Abstract
A new plant commensal Pseudomonas veronii isolate (strain R4) was identified from a Xiphinema index biocontrol screen. Isolated from grapevine roots from vineyards in central Chile, the strain R4 exhibited a slower yet equivalently effective nematicide activity as the well-characterized P. protegens CHA0. Whole genome sequencing of strain R4 and comparative analysis among the available Pseudomonas spp. genomes allowed for the identification of gene clusters that encode putative extracellular proteases and lipase synthesis and secretion systems, which are proposed to mediate—at least in part—the observed nematicidal activity. In addition, R4 strain presented relevant gene clusters related to metal tolerance, which is typical in P. veronii. Bioinformatics analyses also showed gene clusters associated with plant growth promoting activity, such as indole-3-acetic acid synthesis. In addition, the strain R4 genome presented a metabolic gene clusters associated with phosphate and ammonia biotransformation from soil, which could improve their availability for plants.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-016-0198-y) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas veronii, Pseudomonas spp., Xiphinema index, Vitis vinifera L, Exoproteases, Exolipases, Biocontrol
Introduction
Wine and table grape cultivar productions strongly depend on plant root health and physiology. Soil-borne pathogens affecting these systems avoid water and nutrients uptake and lead to several physiological disorders such as root rot and blackening or plant wilt and stunting. In Chile, several genera of plant-parasitic nematodes are limiting factors for grape production, and one of the most damaging is the dagger Xiphinema index [1]. This nematode is also the natural vector of the Grape fan leaf virus, a widespread disease that affects important grape productive areas of the country [2, 3].
Pseudomonas spp. belonging to the fluorescens group are recognized ubiquitous soil nematicidal agents that can also promote plant health [4, 5]. Among Pseudomonas sp. strains exhibiting antagonistic activity against nematodes of agronomic relevance, the P. protegens strain CHA0 [6] has shown an extraordinary capacity against the root-knot nematodes Meloidogyne javanica and M. incognita by producing exoproteases 2,4-DAPG and HCN [7]. The latter was also described to mediate the nematicidal activity of P. chlororaphis O6 over Meloidogyne hapla [8], a broad host-spectrum plant nematode. Nematicidal repertoires in pseudomonads obey to an important degree of genome heterogeneity within the species group; whereas comparison of 16S ribosomal RNA (rRNA) gene sequences have shown a defined clustering for P. protegens strains, the use of antimicrobial secondary metabolites has led to wrong classification in P. fluorescens and P. chlororaphis [9]. In addition, whole genome sequence data from different P. fluorescens strains have highlighted a strain-to-strain variation and diversity [10]; whereas a conserved set of genes forming a core genome represents only 45–52 % of the genome of any individual strain, important variable regions and several hundred genes are unique to each genome [10].
Currently, biocontrol activity has not been described for P. veronii isolates, which have been largely renowned by their biosorption/bioremediation capabilities [11]. In the present work, we report the whole genome sequencing and characterization of a new P. veronii strain R4. This isolate was first identified from a X. index biocontrol panel and presents a highly effective nematicide activity as compared with P. protegens CHA0. Primarily, the strain R4 cell supernatants resulted in important nematode disruption (Fig. 1 a-c), and candidate proteins responsible for this activity have been isolated, partially sequenced, and identified in these extracts [12].
Fig. 1.
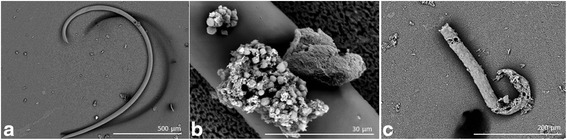
Nematicidal activity of the P. veronii strain R4 over Xiphinema index individuals. Nematicidal activity of strain R4 cell supernatants was obtained from cell cultures grown with milk induction [12], pelleted with acetone, and resuspended using phosphate buffer. Total proteins (20 μg) were added to the wells of 96-well microplates containing 100 μL of buffer and 30 nematodes. The plates were incubated at 24 ± 1 °C for 3 h, and the samples were analysed using scanning electron microscopy (SEM). Initial cuticle degradation in X. index individuals appeared on discrete areas of nematodes’ bodies (a and b), which after challenge led to whole degradation (c)
Organism information
Classification and features
Pseudomonas veronii strain R4 is a motile, Gram-negative, nonsporulating rod in the order Pseudomonadales of the class Gammaproteobacteria. The rod-shaped form varies in size with dimensions of 0.6 μm in width and 2.0 μm in length (Fig. 2a). It is fast growing, forming 2 mm diameter colonies after 48–72 h when grown on KB) [13] at 28 °C. Colonies on KB are white/yellow-opaque, slightly domed, and moderately mucoid with smooth margins (Fig. 2b). The strain R4 was isolated from the roots of healthy nursery-produced grapevine plants in the Maipo valley (Central Chile). It can grow in complex media such as LB [14] or KB as well as in minimal media such as M9 medium [15]. The optimal growth temperature is 28 °C; however, the strain R4 can still replicate at 5 °C in liquid LB and KB. Growth at 37 °C was not observed in these culturing media after 24 h. The bacterium is a colonizer of the grapevine rhizosphere, and it does not cause any deleterious effect on its original host. The strain R4 has natural resistance to carbenicillin (100 mg/L), cefotaxime (300 mg/L), and the mixture of ticarcillin:potassium clavulanate 15:1 (250 mg/L). Minimum Information about the Genome Sequence of P. veronii strain R4 is summarized in Table 1. A phylogenetic tree for the strain R4 and other Pseudomonas spp. was built using a concatenated alignment of 31 universal protein families (Additional file 1: Table S1, Fig. 3).
