplast was made completely of 2′‐O‐methyl modified RNA, its binding affinity was maximized and conversion efficiency was increased. Addition of a 5‐bp GC‐clamp on one side bordering the homology region increased exonuclease resistance and a nick between the 5′ and 3′ ends of the chimeraplast allowed topological unwinding of the molecule. Additionally, when both ends of the duplex were flanked by single‐stranded hairpin loops, concatamerization was prevented. These improvements aided in chemical and thermal stability as well as resistance to nucleases.
Figure 1.
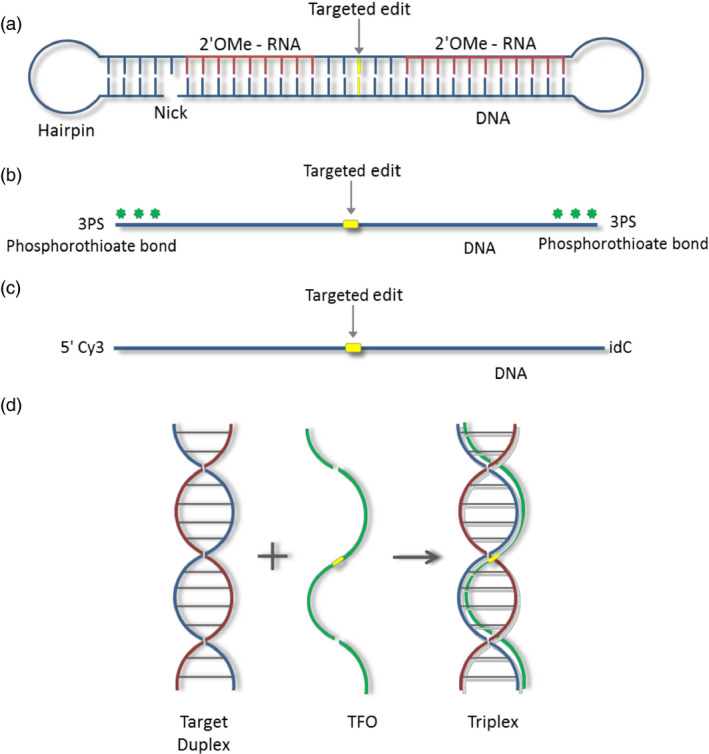
Oligonucleotide designs. (a) chimeraplast schematic showing regions of DNA (blue) and RNA (red; 2′‐O‐methyl modified), a nick and hairpin (total chimeraplast is ~68 nucleobases). (b) A single‐stranded oligonucleotide modified with 3PS (3 phosphorothioate bonds) at both the 5′ and 3′ ends (total oligonucleotide length is 41, 101 or 201 nucleobases). (c) A single‐stranded oligonucleotide modified with a Cy3 dye at the 5′ end and a reverse base (idC) at the 3′ end (total oligonucleotide is 41 nucleobases). (d) Triplex‐forming oligonucleotide (TFO). The target duplex homopurine and homopyrimidine strands are shown in blue and red. The TFO, which binds the homopurine strand, is indicated in green. The location of the targeted nucleotide in all oligonucleotides is shown in yellow.
As the lower strand of the chimeraplast consisted entirely of DNA and because the synthesis of high‐quality, long (generally 68–88mer) oligonucleotides at the time was inefficient, shorter single‐strand oligonucleotide designs mimicking the information strand were tested. Early efforts focused on interrogating thousands of these single‐strand designs in various bacterial, yeast, mammalian and plant systems to identify the design that was most efficacious (Metz, R., DiCola, M., Bailey, A., Metz, R., Kurihara, T., Frank, B. and Walther, D. unpublished data). Oligonucleotide structure and chemistries were designed so that they could be used as DNA templates to promote gene conversion through ODM technology. Oligonucleotides may be short in length and contain a centrally located mismatch in one or a few bases to the target sequence. The oligonucleotides were chemically synthesized and consisted of both DNA and modified nucleotides or other end‐protective chemistries. These modifications prevented the oligonucleotides from undergoing recombination, but still allowed them to act as a mutagen and DNA template. For this reason, ODM is considered a targeted mutagenesis system (Dong et al., 2006; Gocal et al., 2015). Based on peer‐reviewed literature, the most frequently used designs were modified with phosphorothioate linkages on the terminal bases (Figure 1b; Andrieu‐Soler et al., 2005; De Piédoue et al., 2007; Radecke et al., 2006). In mammalian cells, these end linkages can be toxic, with the degree of toxicity being correlated to the number of phosphorothioate linkages as measured as fractional cell survival or γ‐H2AX phosphorylation (Olsen et al., 2005; Rios et al., 2012). As the number of phosphorothioate linkages decreased, the corrected portion of the population increased (Rios et al., 2012). An alternative single‐strand design that has been used successfully to mediate conversion of an inactive GFP target in wheat and AHAS in canola contains a 5′Cy3 label and a 3′idC reverse base (Figure 1c; Dong et al., 2006; Gocal et al., 2015). The Cy3 label allows for the visualization of oligonucleotides within cells and in most plant varieties is less toxic than the phosphorothioate end‐protective linkages (C. Schöpke unpublished data). Triplex‐forming oligonucleotides have also been used for gene correction, but generally they require homopurine or homopyrimidine stretches for triplex formation and thus have been less used (Figure 1d; Havre and Glazer, 1993; Wang et al., 1996).
