Abstract
Genomic analyses are yielding a host of new information on the multiple genetic abnormalities associated with specific types of cancer. A comprehensive description of cancer-associated genetic abnormalities can improve our ability to classify tumors into clinically relevant subgroups, and, on occasion, identify mutant genes that drive the cancer phenotype (“drivers”). More often, though, the functional significance of cancer-associated mutations is difficult to discern. Genome-wide pooled shRNA screens enable global identification of the genes essential for cancer cell survival and proliferation, providing a “functional genomic” map of human cancer to complement genomic studies. Using a lentiviral shRNA library targeting ~16,000 genes and a newly developed, dynamic scoring approach, we identified essential gene profiles in 72 breast, pancreatic, and ovarian cancer cell lines. Integrating our results with current and future genomic data should facilitate the systematic identification of drivers, unanticipated synthetic lethal relationships, and functional vulnerabilities of these tumor types.
Keywords: cancer, shRNA, functional genetics, pooled screens
INTRODUCTION
Recent technological advances have revolutionized our understanding of cancer genetics. Transcriptional profiling, copy number variation (CNV) and deep sequencing have revised the classification of many tumors into molecular subtypes that provide improved prognostic information compared with conventional clinical and histopathological classification schemes (1, 2). Yet often, these subtypes provide little functional information about the molecular events that drive cancer cell behavior. Genome-wide sequencing studies have identified hundreds to thousands of mutations in individual tumors (3-6), yet it often is difficult to know which of these are essential for pathogenesis (i.e., “drivers”), as opposed to “passenger” mutations. Even when a driver oncogene (e.g., KRAS, MYC) or tumor suppressor gene (e.g. TP53, BRCA1/2) is known, these can be poor targets for drug development. In addition, unanticipated gene/pathway dependencies (“synthetic lethality”) can arise as a consequence of the genetic abnormalities in cancer cells, as illustrated by the sensitivity of BRCA1/2-mutant breast cancer cells to PARP inhibitors (7, 8). The systematic identification of such synthetic lethal relationships might suggest new drug targets (9). Comparison of the genetic abnormalities and functional vulnerabilities of cancer cells should help identify new drivers and provide insight into the complex systems biology of cancer.
RNA interference technology has enabled genome-wide loss-of-function screens in mammalian cells. Most screens have used siRNAs, usually arrayed in multi-well plates. Arrayed screens have focused mainly on specific gene families, such as kinases, phosphatases, or selected candidate genes, and have yielded new insights into mechanisms of cancer cell signal transduction, cell division and cell death (10, 11). Cell proliferation assays in multi-well plates are usually constrained to a few population doublings, and gene “knock-downs” in these conditions typically last for at most a week. Therefore, siRNA screens are, by nature, transient, and might underestimate the roles of long-lived proteins on a given phenotype. Moreover, given their cost and the need for extensive automation to interrogate most of the genome, siRNA screens usually are performed on only limited numbers of cell lines, and might fail to capture the genetic heterogeneity in cancer. These properties make it difficult to use arrayed screening approaches to construct extensive functional genomic maps of cancer cells.
The more recent development of large retroviral- or lentiviral-based shRNA libraries facilitates genome-wide screening of cultured cancer cells in a pooled format (12-14), providing a potential solution to the limitations of arrayed screens. Cells are infected with these libraries at a low multiplicity of infection (MOI) and allowed to proliferate for 3-4 weeks, after which shRNAs that have been selectively depleted (referred to as “dropouts”) or enriched are identified on custom microarrays or by deep sequencing. Pooled screens have been used to define genes necessary for cancer cell proliferation/survival in cell culture (12-14), genes that enhance or interfere with the action of specific oncogenes (15) or genes that enhance the effects of anti-neoplastic drugs, suggesting potential new combination therapies (16, 17).
Most large-scale pooled shRNA screens have surveyed cancer cell lines representing multiple histotypes but usually with few representatives of any one tumor type, or have focused on cell lines from different histotypes bearing the same genetic abnormality (e.g., KRAS mutations) (15, 18). As an initial step towards a more comprehensive understanding of the vulnerabilities of breast cancer (BrCa), pancreatic ductal adenocarcinoma (PDAC) and high-grade serous ovarian carcinoma (HGS-OvCa), we performed near genome-wide pooled shRNA screens on 72 cancer cell lines, and established a unique informatics approach to monitor the dynamic evolution of cancer cell populations. We chose breast cancer because the extensive genomic information and subtype classification schemes that exist for this tumor type facilitate integrated genomic/functional genomic analysis. Ongoing genomic efforts should provide similar information for PDAC and HGS-OvCa, but we focused on these malignancies primarily because they typically are detected at an advanced stage, their prognosis remains dismal, and there is therefore an urgent need to define new therapeutic targets. Our large functional genomic dataset can be used in conjunction with orthogonal efforts to map the structural variation within cancer genomes, such as The Cancer Genome Atlas (TCGA) or the International Cancer Genome Consortium (ICGC) (19), to accelerate the identification of drivers. Initial analysis reveals only partial overlap between genomic and functional genomic classifications of cancer, and uncovers novel, unanticipated, cancer cell-specific dependencies in these three major types of cancer, some of which could be amenable to targeted therapies.
RESULTS
Classifying shRNA activity across a compendium of pooled shRNA screens
To catalogue essential genes across a defined set of cancer types, we performed genome-wide pooled screens using a library of 78,432 shRNAs targeting 16,056 unique Refseq genes (“80K” library, Supplemental Table 1), developed by The RNAi Consortium (TRC) (20-22). A total of 72 cell lines were screened, including 29 breast, 28 pancreatic, and 15 ovarian cancer lines (Figure 1A and Supplemental Table 2). Each line was screened in triplicate, and at least three time points were assessed for overall shRNA abundance during population outgrowth. The screens were highly reproducible between replicate biological populations for all of the cell lines (Rav g(BrCa)=0.9, Rav g(PDAC)=0.92, Rav g(HGS-OvCa) =0.87). The result was a dataset containing over 50 million data points from more than 200 independent cell populations.
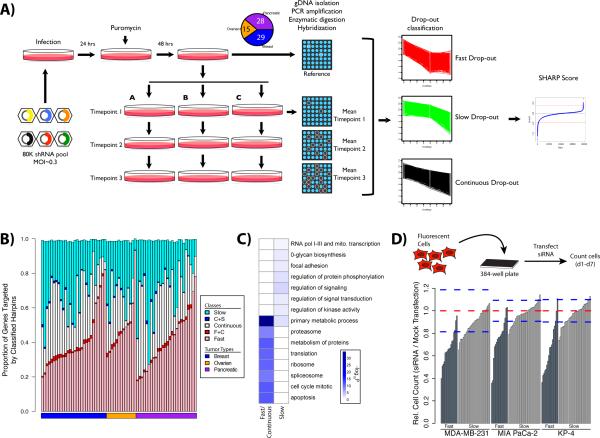
Figure 1.
Outline of Procedure for Timecourse shRNA Screening. (A) Schematic representing the steps that are involved in the shRNA functional screening. (B) Hairpins were classified based on heuristic rules (see Supplemental Table 3). The proportion of genes falling into each category is shown as a stacked bar chart. The pink, white and cyan bar segments detail genes that are targeted by a single hairpin class, while the red and blue segments indicate the sparse overlap between classes. Stacked bars do not reach a value of 1.0 as the minimal overlap observed between fast and slow, as well as all three classes is omitted. The full classification results are shown in Supplemental Table 3. (C) Enrichment plot of GO ‘Biological Process’ terms and MSigDB pathways within classified hairpins. Darker blue colours indicate more significant p-values corresponding to the enrichment. The full set of enriched terms is shown in Supplemental Figure 1. (D) Validation of selected shRNA hits by siRNA. Growth inhibition due to target gene knock-down was determined relative to mock transfected controls in MDA-MB-231, MIA PaCa-2, and KP4 cell lines. Red dashed lines indicate the mean of 60 replicate mock transfections, while the blue dashed lines indicate mean ± 3 standard deviations.
Current scoring algorithms for shRNA and siRNA screens assess dropouts at only a single time point. We reasoned that adding additional time points would provide a detailed history of individual shRNA performance, allow us to model shRNA kinetics during population outgrowth, and increase our confidence in the essentiality score derived for each gene. We also developed a set of heuristics to classify shRNAs as fast, continuous or slow dropouts, based on the rate at which an shRNA disappeared from the bulk population of cells during the screen (see Methods & Supplemental Table 3). Examples of these profiles are shown at the right of Figure 1A. Using heuristics designed to identify the most potent shRNAs in the fast, continuous, or slow classes resulted in the classification of ~2% of the shRNAs in the library into one of these categories, with 40% being fast, 30% continuous, and 30% slow dropouts. These classification criteria largely restricted hairpins to a single class. Moreover, dropout behavior largely appeared to be characteristic of the gene targeted by the hairpin rather than the shRNA itself: within any cell line, a given gene almost always fell into a single dropout class (Figure 1B). Altering our heuristics would allow us to classify more hairpins, but would result in greater overlap between dropout classes, and also would lead to less potent hairpins being classified. On average, ~0.4% of shRNAs were enriched in any given cell line; due to our shRNA barcode detection procedures (see Supplemental Table 4 and Methods), this is almost certainly a substantial underestimate of the true number of enriched hairpins (see Discussion).