Fig. 2.
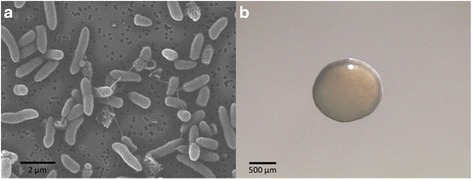
Microscopy analyses of the P. veronii strain R4. Images recording the morphological aspect of the strain R4 cells (a) or an individual colony (b) were acquired using SEM and a light microscope, respectively. Images were acquired to samples grown for 24 h in KB agar medium at 28 °C
Table 1.
Classification and general features of the Pseudomonas veronii strain R4 [39]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain: Bacteria | TAS [40] | |
| Phylum: Proteobacteria | TAS [41] | ||
| Class: Gammaproteobacteria | TAS [42] | ||
| Order: Pseudomonadales | TAS [43] | ||
| Family Pseudomonadaceae | TAS [44] | ||
| Genus Pseudomonas | TAS [11] | ||
| Species Pseudomonas veronii | TAS [12] | ||
| strain: R4 | |||
| Gram stain | Negative | TAS [45] | |
| Cell shape | Rod-shaped | TAS [45] | |
| Motility | Motile | TAS [45 | |
| Sporulation | Not reported | NAS | |
| Temperature range | 5-37 °C | TAS [12] | |
| Optimum temperature | 28 °C | TAS [12] | |
| pH range; Optimum | neutral pH | TAS [12] | |
| Carbon source | Heterotrophic | TAS [12] | |
| MIGS-6 | Habitat | Soil, vine root-associated | TAS |
| MIGS-6.3 | Salinity | 0.85 % NaCl (w/v) | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Rizosphere | NAS |
| MIGS-14 | Pathogenicity | Non-pathogen | IDA |
| MIGS-4 | Geographic location | Chile/Los Andes Province | NAS |
| MIGS-5 | Sample collection | 2009 | NAS |
| MIGS-4.1 | Latitude | S 32° 50′ 42″ | NAS |
| MIGS-4.2 | Longitude | W 70° 36′ 57.599″ | NAS |
| MIGS-4.4 | Altitude | 830 M | NAS |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample but based on a generally accepted property for the species or anecdotal evidence). These evidence codes are from the Gene Ontology project [46]
Fig. 3.
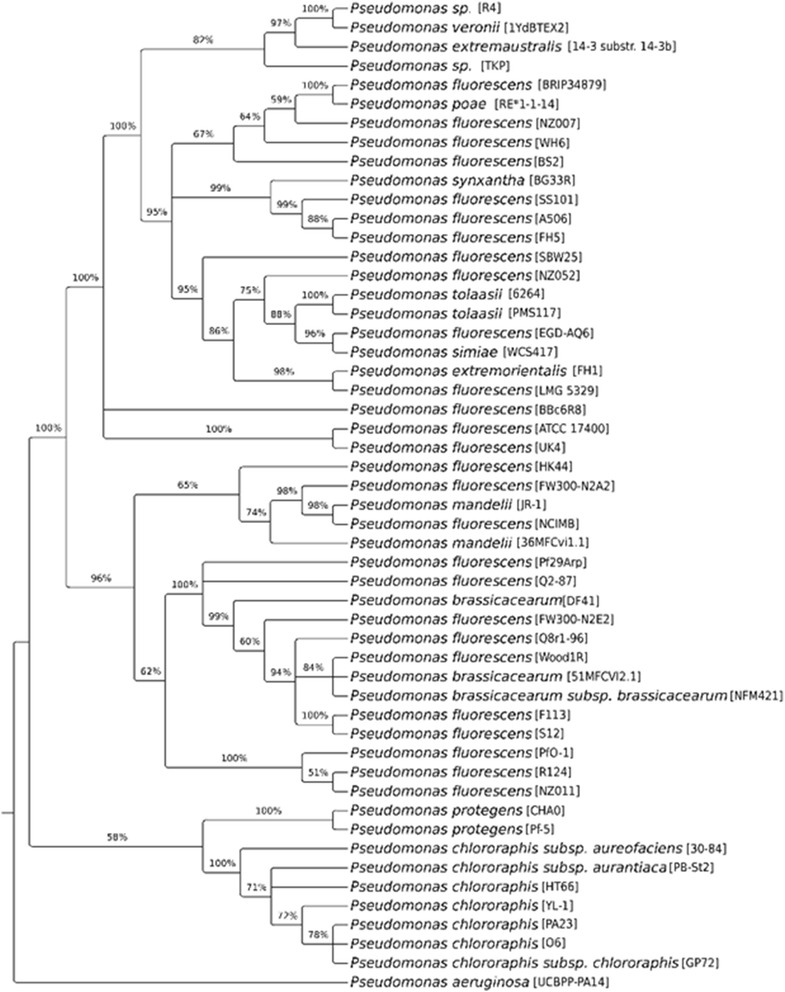
Concatenated alignments of the 31 highly conserved COGs for the 50 members of the Pseudomonas fluorescens group described by [31], which presents a sequenced genome (draft or final) and R4 strain were made with MUSCLE [47]. The poorly aligned and divergent regions were eliminated using Gblocks [48], as described by [49]. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML v3.0 program [50] using the Dayhoff substitution model. Reliability for internal branches was assessed using the Shimodaira-Hasegawa-like Approximate-Likelihood [51]. The resulting tree was visualized using TreeGraph 2 [52]
Genome sequencing information
Genome project history