Precision in gene conversion
ODM employing the chimeraplast design was first used to correct the alkaline phosphatase gene on episomal DNA in mammalian CHO cells, yielding a correction frequency of 30% (Yoon et al., 1996). Shortly thereafter, this technique was successfully used to correct a mutation in the β‐globin gene that is responsible for sickle‐cell anemia, while at the same time the highly (90%) homologous locus δ‐globin remained unaltered, proving that the base change occurred specifically in the targeted gene (Cole‐Strauss et al., 1996). Although an overall conversion frequency was not reported by the authors, the correction of the mutated β‐globin gene was shown to be dose dependent. Oligonucleotide‐directed mutagenesis alterations in the X chromosome gene, hypoxanthine‐guanine phosphoribosyl‐transferase (HPRT), were measured using phosphorothioate‐protected single‐strand oligonucleotides targeting single point loss‐of‐function mutations in the HPRT gene previously generated by ethyl methanesulfonate (EMS) in the V79 male Chinese hamster lung cell line (Kenner et al., 2002). Hypoxanthine‐guanine phosphoribosyl‐transferase encodes an enzyme in the purine salvage pathway, which when mutated becomes resistant to 6‐thioguanine. Because there are many possible loss‐of‐function HPRT alleles that will lead to 6‐thioguanine resistance and because it is an X‐linked gene, it is possible to measure both conversion and random mutagenesis events of a single allele. The study found no significant difference in sensitivity to 6‐thioguanine between the wild‐type control cell populations and the converted cell populations treated with the oligonucleotides, suggesting no detectable random mutagenesis occurred at this locus. Additional work targeting the β‐globin locus in CD34+‐enriched cell population showed a targeting efficiency range from 5% to 13%, while the closely related homologue δ‐globin remained unaltered, thereby providing additional evidence for the specificity of targeting (Xiang et al., 1997). In summary, the data from Cole‐Strauss et al. (1996), Kenner et al. (2002) and Xiang et al. (1997) provide proof that ODM is capable of precisely targeting a single‐base pair mutation in genomic DNA at a reasonable frequency and in a highly specific manner.
Conversion is independent of the transcriptional state of the gene
Additional studies using chimeraplasts have been carried out on target genes with different transcriptional states. For example, Kren et al. (1997) converted the functional, transcriptionally active alkaline phosphatase gene in the human hepatoma cell line HuH‐7 to a mutant form. This was accomplished with a relative frequency of approximately 11.9%, which, when corrected for transfection efficiency, approached nearly 43%. In this report, an alternate chimeraplast design where the mismatch is present only in the DNA strand was also examined. This design differed from the original chimeraplast design structure wherein the mismatch is on both the DNA and the RNA/DNA strands of the chimeraplast. The efficiency of conversion using this altered chimeraplast was only about 2%, which is substantially less than the original design. While previous studies reported success in editing transcriptionally active targets, Kren et al. (1997) were able to show that a chimeraplast was also capable of altering the genomic sequence in a transcriptionally inactive gene. For this work, they targeted the β‐globin locus, which is silent in HuH‐7 cells, and converted it from the wild‐type allele to the sickle‐cell allele, demonstrating that the activity of the chimeraplast is independent of the transcriptional state of the target gene (Kren et al., 1997). Similarly, Bertoni et al. (2005) showed successful targeting of the dystrophin gene proliferating myoblasts, where it is transcriptionally silent.
Stability and heritability of the nucleotide change
Alexeev and Yoon (1998) were the first to demonstrate the stability and heritability of a nucleotide change induced by a chimeraplast. They targeted the tyrosinase gene, which encodes a key enzyme for melanin synthesis and pigmentation in melanocytes derived from albino mice. These melanocytes harboured a point mutation in the tyrosinase gene, resulting in an amino acid change that abolished enzymatic activity. Correction of the point mutation restores the enzymatic activity, enabling cell pigmentation by melanin synthesis. Transfection of albino melanocytes with a chimeraplast resulted in black‐pigmented cells, which were cloned and continued to exhibit the pigmentation throughout several generations. Analysis of the tyrosinase alleles in the clones demonstrated that at least one locus had been corrected, thereby restoring the full‐length enzymatically active protein in these clones. Thus, the phenotypic and genotypic changes effected by chimeraplasts were shown to be permanent and stable. The permanence, stability and heritability of precise changes were later demonstrated, as will be discussed below, in various plant systems for both transgenic marker genes as well as the acetohydroxy acid synthase (AHAS) gene target conferring resistance to various herbicides (Beetham et al., 1999; Gocal et al., 2015; Kochevenko and Willmitzer, 2003; Okuzaki and Toriyama, 2004; Zhu et al., 1999, 2000).
