Abstract
Microchip-based microfluidic electrochemical arrays hold great promise for fast, high-throughput multiplexed detection of cancer biomarker proteins at low cost per assay using relatively simple instrumentation. Here we describe an inexpensive high-throughput electrochemical array featuring 32 individually addressable microelectrodes that is further multiplexed with an 8-port manifold to provide 256 sensors. The gold electrode arrays were fabricated by wet-etching commercial gold compact discs (CD-R) followed by patterned insulation. A print-and-peel method was used to create sub-microliter hydrophobic wells surrounding each sensor to eliminate cross contamination during immobilization of capture antibodies. High-throughput analyses were realized using eight 32-sensor immunoarrays connected to the miniaturized 8-port manifold, allowing 256 measurements in <1 h. This system was used to determine prostate cancer biomarker proteins prostate specific antigen (PSA), prostate specific membrane antigen (PSMA), interleukin-6 (IL-6), and platelet factor-4 (PF-4) in serum. Clinically relevant detection limits (0.05 to 2 pg mL−1) and 5-decade dynamic ranges (sub pg mL−1 to well above ng mL−1) were achieved for these proteins utilizing precapture of analyte proteins on magnetic nanoparticles decorated with enzyme labels and antibodies.
Keywords: high-throughput, electrochemical array, sensor fabrication, immunosensors, cancer biomarkers proteins
Graphical abstract
Elevated levels of proteins in blood have great potential as biomarkers for early cancer detection and personalized therapy.1–6 More reliable diagnostics will be realized by measuring panels of biomarker proteins to provide “snapshots” of patient disease status.7 Recently, new immunoarray devices were reported that can measure up to 4 proteins at low cost while offering accuracy, reliability, and, in some cases, automation.8–13 However, there is a need for a much higher throughput, which is the issue addressed in the present paper.
The enzyme-linked immunosorbent assay (ELISA) has served as the gold standard for clinical protein measurements.6,14 Newer commercial techniques such as bead-based optical or electrochemiluminescent (ECL) methods hold great promise for high-throughput and multiplexed assays.15–18 Simoa, a new technology for protein quantification, has sensitivity in the fg mL−1 range.19 However, these systems have limitations in equipment and reagent cost, and do not yet feature a high degree of multiplexing. Some of these limitations can be overcome using microfluidic electrochemical devices and nanomaterial-enhanced detection.5–7,20,21 Kelley et al. developed a sensor chip featuring gold microelectrode nanostructures to achieve multiplexed detection of an ovarian cancer biomarker.22 By combining the same sensor array with solution-based circuits, they multiplexed detection of pathogenic bacteria and antibiotic-resistance markers.23
Screen-printed electrode arrays integrated with microfluidic platforms have been used to detect a panel of biomarker proteins as possible disposable diagnostic tools.24–27 We previously developed a modular amperometric microfluidic system for multiplexed detection that measured four oral cancer biomarker proteins in serum down to 5 fg mL−1.28 We also used this array for oral mucositis risk assessment29 and decreased assay time to 8 min by trading off sensitivity for assay time using an inkjet-printed array.30
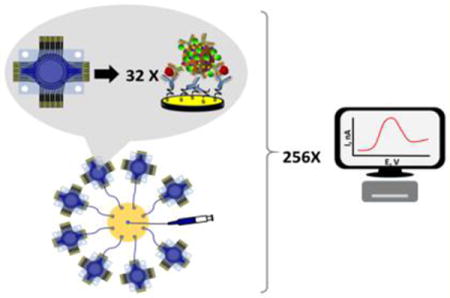
The present paper describes fabrication of a high-throughput microfluidic electrochemical immunoarray system with 256 sensors, and its application to multiplexed protein detection.5,14 Adapting a method previously described, a 32-sensor electrochemical array was fabricated for $0.50 in materials per chip. This new low-cost, high-throughput 32-sensor electrochemical array features on-chip reference and counter electrode and were integrated into simple modular microfluidic chambers for measuring proteins. A miniaturized 8-port manifold was used to connect 8 different devices together allowing 256 individual addressable immunosensors to be monitored simultaneously.