P. veronii strain R4 was selected for sequencing due to the following: its environmental and agricultural potential; its ability to exert in vitro biocontrol against nematode X. index; and its ability to develop a symbiotic relationship with grapevine root tissues. The genome project is deposited in the Genomes OnLine Database, GOLD [16], and the NCBI BioProject database. The draft genome sequence is in GenBank. A summary of the project information is shown in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | High-quality Draft |
| MIGS-28 | Libraries used | 8,000 bp Mate Pair |
| MIGS 29 | Sequencing platforms | 454 GS-FLX Titanium |
| MIGS 31.2 | Fold coverage | 52.0 |
| MIGS 30 | Assemblers | GS De Novo Assembler V2.9 |
| MIGS 32 | Gene calling method | RAST 2.0, GLIMMER 3.0 |
| Locus Tag | SU91 | |
| Genbank ID | JXWQ00000000 | |
| GenBank Date of Release | April 22, 2015 | |
| GOLD ID | Gp0114890 | |
| BIOPROJECT | PRJNA272785 | |
| MIGS 13 | Source Material Identifier | R4 |
| Project relevance | Biotechnological, Agricultural |
Growth conditions and genomic DNA preparation
A 2-mL overnight culture of strain R4 was prepared in a liquid KB medium at 28 °C and 150 rpm. Two hundred microliters from this culture were used as an inoculum for 200 mL of KB medium and incubated for an additional 8 h under the same culture conditions. The bacteria were centrifuged at 3000 × g and subjected to DNA purification using the ZR Fungal/Bacterial DNA MiniPrep™ kit (Zymo Research), according to the manufacturer’s protocol. The concentration and purity of DNA was measured by a BioSpec-Nano spectrophotometer (Shimadzu Corp., Kyoto, Japan). Five micrograms of purified genomic DNA were submitted for the 454 pyrosequencing.
Genome sequencing and assembly
The genome of the strain R4 was sequenced at Macrogen (Macrogen Inc., Seoul, South Korea) using the 454 sequencing platform. The data consisted of a half plate of 454 FLX Titanium from 8 KB mate-paired libraries. A total of 794,931 reads were achieved for this characterization study, yielding 352,645,131 bases and an average read length of 443.618 bases. The GS De Novo Assembler 2.9 (also known as Newbler assembler) developed by 454 Life Sciences (Roche Company, Basel, Switzerland) was used for sequence assembly, quality assessment, and scaffolding.
Genome annotation
The genes in the assembled genome were predicted with Rapid Annotation using Subsystem Technology server databases 2.0 [17] and the gene-caller GLIMMER 3.02 [18]. Clusters of Orthologous Groups of proteins functional classification was based on homology searches using WebMGA [19]. RNAmmer 1.2 [20] and tRNAscan-SE 1.4 [21] were used to identify rRNA genes and tRNA genes, respectively. CRISPR repeats were examined using the CRISPR recognition tool [22]. Signal peptides and transmembrane helices were predicted using SignalP [23] and TMHMM [24], respectively.