Conversion in bacteria
Single‐strand oligonucleotides have also been used to successfully convert genes in bacteria, specifically E. coli (rpsL and rpoB genes, Swingle et al., 2010; various targets, Wang et al., 2009) and in Pseudomonas syringae (rpsL gene, Swingle et al., 2010). In E. coli, both RecA and MutS activities were required for efficient conversion. This was illustrated by the fact that conversion in the recA‐WM1100 strain was indistinguishable from background, and conversion activity was fully restored by complementation with a RecA‐expressing plasmid (Metz et al., 2002). In this same study, similar data was presented for MutS, suggesting a two‐step mechanism of pairing followed by repair. An improvement to the conversion efficiency in bacteria was achieved using the beta protein, a recombinase from phage λ, combined with chemically synthesized single‐strand oligonucleotides to correct the galK restoring auxotrophy (Ellis et al., 2001). The beta protein is encoded by bet, one of the three lambda Red functions. These results were dramatically extended in 2009 by Wang et al. using a technique called multiplex automated genome engineering (MAGE) that enabled a handful of mutations to be obtained within a single E. coli genome. Building on this multiplex approach, Isaacs et al. (2011) showed that using conjugative assembly genome engineering (CAGE) technology, they were able to site specifically replace more than 300 TAG stop codons with synonymous TAA codons to produce a completely recoded E. coli.
Plants
oligonucleotide‐directed mutagenesis (ODM) is a non‐transgenic base pair‐specific precision gene editing platform that has been significantly advanced through RTDS (Gocal et al., 2015) to achieve novel and commercially valuable traits in agriculturally important crops. The RTDS technology harnesses the cell's normal DNA repair system to edit specific targeted bases within the genome through the use of chemically synthesized oligonucleotides. These oligonucleotides are used as repair templates to generate mismatches in the DNA at the target site. Through homology‐directed pairing between the oligonucleotide and the DNA of the target region, the cell's repair machinery is directed to those sites to correct the mismatched base(s) guided by the oligonucleotide sequence. Once the correction process is completed, the oligonucleotide is degraded by the cell through natural processes.
An initial foray of this technology in a plant system involved using cell free extracts to correct a plasmid bearing a non‐sense mutation in the coding sequence of an nptII gene, thereby restoring the active coding sequence and function to confer kanamycin resistance to bacteria into which these corrected plasmids were transformed (Gamper et al., 2000). Using extracts from both mammalian and plant cells, this genetic (phenotypic) readout system in bacteria has been heavily used to dissect the structure/activity relationship of chimeraplasts and later single‐strand oligonucleotide designs (Metz, R., Frank, B., DiCola, M., Kurihara, T., Bailey, A., Rice, M.C., May, G.D., Kipp, P.B., and Kmiec, E.B. unpublished data). The most effective designs were tested for their ability to correct transiently expressed plasmid targets with in vivo readout in various plant cells (Walker, K.A., Avissar, P., Sawycky, X.L., Beetham, P.R. unpublished data).
Several years after the first successful use of ODM in mammalian systems, this gene editing technology was employed in plants. Most applications of ODM in plants to convert endogenous loci have targeted single point mutations in the acetolactate synthase (ALS) gene(s), also known as the acetohydroxy acid synthase (AHAS) gene(s). This enzyme catalyses the first step in the biosynthesis of the essential branched chain amino acids isoleucine, leucine and valine, and mutant enzymes is readily selectable with herbicides that inhibit them; imidazolinones (Imis), sulfonylureas (SUs), chlorsulfuron (CS), pyrimidinylthiobenzoates and bispyribac‐sodium (BS) (Tan et al., 2005). These are Group 2 herbicides based on the nomenclature from the Canadian herbicide classification system – HRAC Group B and Australian Group B. To achieve resistance to the aforementioned herbicide chemistries, one of three amino acid positions was targeted, specifically P197, W574 and S653, with the numbering based on the sequence of the Arabidopsis AHAS protein. The first published study describing the successful application of ODM technology was done in a tobacco cell line known as Nt‐1 (Beetham et al., 1999; Ruiter et al., 2003), followed closely by maize (Zhu et al., 1999, 2000), then Arabidopsis (Kochevenko and Willmitzer, 2003), rice (Okuzaki and Toriyama, 2004) and oil seed rape (Brassica napus) (Gocal et al., 2015; Ruiter et al., 2003). In tobacco, maize, Arabidopsis and rice, chimeraplasts were employed to target the conversions, whereas a 5′Cy3 label and an 3′idC reverse base protected single‐strand oligonucleotide design was used to target conversions in oil seed rape (Gocal et al., 2015). Furthermore, the method used to deliver the oligonucleotides varied between plants, with PEG‐mediated delivery to protoplasts being utilized in Arabidopsis (Kochevenko and Willmitzer, 2003), tobacco (Ruiter et al., 2003) and oil seed rape (Gocal et al., 2015; Ruiter et al., 2003), and biolistics being used as the delivery method for tobacco (Beetham et al., 1999; Ruiter et al., 2003), maize (Zhu et al., 1999, 2000), oil seed rape (Ruiter et al., 2003) and rice (Okuzaki and Toriyama, 2004). In both maize and rice, based on the number of cells receiving oligonucleotides, the biolistics method resulted in a conversion rate of 1 × 10−4, similar to reported conversion frequencies for PEG‐mediated delivery (Okuzaki and Toriyama, 2004; Zhu et al., 1999, 2000). However, it is difficult to compare different oligonucleotide delivery methods because conversion rates varied depending on the crop, the cell biology system, the oligonucleotide type, its concentration, the strand being targeted (coding or non‐coding) and the targeted mutation being made.