We demonstrated performance by amperometric detection of four proteins employing 300 nm magnetic nanoparticles decorated with secondary antibodies (Ab2) and horseradish peroxidase (HRP) labels. The heavily labeled beads are a key approach that achieves high sensitivity. Signals are greatly amplified by the large number of HRPs (8500) per bead.27,28 This method reduces the number of washing and reagent addition steps compared to polyHRP, which we have used previously.31,32 Off-line capture of analyte proteins on the beads minimizes interferences from nonspecific binding before analytes are introduced to the sensors.5 In addition, the multiple antibodies on the magnetic bead conjugates led to cooperative binding facilitating effective capture by sensors.33 The prostate cancer biomarker proteins were prostate specific antigen (PSA),34 prostate specific membrane antigen (PSMA),35 interleukin-6 (IL-6), and platelet factor-4 (PF-4). The full 256-sensor system was used for 6 replicate measurements per sample and standard to achieve full analysis and standardization in <90 min with the possibility to expand the panel to 12 proteins for 8 samples or 96 proteins in 1 sample. The analysis time can also be further reduced to <60 min with a potentiostat capable of simultaneous measurements. A 5-decade wide linear dynamic. range vs log concentration was achieved with clinically relevant detection limits of 0.05 to 2 pg mL−1 for these 4 proteins. Assays on human serum samples from prostate cancer patients confirmed very good correlations with single protein ELISAs.
EXPERIMENTAL SECTION
Chemicals
Mitsu gold compact disc recordable (CD-R) 650 MB were from MAM-A Inc. 1-(3-(Dimethylamino)propyl)-3-ethylcarbo-diimide hydrochloride (EDC), N-hydroxysulfosuccinimide (NHSS), Tween-20, hydroquinone (HQ, ≥99%), and calf serum were from Sigma-Aldrich. Hydrogen peroxide (H2O2, 30%) was from Fisher. Lyophilized 99% bovine serum albumin (BSA) was from Millipore. The polydimethylsiloxane (PDMS) kit was from Dow Corning. Immuno-reagents were dissolved in pH 7.4 PBS buffer (0.01 M in phosphate, 0.14 M NaCl, 2.7 mM KCl) unless otherwise noted. Protein standards were prepared in 150× diluted calf serum in PBS. Streptavidin-coated 300-nm-diameter magnetic nanoparticles (SA-MNP) were from NVigen. Water was purified by a Hydro purification system to 18 MΩ·cm.
Antibodies for prostate specific antigen (PSA) DuoSet (catalog # DY1344), interleukin-6 (IL-6) DuoSet (catalog # DY206), and platelet factor-4 (PF-4) DuoSet (catalog # DY795), monoclonal anti-human prostate specific membrane (PSMA) antibody, biotinylated anti-human PSMA antibody and recombinant human PSMA were from R&D Systems, Inc.
Instrumentation
A Hewlett Packet Laserjet 1020n was used to print the microwell pattern onto glossy paper. A Maxx Press thermal press (Stahls, USA) was used to transfer the pattern onto the gold sensor array. A CHI 1040A multipotentiostat coupled with CHI 685 multiplexer was used to acquire groups of 8 sensor measurements on the 256-sensor array in rapid sequence with 2 s quiet time at 22 ± 2 °C vs Ag/AgCl reference.
The microfluidic device (Figure 1) features a layer of molded, flexible PDMS, sandwiched between two poly(methyl methacrylate) (PMMA) plates to form a circular channel 1.8 mm diameter height 150 μm and volume 40 ± 2 μL (Figure 1c). The top PMMA plate was fitted with 25-gauge stainless steel tubing as inlet and outlet (Figure 1d). PDMS channels were made by precision cutting transparency film using a programmable precision electronic cutter Silhouette CAMEO. In brief, an adhesive-backed transparency film was cut using the electronic cutter and used as a mold for the PDMS when placed onto an aluminum plate. The final detection device is 1 in. × 1 in. × 0.75 in. Disposable reagent reservoirs were made from 0.5 mL microcentrifuge tubes with holes in the bottom for connecting the inlets of each 32 sensor array. Each outlet was connected to a miniaturized 8-port manifold by polystyrene tubing (PE160). A syringe was connected to the outlet of the manifold and used to withdraw reagents from each reservoir into the corresponding arrays (Figure 2b).
Figure 1.