Genome properties
The assembly of the draft genome sequence comprises two scaffolds amounting to 6,649,820 bp (60.8 % average GC content) and a N50 of 6,647,193. In total, 5967 genes were predicted (Table 3, Fig. 4), 5906 of which are protein-coding genes and 61 of which were RNA genes (3 rRNA genes and 58 tRNA genes). The majority of the protein-coding genes (82.9 %) were assigned to a putative function with the remaining annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 6,649,820 | 100.0 |
| DNA coding (bp) | 5,735,337 | 86.2 |
| DNA G + C (bp) | 4,043,431 | 60.8 |
| DNA scaffolds | 2 | – |
| Total genes | 5,967 | 100.0 |
| Protein coding genes | 5,906 | 99.0 |
| RNA genes | 61 | 1.0 |
| Pseudo genes | – | – |
| Genes in internal clusters | – | – |
| Genes with function prediction | 4,894 | 82.9 |
| Genes assigned to COGs | 4,923 | 83.4 |
| Genes with Pfam domains | 4,799 | 80.4 |
| Genes with signal peptides | 557 | 9.4 |
| Genes with transmembrane helices | 1,322 | 22.4 |
| CRISPR repeats | 0 | 0 |
Fig. 4.
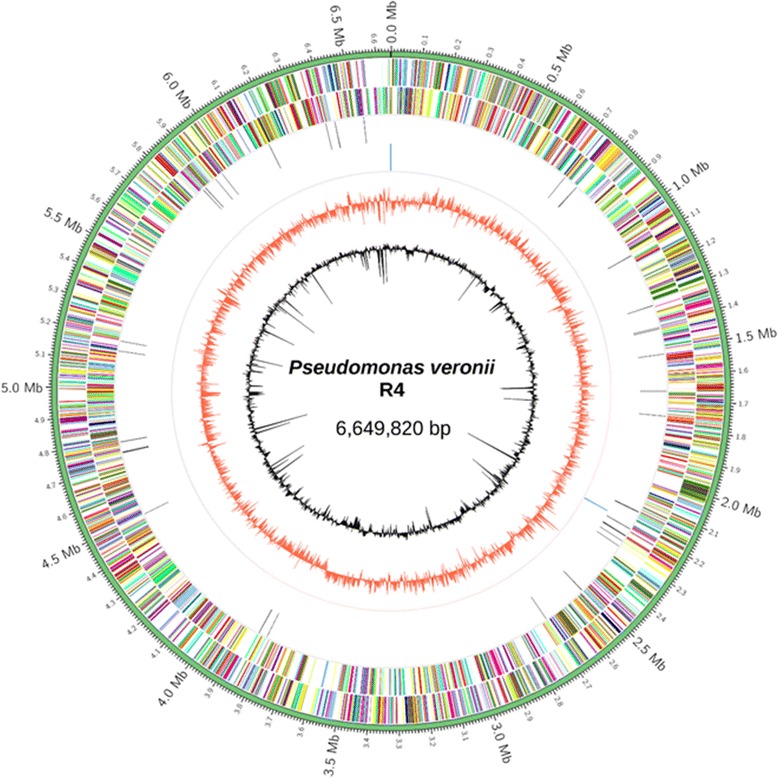
Graphical map of the chromosome. From outside to the centre: genes on forward strand (coloured by COG categories), genes on reverse strand (coloured by COG categories), RNA genes (tRNAs – black, rRNAs – blue, GC skew in red, and G + C in relation to the mean G + C – black), which are both in 2 kb windows. The circular map was generated by Circos [53]
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 190 | 3.22 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.02 | RNA processing and modification |
| K | 523 | 8.86 | Transcription |
| L | 155 | 2.62 | Replication, recombination and repair |
| B | 3 | 0.05 | Chromatin structure and dynamics |
| D | 45 | 0.76 | Cell cycle control, Cell division, chromosome partitioning |
| V | 81 | 1.37 | Defense mechanisms |
| T | 445 | 7.53 | Signal transduction mechanisms |
| M | 271 | 4.59 | Cell wall/membrane biogenesis |
| N | 211 | 3.57 | Cell motility |
| U | 157 | 2.66 | Intracellular trafficking and secretion |
| O | 190 | 3.22 | Posttranslational modification, protein turnover, chaperones |
| C | 340 | 5.76 | Energy production and conversion |
| G | 303 | 5.13 | Carbohydrate transport and metabolism |
| E | 569 | 9.63 | Amino acid transport and metabolism |
| F | 109 | 1.85 | Nucleotide transport and metabolism |
| H | 212 | 3.59 | Coenzyme transport and metabolism |
| I | 235 | 3.98 | Lipid transport and metabolism |
| P | 308 | 5.22 | Inorganic ion transport and metabolism |
| Q | 156 | 2.64 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 611 | 10.35 | General function prediction only |
| S | 470 | 7.96 | Function unknown |
| – | 983 | 16.64 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
In-silico DNA-DNA hybridization was performed using a Genome Blast Distance Phylogeny approach to generate genome based distance measures for phylogenetic inferences and also demonstrated the close relationship between the strain R4 and Pseudomonas veronii. The Genome-to-Genome Distance Calculator [25] revealed a distance of 0.0127 between strain R4 and P. veronii 1YdBTEX2, with a DDH estimate of 89.50 % +/− 2.16. A DDH similarity above 70 % is interpreted as two individuals belonging to the same species, and 79 % is used to discriminate between subspecies [26]. The DDH estimate exceeding the 70 % species threshold was determined from a logistic regression to be 95.63 %. In terms of subspecies relatedness, the probability of exceeding the 79 % threshold was 64.46 % between strain R4 and 1YdBTEX2.
