Abstract
Background
Limonene, a monocyclic monoterpene, is known for its using as an important precursor of many flavoring, pharmaceutical, and biodiesel products. Currently, d-limonene has been produced via fractionation from essential oils or as a byproduct of orange juice production, however, considering the increasing need for limonene and a certain amount of pesticides may exist in the limonene obtained from the citrus industry, some other methods should be explored to produce limonene.
Results
To construct the limonene synthetic pathway in Yarrowia lipolytica, two genes encoding neryl diphosphate synthase 1 (NDPS1) and limonene synthase (LS) were codon-optimized and heterologously expressed in Y. lipolytica. Furthermore, to maximize limonene production, several genes involved in the MVA pathway were overexpressed, either in different copies of the same gene or in combination. Finally with the optimized pyruvic acid and dodecane concentration in flask culture, a maximum limonene titer and content of 23.56 mg/L and 1.36 mg/g DCW were achieved in the final engineered strain Po1f-LN-051, showing approximately 226-fold increase compared with the initial yield 0.006 mg/g DCW.
Conclusions
This is the first report on limonene biosynthesis in oleaginous yeast Y. lipolytica by heterologous expression of codon-optimized tLS and tNDPS1 genes. To our knowledge, the limonene production 23.56 mg/L, is the highest limonene production level reported in yeast. In short, we demonstrate that Y. lipolytica provides a compelling platform for the overproduction of limonene derivatives, and even other monoterpenes.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-016-0626-7) contains supplementary material, which is available to authorized users.
Keywords: Limonene, Yarrowia lipolytica, Neryl diphosphate synthase 1, Limonene synthase
Background
Natural compound monoterpenes are C10 compounds that consist of two isoprene units. Monoterpenes belong to a large family of plant secondary metabolites with valuable applications including using as biofuels, feedstocks for pharmaceutical and other industrial product syntheses, and flavors and fragrances [4, 13, 23]. Limonene, a monocyclic monoterpene, is famous for its citrus-like olfactory properties. Limonene is also an important precursor of many flavor and medicinal compounds such as perillyl alcohol (POH), carvone, and menthol [1, 13]. Limonene has three isomers: d-limonene, dl-limonene, and l-limonene. They all consist in natural plants, while d-limonene is the most widespread [32]. D-limonene is considered as GRAS (generally recognized as safe) material by the US Food and Drug Administration [19]. Furthermore, d-limonene is widely used as a flavoring or fragrance agent in pharmaceuticals, foods, and beverages [27]. Currently, d-limonene has been produced via fractionation from essential oils or as a byproduct of orange juice production, but considering the increasing need for limonene as a polymer and as a jet fuel, the citrus industry may not be able to meet future demands. In the meantime, a certain amount of pesticides may exist in the limonene obtained from the citrus industry, so it is necessary that some other methods should be explored to produce limonene [10, 13].
Metabolic engineering of microorganisms to produce natural products seems to be a good choice. Many terpenoids have been produced by various kinds of microorganisms, such as lycopene produced in Yarrowia lipolytica; miltiradiene and sclareol produced in Saccharomyces cerevisiae; dammarenediol-II produced in Pichia pastoris; β-carotene produced in Escherichia coli and so on [6, 17, 21, 28, 35]. Limonene biosynthesis has been widely studied in other microorganisms including E. coli [1, 11, 33] and S. cerevisiae [4, 13]. In yeast, isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP), the two common building blocks, are the precursors of all terpenoids and they are derived from the mevalonic acid (MVA) pathway, and geranyl diphosphate (GPP) is the direct precursor of monoterpenes catalyzed by geranyl/farnesyl diphosphate synthase (ERG20) [7] (Fig. 1). Limonene can be produced by introducing one key enzyme, limonene synthase (LS) into the pathway based on GPP [4, 13]. LS catalyzes the intramolecular cyclization of GPP to gain limonene.
Fig. 1.
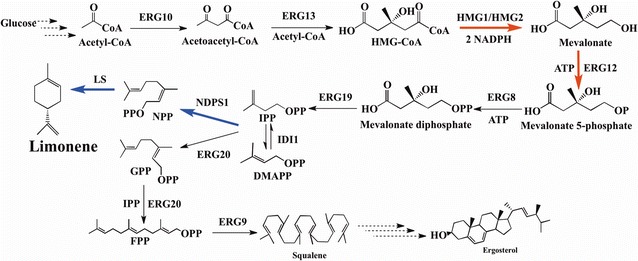
Biosynthesis pathway for limonene production in Y. lipolytica. IPP and DMAPP are converted to NPP by neryl diphosphate synthase 1 (NDPS1) and then NPP is further converted to limonene by limonene synthase (LS). Blue arrows represent that the pathways were exogenously integrated in Y. lipolytica, while red arrows represent that the pathways were overexpressed in Y. lipolytica. Single arrows represent the one-step conversions, while triple arrows represent multiple steps
Yarrowia lipolytica as a non-conventional yeast is considered non-pathogenic and generally regarded as safe (GRAS). The availability of suitable metabolic engineering tools and the full genome sequence of Y. lipolytica make it a favorable host for producing pharmaceutical and food additives, and some studies have verified natural products produced in this strain [3, 21, 34]. Yarrowia lipolytica has the ability to grow to high biomass yield on simple substrates. A wide range of compounds can be produced by Y. lipolytica from glycerol, a side product of biodiesel production [8, 21, 25]. In this study, we aim to construct a limonene biosynthesis pathway in Y. lipolytica and then metabolically engineer this strain for efficient production of limonene. To reach the target, two genes encoding neryl diphosphate synthase 1 (NDPS1) and limonene synthase (LS) were codon-optimized and heterologously expressed in Y. lipolytica. Secondly, to enhance limonene production, several genes involved in the MVA pathway were overexpressed, either in different copies of the same gene or in combination. Lastly, the optimized addition of pyruvic acid and dodecane further improved limonene production. By combining metabolic engineering and optimization of genes expression, an approximately 226-fold increase in limonene productivity was obtained in comparison with the initial strain.