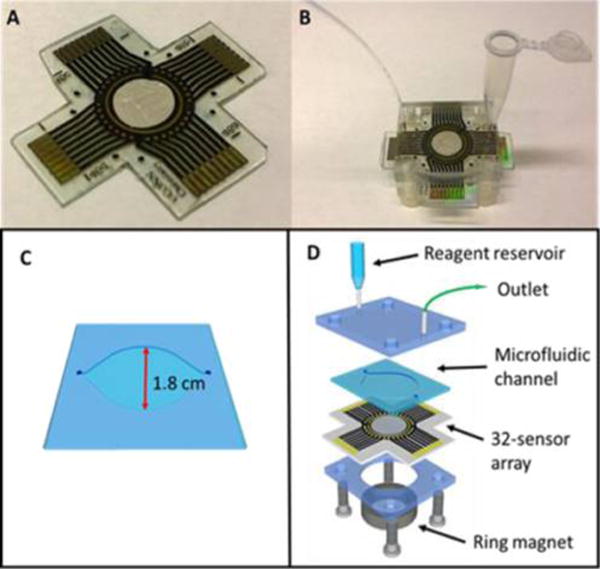
Array configuration: (A) 32-sensor array with microwells of volume ~10 nL that hold 1 μL aqueous droplets. (B) Assembled microfluidic chamber with reagent reservoir. (C) Molded circular PDMS channel. (d) Breakout of one assembly with reagent reservoir, microfluidic channel, 32-sensor electrochemical array, and a ring magnet.
Figure 2.
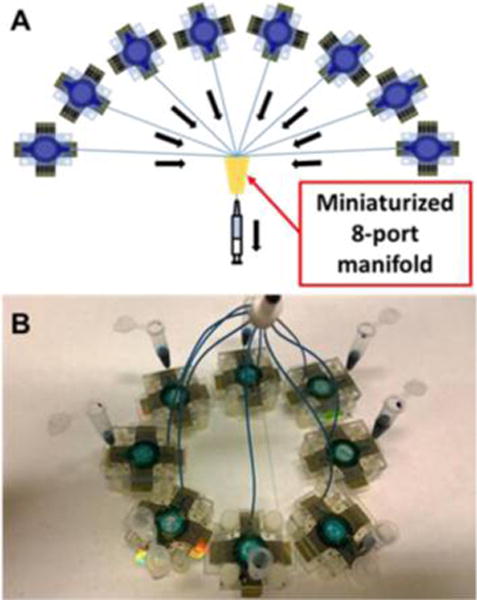
Configuration for 256-sensor system: (a) Schematic representation of 8 microfluidic arrays connected to miniaturized 8-port manifold. (b) 8 devices with built-in reagent reservoirs connected to the manifold (top). Blue dye indicates the flow of reagents into each device, used for all calibrations and patient sample measurements described herein.
Array Fabrication
Sensor arrays were prepared using similar procedures to previous work.31,32 In brief, the sensor pattern was designed using graphic design software Canvas 11 on a 1:1 scale (Figure S1). It was then printed onto a glossy paper (the backing of Avery labels) using HP Laserjet 1022n at 1200 dpi. The pattern was then cut and placed onto a piece of gold CD-R, and then sandwiched in a thermal press at 120 °C for 110 s. The electrical contacts and sensors were manually covered with a Sharpie permanent marker and immersed into a ferricyanide etching solution.36 Sensors were washed with ethanol and water, to expose contact pads and sensors, and then dried under nitrogen. Finally, the reference electrode was made by manually screen-printing Ag/AgCl ink (DuPont 5269) onto the designated area (Scheme S1). Figure 1A shows the completed array. Before capture antibody immobilization, sensors were first cleaned by 10 cyclic potential sweeps between 1.2 V and −0.1 V in 0.18 M sulfuric acid. For 4-protein detection, 8 sensors in each 32-sensor array were assigned to one analyte protein. After cleaning, the arrays were immersed in 4 mM mercaptopropionic acid (MPA) in 20% ethanol for 48 h under N2 to form a self-assembled monolayer (SAM). The outward-facing surface carboxylic groups of MPA on sensors were activated using freshly prepared 400 mM EDC/100 mM NHSS. Arrays were rinsed with water after 10 min and respective capture antibodies (Ab1) were attached by amidization between the activated carboxylic groups of SAM and the primary Ab1 amines using 1.0 μL of 100 μg mL−1 Ab1 per relevant sensor (8 μL total per protein) for 3 h (Scheme 1). Arrays were then washed by gently dipping into 0.05% Tween-20 in PBS, and then PBS 3 times. The antibody-decorated sensor arrays were then placed into the respective microfluidic devices followed by incubation with 1% BSA for 10 min within the microfluidic device (Figure 2b) to block nonspecific binding (NSB) on the sensors and in the PDMS channel. Fresh antibody-decorated arrays were used for each assay and were stable up to 7 days after antibody immobilization and BSA blocking when stored at 4 °C (Figure S3, SI file).