Typical P. veronii elements
Regular genetic elements associated with heavy metal tolerance are found in the strain R4, as expected for a P. veronii isolate. The copABCD operon encodes a multicopper oxidase CopA, which oxidizes cathecol siderophores and generates Cu2+ chelating pigments; in addition, copper-binding proteins (such as CopB, CopC, CopD) that decrease oxidative stress [27, 28] were also found in strain R4 and showed an identical organization to the P. veronii 1YdBTX genome [29] and depicted high identity of encoded proteins showing values >60 %. Also, the strain R4 possesses the gene cluster cznCBA and the cznD gene, encoding for a cation efflux pump of the Resistance-Nodulation-Division family and for a cationic diffusion enhancer, respectively. Both elements confer Zn+2 and Cd+2 resistance in C. metallidurans CH34 [30], and they can be identified in the P. veronii 1YDBTX genome [29].
Genetic elements associated with organic phosphate mineralization from soil were found as extracellular alkaline phosphatase genes PhoD and PhoX. These have been characterized in Pseudomonas fluorescens Pf-0 and allow for soluble phosphate generation and plant absorption [31]. The strain R4 also possesses the amoA gene, which encodes for ammonia mono-oxygenases involved in ammonia-to-nitrite transformation and increases nitrogen availability in soil [32]. Approximately 45 ORFs appeared involved in the denitrification process, which could be organized in three clusters with high homology and with the same organization as in P. fluorescens F113 [33].
Plant-microbe interaction elements
Different plant-bacteria interaction pathways including chemotaxis, adherence, root colonization, nutrients uptake, auxin synthesis, and volatile compound synthesis were deduced from the R4 genome. A two-component chemotaxis system including the kinase sensor (CheA) and the response regulator (CheY) plus potential plant exudate chemoreceptors could activate cell motility into roots, which could be mediated by the flagellar system conformed by almost 77 genes (see Table 5). Root colonization by strain R4 could then be possible by adhesin-like proteins (haemagglutinin and pili types) mediating the plant cell surface association and contact inhibition. Whereas haemagglutining-like genes are distributed throughout the entire genome, Type IV pili genes clustered into two-component systems, signal transduction, and pili structural gene clusters. Also, central carbohydrate metabolism (tricarboxylic acids, Entner-Doudoroff, and pentose cycles) suggests a broad carbon source usage (i.e., D-mannitol, sucrose, trehalose, maltose, xylose, and glucose) derived from plant exudates. They may also use some of the several transporter systems such as regulated by PTS that were also annotated. Supporting these ideas, several GABA catabolic enzymes were found in the strain R4, such as the gadT and gadD genes, which encode for the GABA aminotransferase and succinate semialdehyde dehydrogenase. GABA is a non-proteinogenic amino acid secreted by plants in order to inhibit herbivore, bacterial, and fungal pathogens. The GABA degradative products could be incorporated into the tricarboxylic acids cycle and provide additional carbon provision for the bacteria upon interaction with plants. In addition, the strain R4 presented genes for ABC, MFS, and RND family transporters, enabling nutrient exchange from and into the rhizosphere.
Table 5.