Methods
Strains, vectors, chemicals, and culture media
The vectors and strains used in this study are listed in Tables 1 and 2, respectively. The auxotrophic Y. lipolytica strain Po1f was used as the target gene expression host in this study. This Y. lipolytica strain and the plasmids pINA1269 and pINA1312 have been previously described by Madzak et al. [18]. Yarrowia lipolytica cells were cultured at 30 °C in YPD medium (1 % yeast extract, 2 % peptone, and 2 % glucose) or YNB medium (0.67 % yeast nitrogen base without amino acids, 1 % glucose, and 1.6 % agar). The rich medium YPD was used for strain activation and fermentation, while the synthetic medium YNB, lacking leucine and uracil where appropriate, was used for the screening of transformants. Escherichia coli strain JM109 or DH5α was used as a host for plasmid proliferation and construction of recombinant vectors. The cells were grown in Luria–Bertani (LB) complete medium (0.5 % yeast extract, 1 % tryptone, and 1 % NaCl) at 37 °C for 12 h. Suitable antibiotics or nutrients were added when necessary at the following final concentrations: ampicillin, 100 mg/L; kanamycin 50 mg/L; leucine 0.3 g/L. Plasmid extraction, gel extraction, and purification of DNA were conducted using assay kits from Sangon Biotech Co., Ltd. (Shanghai, China). Primer synthesis was conducted by GeneRay Biotech Co., Ltd. (Shanghai, China). Pyruvic acid and dodecane were purchased from TCI Development Co., Ltd. (Shanghai, China).
Table 1.
Vectors used in this study
| Vectors | Description | Source |
|---|---|---|
| pINA1312 | Y. lipolytica-integrative plasmid, hp4d promoter, XPR2 terminator, ura3d1 selection marker, KmR | [22] |
| pINA1269 | Y. lipolytica-integrative plasmid, hp4d promoter, XPR2 terminator, LEU2 selection marker, AmpR | [22] |
| p1312-tLS | Constitutively expressed truncated LS gene | This study |
| p1312-tNDPS1 | Constitutively expressed truncated NDPS1 gene | This study |
| p1312wLN | Constitutively expressed tLS and tNDPS1 genes | This study |
| p1312LN | Constitutively expressed codon-optimized tLS and codon-optimized tNDPS1 genes | This study |
| p1269tHMG1 | Constitutively expressed tHMG1 gene | This study |
| p1269HMG1 | Constitutively expressed HMG1 gene | This study |
| p1269IDI1 | Constitutively expressed IDI1 gene | This study |
| p1269ERG8 | Constitutively expressed ERG8 gene | This study |
| p1269ERG10 | Constitutively expressed ERG10 gene | This study |
| p1269ERG12 | Constitutively expressed ERG12 gene | This study |
| p1269ERG19 | Constitutively expressed ERG19 gene | This study |
| p1269tLS | Constitutively expressed tLS gene | This study |
| p1269tNPDS1 | Constitutively expressed tNDPS1 gene | This study |
| p1269tHMG1tHMG1 | Constitutively expressed tHMG1 gene of two copies | This study |
| p1269tHMG1tHMG1tHMG1 | Constitutively expressed tHMG1 gene of three copies | This study |
| p1269HMG1HMG1 | Constitutively expressed HMG1 gene of two copies | This study |
| p1269IDI1IDI1 | Constitutively expressed IDI1 gene of two copies | This study |
| p1269IDI1IDI1IDI1 | Constitutively expressed IDI1 gene of three copies | This study |
| p1269HMG1IDI1 | Constitutively expressed HMG1 and IDI1 genes | This study |
| p1269HMG1ERG8 | Constitutively expressed HMG1 and ERG8 genes | This study |
| p1269HMG1ERG10 | Constitutively expressed HMG1 and ERG10 genes | This study |
| p1269HMG1ERG12 | Constitutively expressed HMG1 and ERG12 genes | This study |
| p1269HMG1ERG19 | Constitutively expressed HMG1 and ERG19 genes | This study |
| p1269HMG1tLS | Constitutively expressed HMG1 and tLS genes | This study |
| p1269HMG1tNPDS1 | Constitutively expressed HMG1 and tNDPS1 genes | This study |
Table 2.
Strains used in this study
| Strains | Description | Source |
|---|---|---|
| Escherichia coli | ||
| DH5α, JM109 | For construction of recombinant vectors | Invitrogen |
| Yarrowia lipolytica | ||
| Po1f | leu2−, ura3− | [2] |
| Po1f-LN-000 | Po1f cells harboring p1312LN | This study |
| Po1f-LN-001 | Po1f cells harboring p1312LN and p1269tHMG1 | This study |
| Po1f-LN-002 | Po1f cells harboring p1312LN and p1269tHMG1tHMG1 | This study |
| Po1f-LN-003 | Po1f cells harboring p1312LN and p1269tHMG1tHMG1tHMG1 | This study |
| Po1f-LN-004 | Po1f cells harboring p1312LN and p1269HMG1 | This study |
| Po1f-LN-005 | Po1f cells harboring p1312LN and p1269HMG1HMG1 | This study |
| Po1f-LN-006 | Po1f cells harboring p1312LN and p1269IDI1 | This study |
| Po1f-LN-007 | Po1f cells harboring p1312LN and p1269IDI1IDI1 | This study |
| Po1f-LN-008 | Po1f cells harboring p1312LN and p1269IDI1IDI1IDI1 | This study |
| Po1f-LN-011 | Po1f cells harboring p1312LN and p1269HMG1tLS | This study |
| Po1f-LN-021 | Po1f cells harboring p1312LN and p1269HMG1tNDPS1 | This study |
| Po1f-LN-031 | Po1f cells harboring p1312LN and p1269HMG1ERG8 | This study |
| Po1f-LN-041 | Po1f cells harboring p1312LN and p1269HMG1ERG10 | This study |
| Po1f-LN-051 | Po1f cells harboring p1312LN and p1269HMG1ERG12 | This study |
| Po1f-LN-061 | Po1f cells harboring p1312LN and p1269HMG1ERG19 | This study |
| Po1f-LN-071 | Po1f cells harboring p1312LN and p1269HMG1IDI1 | This study |
Plasmids construction
The gene encoding d-limonene synthase (LS, GenBank ID: AY055214.1) from Agastache rugosa and gene encoding neryl diphosphate synthase 1 (NDPS1, GenBank ID: NM_001247704.1) from Solanum lycopersicum were codon-optimized and synthesized by GeneRay Biotech. The transit peptides of both genes were removed according to the literatures [5, 26]. The truncated genes were named tLS and tNDPS1, respectively. The tLS gene and tNDPS1 gene were cloned into p1312 with primers P1/P2 and P3/P4 to obtain vectors p1312-tLS and p1312-tNDPS1, respectively (Table 3). Then the expression cassette P-tLS-T was cloned into p1312-tNDPS1 with primers P5/P6 to obtain vector p1312LN. The genes HMG1, IDI1, ERG8, ERG10, ERG12, ERG19 amplified from the genome of Po1f with tLS and tNDPS1 were cloned into p1269 with primers P7/P8, P9/P10, P11/P12, P13/P14, P15/P16, P17/P18, P1/P2, and P3/P4 to obtain the respective vectors p1269HMG1, p1269IDI1, p1269ERG8, p1269ERG10, p1269ERG12, p1269ERG19, p1269tLS, p1269tNPDS1. Afterwards, the expression cassettes P-tHMG1-T, P-HMG1-T, P-IDI1-T, P-ERG8-T, P-ERG10-T, P-ERG12-T, P-ERG19-T, P-tLS-T, and P-tNDPS1-T were cloned into p1269tHMG1, p1269HMG1, p1269IDI1 with primers P19/P20 to obtain p1269tHMG1tHMG1, p1269HMG1HMG1, p1269IDI1IDI1, p1269HMG1tLS and p1269HMG1tNPDS1, p1269HMG1ERG8, p1269HMG1ERG10, p1269HMG1ERG12, p1269HMG1ERG19, p1269HMG1IDI1, respectively. Then the expression cassettes P-tHMG1-T and P-IDI1-T were cloned into p1269tHMG1tHMG1 and p1269IDI1IDI1 with primers P21/P22 to obtain p1269tHMG1tHMG1tHMG1 and p1269IDI1IDI1IDI1, respectively. All the plasmids were constructed using the One Step Cloning Kit from Vazyme Biotech Co., Ltd. (Nanjing, China).