To explore the hypothesis that classes of shRNAs are related to functional categories, we compared the biological functions of the gene targets (as assessed by GO categories) of hairpins classified only as fast or continuous dropouts with those classified only as slow dropouts. Fast and continuous dropout shRNAs target genes enriched for the proteasome (e.g., PSMA1, PSMB2), ribosome (e.g., RPS17), splicing machinery (e.g., SNRPD2, SF3B2, AQR, HNRNPC, THOC1), metabolism of proteins (e.g., ARCN1, COPZ1), transcription (e.g., POLR2D, POLR2E, PABPN1) and translation (e.g., EIF3B), all of which are highly conserved, housekeeping functions (Figure 1C and Supplemental Figure 1). Conversely, shRNAs classified as slow dropouts target genes that are enriched for regulation of protein phosphorylation, signaling, signal transduction and kinase activity (e.g., PTPRG, EPGN, PPP1R3B, RNF128, SERPINA3, CSNK2B, ST3GAL3, UBOX5, TNKS2). These results suggest that classifying hairpins on the basis of dropout rate reveals different functional properties of the genes that they target, providing further evidence that hairpin behavior usually reflects the properties of the underlying target gene, rather than the specific hairpin per se.
Validation of fast and slow dropouts defined by hairpin class
To further assess the validity of our results, we performed secondary screens using an orthogonal, siRNA-based assay. Fifty genes (Supplemental Table 5) available in the Dharmacon SMARTpool siRNA library were selected for further analysis. All of the shRNAs for each gene chosen fell into a single dropout class in our original screen, representing either the fast (n=17) or slow (n=33) categories. The chosen siRNAs were transfected into MDA-MB-231, MIA PaCa-2, and KP-4 cell lines, and after 7 days, cells were enumerated and compared with a mock-transfected population. Most (80–93%) of the fast dropout and 29-38% of the slow dropout genes inhibited growth significantly (Supplemental Table 5b, adj. p < 0.05, Wilcoxon rank-sum test) in this assay (Figure 1D and Supplemental Figure 2). These findings again indicate that the shRNA kinetics detected by our pooled shRNA screens reflect the properties of specific genes (i.e., fast dropouts are more likely than slow dropouts to score in this short term assay), rather than the quality/knockdown efficiency of the shRNA reagents targeted against a given gene.
Conversion of shRNA class behavior into an activity score
To convert time-course information from dropout screens into an individual value for each shRNA, we developed the “shARP” (shRNA Activity Rank Profile) score. The shARP score assigns a value to each hairpin by calculating the average slope between the measured microarray expression intensity at each time point and the intensity at time zero (i.e., T0), producing a weighted average of the fold-change across time and accounting for the growth rate of the cell line (see Methods). By integrating information across the time-course, shARP can discriminate between “fast,” “continuous” and “slow” hairpins: in general, slow dropouts have less negative shARP scores than fast or continuous dropouts (Supplemental Figure 3A).
Because each gene represented in the TRC library is targeted by an average of five hairpins, shARP scores for the different shRNAs must be converted into a gene-level score to uncover the behavior of specific genes in a screen. To this end, we defined the “Gene Activity Ranking Profile” score (GARP; see Methods) as the average of the two lowest shARP scores. We then compared the performance of the GARP score with two previously developed scoring metrics, RIGER (12) and RSA (23), for their respective ability to rank the genes in our large panel of screens. These methods differ in how the shRNA sets for a given gene are treated. GARP, RIGER “Weighted Sum” (RIGER_WS), and RIGER “Second best hairpin” (RIGER_SB) consider only the two best hairpins or the second best single hairpin. By contrast, RIGER “Kolmogorov-Smirnov” (RIGER_KS) and RSA consider the behavior of the complete set of hairpins against a given gene.
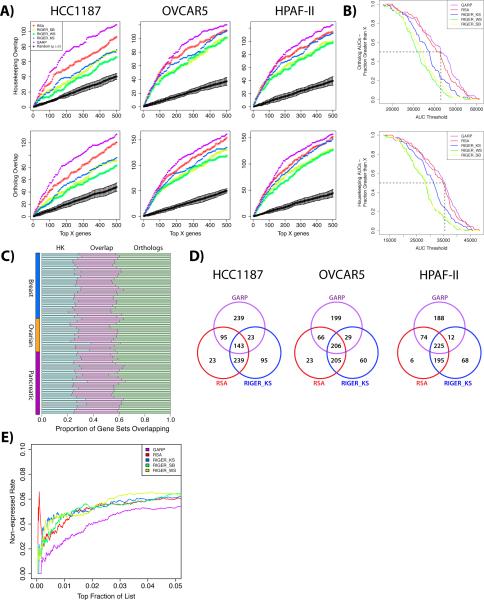
In the absence of a “gold-standard” set of essential human genes, we benchmarked the five scoring approaches using two largely non-overlapping reference sets likely to be enriched for essential genes (see Methods): highly conserved genes from eight diverse species, A. thaliana, B. taurus, C. elegans, C. familiaris, M. mulatta, M. musculus, R. norvegicus, and S. cerevisiae (24), and housekeeping genes (25). If, as seems reasonable, one assumes that housekeeping and ortholog gene sets are enriched for essential genes, then the intersection between the top 500 genes from each scoring method and each of the reference gene sets indicated that GARP outperforms the other scoring approaches in representative breast (HCC1187), ovarian (OVCAR5) and pancreatic (HPAF-II) cancer cell lines (Figure 2A).
Figure 2.
Comparison of shRNA Scoring Approaches. (A) Overlap of genes ranked by GARP, RIGER (‘Kolmogorov-Smirnov’, ‘Second-best hairpin’, and ‘Weighted Sum’), and RSA with two datasets enriched in essential genes: ‘housekeeping genes’ defined by gene expression (see Methods), and human genes conserved in eight eukaryotic species. Representative results for the breast cancer line HCC1187 (left panels), ovarian cancer cell line OVCAR5 (centre panels), and pancreatic cell line HPAF-II (right panels) are shown. Overlaps are included for ten samples of randomly selected genes (black lines, mean ± sd). (B) Cumulative distribution plots of the area under the curve (AUC) for each overlap. At each threshold on the x-axis, the fraction of cell lines with higher AUCs is indicated for each scoring method. (Upper panel) Overlap with highly conserved orthologs. (Lower panel) Overlap with housekeeping genes. (C) Stacked bar chart shows the proportion of genes from the overlap of the top 500 GARP-scored genes and the reference sets (HK and orthologs) that are unique to the HK set, unique to the ortholog set, or common to both. (D) Venn diagrams of the top 500 ranked genes for each scoring method in cell lines HCC1187, OVCAR5, and HPAF-II. (E) GARP shows the lowest rate of targeting non-expressed transcripts. “Non-expression rates” (NERs) were determined by RNA-seq for genes ranked by GARP, RSA and RIGER in seven ovarian shRNA screens. The proportion of non-expressed genes with RPKM = 0 was calculated for the top N ranked genes within each cell line, and the results were averaged for each scoring metric (see Methods).
To examine the performance of each scoring metric more thoroughly, we determined the intersection of scored genes and reference sets across our entire panel of 72 cell lines, and then computed the area under the curve (AUC) for each overlap. The cumulative distribution of AUCs for each scoring method and reference set is shown in Figure 2B, which depicts the fraction of scored gene sets with an AUC greater than a sliding threshold on the X-axis. Again, GARP performed better overall than the other scoring methods, when considering either the conserved gene reference set or the housekeeping gene reference set. Although RSA and RIGER_KS consider all shRNAs targeting each gene, RSA consistently produced higher AUCs, presumably because of its different underlying statistical approach. Importantly, the genes that drive the performance of GARP in each of the ortholog and housekeeping gene sets are largely non-overlapping (Figure 2C). By examining the top 500 hits as defined by each method, we found that GARP identifies a set of genes that are common to RSA and RIGER_KS, as well as a unique subset of genes that are distinct from the overlap between RSA and RIGER_KS in HCC1187, OVCAR5 and HPAF-II cell lines (Figure 2D). In general, the unique subsets of genes identified by GARP in the 72 cell lines account for its superior ability to identify genes in the housekeeping or highly conserved reference sets.