Relevant gene clusters identified on the R4 genome associated with plant-microbe interactions
| Module | Components | Associated genes | Function in strain R4 |
|---|---|---|---|
| Tricarboxylic acids cycle | Catabolic genes | 15 | Carbon metabolism from plant exudates (mannitol, sorbitol, sucrose, trehalose, mannose, arabinose, maltose, xylose and glucose) |
| Entner-Doudoroff pathway | Catabolic genes | 5 | |
| Pentose cycle | Catabolic genes | 14 | |
| Rizosphere nutrients uptake | ABC tranporters regulated by PTS | 23 | |
| Chemotaxis | CheA, CheB, CheR, CheW, CheY and chemoreceptors | 70 | Sensing chemical stimulus and direct motility |
| Motility | Flagello: estructural genes and regulatory genes | 85 | Motility |
| Root colonization | Type IV pili: estructural genes and two components signal transduction proteins | 24 | Host cell surface association and host growth inhibition by contact |
| Haemagglutinin genes | 4 | ||
| Alginate: biosinthetic and regulatoy genes | 24 | Biofilms | |
| Transporters | MFS genes | 32 | Transporters involved in bacterium - rizosphere interaction |
| RND genes | 43 | ||
| ABC genes | 162 | ||
| Acetoin and 2,3-butanediol synthesis | ilvBN; budC; bdh; acoABCX adh | 10 | Plant growth regulators synthesis and catabolism |
| IAA | Two synthetic pathways: from indol-3-acetamide and from indole-3-acetonitrile | 9 | |
| Ethylene | acdS | 1 | ACC catabolism |
| GABA | gadT, gadD, GABA permease gene | 5 | c-aminobutiric acid synthesis |
| Proteases and lipases | Exportable protease (AprA), lipase (LipA) and phopholipase ExoU-like | 8 | biocontrol activity |
| Secondary metabolites | Pyoverdine: estructural genes, and regulatory genes | 19 | |
| Pyochelin: estructural genes and regulatory genes | 30 | ||
| Secretion systems | Type I; Type II; TypeIII and Type VI | 85 | Transport of biocontrol molecules |
Under in vitro conditions, the strain R4 was found to produce IAA from tryptophan; this behaviour was markedly different from other rhizobacteria such as Rhizobium spp. 13, in which differential IAA accumulation has been observed depending on the precursor concentration [34]. The genome in strain R4 contained two potential IAA synthetic pathways from tryptophan: a) the indole-3-acetamide and b) the indole-3-acetonitrile synthetic routes (Table 5). In addition, the R4 genome presents complete synthetic routes for acetoine biosynthesis, i.e., the ilvBN and the acetoine reductase (budC) genes and also the synthetic bdh and the catabolic acoABCX adh genes [10, 35]. These results suggest that strain R4 could transform acetoine into 2,3-butanediol and maybe other PGPs. In addition, the genome data analysis showed the occurrence of a complete catabolic pathway for ethylene, a root elongation inhibitor. The occurrence of the carboxylate-1-aminocyclopropane deaminase (acdS) gene could potentially degrade the ethylene precursor aminocyclopropane into ketobutyric and ammonia, which could synergize the indicated PGP activities.
Biocontrol elements
A gene cluster of 11 Kb in length conserved in P. fluorescens strains SBW25, A506, SS101, F113 [10, 33] and strain R4 included two secretories enzymes, one protease (similar to the metalloprotease Apra very relevant in CHA0 [7]), and one lipase plus an ABC transporter involved in proteases secretion [36]. In CHA0, AprA has been described to inhibit Meloidogyne incognita egg hatching and the death of young nematode individuals [7]. Three other potential exoproteases and two exolipases that have not been described in P. fluorescens were found. A phospholipase (68 kDa) similar in size to the P. aeruginosa ExoU (Acc. No ABJ10150.1) protein, was annotated in the R4 genome. The latter corresponds to an effector protein of the Type III Secretory System in P. aeruginosa, one of the most important virulence factor in that species [37, 38].
Conclusions
The genome analysis allowed for the identification of gene clusters encoding for putative extracellular proteases, lipases, and eventual transport systems that are proposed to mediate, at least in part, the nematicidal activity found in this P. veronii strain in a X. index biocontrol panel. In addition, bioinformatics analyses supported preliminary experimental data that describe plant growth promotion through a putative IAA synthesis pathway.
The phylogenetic relationships between the strain R4 and other sequenced Pseudomonas spp. strains on the basis of concatenated alignment of 31 universal protein families showed the closest relationship with P. veronii strains 1YdBTEX2 and P. extremaustralis 14–3 sbstr. 14-3b. These formed a clade with a high similarity to a group conformed by numerous P. fluorescens isolates.
A predicted R4 genome consisted of 6,678,155 bases in which an assignment of 5840 CDS depicted a coding density of 86.8 %. Using a functional classification of 3796 CDS (65 % of total CDS) by comparing protein sequences from complete genomes and executing a COG, candidate gene sequences revealed several functions such as complete pathways related to carbohydrate central metabolism (i.e., the tricarboxylic acid cycle, the Entner-Doudoroff pathway, and the pentose cycle), and metabolic routes related to plant–bacteria interactions were found. Similarly, metabolic pathways for the synthesis of PGPs such as IAA, acetoin, and 2,3-butanediol were also successfully identified. Moreover, gene groups for chemotaxis, root colonization, rhizosphere nutrient uptake, and volatile compounds were found.
Acknowledgements
The Pseudomonas protegens strain CHA0 was kindly provided by Dr. Dieter Haas from the Department of Fundamental Microbiology at the University of Lausanne in Switzerland. This work was funded by the Biofrutales S.A. Consortium and the Corfo-Chile grant 13CTI- 21520-SP7. HC and FA are Comisión Nacional de Ciencia y Tecnología (CONICYT) Chile doctoral fellows.
Authors’ contributions
CM, FA, ES, JV, PT, and CG developed the bioinformatics processing of the R4 genome. HC, HP, AS, AC, ET, and MM developed the primary functional identification of the strain R4. HP conceived of the study and participated in its design and coordination. CM, FA, and HP helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
Authors declare have no competing interests as defined by the Journal, or other interests that might be perceived to influence the results and/or discussion reported in this article.
