Introduction
In the past decade, our laboratory has been greatly interested in studying the Fanconi Anemia (FA) signaling pathway [1–5]. This interest stems from the ability of the FA pathway to maintain genome stability and suppress tumorigenesis. Oncogenic proliferation occurs without sufficient nucleotides to support DNA replication, resulting in replication failure at common fragile sites and leads to the formation of double-strand DNA breaks [6]. The failure in correctly sensing or rescuing stalled replication intermediates is one of underlying causes of several chromosomal instability syndromes such as FA. It is an inherited disorder characterized by congenital abnormalities, bone marrow failure, and cancer proneness [7]. However, much remains unknown regarding how the FA pathway acts under normal conditions and about the manners by which FA proteins contribute a proper progression among individual phases of cell cycle. Moreover, any insight into the FA pathway is important because such insight could lead to understanding of how genomic instability occurs [8]. We expect that the knowledge from understanding this pathway will provide valuable information leading to building up effective tools for cancer prevention as well as therapy.
Successful DNA replication is essential for ongoing cellular life; however, this process poses the threat of DNA damage [9]. Considering the fact that FANCD2 is only monoubiquitinated in the S-phase of each normal cell cycle, our study explored how monoubiquitinated FANCD2 acts during the S-phase of cell cycle [10]. The chromatin binding nature of FANCD2 protein prompted us to reveal that monoubiquitinated FANCD2 has a stronger binding capacity to the replication origins than the non-monoubiquitinated one. Importantly, we observed that the fired origins are reduced in cells deficient in the basal level of monoubiquitinated FANCD2 in comparison with cells carrying a normal level of FANCD2 monoubiquitination. All of these pushed us to go a step further into the mechanisms underlying the functions of monoubiquitinated FANCD2 at replication origins. Through mass spectrometric analysis, we found the interaction between the monoubiquitinated FANCD2 and minichromosome maintenance protein 3 (MCM3). This interaction affects the licensed origin firing of replication at the replication origins. Monoubiquitinated FANCD2 has been studied for a long time for its relationship with DNA damage; but little is known regarding its roles in DNA replication. Our study allow us to first propose a work model regarding how monoubiquitinated FANCD2 performs in DNA replication origin firing and, thus, participates in the control of S-phase progression.
Replication origin firing marks the initiation of DNA replication. This process is regulated through a number of binding-factors, which include the origin recognition complex (ORC) providing a platform for Cdt1 and Cdc6 to recruit MCM2-7 proteins. This complex of helicases unwinds DNA for DNA polymerase to synthesize new DNA [11]. MCM3 was found to be a functional dock for the monoubiquitinated FANCD2 to tether a proper amount of MCM helicase complexes at the origins. This binding relationship appeared to be attributed to the arginine finger region (AA 477–480) of MCM3. And the finding was further supported by the molecular docking using the Z-DOCK web tool [12]. As shown in Figure 1, the predicted structure of human MCM3 (blue), based on the crystalized MCM5 of Saccharomyces cerevisiae (PDB: 3JA8.5) using I-TASSER [13–16], adapted well to that of FANCD2 protein (red) (PDB: 3S4W.2). This model also further validated their specific interaction region [MCM3 (magenta) and FANCD2 (cyan)]. Together, our results strongly suggest that this newly recognized role of monoubiquitinated FANCD2 in each S-phase of normally growing cells, will give a post-licensing check on the licensed origins prior to the commencement of firing.
Figure 1.
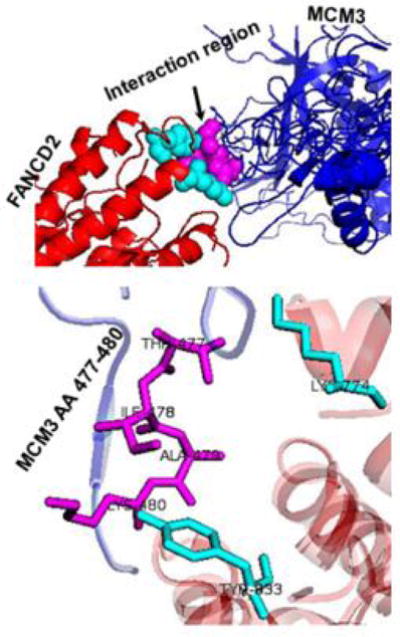
The molecular docking of MCM3 and FANCD2.