Scheme 1.
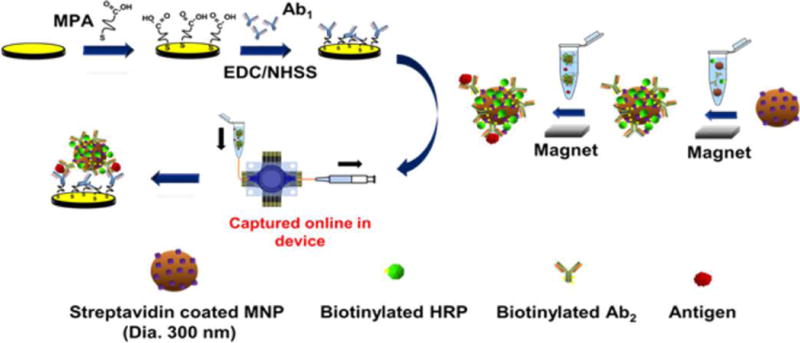
Strategy for Multiplexed Voltammetric Detection with Wide Dynamic Range, Showing One Sensor with Capture Antibodies
Biotinylated secondary antibodies (Ab2) and biotinylated horseradish peroxidase (HRP) labels were chemically linked onto 300-nm-diameter streptavidin-coated magnetic nanoparticles similar to a previously reported procedure for 1 μm streptavidin-coated magnetic beads (Scheme 1).27 Previously reported procedures were followed to capture the antigen of interest (PSA, IL-6, IL-8, or PSMA) from standard solutions prepared using 150× diluted calf serum (Scheme S1; see SI for details). The resulting magnetic nanoparticles containing the analytes were loaded and drawn by syringe into the fluidic chambers using the setup in Figure 2b (see SI for details).
Electrochemical detection was done by drawing in freshly prepared 1 mM hydroquinone (HQ) and 100 μM hydrogen peroxide (H2O2) in PBS purged with purified nitrogen to fill microfluidic detector channels using a syringe from individual reagent reservoirs before each successive measurement to account for the time delay in the multiplexer. Differential pulse voltammetry was done at 4 mV step, 25 mV amplitude, and 0.5 s pulse vs Ag/AgCl reference. Total measurement time of all 256 sensor was 30 min.
RESULTS
Characterization of 32-Sensor Array
The design pattern is printed for 32 sensors with diameter of 850 μm onto glossy paper. Thermal transfer onto the gold CDs causes toner spread which reduced the sensor effective geometric areas. After thermal transfer, the diameter of each sensor was 800 ± 25 μm estimated using fiducial camera images. The hydrophobic wells have heights of 6–14 μm around the sensors and minimum volumes of ~10 nL, but can hold up to 1 μL drops of aqueous reagents due to the high water contact angle of the toner material. The dry resistance of gold array contract pads to working electrodes was 13.6 ± 0.2 ohms.
Sensors were first cleaned by cyclic potential sweeping between −0.1 V to +1.2 V vs Ag/AgCl at 100 mV s−1 in 0.18 M sulfuric acid. Responses of the 32-sensor array were first characterized by assessing reproducibility of the electrochemically addressable area. Cyclic voltammograms (Figure 3a) were completed by filling the microfluidic device with 5 mM ruthenium hexamine chloride [Ru(NH3)6]Cl3 (RuHex) and 0.1 M PBS after cleaning the gold surface. Using the Randles-Sevcik equation and the known diffusion coefficient of ruthenium hexamine chloride, the electro-active surface area was 0.46 ± 0.02 mm2 (4% RSD) from sensor to sensor and 0.50 ± 0.03 mm2 (6% RSD) from array to array, or ~80% of estimated geometric area. Differential pulse voltammograms (Figure S2a) were also measured for all sensor electrodes in the microfluidic device. A linear relation between peak current and RuHex concentration (Figure S2b) with detection limit of 5 μM, demonstrating good analytical performance.
Figure 3.