Abbreviations
- 2,4 DAPG
2,4-diacetylphloroglucinol
- DDH
Digital DNA-DNA hybridization
- GABA
Gamma-aminobutiric acid
- IAA
Indole acetic acid
- KB
King’s agar
- PGPs
Plant growth promoters
- PTS
Phosphotransferase system
Additional file
Table S1.Cluster of Orthologous Genes (COG) considered in the phylogenetic analysis. (DOCX 12 kb)
References
- 1.Aballay E, Mårtensson A, Persson P. Screening of rhizosphere bacteria from grapevine for their suppressive effect on Xiphinema index Thorne & Allen on in vitro grape plants. Plant Soil. 2011;347:313–25. doi: 10.1007/s11104-011-0851-6. [DOI] [Google Scholar]
- 2.Fiore N, Prodan S, Montealegre J, Aballay E, Pino A, Zamorano A. Survey of grapevine viruses in Chile. J Plant Pathol. 2008;90:125–30. [Google Scholar]
- 3.Valenzuela A, Aballay E, Torres M. Identificación y frecuencia de nematodos asociados a la vid en la Región Metropolitana, Chile. Investigación Agrícola (Chile) 1992;12:15–7. [Google Scholar]
- 4.Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–19. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 5.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA, Raaijmakers JM. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 6.Stutz EW, Défago G, Kern H. Naturally occurring fluorescent pseudomonads involved in suppression of Black Root Rot of tobacco. Phytopathology. 1986;76:181–5. doi: 10.1094/Phyto-76-181. [DOI] [Google Scholar]
- 7.Siddiqui IA, Haas D, Heeb S. Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. Appl Environ Microbiol. 2005;71:5646–9. doi: 10.1128/AEM.71.9.5646-5649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, Ko SJ, Ma KC, Kang BR, Kim YC, Kim IS. Nematicidal activity of a nonpathogenic biocontrol bacterium, Pseudomonas chlororaphis O6. Curr Microbiol. 2011;62:746–51. doi: 10.1007/s00284-010-9779-y. [DOI] [PubMed] [Google Scholar]
- 9.Ramette A, Frapolli M, Fischer-Le Saux M, Gruffaz C, Meyer JM, Défago G, Sutra L, Moënne-Loccoz Y. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol. 2011;34:180–8. doi: 10.1016/j.syapm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Loper JE, Hassan KA, Mavrodi DV, Davis EW, II, Lim CK, Shaffer BT, Elbourne L, Stockwell V, Hartney S, Breakwell K, Henkels M, Tetu S, Rangel L, Kidarsa T, Wilson N, van de Mortel J, Song C, Blumhagen R, Radune D, Hostetler J, Brinkac L, Durkin A, Kluepfel D, Wechter P, Anderson A, Kim Y, Pierson L, III, Pierson E, Lindow S, Kobayashi D, Raaijmakers J, Weller D, Thomashow L, Allen A, Paulsen I. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elomari M, Coroler L, Hoste B, Gillis M, Izard D, Leclerc H. DNA relatedness among Pseudomonas strains isolated from natural mineral waters and proposal of Pseudomonas veronii sp. nov. Int J Syst Bacteriol. 1996;46:1138–44. doi: 10.1099/00207713-46-4-1138. [DOI] [PubMed] [Google Scholar]
- 12.Canchignia H, Altimira F, Montes C, Sánchez E, Tapia E, Miccono MA, Espinoza D, Aguirre C, Michael S, Prieto H. Candidate nematicidal proteins in a new Pseudomonas veronii isolate identified by its antagonic properties against Pseudomonas veronii. J. Gen. Appl. Microbiol. 2016. doi: 10.2323/jgam.2016.07.001. [DOI] [PubMed]
- 13.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–7. [PubMed] [Google Scholar]
- 14.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harwood CR, Cutting SM. Chemically defined growth media and supplements. In: Harwood CR, Cutting SM, editors. Molecular biological methods for Bacillus. Chichester: Wiley; 1990. p. 548. [Google Scholar]
- 16.Reddy TBK, Thomas A, Stamatis D, Bertsch J, Isbandi M, Jansson J, Mallajosyula J, Pagani I, Lobos E, Kyrpides N. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta) genome project classification. Nucleic Acids Res. 2015;43:D1099–106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444–52. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagesen K, Hallin PF, Rødland E, Stærfeldt HH, Rognes T, Ussery DW. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinf. 2007;8:209–16. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 24.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 25.Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–34. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier-Kolthoff JP, Hahnke RL, Petersen JP, Scheuner CS, Michael VM, Fiebig AF, Rohde CR, Rohde MR, Fartmann BF, Goodwin LA, Chertkov OC, Reddy TR, Pati AP, Ivanova NN, Markowitz VM, Kyrpides NC, Woyke TW, Klenk HP, Göker M. Complete genome sequence of DSM 30083 T, the type strain (U5/41 T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha JS, Cooksey DA. Copper resistance in Pseudomonas syringae mediated by perimasmic and outer membrane proteins. Proc Natl Acad Sci U S A. 1991;88:8915–9. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magnani D, Solioz M. How bacteria handle copper. In: Nies D, Silver S, editors. Bacterial transition metal homeostasis. Germany: Springer, Heidelberg; 2007. pp. 259–85. [Google Scholar]