RTDS technology applied to the conversion of BFP to GFP in Arabidopsis
Oligonucleotide‐mediated conversions have been improved in many ways by utilizing a fluorescence conversion system wherein a blue fluorescent protein (BFP) can be changed to a green fluorescent protein (GFP) by editing a single nucleotide of the BFP gene. For instance, optimization of oligonucleotide length and end‐protective chemistries has shown promise in enhancing conversion efficiency. To illustrate the effectiveness of oligonucleotide‐mediated conversions in Arabidopsis, protoplasts derived from a BFP transgenic line were tested for BFP to GFP gene editing. Protoplasts were transfected with either a 41, 101, or 201 nucleobase (nb) oligonucleotide (BFP/41, BFP/101, BFP/201), each containing the C→T edit required to convert BFP to GFP, and monitored for GFP fluorescence 72 h after oligonucleotide introduction using cytometry. All three oligonucleotide lengths tested resulted in a notably higher percentage of GFP‐positive cells when compared to the control treatment (Figure 2; Warburg, Z.J., Miller, R., Mozoruk, J. and Sauer, N.J. unpublished data). Oligonucleotide length had a positive correlation with respect to GFP fluorescing cells, with oligonucleotide BFP/201 resulting in nearly five times more GFP‐positive cells than oligonucleotide BFP/41 (Figure 2). This result demonstrates that oligonucleotide‐mediated conversions are an effective method to make precise changes in Arabidopsis, and further that oligonucleotide optimization can play an important role with respect to the frequency of targeted edits.
Figure 2.
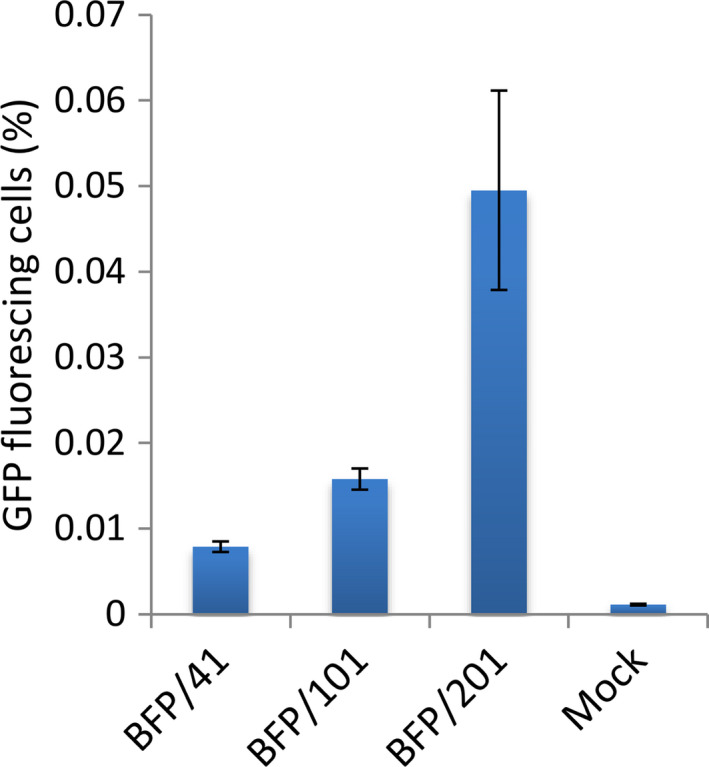
Frequency of transgene editing using RTDS technology in Arabidopsis protoplasts. Oligonucleotides of three different lengths (nb) each possessing the C→T edit in codon H66 (H66Y; CAC→TAC) required to convert blue fluorescent protein (BFP) to green fluorescent protein (GFP) were delivered into Arabidopsis protoplasts by the PEG method. Percentage of GFP fluorescing cells was measure 72 h after delivery by cytometry. Error bars are mean ± SEM (n = 3).
Regulatory view on ODM
With an ability to precisely change sequences in genomes, ODM is one of several new breeding techniques that is leading to the commercialization of crop plants. In the United States, the Animal Plant Health Inspection Service (APHIS) of the U.S. Department of Agriculture indicated that a herbicide tolerant canola developed by Cibus through ODM is not subject to their regulation and that is it is exempt from biotechnology regulation under 7 CFR Part 340. In Canada, the approval for release of a plant variety with a new trait does not take into account the method with which it was produced (Shearer, 2015). Therefore, crops developed through ODM are evaluated the same way as crops developed through other breeding methods. In 2007, the European Commission set up a New Plant Breeding Techniques working group with the task to assess whether eight new plant breeding techniques, among them ODM, would fall under the scope of European GMO legislation. Based on the final report to the European Commission (discussed in Schiemann and Hartung, 2015, p 202), a majority of the members of the working group deemed that ODM is a mutagenesis technique and, therefore, would fall outside the scope of the directives governing GMOs. Similar conclusions were reached in a commentary article by a group of scientists, in which regulatory aspects of ODM are discussed (Breyer et al., 2009). This decision will allow for the commercialization of crop plants with traits developed by ODM to occur in a timely manner.
Improving conversion efficiency and future prospects
While significant and practical gene editing frequency has been demonstrated with ODM techniques in many plant species, correction rates have been relatively low and editing has depended on large amounts of the introduced oligonucleotide.