Table 3.
Primers used in this study
| Primer | Primer sequence (5′–3′) |
|---|---|
| P1 | ACAACCACACACATCCACGTGATGCGACGATCCGGTAACTACTCCCCTT (PmlI) |
| P2 | TTAGTTTCGGGTTCCCACGTGCTAGGCGAAAGGCTGGAACAGGCAA (PmlI) |
| P3 | ACAACCACACACATCCACGTGATGTCCGCCCGAGGTCTCAACAAAA (PmlI) |
| P4 | TTAGTTTCGGGTTCCCACGTGCTAGTAGGTGTGGCCACCGAATCGT (PmlI) |
| P5 | AGATAGAGTCGACAAAGGCCTGCTAGCTTATCGATACGCGTGCATG (StuI) |
| P6 | TGTACACCGAGAAACAGGCCTCATCTCACTTGCGTATGTATGGAAA (StuI) |
| P7 | ACAACCACACACATCCACGTGATGCTACAAGCAGCTATTGG (PmlI) |
| P8 | TTAGTTTCGGGTTCCCACGTGCTATGACCGTATGCAAATATT (PmlI) |
| P9 | ACAACCACACACATCCACGTGATGACGACGTCTTACAGCGA (PmlI) |
| P10 | TTAGTTTCGGGTTCCCACGTGCTACTTGATCCACCGCCGAA (PmlI) |
| P11 | ACAACCACACACATCCACGTGATGACCACCTATTCGGCTCC (PmlI) |
| P12 | TTAGTTTCGGGTTCCCACGTGCTACTTGAACCCCTTCTCGA (PmlI) |
| P13 | ACAACCACACACATCCACGTGATGCGACTCACTCTGCCCCG (PmlI) |
| P14 | TTAGTTTCGGGTTCCCACGTGCTACTCGACAGAAGAGACCT (PmlI) |
| P15 | ACAACCACACACATCCACGTGATGGACTACATCATTTCGGC (PmlI) |
| P16 | TTAGTTTCGGGTTCCCACGTGCTAATGGGTCCAGGGACCGA (PmlI) |
| P17 | ACAACCACACACATCCACGTGATGATCCACCAGGCCTCCAC (PmlI) |
| P18 | TTAGTTTCGGGTTCCCACGTGCTACTTGCTGTTCTTCAGAG (PmlI) |
| P19 | CGAGGCAGCAGATCCACTAGTAGCACCGCCGCCGCAAGGAATGG (SpeI) |
| P20 | GCGGCCGCATAGGCCACTAGTCTGTCAAACATGAGAATTCGG (SpeI) |
| P21 | CTCTCAAGGGCATCGGTCGACAGCACCGCCGCCGCAAGGAATGG (SalI) |
| P22 | CGCATAAGGGAGAGCGTCGACCTGTCAAACATGAGAATTCGG (SalI) |
| P23 | CGGCATCCGCTTACAGAC |
| P24 | GGAGGCATCAGTGACCAAA |
| P25 | CATTAGGAAGCAGCCCAGTA |
| P26 | GAGATCGTCAAGGGTTTG |
| P27 | CATAAGTGCGGCGACGAT |
| P28 | CTACTACTGGGCTGCTTCCTA |
Restriction sites are underlined
Strain construction
p1312LN was linearized with NotI and then was transformed into competent Po1f cells using the kit, Frozen-EZ yeast transformation II. After transformation, cells were cultured on YNB medium plates with leucine added. The right colonies verified with primers P1/P2 and P3/P4 were named Po1f-LN-000. Then, p1269tHMG1, p1269tHMG1tHMG1, and p1269tHMG1tHMG1tHMG1 were linearized with ApaI while p1269HMG1, p1269HMG1HMG1, p1269IDI1, p1269IDI1IDI1, and p1269IDI1IDI1IDI1 were linearized with BsrGI. All these linearized plasmids were transformed into competent Po1f-LN-000 cells, respectively. These cells were cultured on YNB medium plates and the right colonies, verified with primers P23/P24, P25/P26, and P27/P28 were named Po1f-LN-001, Po1f-LN-002, Po1f-LN-003, Po1f-LN-004, Po1f-LN-005, Po1f-LN-006, Po1f-LN-007, and Po1f-LN-008, respectively. Afterwards, p1269HMG1tLS, p1269HMG1tNPDS1, p1269HMG1ERG8, p1269HMG1ERG10, p1269HMG1ERG12, p1269HMG1ERG19, and p1269HMG1IDI1 were linearized separately with BsrGI and transformed into Po1f-LN-000. After verification, the right colonies obtained were therefore named Po1f-LN-011, Po1f-LN-021, Po1f-LN-031, Po1f-LN-041, Po1f-LN-051, Po1f-LN-061, and Po1f-LN-071, respectively.
Yeast cultivation
YPD medium was used to cultivate the engineered strains. All strains were firstly inoculated into 10 mL culture tubes containing 2 mL medium, and grown at 30 °C at 220 rpm to reach an OD600 of about 1.0. The cultures were started by inoculating 50 mL medium containing 1 mL dodecane as organic extractant phase with the preculture [30] and the initial OD600 was 0.01. Strains were cultured at 30 °C and 220 rpm for 3 days. All the flask fermentation results represented the mean ± S.D. of three independent experiments. At the same time, to investigate the influence of the addition of pyruvic acid in vitro, pyruvic acid of different concentrations 2, 4, and 8 g/L were added to the medium of strain Po1f-LN-051, respectively. Furthermore, effects of the amount of dodecane on limonene synthesis by strain Po1f-LN-051 were also surveyed, with dodecane varying from 2 to 10 % (volume/volume) and constant pyruvic acid concentration of 4 g/L.