Correlating gene activity scores with target expression levels
All RNAi screens are susceptible to off-target effects (11, 26). Although it is difficult to quantify such effects from screening data alone, if off-target effects are common, one might expect hairpins directed against non-expressed genes to “score” as frequently as those targeting expressed genes. We performed RNA-seq on seven ovarian cancer cell lines and determined the fraction of non-expressed genes that were adjudged “essential” by GARP and the other published scoring metrics (Figure 2E). Regardless of the scoring method, the vast majority of “hits” reflected genes that were expressed (RPKM >0). Compared to the other scoring systems, though, GARP identified fewer non-expressed genes as “hits”.
We also used Receiver Operating Characteristic (ROC) curves to compare the relative ability of each score to “call” essential transcripts that also were expressed. Notably, the AUCs computed by GARP (μ=61.8±5.6%) were higher than those computed by RSA (μ=56.1±3.5%; p=0.013), RIGER_KS (μ=56.4±4.3%; p=0.006), and RIGER_SB (μ=55.3±4.2%; p=0.016), using a paired t-test with unequal variances. The difference between GARP and RIGER_WS (μ=59.6±5.4%; p=0.079) was not significant, although GARP still outperformed RIGER_WS in 5/7 (71%) cell lines. Taken together, these results suggest that GARP performs at least as well, and by many metrics superior to, previous scoring systems for shRNA screens (see Discussion).
A snapshot of gene essentiality across 3 major tumor types
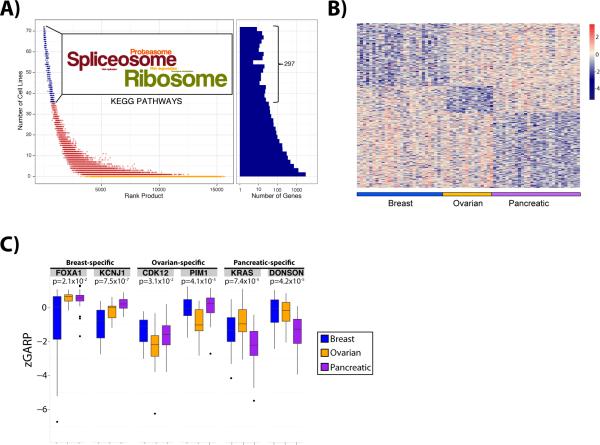
Next, we looked for essential genes across all of the cell lines. We identified 285 genes with significant GARP scores (p-value ≤ 0.05) in at least 50% of the 72 screens (Supplemental Table 6). These “general essential” genes were enriched in housekeeping functions involving the ribosome (p = 4.6e-48, Fisher's exact test), proteasome (p = 9.4e-12, Fisher's exact test), spliceosome (p = 2.9e-32, Fisher's exact test), DNA replication (p = 4e-3, Fisher's exact test), protein metabolism (p = 5.2e-20, Fisher's exact test) or mRNA processing (p = 3.3e-17, Fisher's exact test) (Figure 3A), consistent with what was observed in the fast hairpin dropout class (Figure 1C). Not surprisingly, general essential genes displayed potent inhibitory effects on cell growth; all general essentials were “fast dropouts” in at least one cell line, and behaved as “fast dropouts” in an average of 14 cell lines (Supplemental Figure 3B). Notably, there was significant overlap (p < 1.1e-112, Fisher's exact text) between general essentials identified in our study and in a recent report (18) that also used pooled shRNA screening but tested a smaller (~54,000 versus ~78,000) set of shRNAs (Supplemental Figure 4), and an in-depth cell line-by-cell line comparison showed significant overlap in 11 out of 15 cell lines in common between the screens (Supplemental Table 7).
Figure 3.
Identification of general and tissue-specific essential genes. (A) Representation of the selection criteria underlying the definition of the 285 general essential genes. Histogram at right shows the number of cell lines in which each gene is found essential. The ‘Wordle’ (inset) depicts the most frequently occurring KEGG pathways among the general essential genes, where the most frequently occurring terms are illustrated in larger font. (B) Heatmap of differentially essential genes in the three tumor types. 151 genes were identified as specific to breast cell lines, 72 to ovarian cell lines, and 175 to pancreatic cell lines. The complete list of genes is available in Supplemental Table 8. (C) The range of zGARP scores for six tissue-specific genes. P-values comparing the range of scores were determined using the Kruskal-Wallis test.
Next, we asked if the 72 cell lines could be classified on the basis of “functional genomics” (i.e., their relative sensitivity to shRNAs in the 80K library), and if so, how such groups would compare with their respective histotypes. Of the genes with a significant normalized GARP score (p < 0.05) in at least two cell lines (~5,510 genes), we identified the 10% most variable across all of the lines. These genes (n = 551) were subjected to unsupervised complete linkage hierarchical clustering using the Pearson correlation, which divided the cell lines into two major groups (Supplemental Figure 5). Notably, cell lines derived from the same tissue did not cluster into the same groups, although one of the major clusters was markedly enriched (p = 8.7e-5) for breast cancer cell lines: 23/29 breast cancer cell lines appear in this cluster, which contains a total of 36 cell lines. Accordingly, this cluster included genes that we classified as breast-specific (FOXA1, CDK4, SNW1, ATP5B, CLYBL, NDUFS7, CAD, INTS10,) or luminal/HER2-specific (TFAPC2, AF3GL2) in subsequent analyses (see below). Interestingly, even though all of the PDAC lines contained a KRAS mutation, these lines scattered throughout the two major clusters, with10 PDAC lines in the breast-enriched set and 18 in the other major cluster (see Discussion).
Besides “general essential” genes, we performed a supervised analysis of the 72 cell line dataset to identify two other types of essential genes: (1) tissue-specific and (2) subtype-specific essentials; the latter can be classified further into essential genes found in a subset of cell lines from different tumor types or essentials found in selected cell lines within a single tumor type. Non-parametric statistical testing identified 72 ovarian-, 175 pancreatic-, and 151 breast-specific essential genes, distinguishable by a significantly lower normalized GARP score (see Methods) in that tumor type (Wilcoxon Rank Sum test p<0.01) relative to the other two tumor types. A heat map of these tissue-specific essential genes is shown in Figure 3B. We examined each list of tissue-specific essentials for gene set enrichment using the Molecular Signatures Database (27). The breast cancer-specific essentials were enriched for a broad array of functions (e.g., cell cycle [q = 7.1e-10], ubiquitin-mediated proteolysis [ q = 1.1e-3], spliceosome [q = 9.3e-4], oxidative phosphorylation [q = 4.4e-3], snRNP assembly [q = 2.1e-3], pyrimidine metabolism [q = 3.5e-4]), as well as more specific processes, such as nucleotide excision repair (NER; q = 1.9e-2) and transcription-coupled NER (q = 1.7e-2). Ovarian cancer-specific essentials were mildly enriched for G-protein activation (q = 2.1e-2) and downstream events in GPCR signaling (q = 7.8e-2), whereas pancreatic tissue-specific essential genes were enriched for signaling in the immune system (q = 2e-3), the ETS pathway (q = 2.2e-2), and the LAIR pathway (q = 2.2e-2) (Supplemental Figure 6).
More detailed examination of the tissue-specific essentials revealed that the pancreatic cancer-specific essentials included KRAS and the cell cycle regulator CDK6 (Figure 3C and Supplemental Table 8). KRAS is mutated in most pancreatic tumors and promotes ETS-mediated transcription (28), which could account for the enrichment for ETS pathway genes noted above. Most pancreatic cancer lines fail to express the CDK inhibitor CDKN2A (data not shown), which might sensitize them to a reduction in CDK activity. Novel pancreatic cancer-specific essential genes included DONSON (Figure 3C), a centrosomal protein involved in DNA-damage response signaling and genomic integrity (29), the histone acetyltransferase MYST3 (30), and the transcriptional repressor SSX4 (Supplemental Table 8), although the latter gene has been implicated in other tumor types (NSCLC, endometrial, cervical) (31, 32). Also of note are PRKAA1 (encoding the catalytic subunit of PKA) and TRAF6, which, along with TLR4 (another hit in the screen) is involved in autophagy and adaptive and innate immune responses (33) (Supplemental Table 8).
Ovarian cancer-specific essentials included the cyclin-dependent kinase CDK12, involved in RNA splicing and recently found to be mutated somatically in 3% of HGS-OvCa cases (34). Two other ovarian cancer-specific essentials, PIM1 and CARD11, have not been linked to HGS-OvCa, but are involved in promoting cell cycle progression and in NF-κB activation, respectively (35, 36), both of which are broadly implicated in oncogenesis.
Breast cancer-specific essentials included known mammary oncogenes such as AKT1, a downstream effector of another mammary oncogene, PIK3CA (37), and CDK4, which encodes a binding partner of CyclinD1; the latter is amplified in a substantial percentage of breast cancers (38). Other breast cancer-specific essentials reportedly are over-expressed in breast cancer, including ERH, GRN, and KDM1A (39-41), co-activate ERα(SNW1), or are involved in tamoxifen resistance (ABCC2) (17, 42, 43). FOXA1 knockdown was particularly deleterious to ER+ breast cancer cell lines (see below and Figure 3C), consistent with its well-established role in estrogen receptor action (44, 45), and its prognostic significance (46) in breast cancer.