Various treatments have been explored to enhance conversion efficiency. In mammalian systems, recognition by the mismatch repair machinery or components of the non‐homologous end‐joining (NHEJ) DNA repair pathway have had a negative effect on conversion frequency, and consequently conversion frequencies have increased when such components are knocked down or inactivated (Dekker et al., 2003, 2006; Morozov and Wawrousek, 2008). By comparison, chemicals or treatments that augment homology‐based DNA repair also increased conversion efficiency (Morozov and Wawrousek, 2008; Olsen et al., 2005 Parekh‐Olmedo et al., 2005).
Another such improvement has been achieved through the use of DNA double‐strand breakers. When oligonucleotides were combined with chemicals or antibiotics that generate DNA double‐strand breaks, significant enhancement in the frequency of gene targeting was observed (Ferrara et al., 2004; Parekh‐Olmedo et al., 2005). Similarly, engineered nucleases have been shown to enhance the efficacy and precision of gene editing in combination with oligonucleotides, primarily in eukaryotic systems (Chen et al., 2011; Connelly et al., 2010; DiCarlo et al., 2013; Gratz et al., 2013; Strouse et al., 2014; Suzuki et al., 2003; Svitashev et al., 2015; Voytas, 2013; Wang et al., 2015; Wu et al., 2013; Zhao et al., 2014). These nucleases were engineered to cleave DNA in a target‐specific manner. They include meganucleases, zinc finger nucleases (ZFN), TAL effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)‐associated endonuclease Cas9 (CRISPR‐Cas9) (Bortesi and Fischer, 2015; Carroll, 2014; Rousseau et al., 2011).
While numerous studies using the combination of engineered nucleases and oligonucleotides have been reported in fish and mammals (Ding et al., 2013; Hwang et al., 2013; Strouse et al., 2014; Wefers et al., 2013; Yang et al., 2013), the published data for plant systems is very limited. To date, we are aware of only five examples. In one study using rice protoplasts, Shan et al. (2015) introduced two restriction enzyme sites into the sequence of the OsPDS gene in a partial transgenic approach using an integrated CRISPR/Cas9 and single‐stranded oligonucleotides. In this publication, the authors did not report successful regeneration of plants from edited protoplasts. In another study, Wang et al. (2015) targeted conversion of the enolpyruvylshikimate‐3‐phosphate synthase (EPSPS) locus in rice by employing TALENs transgenically integrated into the genome combined with a chimeraplast. EPSPS encodes an enzyme in the shikimate pathway found in plants but not mammalian systems, which is a key step in the biosynthesis of the aromatic amino acids phenylalanine, tyrosine and tryptophan. In plants, EPSPS is a target for the herbicide, glyphosate, where it acts as a competitive inhibitor of the binding site for phosphoenolpyruvate (Schönbrunn et al., 2001). They reported detecting 1 of 25 transgenic lines with the intended EPSPS gene edit. However, this line, in addition to the single base edit, also contained an imprecise NHEJ event, sending the coding sequence with the intended edit out of frame (Wang et al., 2015).
In a recent preprint (Svitashev et al., 2015), CRISPR/Cas9 and a phosphinothricin acetyltransferase (PAT) selectable marker to confer resistance to bialaphos were delivered into immature corn embryos using bombardment, in combination with either a double‐stranded PCR product (794 bp) or two different single‐stranded 127mer oligonucleotides, which served as template DNA targeting the P197 locus in the AHAS gene. After selection with bialaphos and/or chlorsulfuron, an editing frequency of three or four events in 1000 was reported for oligonucleotides, compared with two events in 1000 for double‐stranded DNA. In a fourth example, a double‐strand DNA donor was also used as the repair template for a CRISPR/Cas9‐induced DSB in the work of Li et al. (2013) to target a new restriction site in the PDS locus of Nicotinia benthamiana. The converted protoplasts in this work were not regenerated into whole plants. In our unpublished data (Woodward, M. and Narvaez‐Vasquez, J.), a study was carried out targeting the two EPSPS loci in flax using a combination of TALEN and oligonucleotide. In this work, we detected 0.19% precise and scarless EPSPS edits in both loci in 7‐day‐old microcolonies. Collectively, these studies, as well as work performed by our group, demonstrate that while significant precise gene editing events in plants can be achieved using oligonucleotides alone, this effect can be enhanced by a variety of reagents that cause DNA double‐strand breaks.
The promise of ODM and RTDS to precisely deliver predictable targeted edits to specific targets within the nuclear genome, as directed by exogenously supplied chemically synthesized oligonucleotides, remains strong. For more than a decade, Cibus has focused on improving this process and extending it from model systems to develop traits in commercial crops. As was the case in E. coli, by applying treatments that improve conversion efficiency, the range of possibilities will expand to allow multiple conversions in single gene targets as well as simultaneous conversion of multiple targets in a single cell. Together, improvements to this non‐transgenic breeding technology will deliver traits more rapidly and offer the promise of accelerating and completely revolutionizing the breeding process.
Acknowledgements
We would like to thank all members of the Cibus team, current and past, for their many contributions to the exciting field of precision genome editing technologies in plants. All authors are employees of Cibus, an industry leader in developing and applying precision gene editing tools to meet agricultural, industrial and human health needs. Cibus is a global company with offices in Europe and North America, including its state‐of‐the‐art research and development campus in San Diego, California.