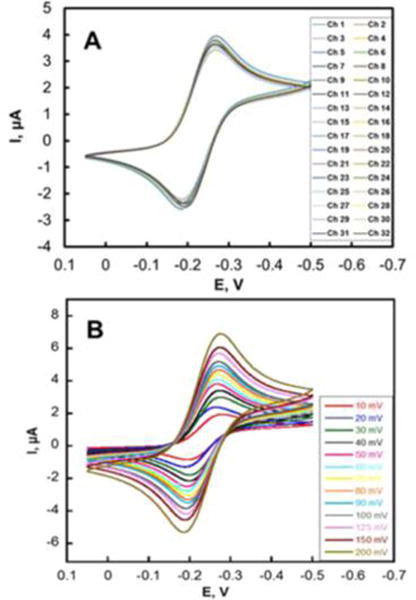
Array reproducibility. (a) CVs at 50 mV s−1 on 32 electrodes in 5 mM ([Ru(NH3)6]-Cl3) in aqueous 0.1 M PBS in the microfluidic device. (b) CVs at various scan rates on 32 electrodes in the same solution.
Multiplexed Biomarker Detection
Microfluidic devices with sensors having relevant attached Ab1 were filled with magnetic bead dispersion that have captured standard or sample proteins, as described in the Experimental Section. Hydroquinone mediator/hydrogen peroxide solution was drawn into the sensor arrays to activate the HRP to its ferryloxy form that is reduced in the mediated electrode reaction. Total assay time for six replicate measures of eight concentrations included in the calibration curves (from offline capture of antigens to quantitative results) was <60 min for four proteins. Control experiments included the full immunoassay procedure without antigens reflecting the sum of residual nonspecific binding and direct reduction of hydrogen peroxide.
Each 32-sensor array was divided into 4 sections of 8 sensors, each designated for PSA, IL-6, PF4, and PSMA, respectively, 2 sensors in each section were assigned BSA to act as a negative control reflecting responses from any nonspecific absorbed proteins on the sensor surface. For simultaneous detection of PSA, PSMA, IL-6, and PF-4, the 4 target protein standards were dissolved in calf serum diluted 150× in PBS. Ab2-MNP-HRP beads for the 4 proteins were combined and reconstituted in PBS and incubated with the protein standard mixtures to capture the analyte proteins. The ratio of the HRP and Ab2 was optimized using a method similar to those previously reported (Figure S4, see SI file). From enzyme activity assays, the number of HRP labels per optimized MNP conjugate was 8500 ± 1500. The average number of Ab2’s was 1800 ± 350 measured using a bicinchoninic acid (BCA) assay kit. The magnetic bead conjugates were washed, and then loaded into the full 256-sensor microfluidic system (Figure 2b). After incubation of Ab2-MNP-HRP beads to allow the Ab1-decorated sensors to capture beads bearing the corresponding analyte protein, the sensors were washed with PBS-T20 and PBS, followed by detection using differential pulse voltammetry (DPV). The peak currents increased linearly with log C from 2 pg mL−1 to 200 ng mL−1 for PSA, 0.05 pg mL−1 to 5 ng mL−1 for IL-6, 0.1 pg mL−1 to 10 pg mL−1 for PF-4, and 0.15 pg mL−1 to 15 ng mL−1 for PSMA (Figure 4). The lower concentrations are detection limits (DLs), as the zero protein control plus three times the average standard deviation (Figure 5). Variation in peak currents was ≤10% for sensor-to-sensor (n = 16) and ≤8% for array-to-array (n = 3). Full calibrations for 4 proteins were achieved within <60 min due to the high-throughput capability of the device. Fresh arrays were used for each assay, and crosstalk was minimized by a symmetric arrangement of reference and counter with respect to the sensor electrodes.26 Essentially no crosstalk was observed as indicated by reproducibility of calibration data using different electrodes in the array (Figure 5), good agreement of patient sample results with ELISA (Figure 7), and the ability to distinguish small concentrations of protein analytes in the presence of high concentration of other proteins (Figure S3, see SI file).
Figure 4.