Functional screening results partially recapitulate breast cancer subtypes
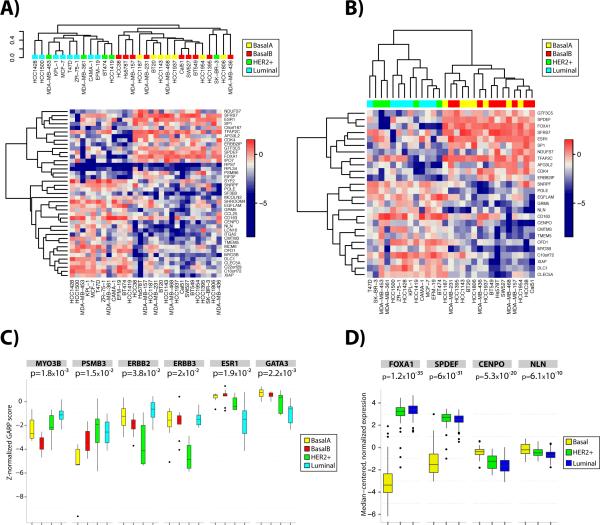
Breast tumors can be classified by transcriptional profiling into multiple subtypes with different prognostic significance (2, 47). By contrast, unsupervised analysis clusters breast cancer cell lines into two major subgroups, basal and luminal/HER2. The basal cluster can be divided further (by unsupervised clustering) into Basal A and Basal B, with the Basal A subgroup being most reminiscent of classical triple-negative breast tumors and Basal B being enriched for the recently described claudin-low tumor subtype (48). The luminal/HER2 cluster can be sub-divided further on the basis of HER2 amplification.
We asked whether breast cancer cell lines also could be classified on the basis of functional genomics, and if so, how such groups would compare with genomic subclasses. Of the genes with a significant normalized GARP score (p <0.05) in at least two breast cancer cell lines (~3,500 genes), we identified the 10% that varied the most across all of the lines. Subjecting these genes (n = 348) to unsupervised hierarchical clustering using Pearson correlation and complete linkage clustering divided the cell lines into two major groups. A heat map representation of the genes (n = 41; Supplemental Table 9) that most significantly discriminated between these two major clusters (t test, FDR < 0.1, Benjamini-Hochberg correction) is shown in Figure 4A. Notably, the MCF-7 and KPL-1 cell lines, which are derived from the same tumor, clustered most closely in this analysis. Moreover, the two functional genomic clusters corresponded almost exactly with the basal and HER2/luminal subtypes defined by expression profiling of the same cell lines. The single outlier, SkBr3, is classified as HER2/luminal by microarray analysis but behaved like a basal cell line in our functional genomic screen. This unsupervised clustering approach robustly distinguished Basal and Luminal/HER2+ cell lines even if we used the top 50% most variable genes for analysis (Supplemental Figure 7). By contrast, unsupervised analysis of our functional genomic data did not segregate the breast cancer cell lines further into, for example, Basal A and Basal B, or HER2 versus luminal.
Figure 4.
Subtype classification of breast functional screening results. (A) Unsupervised hierarchical clustering of functional screening data classifies the breast cancer cell lines into the known breast cancer subtypes. Seventeen genes correspond to the Luminal/HER2+ subtypes, while 24 were specific to the Basal subtype. (B) Supervised clustering of the screening results according to known breast cancer subtypes. (C) Normalized zGARP scores for genes displaying differential essentiality in specific breast subtypes. Only MYO3B and ESR1 had signals strong enough to be included in the unsupervised analysis at the FDR threshold used. p-values comparing the range of scores were determined using the Kruskal-Wallis test. (D) The normalized gene expression level of four genes identified as luminal/HER2+ specific (FOXA1, SPDEF) or basal subtype-specific (CENPO, NLN) was obtained from the TCGA data portal. The expression data included 42 basal subtype, 63 HER2+ subtype, and 223 luminal subtype tumor samples. p-values comparing the range of expression values were determined using the Kruskal-Wallis test.
We also carried out a supervised analysis on the same dataset, asking for the genes that best distinguished basal from HER2/luminal cell lines (Figure 4B). Remarkably, all 26 of the genes that met our significance criteria (t test, FDR < 0.1) also were identified by unsupervised clustering (Figure 4A) and included well-known determinants of the luminal/HER2 subtype, such as ESR1, FOXA1, SPDEF, and TFAP2C. Although known drivers of the HER2 subtype (e.g., HER2 and HER3) did not achieve scores significant enough to appear in the unsupervised or supervised clustering panels, they clearly were more essential to the HER2 subtype (Figure 4C). Finally, we interrogated the TCGA database for the levels of expression of these 41 genes in breast cancer subtypes. Overall, expression did not correlate with functional subtype specificity, except for the few examples depicted in Figure 4D.
Identification of putative oncogenic drivers through integrative analyses
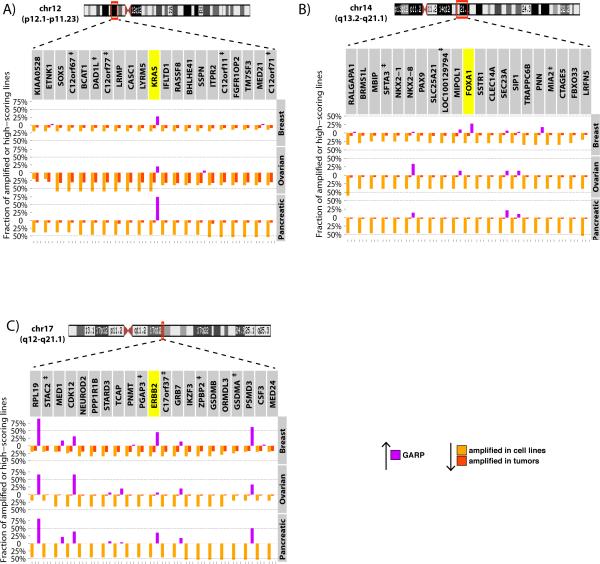
Genome-scale functional screens can be coupled with genomic information to identify potential cancer driver genes. To facilitate such analyses, we developed a representation (“query plot”) that permits simultaneous visualization of copy number information from publicly available data (see Methods) and GARP scores from functional screens. Each query plot displays amplification data (from tumors and cell lines) for each gene (as a percentage of total tumors and cell lines) as downwardly projecting bars, and the percentage of cell lines in which the same gene scored as essential (GARP p < 0.05) as upwardly projecting bars (for details, see Methods).
Initial query plots were generated for known oncogenes, such as KRAS, FOXA1, and ERBB2. Notably, compared with the other tumor types, KRAS was particularly essential in pancreatic cancer cell lines (Figure 5A), consistent with the known role of KRAS mutations in PDAC. Although FOXA1 is amplified in breast, ovarian and pancreatic cancer cell lines, it was essential only in luminal/HER+ breast cancer cells (Figures 5B). As expected, ERBB2 is clearly the focal point of the ERBB2 amplicon in primary tumors and cell lines, and was essential in a subset of breast cancer cell lines (Figure 5C). The ERBB2 locus also was amplified in a subset of PDAC and HGS-OvCa cell lines and tumors, but was essential only in selected PDAC cell lines. Scanning across the ERBB2 amplicon, we see general essential genes (e.g. RPL19, PSMD3), as well as CDK12, recently implicated as a driver in HGS-OvCa (34). Taken together (also see discussion of ovarian cancer-specific essentials above), these data suggest that CDK12 may be the key driver within the ERBB2 amplicon in HGS-OvCa.
Figure 5.
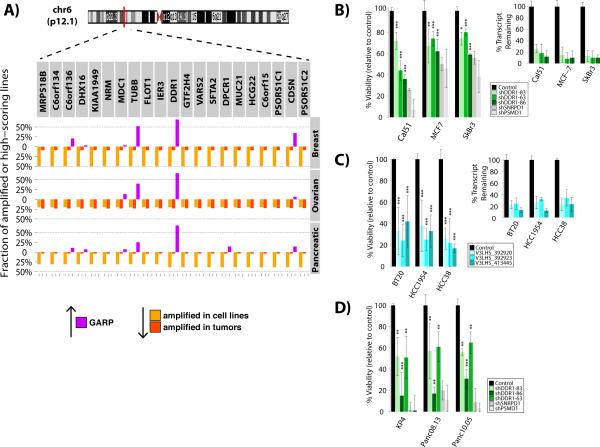
Pattern of functional screening data in amplifications with known drivers. Chromosome ideograms indicate known regions of amplification in tumors. Barplots above the X-axis depict frequency of cell lines in which each indicated gene is essential according to GARP scores (p<0.05), while below the X-axis depict the frequency of observed amplifications in cell lines (orange) or tumors (red). For each plot, the ten genes upstream and downstream of the suspected oncogenic driver gene (yellow) are shown.