Analysis
Optical densities at 600 nm (OD600) were measured using a Shimadzu UV-1800 spectrophotometer (Shimadzu Co., Kyoto, Japan). 5 mL of wet cell culture was harvested, washed twice with distilled water, and centrifuged at 12,000 rpm for 5 min. The upper layer was discarded and dry cell weight was determined by measuring the weight of cell pellets which were dried at 105 °C for 48 h.
For limonene detection, the dodecane layers were collected and myrcene was used as an internal standard. The dodecane overlay samples were mixed with myrcene by 4:1 (v/v). Then the mixture (1 μL) was analyzed by GC/MS using an Agilent System 6890 gas chromatograph coupled to an Agilent 5975 quadrupole mass selective detector (EI) (Agilent Technologies, Santa Clara, CA), equipped with a HP-5 (30 m × 0.25 mm, 0.25 µm film thickness) GC column. The GC oven temperature program was as follows: 100 °C for 1 min, a ramp of 0.5 °C/min to 102 °C, and then a rise to 280 °C in 5 min. The split ratio was 20:1. Limonene and myrcene standards (purchased from Sigma-Aldrich) were used for quantification.
For squalene extraction, 1.8 mL of fermentation broth was collected and resuspended with 600 μL of 20 % KOH/50 % ethanol solution then voltexed at once using a multi holder for 5 min at the fastest rate. After that, the tubes were placed into boiling water for 5 min then cooled in ice water. HPLC grade hexane 600 μL was added to the tubes, which was then voltexed using a multi holder for 5 min. The two phases were separated by centrifugation at 12,000 rpm for 5 min at 4 °C. After centrifugation, 400 μL of top hexane layer was transferred into the new 1.5 mL eppendorf (EP) tubes and evaporated using a vacuum dryer for 15 min at a low dry rate, after that 50 μL of HPLC grade ethanol and 450 μL of HPLC grade acetonitrile were added to the tubes. Squalene was quantified with the Shimadzu LC-20A (Shimadzu Co., Kyoto, Japan) equipped with C18 column (Phenomenex Kinetex 5 μm C18) and a UV detector. The wavelength was 195 nm and mobile phase was 100 % acetonitrile with 2 mL/min flow rate and a column temperature of 35 °C. Squalene standard (purchased from TCI Biotech) was used for quantification.
Inhibitory effects of d-limonene on Y. lipolytica Po1f
A fixed volume of serially diluted d-limonene in ethanol ranging from 100 to 3000 mg/L was added to YPD medium with 0.5 % Tween 80 v/v. Tween 80 was dissolved into the medium to increase the solubility of d-limonene. YPD medium contained 0.5 % Tween 80v/v and ethanol was regarded as control.
Results and discussion
Construction of the limonene synthetic pathway in Y. lipolytica
Though Y. lipolytica possesses a native MVA pathway which can supply the intermediates DMAPP and IPP (Fig. 1), it cannot produce limonene because of the absence of limonene synthase. Consequently, the gene encoding d-limonene synthase (LS) was first amplified from the cDNAs of Agastache rugosa. Meanwhile, to overcome the possible expression problem a codon-optimized artificial LS gene was also designed and amplified. The integrative vector pINA1312 carrying either the wild-type LS gene or the codon-optimized artificial LS gene was successfully integrated into the chromosome of Po1f strain, respectively. Unfortunately, after three-day cultivation of these two engineered strains, no limonene could be detected in the dodecane layers by GC–MS (data not shown), based on the relative retention time and total ion mass spectral comparison with the external standard.
Geranyl diphosphate (GPP) has been generally regarded as the substrate for monoterpenes, while very little was reported about NPP, the isomer of GPP. Recently, monoterpenes in the glandular trichomes of tomato were found to be synthesized from NPP precursor rather than GPP [26]. Another study showed that NPP instead of GPP was the substrate of limonene in S. cerevisiae [16]. Therefore, we hypothesized that NPP might also be the major substrate for limonene biosynthesis in Y. lipolytica. To address the precursor problem, codon-optimized NDPS1 gene, which encodes NDPS1 catalyzing the conversion of DMAPP and IPP to NPP in Solanum lycopersicum, were integrated into the chromosome of Y. lipolytica Po1f together with the codon-optimized LS gene to obtain strain Po1f-LN-000. Dodecane layers were collected on the third day and tested by GC/MS. Limonene (m/z 68.1, 93.1, and 136.5) was monitored at 3.9 min (Fig. 2). The engineered strain Po1f-LN-000 produced limonene in detectable quantities at 0.006 mg/g DCW and was therefore used as the initial limonene-producing strain in this study (Fig. 3). Thus, using codon-optimized gene tLS from Agastache rugosa and tNDPS1 from S. lycopersicum, the biosynthetic pathway for limonene production was successfully constructed in Y. lipolytica. The results also demonstrated that introduction of NPP pathway is essential for limonene production and the genes derived from plants were necessary to be codon-optimized when expressed in Y. lipolytica. After this initial success in metabolic engineering of Y. lipolytica for limonene production, further stepwise improvements were attempted using this Po1f-LN-000 strain. The optimized nucleotide sequences of truncated LS gene and truncated NDPS1 gene were provided in Additional file 1.
Fig. 2.
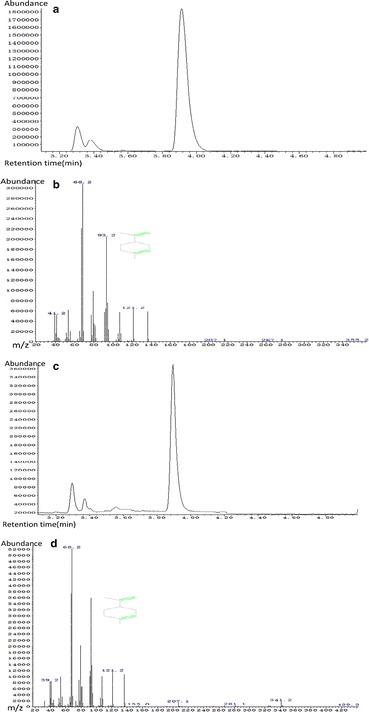
GC–MS analysis of limonene from the dodecane phase of the cultures in engineered Y. lipolytica. The strain was cultivated in YPD medium for 72 h. a Limonene standard; b Mass spectrum of limonene standard; c Limonene obtained in YPD medium containing 4 g/L pyruvic acid of Po1f-LN-051; d Mass spectrum of limonene obtained in YPD medium containing 4 g/L pyruvic acid of Po1f-LN-051
Fig. 3.