- 29.de Lima-Morales D, Chaves-Moreno D, Jarek M, Vilchez-Vargas R, Jauregui R, Pieper DH. Draft genome sequence of Pseudomonas veronii strain 1YdBTEX2. Genome Announc. 2013;1:e00258–13. doi: 10.1128/genomeA.00258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monchy S, Benotmane M, Jassen P, Vallaey T, Taghavi S, van der Lelie D, Mergeay M. Plasmids pMol28 and pMol30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J Bacterial. 2007;189:7417–7125. doi: 10.1128/JB.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monds RD, Newell PD, Schwartzman JA, O’Toole GA. Conservation of the Pho regulon in Pseudomonas fluorescens p f0–1. Appl Environ Microbiol. 2006;72:1910–24. doi: 10.1128/AEM.72.3.1910-1924.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daum M, Zimmer W, Papen H, Kloos K, Nawrath K, Bothe H. Physiological and molecular biological characterization of ammonia oxidation of heterotrophicnitrifier Pseudomonas putifda. Curr Microbiol. 1998;37:281–8. doi: 10.1007/s002849900379. [DOI] [PubMed] [Google Scholar]
- 33.Redondo-Nieto M, Barret M, Morrissey J, Germaine K, Martínez-Granero F, Barahona E, Navazo A, Sánchez-Contreras M, Moynihan JA, Muriel C, Dowling D, Fergal O’Gara F, Martín M, Rivilla R. Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genomics. 2013;14:54–70. doi: 10.1186/1471-2164-14-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sridevi M, Mallaiah K. Bioproduction of indole acetic acid by Rhizobium strains isolated from root nodules of green manure crop, Sesbania sesban (L.) Merr. Iran J Biotechnol. 2007;5:178–82. [Google Scholar]
- 35.Taghavi S, van der Lelie D, Hoffman A, Zhang Y, Walla M, Vangronsveld J, Newman L, Monchy S. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 2010;6:e1000943. doi: 10.1371/journal.pgen.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn J, Pan J, Shick Rhee J. Identification of the tliDEF ABC transporter specific for lipase in Pseudomonas fluorescens SIK W1. J Bacteriol. 1999;181:1847–52. doi: 10.1128/jb.181.6.1847-1852.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abd H, Wretlind B, Saeed A, Idsund E, Hultenby K, Sandström G. Pseudomonas aeruginosa utilises its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J Eukaryot Microbiol. 2008;55:235–43. doi: 10.1111/j.1550-7408.2008.00311.x. [DOI] [PubMed] [Google Scholar]
- 38.Matz C, Moreno AM, Alhede M, Manefield M, Hauser AR, Givskoy M, Kjelleberg S. Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2008;2:843–52. doi: 10.1038/ismej.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, Hermjakob H, Hertz-Fowler C, Hugenholtz P, Joint I, Kagan L, Kane M, Kennedy J, Kowalchuk G, Kottmann R, Kolker E, Kravitz S, Kyrpides N, Leebens-Mack J, Lewis SE, Li K, Lister AL, Lord P, Maltsev N, Markowitz V, Martiny J, Methe B, Mizrachi I, Moxon R, Nelson K, Parkhill J, Proctor L, White O, Sansone SA, Spiers A, Stevens R, Swift P, Taylor C, Tateno Y, Tett A, Turner S, Ussery D, Vaughan B, Ward N, Whetzel T, San Gil I, Wilson G, Wipat A. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woese CR. Towards a natural system of organisms: proposal for the domains archea, bacteria and eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrity GM, Bell JA. Phylum Proteobacteria, XIV: phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriol-ogy. Volume 2. Part B. 2. New York: Springer; 2005. p. 1. [Google Scholar]
- 42.Garrity GM, Bell JA. Class Gammaproetobacteria III: class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. Volume 2. Part B. 2. New York: Springer; 2005. p. 1. [Google Scholar]
- 43.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 44.Garrity G, Bell J, Lilburn T. In: Bergey’s manual of systematic bacteriology. Volume 2, Part B. 2. Garrity G, Brenner D, Krieg N, Staley J, editors. New York: Springer; 2005. p. 323. [Google Scholar]
- 45.Palleroni NJ. Pseudomonadaceae. In: Krieg NR, Holt JG, editors. Bergey’s manual of systematic bacteriology. Baltimore: The Williams and Wilkins Co; 1984. pp. 141–99. [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 49.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 51.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–52. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 52.Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krzywinski M, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones S, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]