To survey the landscape of potential drivers for breast, pancreatic, and ovarian cancer, we examined genes with measurable amplification in multiple cell lines that also scored as essential (p-value<0.05, GARP) in multiple cell lines of a given tumor type. Several general essential genes identified by GARP are involved in cell signaling and represent putative therapeutic targets. For example, the gene encoding the receptor tyrosine kinase DDR1 hit in 49 out of 72 cell lines (GARP p<0.05). To further explore the role of DDR1, three shRNAs from the TRC library, as well as three hairpins from an alternative shRNA library (see Methods), were used for secondary assays in the breast cancer cell lines Cal51, MCF7, SkBr3, BT20, HCC1954 and HCC38. As positive controls for cell killing, we used two shRNAs that were lethal in almost all cell lines, targeting small nuclear ribonucleoprotein D1 polypeptide (SNRPD1) and the 26S proteasomal subunit non-ATPase 1 (PSMD1), respectively (Supplemental Table 6). All six DDR1-directed hairpins efficiently lowered DDR1 transcript levels and inhibited proliferation in the six breast cancer cell lines (Figure 6B-C). DDR1 depletion had similar effects on the three pancreatic cancer cell lines tested, KP4, Panc08.13 and Panc10.05 (Figure 6D), confirming that it is essential for cancer cell proliferation. Notably, the most efficacious DDR1 shRNAs were as effective at inhibiting proliferation as the SNRPD1 and PSMD1 positive controls.
Figure 6.
Confirmation of the essential role of DDR1 in breast and pancreatic cell lines. (A) As described in Figure 4, but for DDR1. (B) Relative number of cells remaining six days post-infection with three different DDR1 shRNAs. SNRPD1 and PSMD1 shRNA were used as positive controls, as these were the two most potent shRNA in pooled screening. Control shRNA is the average of a GFP, a LacZ, and a Luciferase shRNA. DDR1 shRNA viability/cell number is depicted relative to control. Experiments were performed in triplicate. p-value was derived using a one-tailed Student's T-test. (shDDR1-83: TRCN0000121083, shDDR1-86: TRCN0000121086, shDDR1-63: TRCN0000121163). Small graph insert, Percentage transcript remaining determined by qPCR relative to GFP shRNA control. (C) Relative number of cells remaining nine days post-infection with three different pGIPZ DDR1 shRNAs. Negative control was a non-silencing shRNA. Small graph insert, Percentage transcript remaining determined by qPCR relative to GFP shRNA control. (D) Relative cell number determined as in (B), in pancreatic cell lines. *p<0.05, **p<0.01, ***p<0.001
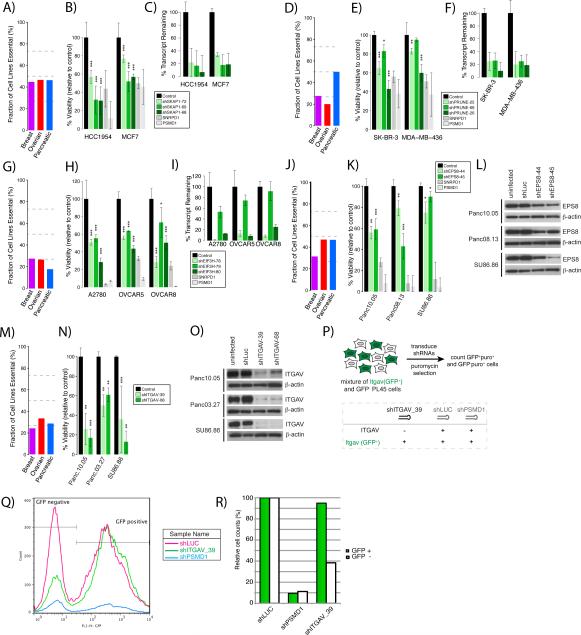
We selected for further analysis an additional six genes (SKAP1, PRUNE, EIF3H, EPS8 and ITGAV) that mapped within amplicons in at least one of the three tumor types (Supplemental Figure 8) and scored as essential in our screens. SKAP1, which scored in multiple breast (13), PDAC (13) and HGS-OvCa (7) cell lines (total of 33 lines), is a SRC kinase-associated phosphoprotein thought to function only in T-cells (49). We confirmed that SKAP1 is expressed in breast cancer cell lines, and found that SKAP1 knockdown significantly reduced proliferation in HCC1954 and MCF7 breast cancer cells (Figure 7A-C). PRUNE encodes a phosphodiesterase reportedly involved in cell migration and was essential in 23 of our cell lines (PDAC (13), BrCa (7), and HGS-OvCa (3)) (50). PRUNE knockdown (by either of two shRNAs) significantly reduced the proliferation of SkBr3 and MDA-MB-436 cells (Figure 7D-F). Knockdown of the translation initiation factor EIF3H, which scored as essential in 8 BrCa, 5 PDAC, and 3 HGS-OvCa cell lines, by any of three independent shRNAs significantly reduced cell proliferation in the HGS-OvCa cell lines tested (OVCAR5, OVCAR8 and A2780) (Figure 7G-I). Furthermore, knocking down EPS8, which encodes an adaptor protein involved in endocytosis and was essential in 29 of our cell lines (PDAC (12), breast (10), and HGS-Ov (7)), also significantly reduced proliferation in certain PDAC-derived cancer cell lines (Figure 7J-L). ITGAV, which encodes integrin alpha V, a component of the vitronectin receptor, was essential in 20 of our cell lines (PDAC (8), breast (7), and HGS-Ov (5)). Accordingly, knockdown of ITGAV significantly inhibited the proliferation of Panc10.05, SU86.86 and Panc08.13 cells (Figure 7M-O).
Figure 7.
Validation of genes that are overexpressed or in regions of copy number gains in cancer cells. (A) Percentage of cell lines in which SKAP1 was in the top 5% of essential genes according to GARP. (B) Relative number of breast cancer cells remaining six days post-infection with three different SKAP1 shRNAs. SNRPD1 and PSMD1 shRNA were included as positive controls. Control shRNA is the average of a GFP, a LacZ, and a Luciferase shRNA. Viability/cell number is depicted relative to control. p-value was derived using a one-tailed Student's T-test from triplicate experiments. (C) Percentage transcript remaining determined by qPCR relative to GFP shRNA control. (D-F) Same as (A-C) for PRUNE. (G-I) Same as (A-C) for EIF3H in ovarian cell lines. (J-K) Same as (A-B) for EPS8 in pancreatic cell lines. (L) Western blot of EPS8 knockdown in pancreatic cell lines. (M-N) Same as (A-B) for ITGAV in pancreatic cell lines. (O) Western blot showing effects of ITGAV knockdown in pancreatic cell lines. (P) Experimental scheme for ITGAV rescue experiment. (Q) Flow cytometry results for ITGAV rescue experiment. (R) Quantification of results in Q. *p<0.05, **p<0.01, ***p<0.001.
Finally, we tested whether the effects of knocking down a gene identified in our screens could be rescued by re-expressing an shRNA resistant form of that gene. We developed a competition assay using PL45 cells expressing (GFP+) or not expressing (GFP−) mouse Itgav (Figure 7P), and then monitored the effect on proliferation of an shRNA targeting the 3’UTR of human ITGAV. Indeed, cells expressing Itgav were resistant to the effects of the human shRNA, confirming that the inhibitory effect of ITGAV knockout on proliferation were specific(Figure 7Q-R).
DISCUSSION
Functional genomic approaches can complement genomic analyses, providing a more comprehensive view of cancer cell biology and suggesting new therapeutic strategies. Earlier work using large-scale pooled shRNA screens to investigate gene essentiality in cancer cell lines surveyed multiple histotypes, but examined relatively few examples of each type (18) or sought synthetic lethal relationships with the same oncogene expressed in different types of cancer (15). Our study represents an extensive functional genetic survey of three major cancers, BrCa, PDAC, and HGS-OvCa, establishes a new metric for scoring shRNA dropout screens, uncovers complexity in the relationship between genomic and functional genomic classification schemes, and reveals unexpected gene dependencies and new potential therapeutic targets in these malignancies.