References
- Aarts, M. , Dekker, M. , de Vries, S. , van der Wal, A. and te Riele, H. (2006) Generation of a mouse mutant by oligonucleotide‐mediated gene modification in ES cells. Nucleic Acids Res. 34, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeev, V. and Yoon, K. (1998) Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA‐DNA oligonucleotide. Nat. Biotechnol. 16, 1343–1346. [DOI] [PubMed] [Google Scholar]
- Andrieu‐Soler, C. , Casas, M. , Faussat, A.M. , Gandolphe, C. , Doat, M. , Tempé, D. , Giovannangeli, C. et al. (2005) Stable transmission of targeted gene modification using single‐stranded oligonucleotides with flanking LNAs. Nucleic Acids Res. 33, 3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetham, P.R. , Kipp, P.B. , Sawycky, X.L. , Arntzen, C.J. and May, G.D. (1999) A tool for functional plant genomics: chimeric RNA/DNA oligonucleotides cause in vivo gene‐specific mutations. Proc. Natl Acad. Sci. USA, 96, 8774–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni, C. , Morris, G.E. and Rando, T.A. (2005) Strand bias in oligonucleotide‐mediated dystrophin gene editing. Hum. Mol. Genet. 14, 221–233. [DOI] [PubMed] [Google Scholar]
- Bortesi, L. and Fischer, R. (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. [DOI] [PubMed] [Google Scholar]
- Breyer, D. , Herman, P. , Brandenburger, A. , Gheysen, G. , Remaut, E. , Soumillion, P. , Van Doorsselaere, J. et al. (2009) Genetic modification through oligonucleotide‐mediated mutagenesis. A GMO regulatory challenge? Environ. Biosafety Res. 8, 57–64. [DOI] [PubMed] [Google Scholar]
- Carroll, D. (2014) Genome engineering with targetable nucleases. Methods Mol. Biol. 83, 409–439. [DOI] [PubMed] [Google Scholar]
- Chen, F. , Pruett‐Miller, S.M. , Huang, Y. , Gjoka, M. , Duda, K. , Taunton, J. , Collingwood, T.N. et al. (2011) High‐frequency genome editing using ssDNA oligonucleotides with zinc‐finger nucleases. Nat. Methods, 8, 753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole‐Strauss, A. , Yoon, K. , Xiang, Y. , Byrne, B.C. , Rice, M.C. , Gryn, J. , Holloman, W.K. et al. (1996) Correction of the mutations responsible for sickle cell anemia by an RNA‐DNA oligonucleotide. Science, 273, 1386–1389. [DOI] [PubMed] [Google Scholar]
- Connelly, J.P. , Barker, J.C. , Pruett‐Miller, S. and Porteus, M.H. (2010) Gene correction by homologous recombination with zinc finger nucleases in primary cells from a mouse model of a generic recessive genetic disease. Mol. Ther. 18, 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piédoue, G. , Andrieu‐Soler, C. , Concordet, J.P. , Sun, J.S. , Lopez, B. , Kuzniak, I. , Leboulch, P. et al. (2007) Targeted gene correction with 5′ acridine‐oligonucleotide conjugates. Oligonucleotides, 17, 258–263. [DOI] [PubMed] [Google Scholar]
- Dekker, M. , Brouwers, C. and Te Riele, H. (2003) Targeted gene modification in mismatch‐repair‐deficient embryonic stem cells by single‐stranded DNA oligonucleotides. Nucleic Acids Res. 31, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, M. , Brouwers, C. , Aarts, M. , van der Torre, J. , de Vries, S. , van de Vrugt, H. and Te Riele, H. (2006) Effective oligonucleotide‐mediated gene disruption in ES cells lacking the mismatch repair protein MSH3. Gene Ther. 13, 686–694. [DOI] [PubMed] [Google Scholar]
- DiCarlo, J.E. , Norville, J.E. , Mali, P. , Rios, X. , Aach, J. and Church, G.M. (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR‐Cas systems. Nucleic Acids Res. 41, 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Q. , Lee, Y.‐K. , Schaefer, E.A.K. , Peters, D.T. , Veres, A. , Kim, K. , Kuperwasser, N. et al. (2013) A TALEN genome‐editing system for generating human stem cell‐based disease models. Cell Stem Cell, 12, 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C. , Beetham, P. , Vincent, K. and Sharp, P. (2006) Oligonucleotide‐directed gene repair in wheat using a transient plasmid gene repair assay system. Plant Cell Rep. 25, 457–465. [DOI] [PubMed] [Google Scholar]
- Ellis, H.M. , Yu, D. and DiTizio, T. (2001) High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single‐stranded oligonucleotides. Proc. Natl Acad. Sci. USA, 98, 6742–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara, L. , Parekh‐Olmedo, H. and Kmiec, E.B. (2004) Enhanced oligonucleotide‐directed gene targeting in mammalian cells following treatment with DNA damaging agents. Cell Res. 300, 170–179. [DOI] [PubMed] [Google Scholar]
- Gamper, H.B. , Parekh, H. , Rice, M.C. , Bruner, M. , Youkey, H. and Kmiec, E.B. (2000) The DNA strand of chimeric RNA/DNA oligonucleotides can direct gene repair/conversion activity in mammalian and plant cell‐free extracts. Nucleic Acids Res. 28, 4332–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal, G.F.W. , Schöpke, C. and Beetham, P.R. (2015) Oligo‐mediated targeted gene editing. In Advances in New Technology for Targeted Modification of Plant Genomes Chapter 5 ( Zhang, F. , Puchta, H. and Thomson, J.G. , eds), pp. 73–90. Berlin, Heidelberg: Springer Verlag. [Google Scholar]