Our experiments demonstrate that the basal level of monoubiquitinated FANCD2 is tasked with an active role in replisome surveillance. As shown in Figure 2, the assembling an origin of DNA replication and the normally monoubiquitinated FANCD2 both act in DNA replication initiation. Eukaryotic cells initiate their DNA replication at origins via the interaction between DNA elements (replicators and origins) and protein elements (initiator and other initiation proteins) [17]. Monoubiquitinated FANCD2 in each S-phase interacts with replication origins as well as the member(s) of MCM complex, such as MCM3, to provide “a temporal and spatial check” for securing an adequate amount of licensed-replication origins to fire. Without the normal basal level of FANCD2 monoubiquitination, cells would not have an enough number of licensed origins to initiate a normal replication, thereby overtime rendering genomic instability, aging and cancer, as shown in FA. Our long-term goal is to use the FA pathway as a unique genetic model system to expand our knowledge of tumorigenesis and to use this knowledge to enable preventive and therapeutic approaches for cancer patient care. This has guided us to conduct many studies leading the field of investigation [18,19].
Figure 2.
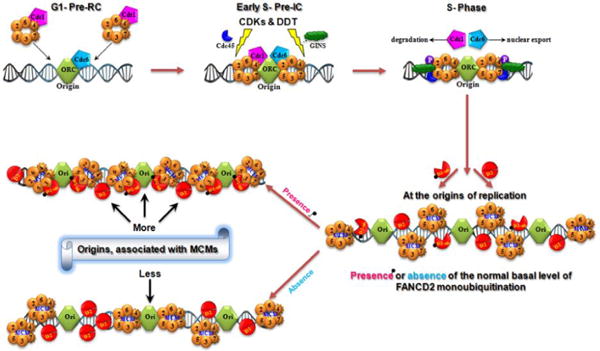
FANCD2-monoubiquitination Role in Replication Initiation.
Conclusion
In conclusion, we believe that investigating the FA pathway will allow us to not only understand basic mechanisms underlying DNA repair and response to the cancer treatment but also will be helpful for generating new effective tools in the fight against human cancer. For example, our study would suggest a promising chemotherapeutic approach of targeting specific parts of MCMs to lower down cancer cell progression by screening our Hawaii or other natural products or chemical libraries. The resultant lead compounds would effectively modulate the origin firing of cancer cells and, thus, stop their malignant growth.
Acknowledgments
The work is supported partly by R01CA188251 & R01CA136532 to PF. Our studies are also partly supported by the University of Hawaii Cancer Center, Honolulu, HI.
References
- 1.Fei P, Yin J, Wang W. New advances in the DNA damage response network of Fanconi anemia and BRCA proteins. FAAP95 replaces BRCA2 as the true FANCB protein. Cell Cycle. 2005;4:80–86. doi: 10.4161/cc.4.1.1358. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Zhao D, Wang H, Lin CJ, Fei P. FANCD2 monoubiquitination provides a link between the HHR6 and FA-BRCA pathways. Cell Cycle. 2008;7:407–413. doi: 10.4161/cc.7.3.5156. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Zhao D, Park HK, Wang H, Dyer RB, et al. FAVL elevation in human tumors disrupts Fanconi anemia pathway signaling and promotes genomic instability and tumor growth. J Clin Invest. 2010;120:1524–1534. doi: 10.1172/JCI40908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu D, Dudimah FD, Zhang J, Pickering A, Paneerselvam J, et al. Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle. 2013;12:803–809. doi: 10.4161/cc.23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Y, Zhang J, Yu H, Fei P. Advances in the understanding of Fanconi Anemia Complementation Group D2 Protein (FANCD2) in human cancer. Cancer Cell Microenviron. 2015;2 doi: 10.14800/ccm.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aird KM, Zhang G, Li H, Tu Z, Bitler BG, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panneerselvam J, Xie G, Che R, Su M, Zhang J, et al. Distinct Metabolic Signature of Human Bladder Cancer Cells Carrying an Impaired Fanconi Anemia Tumor-Suppressor Signaling Pathway. J Proteome Res. 2016;15:1333–1341. doi: 10.1021/acs.jproteome.6b00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longerich S, Kwon Y, Tsai MS, Hlaing AS, Kupfer GM, et al. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014;42:5657–5670. doi: 10.1093/nar/gku198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koprinarova M, Schnekenburger M, Diederich M. Role of Histone Acetylation in Cell Cycle Regulation. Curr Top Med Chem. 2016;16:732–744. doi: 10.2174/1568026615666150825140822. [DOI] [PubMed] [Google Scholar]
- 10.Panneerselvam J, Pickering A, Han B, Li L, Zheng J, et al. Basal level of FANCD2 monoubiquitination is required for the maintenance of a sufficient number of licensed-replication origins to fire at a normal rate. Oncotarget. 2014;5:1326–1337. doi: 10.18632/oncotarget.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, et al. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Yan R, Roy A, Xu D, Poisson J, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:174–181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méchali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Lee YH, Panneerselvam J, Zhang J, Loo LW, et al. Mutated Fanconi anemia pathway in non-Fanconi anemia cancers. Oncotarget. 2015;6:20396–20403. doi: 10.18632/oncotarget.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panneerselvam J, Wang H, Zhang J, Che R. BLM promotes the activation of Fanconi Anemia signaling pathway. Oncotarget. 2016 doi: 10.18632/oncotarget.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]