Previous studies quantified hairpin dropout at a single (usually fairly long) time point. Dynamic measurements provide additional power for tracking shRNA abundance in a given population of cells, and we found that this helps to group shRNAs into functional classes that reflect the intrinsic properties of their corresponding target genes (Figure 1C). For example, shRNAs that drop out early are more likely to target housekeeping genes. We developed a new scoring approach (shARP) that captures these dynamic properties, as well as a gene-level metric, termed GARP. Importantly, if one considers “housekeeping” or “highly conserved” gene subsets (which are largely non-overlapping; see Figure 2C) as reasonable surrogates for essentiality, GARP outperforms previous scoring metrics in its ability to “call” general essential genes (Figures 2A-B)
Analysis of our 72 screens allowed us to define three types of essential genes: (i) general essentials, (ii) tissue-specific essentials, and (iii) subtype-specific essentials. General essentials are, as expected, enriched for highly conserved, housekeeping functions, such as those associated with transcription, translation, splicing and the proteasome (Figure 3A, Supplemental Table 6). Moreover, the general essentials identified in our screen showed considerable (although not complete) overlap with those defined in an earlier study (18) (Supplemental Figure 4). Such similarity in experimental results across a large number of experiments in different institutions argues for the robustness of dropout screening methodology, and strongly suggests that the genes identified as essential in all of these screens are, in fact, generally required for cancer cell proliferation/survival. Several possible explanations likely contribute to the lack of complete overlap in general essentials defined in previous work and our study. For example, we observed, retrospectively, that calculating fold-changes in shRNA barcode abundance for different endpoints within the same screen yields rank-ordered gene lists that do not overlap perfectly. Our screening approach, which employs multiple time points, helps to buffer some of the noise introduced by endpoint assays, obviates the need to terminate a screen at a precise number of generations, and permits comparison of shRNA dropout profiles across multiple cell lines. Thus, we believe that dynamic shRNA profiles provide a more robust metric than fold-change measurements for ranking shRNA dropouts, and hence essential genes.
Although we are confident that genes that score as general essentials in our screen (and particularly those that also are classified as general essentials by the other scoring metrics) are, in fact, required for cancer cell proliferation, inherent limitations of shRNA screening methodology make it likely that we are underestimating the actual number of general essential genes. Indeed, we have noted that some genes with GARP p>0.05 are, in fact, required for proliferation in secondary assays. Because biologically significant results can be obtained for hairpins that lie outside of the statistically significant range, future users of our resource should not a priori exclude from further investigation genes with GARP scores lying outside of a stringent statistical cut-off. Also, it should be noted that the shRNA detection strategy that we used in our screens precludes efficient identification of genes whose depletion enhances cell proliferation: in order to detect dropouts, hybridizations are carried out with a standard amount of excess probe, which limits the detection of “enhancing” shRNAs.
Interestingly, some genes that qualified as general essentials in our screen do not serve obvious housekeeping roles. Perhaps the most intriguing is DDR1, which encodes a receptor tyrosine kinase (RTK) that binds to collagens (51). Secondary screens using shRNAs from two independent libraries confirmed that DDR1 is required in the six breast and four pancreatic cancer lines tested. Also, several previous studies reported increased DDR1 expression in various human tumors, including breast, ovarian, lung, esophageal, and pediatric brain cancers (reviewed in (52)), and DDR1 knockdown suppresses tumorigenicity in HCT116 cells (53). Furthermore, the DDR1 locus is amplified in a significant number of breast, ovarian and pancreatic tumors and cell lines (Figure 6A). Notably, although they are smaller than control, Ddr1 knockout mice are viable (51), arguing for a selective requirement in malignant cells. These data suggest that DDR1 might be an attractive target for anti-neoplastic drug development, although it will be important to confirm its requirement for tumor maintenance, not just for cancer cell proliferation in tissue culture. Recently, DDR2, which shares substantial sequence similarity with DDR1 and also encodes a collagen-responsive RTK, was found to be mutated and required in about 5% of squamous cell carcinomas of the lung (54). We found that DDR2 also was essential in 37 of our cancer cell lines. Taken together, these results raise the possibility that both DDRs play a broader role in oncogenesis than previously appreciated.
Supervised analysis of our screening results identified tissue-specific genes, which are potentially responsible for enrichment in different biological processes and may reveal emergent properties of cancer cells from tissues of different origin. However, these tissue-specific genes did not drive (unsupervised) clustering of the lines into histotype-specific groups. Notably, one major cluster was enriched in breast cancer cell lines, including 13/14 lines of luminal/HER2 subtype, and this grouping was driven mainly by breast cancer-specific genes and luminal/HER2-specific genes. Most HGS-OvCa and PDAC cell lines segregated into the same group, even though breast and ovarian cancer share some oncogenic drivers (BRCA1/2, HER2, HER3) and unlike PDAC, usually lack KRAS mutations. Moreover, while nearly all of the PDAC lines tested have a KRAS mutation, these lines did not co-segregate by unsupervised clustering analysis. These data indicate that additional genetic events in PDAC modulate the gene essentiality landscape imposed by KRAS mutations. Furthermore, they suggest that, in contrast to previous reports (15, 55-57), it might be difficult to identify a universal set of “KRAS” synthetic lethal genes; we were not able to uncover a set of essential genes specific to mutant KRAS. Rather, there might be context-dependent KRAS synthetic lethality imposed by the cell of origin of the malignancy and/or its other underlying genetic abnormalities.
Because the breast cancer cell lines we studied have been analyzed by expression profiling (and CNV analysis), we could explore the relationship between genomics and functional genomics in these lines. Remarkably, applying an unsupervised clustering algorithm to these screening data resulted in clustering of the breast cancer cell lines into functional subsets that are essentially identical to the classical HER2/luminal and basal subgroups (58, 59). Reassuringly, several of the genes responsible for the clustering of the HER2/luminal cell lines are well-known drivers of these subtypes (e.g., ESR1, FOXA1, SPDEF). The genes whose knockdown preferentially impaired basal breast cancer cell proliferation are less well studied in this subtype or in breast cancer in general. However, they include genes encoding proteins involved in DNA repair (POLE) and with anti-apoptotic function (XIAP). Our functional genomic classification does not separate the HER2/luminal or basal subgroups further (i.e., into HER2-specific, luminal-specific, or Basal A or Basal B specific) by unsupervised clustering. For the HER2/luminal breast cancer lines, this is not surprising, as HER2 and luminal subgroups also are not distinguishable by expression profiling (59). Basal A and Basal B cell lines can, however, be discerned by transcriptional differences (59). Failure to separate these subgroups by functional genomic clustering could reflect insufficient numbers of cell lines in our screen or biological nuances that are not completely captured at the mRNA expression level. Notably, genes specific for each subclass (i.e., luminal, HER2, Basal A, Basal B) can be identified by supervised methods (Figure 4C); such genes, if validated, could yield new insights into subgroup-specific biological differences and suggest subgroup specific therapeutic targets.
Recently, PDAC and HGS-OvCa were classified into subgroups based on expression differences (34, 60). However, unsupervised clustering did not reveal subgroup-specific essential gene maps for our pancreatic or ovarian cancer cell lines. Conceivably, the number of cell lines that we screened did not provide sufficient predictive power. Alternatively, our cell lines might not adequately represent the range of transcriptional subclasses seen in tumors. It also is possible that the transcriptional subclasses themselves are not predictive of gene sets that are essential for viability. For example, our PDAC collection contains cell lines that conform to both the classical and quasi-mesenchymal pancreatic subtypes (60), yet these lines did not segregate from each other by unsupervised functional genomic clustering. Additional genomic data and further analyses are needed to explore the relationship between functional genetic screening data and genomic data to identify better prognostic and therapeutic factors for pancreatic and ovarian cancers.
Finally, by combining copy number information with the results of our dropout screens, we identified—and confirmed—several unexpected vulnerabilities in breast, pancreatic and ovarian cancer cell lines. For example, SKAP1 encodes an adaptor that is generally thought to function only in T cells, where it reportedly modulates T cell antigen receptor-induced activation of the Ras-ERK-AP1 pathway (49, 61), as well as integrin clustering and adhesion (62). Recently, SKAP1 was identified in genome-wide association studies as a susceptibility locus for ovarian (63) and prostate cancer (64). Although this could indicate a role for SKAP1 in immune surveillance, our data suggest that these alleles might have cell-autonomous effects on tumorigenesis. We identified PRUNE as a gene that was preferentially essential in breast and pancreatic cancer cell lines (p<0.028) and confirmed this in several breast cell lines (Figure 7). PRUNE encodes a phosphodiesterase belonging to the DHH superfamily, which reportedly binds NM23-H1 to promote metastasis (65). PRUNE also binds to glycogen synthase kinase and reportedly regulates cell migration by modulating focal adhesions (50). Moreover, amplification and over-expression of PRUNE reportedly correlates with advanced breast carcinomas (66, 67). EIF3H encodes a translation initiation factor, and is amplified and over-expressed in a variety of cancer types including colon (68), NSCLC (69), prostate (70), breast (71), and HCC (72). Notably, several other members of the eIF-3 translation initiation complex scored as essential in our screens, including EIF3A (67 lines), EIF3B (70 lines), EIF3C (66 lines), EIF3D (64 lines), EIF3G (57 lines), and EIF3I (54 lines). EPS8 is an EGFR substrate that mediates RAC1 activation and trafficking of EGFR in a RAB5-dependent manner (73). EPS8 also has been implicated in cell migration and ovarian cancer metastasis (74), and EPS8 over-expression has been observed in PDAC (75) and oral squamous cell carcinoma (76). Lastly, ITGAV encodes integrin alpha chain V, a component of the vitronectin receptor, which has been associated with multiple different cancers (77).