- Gratz, S.J. , Cummings, A.M. , Nguyen, J.N. , Hamm, D.C. , Donohue, L.K. , Harrison, M.M. , Wildonger, J. et al. (2013) Genome engineering of Drosophila with the CRISPR RNA‐guided Cas9 nuclease. Genetics, 194, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havre, P.A. and Glazer, P.M. (1993) Targeted mutagenesis of simian virus 40 DNA mediated by a triple helix‐forming oligonucleotide. J. Virol. 67, 7324–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, C.A. 3rd , Phillips, S. , Edgell, M.H. , Gillam, S. , Jahnke, P. and Smith, M. (1978) Mutagenesis at a specific position in a DNA sequence. J. Biol. Chem. 253, 6551–6560. [PubMed] [Google Scholar]
- Hwang, W.Y. , Fu, Y. , Reyon, D. , Maeder, M.L. , Tsai, S.Q. , Sander, J.D. , Peterson, R.T. et al. (2013) Efficient genome editing in zebrafish using a CRISPR–Cas system. Nat. Biotechnol. 31, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs, F.J. , Carr, P.A. , Wang, H.H. , Lajoie, M.J. , Sterling, B. , Kraal, L. , Tolonen, A.C. et al. (2011) Precise manipulation of chromosomes in vivo enables genome‐wide codon replacement. Science, 333, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner, O. , Kneisel, A. , Klingler, J. , Bartelt, B. , Speit, G. , Vogel, W. and Kaufmann, D. (2002) Targeted gene correction of hprt mutations by 45 base single‐stranded oligonucleotides. Biochem. Biophys. Res. Comm. 299, 787–792. [DOI] [PubMed] [Google Scholar]
- Kochevenko, A.J. and Willmitzer, L. (2003) Chimeric RNA/DNA oligonuclelotide‐based site specific modification of the tobacco acetolactate synthase gene. Plant Physiol. 132, 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren, B.T. , Cole‐Strauss, A. , Kmiec, E.B. and Steer, C.J. (1997) Targeted nucleotide exchange in the alkaline phosphatase gene of HuH‐7 cells mediated by a chimeric RNA/DNA oligonucleotide. Hepatology, 25, 1462–1468. [DOI] [PubMed] [Google Scholar]
- Li, J.F. , Aach, J. , Norville, J.E. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. et al. (2013) Multiplex and homologous recombination‐mediated plant genome editing via guide RNA/Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz, R. , DiCola, M. , Kurihara, T. , Bailey, A. , Frank, B. , Roecklein, B. and Blaese, M. (2002) Mode of action of RNA/DNA oligonucleotides: progress in the development of gene repair as a therapy for alpha(1)‐antitrypsin deficiency. Chest, 121, 91S–97S. [DOI] [PubMed] [Google Scholar]
- Moerschell, R.P. , Tsunasawa, S. and Sherman, F. (1988) Transformation of yeast with synthetic oligonucleotides. Proc. Natl Acad. Sci. USA, 85, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov, V. and Wawrousek, E.F. (2008) Single‐strand DNA‐mediated targeted mutagenesis of genomic DNA in early mouse embryos is stimulated by Rad51/54 and by Ku70/86 inhibition. Gene Ther. 15, 468–472. [DOI] [PubMed] [Google Scholar]
- Okuzaki, A. and Toriyama, K. (2004) Chimeric RNA/DNA oligonucleotide‐directed gene targeting in rice. Plant Cell Rep. 22, 509–512. [DOI] [PubMed] [Google Scholar]
- Olsen, P.A. , Randol, M. and Krauss, S. (2005) Implications of cell cycle progression on functional sequence correction by short single‐stranded DNA oligonucleotides. Gene Ther. 12, 546–551. [DOI] [PubMed] [Google Scholar]
- Parekh‐Olmedo, H. , Ferrara, L. , Brachman, E. and Kmiec, E.B. (2005) Gene therapy progress and prospects: targeted gene repair. Gene Ther. 12, 639–646. [DOI] [PubMed] [Google Scholar]
- Radecke, S. , Radecke, F. , Peter, I. and Schwarz, K. (2006) Physical incorporation of a single‐stranded oligodeoxynucleotide during targeted repair of a human chromosomal locus. J. Gene Med. 8, 217–228. [DOI] [PubMed] [Google Scholar]
- Rios, X. , Briggs, A.W. , Christodoulou, D. , Gorham, J.M. , Seidman, J.G. and Church, G.M. (2012) Stable gene targeting in human cells using single‐strand oligonucleotides with modified bases. PLoS ONE, 7, e36697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau, J. , Chapdelaine, P. , Boisvert, S. , Almeida, L.P. , Corbeil, J. , Montpetit, A. and Tremblay, J.P. (2011) Endonucleases: tools to correct the dystrophin gene. J. Gene Med. 13, 522–537. [DOI] [PubMed] [Google Scholar]
- Ruiter, R. , van den Brande, I. , Stals, E. , Delauré, S. , Cornelissen, M. and D'Halluin, K. (2003) Spontaneous mutation frequency in plants obscures the effect of chimeraplasty. Plant Mol. Biol. 53, 675–689. [DOI] [PubMed] [Google Scholar]
- Schiemann, J. and Hartung, F. (2015) EU perspectives on new plant‐breeding techniques. In NABC Report 26. New DNA‐editing Approaches: Methods, Applications and Policy for Agriculture ( Eaglesham, H. and Hardy, R.W.F. , eds), pp. 201–210. Ithaca, NY: Boyce Thompson Institute. [Google Scholar]