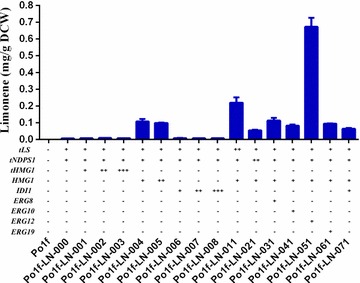
Quantitative analysis of limonene production in engineered Y. lipolytica. Limonene productions in the engineered strains Po1f-LN-000, Po1f-LN-001, Po1f-LN-002, Po1f-LN-003, Po1f-LN-004, Po1f-LN-005, Po1f-LN-006, Po1f-LN-007, Po1f-LN-008, Po1f-LN-011, Po1f-LN-021, Po1f-LN-031, Po1f-LN-041, Po1f-LN-051, Po1f-LN-061, Po1f-LN-071. All strains were cultured in YPD medium for 3 days. Three repeats were performed for each strain, and error bars represent standard deviations
Improving limonene production by overexpressing HMG-CoA reductase gene
The enzyme 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase is well known as the major rate-limiting enzyme of the mevalonate (MVA) pathway in many organisms, including yeasts. Overexpression of the catalytic domain of the HMGR (producing a form of the enzyme that lacks the membrane-binding region, namely truncated HMG-CoA reductase gene or tHMG1) in yeasts has been shown to boost production of isoprenoid biosynthesis [9, 24, 29]. Furthermore, in the oleaginous yeast Y. lipolytica, it was also reported that the integration of HMG1 into the engineered strain led to a 6.9-fold increase in lycopene content after 96 h of cultivation, probably attributable to the increased supply of precursors [21]. Thus, it was determined whether overexpression of HMG1 could increase limonene production in limonene-producing Y. lipolytica cells. For this purpose, heterologous tHMG1 gene from S. cerevisiae and homologous HMG1 gene from Y. lipolytica were overexpressed under the control of hy4d promoter in the limonene-producing strain Po1f-LN-000, as showed in Fig. 3, resulting in the recombinant strains Po1f-LN-001 and Po1f-LN-004, respectively. Limonene production was only improved around 30 % in the heterologous recombinant strain Po1f-LN-001, while homologous recombination of HMG1 significantly enhanced the limonene-producing level. In the fermentation broths of the strain Po1f-LN-004, the limonene titer (or content) increased from 0.006 mg/g DCW to 0.11 ± 0.01 mg/g DCW, 18-fold higher than the control strain Po1f-LN-000 (Fig. 3). These results did confirm that the production of limonene was accumulated by the duplication of HMG-CoA reductase gene and, more importantly, the overexpression of homologous HMG1 was more effective to increase the biosynthesis of limonene in Y. lipolytica.
Further enhancing the HMG1 copy number might lead to further improvement of the limonene production. To validate the hypothesis, the plasmids carrying extra gene copies were introduced into Po1f-LN-000, yielding another type of recombinant strains (Po1f-LN-005) that contain HMG1 of two copies. We found that the two copies of HMG1 did not necessarily improve limonene production, instead a slight decrease in limonene titer was observed. Similar phenomenon also happened for strains Po1f-LN-002 harboring two copies of tHMG1 and Po1f-LN-003 with three copies of tHMG1 (Fig. 3). These results implied that it was not necessary to increase the copy number of HMG-CoA reductase gene, and single copy of homologous HMG1 was good enough for limonene overproduction in Y. lipolytica.
Effects of re-engineering of limonene synthesis on limonene production
Since there was only a single copy of tLS and tNDPS1 gene in chromosome with relatively low expression strength, limonene synthase and neryl diphosphate synthase enzymes might be rate limiting for limonene production. In order to investigate the effects of increased activities of these enzymes on limonene production, plasmids p1269HMG1tLS and p1269HMG1tNDPS were transformed into strain Po1f-LN-000 (Po1f cells harboring p1312LN and p1269HMG1) to generate strain Po1f-LN-011 with two copies of tLS and strain Po1f-LN-021 with two copies of tNDPS1, respectively. Shake flask cultures of strain Po1f-LN-004 and the engineered strains of Po1f-LN-011and Po1f-LN-021 were carried out and the limonene productions were compared (Fig. 3). In contrast to the slight decrease in limonene production in strain Po1f-LN-021, the final limonene concentration by Po1f-LN-011 doubled in comparison with that of the engineered strain Po1f-LN-004, suggesting that strengthening the conversion of precursor NPP to limonene plays a more important role in improving the production. In contrast to the effectiveness of enhancing tLS expression, strain with extra copy of tNDPS1 produced less amount of limonene than that of Po1f-LN-004 strain, indicating that single expression of tNDPS1 was more efficient in converting IPP and DMAPP into NPP.
Effects of overexpression of gene encoding isopentenyl diphosphate isomerase on limonene production
The gene IDI1 (Fig. 1) encodes for an isopentenyl diphosphate isomerase that catalyzes an essential step in the sterol pathway, the isomerization of IPP to DMAPP. A number of works focused on the overexpression of IDI gene to improve isoprenoid accumulation in E. coli [14, 20, 31]. In S. cerevisiae, an extra copy of IDI1 was introduced into the genome leading to a 50 % increase in geraniol yield. Furthermore, overexpression of IDI1 using a multi-copy plasmid in the engineered strains increased geraniol yield 1.45-fold [15]. In another study, overexpression of IDI1 increased the monoterpene cineole production by twofold, and approximately fivefold increase was obtained when IDI1 was expressed from a high copy plasmid in the engineered S. cerevisiae strains [12]. In order to investigate the effects of overexpressing homologous IDI on limonene production, the plasmids carrying different copies of IDI were transformed into Po1f-LN-000 strain resulting in strains Po1f-LN-006 (single copy), Po1f-LN-007 (two copies), and Po1f-LN-008 (three copies), respectively. The data showed that single copy of IDI1 gene could only increase the limonene concentration by 30 %. However, there was no obvious difference in limonene production with the increase in copy number of the IDI1 gene. Although one extract IDI1 integration could enhance the limonene biosynthesis in Y. lipolytica, the improvement was limited compared with the overexpression of HMG1 (Fig. 3). When both HMG1 and IDI1 genes were overexpressed in Po1f-LN-000 (resulting in strain Po1f-LN-071), interestingly, we observed a slight decrease in limonene production instead of the production enhancement in comparison with Po1f-LN-004, suggesting that IDI1 might exert limited influence on limonene accumulation in Y. lipolytica.