Taken together, our results have interesting implications for the systems biology of cancer. Our finding that functional genomic and genomic classification schemes yield only partially overlapping results implies that functional genomic studies reveal nuances in cancer cell biology that have not been captured by existing genomic analyses. By (re)-analyzing genomic data in cancers cells grouped by similar gene essentiality profiles, it is likely that new drivers and synthetic lethal relationships will emerge. Future exploitation of our functional genomic resource will require validation of the multiple types of essential genes that we have identified, along with integrative analysis of functional genomic and detailed genomic information from these cell lines.
METHODS
Cell Lines
A full description of each cell line used in this study, where the cell lines were obtained, and the method and date of cell line authentication are detailed in Supplemental Table 2.
shRNA Dropout Screens
Each cell line was grown to a population size of ~2×108 cells in the requisite media (see Supplemental Table 2). Cells were washed with warm PBS, trypsinized, resuspended in warm media and counted. An aliquot of the 78k human shRNA lentivirus pool (22) and either polybrene (4-8μg/ml) or protamine sulphate (5μg/ml) were added such that a multiplicity of infection of 0.3-0.4 would be achieved (determined by prior testing of each cell line). Cell-lentivirus mixtures were plated into 15cm diameter culture dishes and incubated at 37°C with 5% CO2. Twenty-four hours post-infection, the media was replaced with fresh media containing puromycin (puromycin concentration determined by prior tests on each cell line) and cells were incubated for an additional 48 hours. Culture dishes were washed with warm PBS to remove dead cells, and surviving cells were collected by trypsinization and resuspended in warm media. Cell populations were quantified, aliquots of 2×107 cells were removed and pelleted by centrifugation, and three replicate populations of 2×107 cells were plated at appropriate density into 15cm culture dishes. When replicate populations reached 80%-90% confluency, cells from each replicate were collected by trypsinization and mixed (cells from different replicates were not co-mingled). From these mixtures, two aliquots of 2×107 cells were removed, pelleted, and frozen down, while one aliquot of 2×107 cells was re-plated for further growth. This step was repeated until a minimum of six to eight doublings for each replicate cell population was obtained. Genomic DNA was prepared from cell pellets using the QIAmp Blood Maxi kit (Qiagen #51194). Genomic DNA was precipitated using ethanol and sodium chloride, and resuspended at 400ng/μl in 10mM Tris-HCl pH 7.5. shRNA populations from cell lines were amplified via PCR and prepared and applied to GMAP arrays as described previously (22).
Identification of Hairpin Classes
Hairpins were segregated into classes using rules based on a boolean combination of features that describe the hairpin behaviour through the timecourse. The rules involved features describing the rate of dropout over the first and second time intervals (i.e. the slope of the expression intensity change between timepoints), the ratio of the dropout rates, the fold-change at the end-point relative to T0 (FC2), and the initial expression intensity at T0. The classes include: K – fast dropouts, which lead to rapid hairpin depletion; S – slow dropouts, displaying a lag prior to hairpin depletion; E – enhancers, or hairpins that show an increase in abundance over time; and C – continuous dropouts, or hairpins that show a constant rate of depletion over time.. These rules are summarized in Supplemental Table 3.
Scoring of shRNA Screens
In order to incorporate measurements from multiple timepoints in a shRNA screen, we developed the shRNA Activity Ranking Profile score (shARP), as shown in Equation 1:
| Eq. 1 |
where n is the number of timepoints, Δy is the change in expression intensity at ti relative to t0, and Δx is the number of doublings for the cell line at ti relative to t0. The shARP scores are determined for each of the 78,432 hairpins in the library, and are then used to calculate the Gene Activity Ranking Profile score (GARP) by averaging the two lowest shARP scores. A significance value is assigned to each GARP score through bootstrapping, where the shARP scores are randomly permuted 1000 times, GARP scores recomputed, and a p-value is determined by the frequency with which the actual GARP score is lower than the permuted GARP scores. To facilitate comparisons between screens, GARP scores were Z-score normalized.
In addition to the GARP score, we also applied two previously published shRNA scoring methods to rank the normalized shARP scores from our screens. First, the Redundant siRNA Activity (RSA) (23) was applied using the R package provided by the original authors. Briefly, all shARP scores were normalized using a robust Z-score before applying RSA iteratively to each screen, setting the UB parameter to 0, the LB parameter to −3, and using the Entrez GeneID as the unique identifier for each gene. Second, we ranked the normalized shARP scores by RIGER (12), as implemented in the GENE-E software package. Genes were ranked by each of the three RIGER methods to collapse hairpins to genes, namely “Weighted sum of the first two ranks of hairpins”, “Second best rank”, and “Kolmogorov-Smirnov”. To avoid RIGER scoring dropouts and enhancers separately, the shARP score distribution was shifted below zero prior to applying Kolmogorov-Smirnov scoring by subtracting the maximum value from each distribution.
Benchmarking Scoring Methods
In order to benchmark our scoring method against existing approaches such as RIGER and RSA, the top ranked genes from each screen were overlapped against two datasets likely enriched in essential genes. First, housekeeping (HK) genes are genes universally expressed to maintain cellular function: the more tissues in which a gene is expressed, the higher the likelihood that it will be essential (78). Second, highly conserved orthologs are genes that are shared among species, which have a higher likelihood of being essential (79, 80). HK genes (n = 1722) were identified as genes expressed in at least 73/79 tissues in a human expression compendium (25). Highly conserved orthologs (n = 1617) were identified as human genes with orthologs in eight different species (A. thaliana, B. taurus, C. elegans, C. familiaris, M. mulatta, M. musculus, R. norvegicus, and S. cerevisiae), as determined by InParanoid (24). There are 315 genes in common between the housekeeping genes and conserved orthologs.
The top 500 genes ranked by each scoring method in each screen were selected and overlapped with the reference sets, starting with the top five genes up to the top 500 in five gene steps. The overlap was plotted, and the area under the curve (AUC) was calculated for each overlapping gene set.
Supplementary Material
SIGNIFICANCE.
This study presents a resource of genome-scale, pooled shRNA screens for 72 breast, pancreatic, and ovarian cancer cell lines that will serve as a functional complement to genomics data, facilitate construction of essential gene profiles, help uncover synthetic lethal relationships, and identify uncharacterized genetic vulnerabilities in these tumor types.
ACKNOWLEDGEMENTS
We thank Corey Nislow and Guri Giaever for use of the equipment, Real Franciscio (Madrid, Spain), Ann Marie Mes-Masson (Montreal, Canada), Patricia Tonin (Montreal, Canada), Graham Fletcher (Toronto, Canada), Gordon Mills (MD Anderson), and Robert Bast (MD Anderson) for cell lines and Luigi Naldini for plasmids.
Financial Support: This work was supported by the Ontario Institute for Cancer Research and Terry Fox Research Institute through the Selective Therapies Program (R.R., B.G.N., J.W. and J.M.), the Canadian Institutes for Health Research (J.W., PRG-82679, B.G.N., and J.M.), the Canadian Foundation for Innovation (J.M.), and the Ontario Research Fund (B.G.N., J.M. and R.R.). Work in the Neel and Rottapel laboratories is partially supported by the Ontario Ministry of Health and Long Term Care and the Princess Margaret Hospital Foundation. JM holds a CIHR New Investigator Award. B.G.N. is a Canada Research Chair, Tier 1 and the recipient of the Premier of Ontario's Summit Award. RM is the recipient of a postdoctoral fellowship from the Canadian Breast Cancer Foundation.
Footnotes
Author Contributions: RM, KRB, TK, RR, BGN and JM designed the research. RM, MH, YF, ML, BF, PM, MM, KK, DVD, FJV, FSV, AD, JW, TK, CM, DK and JN contributed to and/or performed experiments. RM, KRB, FS, AS, JLK, PMK, FS, GCB, TK, RR, BGN and JM analyzed the data. RR, BGN and JM supervised the research. RM, KRB, BGN and JM wrote the manuscript.