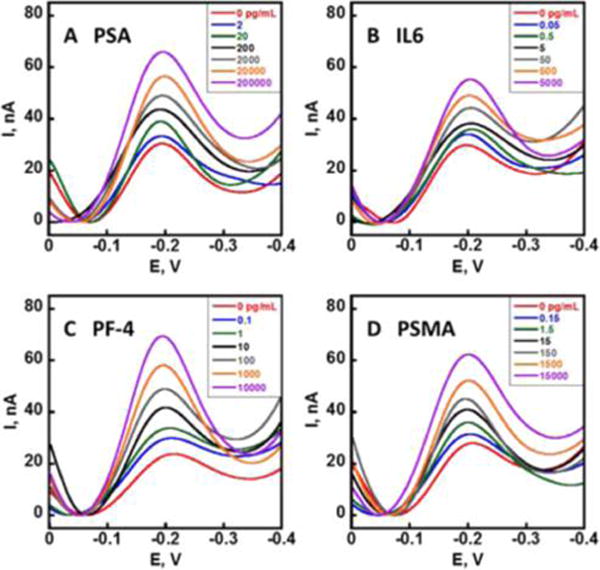
Multiplexed biomarker protein detection. DPV peaks developed after the capture of the Ag-Ab2-MNP-HRP conjugates on the immunoarrays using 1 mM hydroquinone and 100 μM H2O2 for (a) PSA, (b) IL-6, (c) PF-4, and (d) PSMA in 150× diluted calf serum. DPV was done at 4 mV step, 25 mV pulse amplitude, and 15 Hz frequency vs Ag/AgCl from 0.0 V to −0.4 V.
Figure 5.
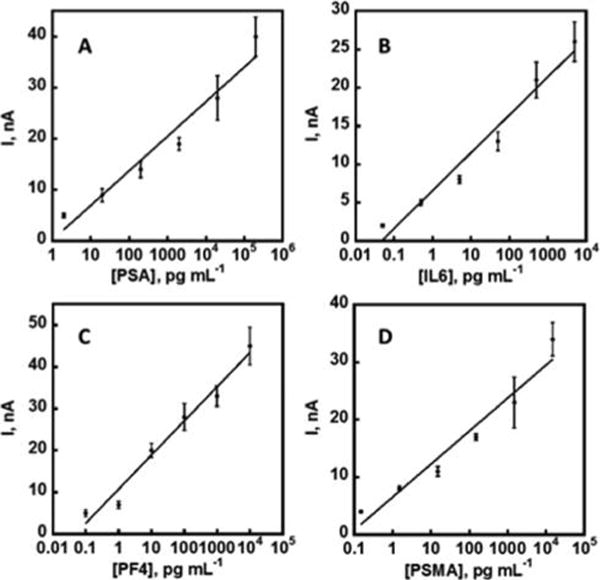
Calibration plots for multiplexed biomarker protein detection developed using protein standard mixtures in 150× diluted calf serum using the described microfluidic setup housing 8 arrays of 32 sensors (256 total sensors). All 4 calibration plots were generated simultaneously in <60 min for (a) PSA, (b) IL-6, (c) PF-4, and (d) PSMA. Control subtracted, n = 6.
Figure 7.
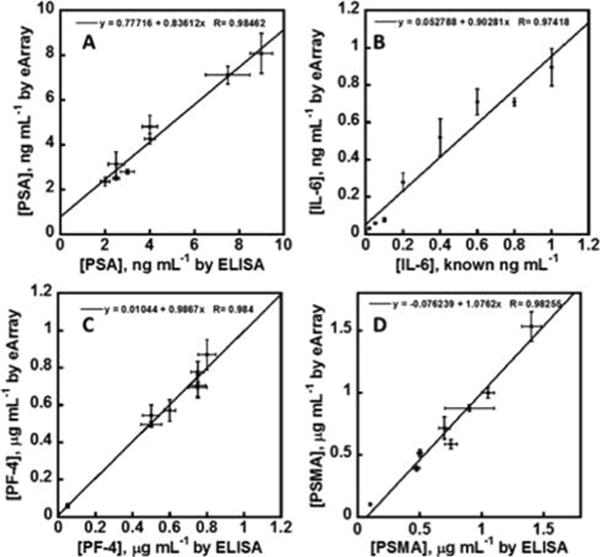
Linear correlation plots of electrochemical immunoarray vs ELISA results for 8 human serum samples for (A) PSA, (B) Spiked IL-6, (C) PF-4, and (D) PSMA. Error bars are standard deviations for the 32-sensor arrays (n = 6) and ELISA (n = 3). Four biomarkers for all 8 samples were measured simultaneously using the 256-sensor array.
Accuracy Validation
Seven serum samples from prostate cancer patients and one sample from a cancer-free patient were analyzed and compared with results from single-protein ELISA.13 These samples were diluted 150-fold in PBS to bring the electrochemical response into the linear ranges of the calibrations. Concentrations of PSA, IL-6, PF-4, and PSMA were found to be within the detection limits of their respective ELISAs, but IL-6 concentrations in serum samples were well below the detection limit of ELISA. For each array, 32 data points were obtained for measuring 4 different proteins in one sample (Figure 6) and 256 data points were realized simultaneously using the described microfluidic setup. For this validation study, each sample was spiked with 20 to 1000 pg mL−1 of IL-6.