Optimization of mevalonate pathway improved limonene production
Our previous work has already shown that the homologous overexpression of HMG1 was efficient in boosting limonene production in Y. lipolytica. In addition to the overexpression of the HMG1 gene alone, we also emphasized on the co-overexpression of the HMG1 gene together with other genes involved in the MVA pathway (e.g., ERG8 encoding phosphomevalonate kinase, ERG10 encoding acetoacetyl-CoA thiolase, ERG12 encoding mevalonate kinase, and ERG19 encoding mevalonate diphosphate decarboxylase), in order to further explore potential and efficient strategies for improving limonene productivity. The plasmids p1269HMG1ERG8, p1269HMG1ERG10, p1269HMG1ERG12, and p1269HMG1ERG19 were integrated into Po1f-LN-000 separately, resulting strains Po1f-LN-031, Po1f-LN-041, Po1f-LN-051, and Po1f-LN-061, respectively. As shown in Fig. 3, among all of the above recombinant strains, the improved limonene production could only be observed for the strain Po1f-LN-051 in which ERG12 and HMG1 were co-overexpressed. One hundred and 12- and 6-fold increases in limonene content were obtained in Po1f-LN-051 in comparison with those obtained by the control strains of Po1f-LN-000 and Po1f-LN-004, respectively. The results suggested the significant importance of mevalonate biosynthesis and its subsequent phosphorylation in supplying an enhanced carbon flux for the improved production of MVA pathway-related metabolites. It has not yet been reported that the overexpression of the ERG12 gene is favorable to the synthesis of monoterpene. Actually, when both the HMG1 and ERG12 genes were overexpressed, we also observed a 2.5-fold increase in the content of squalene, a shunt biosynthetic product (i.e., 0.36 mg/g DCW and 0.96 mg/g DCW in strains Po1f-LN-000 and Po1f-LN-051, respectively), while the growth rates of these strains did not vary too much (Fig. 4).
Fig. 4.
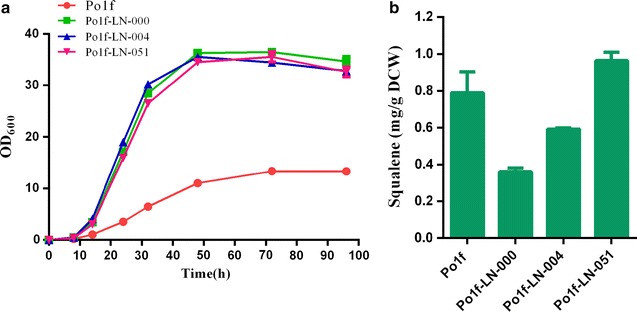
a The OD600 values of strains Po1f, Po1f-LN-000, Po1f-LN-004, and Po1f-LN-051 cultured in YPD medium, measured at 0, 8, 14, 24, 32, 48, 72, and 96 h. b Squalene production in strains Po1f, Po1f-LN-000, Po1f-LN-004, Po1f-LN-051 cultured in YPD medium for 5 days
Effects of pyruvic acid and dodecane addition
Based on the successes in genetic modification of Y. lipolytica for improving limonene biosynthesis, extensive efforts have been made in the optimization of culture medium to further increase limonene productivity. Among several key influential factors, effects of pyruvic acid and dodecane were investigated in this study. Pyruvic acid is a substrate of the DXP pathway and previous studies indicated that the addition of pyruvic acid as an auxiliary carbon source could increase amorphadiene and limonene accumulation in E. coli [10, 36]. In order to evaluate the effect of the amount of pyruvic acid on the growth of Y. lipolytica and limonene production, shake flask cultures with initial pyruvic acid concentrations varying from 0 to 8 g/L in YPD medium were conducted and compared. Although no obvious differences were observed for cell growth, the limonene contents were increased and 4 g/L of initial pyruvic acid seemed to favor the limonene production best (Fig. 5b). We also tested the use of glycerol as a sole carbon source or auxiliary carbon source. However, limonene productions were decreased apparently only as a sole carbon source in comparison with other combinations (Fig. 5a). The reason may be that the promoter when glycerol was used as substrate resulted in the lower expression of genes and reduced the production of limonene.
Fig. 5.
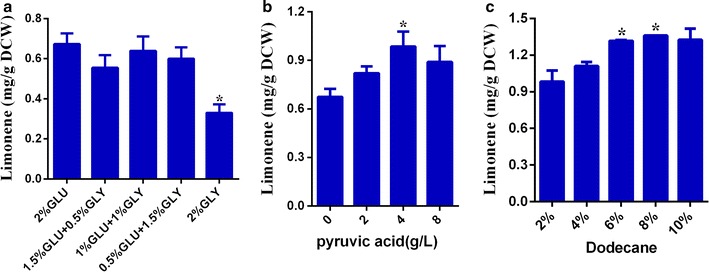
Effects of different combinations of glucose and glycerol as substrate and heterologous addition of pyruvic acid and dodecane on the production of limonene. a Glucose of 2, 1.5, 1, 0.5, 0 % were respectively combined with glycerol of 0, 0.5, 1, 1.5, 2 % as substrate in the YP medium of strain Po1f-LN-051. b Pyruvic acid was added to the YPD medium of strain Po1f-LN-051 to a final concentration of 0, 2 , 4, or 8 g/L. c Dodecane was added to the YPD medium of strain Po1f-LN-051 with 4 g/L pyruvic acid as the auxiliary carbon source and the proportion of dodecane was from 2 to 10 %. Three repeats were performed for each medium, and error bars represent standard deviations. t tests were conducted to evaluate statistical significance at p < 0.05. Particularly, for (a) the asterisk shows statistical significance between 2 % glucose and other combinations of glucose and glycerol, for (b) the asterisk shows statistical significance between YPD medium containing 0 g/L pyruvic acid and other concentrations of pyruvic acid, and for (c) the asterisk shows statistical significance between YPD medium containing 2 % dodecane and other proportions of dodecane
Limonene is produced and secreted by Y. lipolytica into the medium. Usually dodecane is added into the culture medium to enrich limonene. To investigate the amount of dodecane on limonene production, the cultures were performed with different initial dodecane concentrations varying from 2 to 10 % in YPD medium with 4 g/L pyruvic acid as the auxiliary carbon source. Maximum production of limonene in recombinant cells was observed when dodecane concentration was 8 %, and no significant differences on cell growth were found in all cases. With the optimized pyruvic acid and dodecane concentrations, a limonene production of 23.56 mg/L and 1.36 mg/g DCW was achieved, approximately 226-fold higher than those of the control strain Po1f-LN-000 (with only codon-optimized LS and NDPS1 genes overexpressed) (Fig. 5c). These results demonstrated that both the addition of pyruvic acid and the appropriate dodecane concentration were important in enhancing limonene production by Y. lipolytica.