REFERENCES
- 1.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–90. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 8.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 9.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–8. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Moffat J, Sabatini DM. Building mammalian signalling pathways with RNAi screens. Nat Rev Mol Cell Biol. 2006;7:177–87. doi: 10.1038/nrm1860. [DOI] [PubMed] [Google Scholar]
- 11.Sigoillot FD, King RW. Vigilance and validation: Keys to success in RNAi screening. ACS Chem Biol. 2011;6:47–60. doi: 10.1021/cb100358f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–4. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–20. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF- kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–6. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendes-Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I, et al. Breast Cancer Special Feature: Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1018872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, et al. International network of cancer genome projects. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 21.Root DD, Wang K. High-affinity actin-binding nebulin fragments influence the actoS1 complex. Biochemistry. 2001;40:1171–86. doi: 10.1021/bi0015010. [DOI] [PubMed] [Google Scholar]
- 22.Ketela T, Heisler LE, Brown KR, Ammar R, Kasimer D, Surendra A, et al. A comprehensive platform for highly multiplexed mammalian functional genetic screens. BMC Genomics. 2011;12:213. doi: 10.1186/1471-2164-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konig R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nature methods. 2007;4:847–9. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–80. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moffat J, Reiling JH, Sabatini DM. Off-target effects associated with long dsRNAs in Drosophila RNAi screens. Trends Pharmacol Sci. 2007;28:149–51. doi: 10.1016/j.tips.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang JS, Koenig A, Harrison A, Ugolkov AV, Fernandez-Zapico ME, Couch FJ, et al. Mutant K-Ras increases GSK-3beta gene expression via an ETS-p300 transcriptional complex in pancreatic cancer. Oncogene. 2011 doi: 10.1038/onc.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs F, Pau G, Kranz D, Sklyar O, Budjan C, Steinbrink S, et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol Syst Biol. 2010;6:370. doi: 10.1038/msb.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katsumoto T, Yoshida N, Kitabayashi I. Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci. 2008;99:1523–7. doi: 10.1111/j.1349-7006.2008.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasegawa K, Koizumi F, Noguchi Y, Hongo A, Mizutani Y, Kodama J, et al. SSX expression in gynecological cancers and antibody response in patients. Cancer Immun. 2004;4:16. [PubMed] [Google Scholar]
- 32.Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res. 2005;11:8055–62. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- 33.Shi CS, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy. 2010;6:986–7. doi: 10.4161/auto.6.7.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37:726–30. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Lamason RL, McCully RR, Lew SM, Pomerantz JL. Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry. 2010;49:8240–50. doi: 10.1021/bi101052d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 38.Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003;78:323–35. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- 39.Zafrakas M, Losen I, Knuchel R, Dahl E. Enhancer of the rudimentary gene homologue (ERH) expression pattern in sporadic human breast cancer and normal breast tissue. BMC Cancer. 2008;8:145. doi: 10.1186/1471-2407-8-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrero G. Autocrine growth factor revisited: PC-cell-derived growth factor (progranulin), a critical player in breast cancer tumorigenesis. Biochem Biophys Res Commun. 2003;308:409–13. doi: 10.1016/s0006-291x(03)01452-9. [DOI] [PubMed] [Google Scholar]
- 41.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–20. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 42.Lu S, Becker KA, Hagen MJ, Yan H, Roberts AL, Mathews LA, et al. Transcriptional responses to estrogen and progesterone in mammary gland identify networks regulating p53 activity. Endocrinology. 2008;149:4809–20. doi: 10.1210/en.2008-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287–93. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, et al. FOXA1 expression in breast cancer--correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13:4415–21. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 47.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene- expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 48.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosco KA, Cerignoli F, Williams S, Abraham RT, Mustelin T. SKAP55 modulates T cell antigen receptor-induced activation of the Ras-Erk-AP1 pathway by binding RasGRP1. Mol Immunol. 2008;45:510–22. doi: 10.1016/j.molimm.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi T, Hino S, Oue N, Asahara T, Zollo M, Yasui W, et al. Glycogen synthase kinase 3 and h-prune regulate cell migration by modulating focal adhesions. Mol Cell Biol. 2006;26:898–911. doi: 10.1128/MCB.26.3.898-911.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogel WF, Aszodi A, Alves F, Pawson T. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol Cell Biol. 2001;21:2906–17. doi: 10.1128/MCB.21.8.2906-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel WF, Abdulhussein R, Ford CE. Sensing extracellular matrix: an update on discoidin domain receptor function. Cell Signal. 2006;18:1108–16. doi: 10.1016/j.cellsig.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 53.Kim HG, Hwang SY, Aaronson SA, Mandinova A, Lee SW. DDR1 receptor tyrosine kinase promotes prosurvival pathway through Notch1 activation. J Biol Chem. 2011;286:17672–81. doi: 10.1074/jbc.M111.236612. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Peter S, Hammerman, Sos Martin L., Ramos Alex H., Xu Chunxiao, Dutt Amit, Zhou Wenjun, et al. Mutations in the DDR2 Kinase Gene Identify a Novel Therapeutic Target in Squamous Cell Lung Cancer. Cancer Discovery. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–34. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 57.Singh A, Settleman J. Oncogenic K-ras “addiction” and synthetic lethality. Cell Cycle. 2009;8:2676–7. doi: 10.4161/cc.8.17.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lehmann R, Meyer J, Schuemann M, Krause E, Freund C. A novel S3S-TAP-tag for the isolation of T-cell interaction partners of adhesion and degranulation promoting adaptor protein. Proteomics. 2009;9:5288–95. doi: 10.1002/pmic.200900294. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, Liu H, Lu Y, Lovatt M, Wei B, Rudd CE. Functional defects of SKAP-55-deficient T cells identify a regulatory role for the adaptor in LFA-1 adhesion. Mol Cell Biol. 2007;27:6863–75. doi: 10.1128/MCB.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–9. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang CN, Huang SP, Pao JB, Chang TY, Lan YH, Lu TL, et al. Genetic polymorphisms in androgen receptor-binding sites predict survival in prostate cancer patients receiving androgen-deprivation therapy. Ann Oncol. 2011 doi: 10.1093/annonc/mdr264. [DOI] [PubMed] [Google Scholar]
- 65.D'Angelo A, Garzia L, Andre A, Carotenuto P, Aglio V, Guardiola O, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5:137–49. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 66.Zollo M, Andre A, Cossu A, Sini MC, D'Angelo A, Marino N, et al. Overexpression of h- prune in breast cancer is correlated with advanced disease status. Clin Cancer Res. 2005;11:199–205. [PubMed] [Google Scholar]
- 67.Forus A, D'Angelo A, Henriksen J, Merla G, Maelandsmo GM, Florenes VA, et al. Amplification and overexpression of PRUNE in human sarcomas and breast carcinomas-a possible mechanism for altering the nm23-H1 activity. Oncogene. 2001;20:6881–90. doi: 10.1038/sj.onc.1204874. [DOI] [PubMed] [Google Scholar]
- 68.Pittman AM, Naranjo S, Jalava SE, Twiss P, Ma Y, Olver B, et al. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cappuzzo F, Varella-Garcia M, Rossi E, Gajapathy S, Valente M, Drabkin H, et al. MYC and EIF3H Coamplification significantly improve response and survival of non-small cell lung cancer patients (NSCLC) treated with gefitinib. J Thorac Oncol. 2009;4:472–8. doi: 10.1097/JTO.0b013e31819a5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saramaki O, Willi N, Bratt O, Gasser TC, Koivisto P, Nupponen NN, et al. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am J Pathol. 2001;159:2089–94. doi: 10.1016/S0002-9440(10)63060-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Savinainen KJ, Linja MJ, Saramaki OR, Tammela TL, Chang GT, Brinkmann AO, et al. Expression and copy number analysis of TRPS1, EIF3S3 and MYC genes in breast and prostate cancer. Br J Cancer. 2004;90:1041–6. doi: 10.1038/sj.bjc.6601648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okamoto H, Yasui K, Zhao C, Arii S, Inazawa J. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology. 2003;38:1242–9. doi: 10.1053/jhep.2003.50457. [DOI] [PubMed] [Google Scholar]
- 73.Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, et al. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–7. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 74.Chen H, Wu X, Pan ZK, Huang S. Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer metastasis. Cancer Res. 2010;70:9979–90. doi: 10.1158/0008-5472.CAN-10-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Welsch T, Endlich K, Giese T, Buchler MW, Schmidt J. Eps8 is increased in pancreatic cancer and required for dynamic actin-based cell protrusions and intercellular cytoskeletal organization. Cancer Lett. 2007;255:205–18. doi: 10.1016/j.canlet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 76.Yap LF, Jenei V, Robinson CM, Moutasim K, Benn TM, Threadgold SP, et al. Upregulation of Eps8 in oral squamous cell carcinoma promotes cell migration and invasion through integrin-dependent Rac1 activation. Oncogene. 2009;28:2524–34. doi: 10.1038/onc.2009.105. [DOI] [PubMed] [Google Scholar]
- 77.Cooper CR, Chay CH, Pienta KJ. The role of alpha(v)beta(3) in prostate cancer progression. Neoplasia. 2002;4:191–4. doi: 10.1038/sj.neo.7900224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doyle MA, Gasser RB, Woodcroft BJ, Hall RS, Ralph SA. Drug target prediction and prioritization: using orthology to predict essentiality in parasite genomes. BMC Genomics. 2010;11:222. doi: 10.1186/1471-2164-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.