- Schönbrunn, E. , Eschenburg, S. , Shuttleworth, W.A. , Schloss, J.V. , Amrhein, N. and Evans, J.N.S. (2001) Interaction of the herbicide glyphosate with its target enzyme 5‐enolpyruvylshikimate 3‐phosphate synthase in atomic detail. Proc. Natl Acad. Sci. USA, 98, 1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Zhang, Y. , Chen, K. , Zhang, K. and Gao, C. (2015) Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13, 791–800. [DOI] [PubMed] [Google Scholar]
- Shearer, H. (2015) Regulation of plants with novel traits: Canadian perspectives on the “novelty” trigger. In NABC Report 26. New DNA‐editing Approaches: Methods, Applications and Policy for Agriculture ( Eaglesham, H. and Hardy, R.W.F. , eds), pp. 193–200. Ithaca, NY: Boyce Thompson Institute. [Google Scholar]
- Strouse, B. , Bialk, P. , Niamat, R. , Rivera‐Torres, N. and Kmiec, E.B. (2014) Combinatorial gene editing in mammalian cells using ssODNs and TALENs. Sci. Rep. 4, 3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T. , Murai, A. and Muramatsu, T. (2003) Low‐dose bleomycin induces targeted gene repair frequency in cultured melan‐c cells using chimeric RNA/DNA oligonucleotide transfection. Int. J. Mol. Med. 12, 109–114. [PubMed] [Google Scholar]
- Svitashev, S. , Young, J.K. , Schwartz, C. , Gao, H. , Falco, S.C. and Cigan, A.M. (2015) Targeted mutagenesis, precise gene editing and site‐specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingle, B. , Markel, E. , Costantino, N. , Bubunenko, M.G. , Cartinhour, S. and Court, D.L. (2010) Oligonucleotide recombination in Gram‐negative bacteria. Mol. Microbiol. 75, 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, S. , Evans, R.R. , Dahmer, M.L. , Singh, B.K. and Shaner, D.L. (2005) Imidazolinone‐tolerant crops: history, current status and future. Pest Manag. Sci. 61, 246–257. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F. (2013) Plant genome engineering with sequence‐specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Seidman, M.M. and Glazer, P.M. (1996) Mutagenesis in mammalian cells induced by triple helix formation and transcription‐coupled repair. Science, 271, 802–805. [DOI] [PubMed] [Google Scholar]
- Wang, H.H. , Isaacs, F.J. , Carr, P.A. , Sun, Z.Z. , Xu, G. , Forest, C.R. and Church, G.M. (2009) Programming cells by multiplex genome engineering and accelerated evolution. Nature, 460, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Liu, Y. , Zhang, C. , Liu, J. , Liu, X. , Wang, L. , Wang, W. et al. (2015) Gene editing by co‐transformation of TALEN and chimeric RNA/DNA oligonucleotides on the rice OsEPSPS Gene and the inheritance of mutations. PLoS ONE, 10(4), e0122755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefers, B. , Meyer, M. , Ortiz, O. , Hrabé de Angelis, M. , Hansen, J. , Wurst, W. and Kühn, R. (2013) Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc. Natl Acad. Sci. USA, 110, 3782–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Liang, D. , Wang, Y. , Bai, M. , Tang, W. , Bao, S. , Yan, Z. et al. (2013) Correction of a genetic disease in mouse via use of CRISPR‐Cas9. Cell Stem Cell, 13, 659–662. [DOI] [PubMed] [Google Scholar]
- Xiang, Y. , Cole‐Strauss, A. , Yoon, K. , Gryn, J. and Kmiec, E.B. (1997) Targeted gene conversion in a mammalian CD34+‐enriched cell population using a chimeric RNA/DNA oligonucleotide. J. Mol. Med. 75, 829–835. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T. , Moerschell, R.P. , Wakem, L.P. , Komar‐Panicucci, S. and Sherman, F. (1992) Strand‐specificity in the transformation of yeast with synthetic oligonucleotides. Genetics, 131, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Guell, M. , Byrne, S. , Yang, J.L. , De Los Angeles, A. , Mali, P. , Aach, J. et al. (2013) Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 41, 9049–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, K. , Cole‐Strauss, A. and Kmiec, E.B. (1996) Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA. DNA oligonucleotide. Proc. Natl Acad. Sci. USA, 93, 2071–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, P. , Zhang, Z. , K.E., H. , Yue, Y. and Xue, D. (2014) Oligonucleotide‐based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24, 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T. , Peterson, D.J. , Tagliani, L. , St. Clair, G. , Baszczynski, C.L. and Bowen, B. (1999) Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc. Natl Acad. Sci. USA, 96, 8768–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T. , Mettenburg, K. , Peterson, D.J. , Tagliani, L. and Baszczynski, C. (2000) Engineering herbicide‐resistant maize using chimeric RNA/DNA oligonucleotides. Nat. Biotechnol. 18, 555–558. [DOI] [PubMed] [Google Scholar]