Inhibitory effects of d-limonene on Y. lipolytica
As seen from the Fig. 6, yeast growth was similar to the control when the concentration of limonene in the medium was lower than 500 mg/L. The cell growth was significantly inhibited when limonene concentration was higher than 500 mg/L.
Fig. 6.
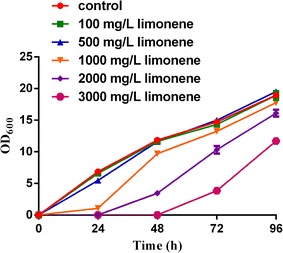
Inhibitory effects of d-limonene on Y. lipolytica. A fixed volume of serially diluted d-limonene in ethanol ranging from 100 to 3000 mg/L was added to YPD medium which contained 0.5 % Tween 80 (v v-1), YPD medium contained 0.5 % Tween 80 (v v-1) and ethanol was regarded as control. Po1f was used as the trial strain
Limonene biosynthesis has been widely studied in E. coli and S. cerevisiae. In E. coli, the highest limonene production was 435 mg/L from simple sugars using a heterologous mevalonate pathway. Furthermore, the purification of limonene produced in E. coli was also explored recently [1, 4, 13, 33]. In S. cerevisiae, limonene trapped in a dodecane phase resulted in the recovery of 0.028 mg/L (+)-limonene and 0.060 mg/L (−)-limonene in strains expressing a mutant ERG20 enzyme and the truncated Citrus and Perilla synthases, respectively. Another study showed that the best limonene titer obtained in S. cerevisiae was 1.48 ± 0.22 mg/L when a truncated HMG1 and upc2-1 were expressed with codon-optimized citrus (+)-limonene synthase in supplemented YP medium. Although the yield of limonene obtained in our study is lower than that in E. coli, considering the non-GRAS status of E. coli, it is of great commercial interest for GRAS status organism Y. lipolytica to produce limonene. In addition, the limonene production of the engineered strain in this study is the highest reported to date in yeasts, which might prompt further research on the biosynthesis of various terpenoids in Y. lipolytica.
Conclusion
Limonene, an important monoterpene, is used as a precursor of many flavoring, pharmaceutical, and biodiesel products and majorly supplied limitedly in plant source. Recently, microbial production of valuable chemicals by economically efficient bioprocesses has emerged as an attractive alternative way. Here, our work presented the first report of the limonene biosynthetic pathway in oleaginous yeast Y. lipolytica by heterologous expression of codon-optimized tLS and tNDPS1 genes. Specifically, we optimized the MVA pathway and re-engineered the limonene biosynthetic pathway for further stepwise improvement of limonene production capacity. Combining this metabolic engineering strategy with the optimization of medium in flask culture, a maximum limonene titer and content of 23.56 mg/L and 1.36 mg/g DCW (the highest reported in yeasts) was achieved in this study, showing approximately 226-fold increase compared with the initial yield of 0.006 mg/g DCW. As a conclusion, we demonstrated that Y. lipolytica could be a compelling platform for a feasible, scalable, and economic route to the overproduction of limonene derivatives, and even other monoterpenes. In our next experiments, fermentation strategies will be developed based on the engineered strains to obtain enhanced production. We will be focusing on several issues, such as optimizing the medium compositions and fermentation parameters, selecting suitable solvent and antifoam.
Authors’ contributions
QH, JW, and XC conceived and designed the experiments. XC, BL, and JC did the lab work, reagent purchasing, plasmid construction, strain screening and cultivation, fermentation, and product detection. XC did the literature review and prepared the manuscript. QH, JW, and TI helped to revise the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Prof. Catherine Madzak (INRA, UMR1319 Micalis, Domair e de Vilvert, Jouy-en-Josas, France) for the auxotrophic Y. lipolytica strain Po1f and the plasmids pINA1269 and pINA1312.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was financially supported by National Basic Research Program of China (973 Program) (2012CB721101), and National Natural Science Foundation of China (21576089).
Abbreviations
- DMAPP
dimethylallyl pyrophosphate
- IPP
isopentenyl pyrophosphate
- GPP
geranyl diphosphate
- NPP
neryl diphosphate
- MVA
mevalonate
- NDPS1
neryl diphosphate synthase
- LS
limonene synthase
Additional file
10.1186/s13068-016-0626-7 The optimized nucleotide sequences of truncated LS gene and truncated NDPS1 gene.
Contributor Information
Xuan Cao, Email: caoxuanflora@live.com.
Yu-Bei Lv, Email: zyx_ssb@163.com.
Jun Chen, Email: junchen@ecust.edu.cn.
Tadayuki Imanaka, Email: imanaka@sk.ritsumei.ac.jp.
Liu-Jing Wei, Phone: +86-021-64250972, Email: weiliujing@ecust.edu.cn.
Qiang Hua, Phone: +86-021-64250972, Email: qhua@ecust.edu.cn.
References
- 1.Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Barth G, Gaillardin C. 1996. Yarrowia lipolytica. Nonconventional yeasts in biotechnology. pp. 313–88.
- 3.Barth G, Gaillardin C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev. 1997;19:219–237. doi: 10.1111/j.1574-6976.1997.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 4.Behrendorff JB, Vickers CE, Chrysanthopoulos P, Nielsen LK. 2,2-Diphenyl-1-picrylhydrazyl as a screening tool for recombinant monoterpene biosynthesis. Microb Cell Fact. 2013;12:1. doi: 10.1186/1475-2859-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colby SM, Alonso WR, Katahira EJ, McGarvey DJ, Croteau R. 4S-limonene synthase from the oil glands of spearmint (Mentha spicata) J Biol Chem. 1993;268:23016–23024. [PubMed] [Google Scholar]
- 6.Dai ZB, Liu Y, Huang LQ, Zhang XL. Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109:2845–2853. doi: 10.1002/bit.24547. [DOI] [PubMed] [Google Scholar]
- 7.Dai ZB, Liu Y, Zhang XN, Shi MY, Wang BB, Wang D, Huang LQ, Zhang XL. Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab Eng. 2013;20:146–156. doi: 10.1016/j.ymben.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 8.De Pourcq K, Vervecken W, Dewerte I, Valevska A, Van Hecke A, Callewaert N. Engineering the yeast Yarrowia lipolytica for the production of therapeutic proteins homogeneously glycosylated with Man8GlcNAc2 and Man5GlcNAc2. Microb Cell Fact. 2012;11:1. doi: 10.1186/1475-2859-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donald K, Hampton RY, Fritz IB. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol. 1997;63:3341–3344. doi: 10.1128/aem.63.9.3341-3344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du FL, Yu HL, Xu JH, Li CX. Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E. coli. Bioresour Technol. 2014;1:1. [Google Scholar]
- 11.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ignea C, Cvetkovic I, Loupassaki S, Kefalas P, Johnson CB, Kampranis SC, Makris AM. Improving yeast strains using recyclable integration cassettes, for the production of plant terpenoids. Microb Cell Fact. 2011;10:1. doi: 10.1186/1475-2859-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jongedijk E, Cankar K, Ranzijn J, van der Krol S, Bouwmeester H, Beekwilder J. Capturing of the monoterpene olefin limonene produced in Saccharomyces cerevisiae. Yeast. 2015;32:159–171. doi: 10.1002/yea.3038. [DOI] [PubMed] [Google Scholar]
- 14.Kim SW, Keasling JD. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng. 2001;72:408–415. doi: 10.1002/1097-0290(20000220)72:4<408::AID-BIT1003>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 15.Liu JD, Zhang WP, Du GC, Chen J, Zhou JW. Overproduction of geraniol by enhanced precursor supply in Saccharomyces cerevisiae. J Biotechnol. 2013;168:446–451. doi: 10.1016/j.jbiotec.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Liu JD. 2013. Key issues in the metabolic engineering of Saccharomyces cerevisiae for monoterpene production [D]. Wuxi: Jiangnan University.
- 17.Liu XB, Liu M, Tao XY, Zhang ZX, Wang FQ, Wei DZ. Metabolic engineering of Pichia pastoris for the production of dammarenediol-II. J Biotechnol. 2015;216:47–55. doi: 10.1016/j.jbiotec.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Madzak C, Gaillardin C, Beckerich JM. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J Biotechnol. 2004;109(1–2):63–81. doi: 10.1016/j.jbiotec.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 19.Mamidipally PK, Liu SL. First approach on rice bran oil extraction using limonene. Eur J Lipid Sci Technol. 2004;106:122–125. doi: 10.1002/ejlt.200300891. [DOI] [Google Scholar]
- 20.Matthews PD, Wurtzel ET. Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of de-oxyxylulose phosphate synthase. Appl Microbiol Biotechnol. 2000;53:396–400. doi: 10.1007/s002530051632. [DOI] [PubMed] [Google Scholar]
- 21.Matthäus F, Ketelhot M, Gatter M, Barth G. Production of lycopene in the non-carotenoid producing yeast Yarrowia lipolytica. Appl Environ Microbiol. 2014;80(5):1660–1669. doi: 10.1128/AEM.03167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicaud JM, Madzak C, Broek P, Gysler C, Duboc P, Niederberger P, Gaillardin C. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002;2:371–379. doi: 10.1016/S1567-1356(02)00082-X. [DOI] [PubMed] [Google Scholar]
- 23.Oswald M, Fischer M, Dirninger N, Karst F. Monoterpenoid biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 2007;7:413–421. doi: 10.1111/j.1567-1364.2006.00172.x. [DOI] [PubMed] [Google Scholar]
- 24.Polakowski T, Stahl U, Lang C. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl Microbiol Biotechnol. 1998;49:66–71. doi: 10.1007/s002530051138. [DOI] [PubMed] [Google Scholar]
- 25.Rywińska A, Juszczyk P, Wojtatowicz M, Robak M, Lazar Z, Tomaszewska L, Rymowicz W. Glycerol as a promising substrate for Yarrowia lipolytica biotechnological applications. Biomass Bioenerg. 2013;48:148–166. doi: 10.1016/j.biombioe.2012.11.021. [DOI] [Google Scholar]
- 26.Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. PNAS. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun JD. D-Limonene: safety and clinical applications. Altern Med Rev. 2007;12:259–264. [PubMed] [Google Scholar]
- 28.Trikka FA, Nikolaidis A, Athanasakoglou A, Andreadelli A, Ignea C, Kotta K, Argiriou A, Kampranis SC, Makris AM. Iterative carotenogenic screens identify combinations of yeast gene deletions that enhance sclareol production. Microb Cell Fact. 2015;14:60. doi: 10.1186/s12934-015-0246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verwaal R, Wang J, Meijnen JP, Visser H, Sandmann G, van den Berg JA, van Ooyen AJ. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl Environ Microbiol. 2007;73:4342–4350. doi: 10.1128/AEM.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers CE, Bongers M, Bydder SF, Chrysanthopoulos P, Hodson MP, 2015. Protocols for the production and analysis of isoprenoids in bacteria and yeast. Hydrocarbon and lipid microbiology protocols, Springer protocols handbooks. 1–30.
- 31.Villalóna AR, Gil JP, Concepción MR. Carotenoid accumulation in bacteria with enhanced supply of isoprenoid precursors by upregulation of exogenous or endogenous pathways. J Biotechnol. 2008;135:78–84. doi: 10.1016/j.jbiotec.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 32.Wang WJ. Recent advances on limonene, a natural and active monoterpene (in Chinese) China Food Addit. 2005;1:33–37. [Google Scholar]
- 33.Willrodt C, David C, Cornelissen S, Bühler B, Julsing MK, Schmid A. Engineering the productivity of recombinant Escherichia coli for limonene formation from glycerol in minimal media. Biotechnol J. 2014;9:1000–1012. doi: 10.1002/biot.201400023. [DOI] [PubMed] [Google Scholar]
- 34.Xue ZH, Sharpe PL, Hong SP, Yadav NS, Xie DM, Short DR, Damude HG, Rupert RA, Seip JE, Wang J, Pollak DW, Bostick MW, Bosak MD, Macool DJ, Hollerbach DH, Arcilla DM, Bledsoe SA, Croker K, McCord EF, Tyreus BD, Jackson EN, Zhu Q. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol. 2013;31:734–740. doi: 10.1038/nbt.2622. [DOI] [PubMed] [Google Scholar]
- 35.Yoon SH, Lee SH, Das A, Ryu HK, Jang HJ, Kim JY, Oh DK, Keasling JD, Kim SW. Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. J Biotechnol. 2009;140:218–226. doi: 10.1016/j.jbiotec.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Zhou K, Zou R, Zhang CQ, Stephanopoulos G, Too HP. Optimization of amorphadiene synthesis in Bacillus Subtilis via transcriptional, translational, and media modulation. Biotechnol Bioeng. 2013;110:2556–2561. doi: 10.1002/bit.24900. [DOI] [PubMed] [Google Scholar]