Figure 6.
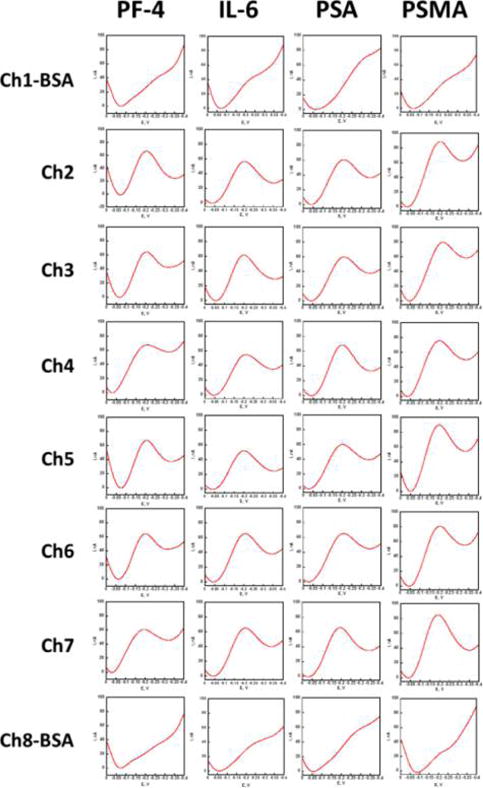
Array sensor results showing individual DPV peaks acquired for patient sample 1 using a 32-sensor immunoarray housed in the modular microfluidic device. For each experiment, 8 32-sensor arrays were assayed simultaneously for 8 different samples leading to 256 DPV curves. Channels 1 and 8 are controls with BSA replacing antibodies.
Linear correlation plots of the ELISA vs immunoarray data gave slopes of for 0.84 ± 0.06 for PSA, 0.90 ± 0.09 for IL-6, 0.98 ± 0.07 for PF-4, and 1.1 ± 0.1 for PSMA (Figure 7). Intercepts of these plots were near zero, i.e., 0.77 ± 0.49 for PSA, 0.053 ± 0.025 for IL-6, 0.010 ± 0.046 for PF-4, and −0.076 ± 0.087 for PSMA (Figure 7). These results demonstrate good correlation of the high-throughput immunoassay with standard ELISA while confirming the high selectivity and specificity of the assay for each of the four proteins in the presence of the hundreds of other proteins in human serum.37
DISCUSSION
Results above demonstrate high-throughput capabilities of a modular microfluidic immunoarray combined with off-line protein capture on magnetic nanobeads. Four proteins in serum were determined with good accuracy. Fabrication of the 32-sensor array required no photolithography or special equipment. A single array costs ~$0.50 US in materials to produce. The nonlithographic fabrication technique is a facile and convenient way to prototype microelectrodes without relying on extensive mask development or expensive fabrication equipment. It uses direct computer printing of simple masks using a laser printer to pattern sensor arrays with good reproducibility (4% RSD). This technique offers a valuable tool to researchers for a variety of applications in low-resource environments where microfabrication facilities are not available.38
Microfluidic channels were made by molding PDMS on an inexpensive precision-cut transparency film. Major advantages of these approaches include low-cost fabrication and ease of modification of printed sensors and microfluidic pattern, which could be customized for virtually any design prototypes within minutes. Good device-to-device sensor area reproducibility is shown by small standard deviations (6% RSD). The sensor arrays were reproducible and disposable, making them suitable for clinical applications. The complete system with 8 microfluidic devices, miniaturized manifold, and tubing cost less than $200.
Antibodies were utilized as the capture agents to facilitate specific capture of analytes because of their wide availabity and excellent performance. While fragmented antibodies (Fab) provide benefits in better protein orientation and reduced nonspecific binding, Fab are not available for a wide variety of proteins.39 Capture antibodies were immobilized onto SAMs with surface exposed carboxyl groups through amine coupling using EDC/NHSS chemistry with a method we have successfully used in the past.27,29,30 Short-chain alkanethiol mercaptopropionic acid (MPA) was used for the SAMs to ensure efficient electron transfer.40,41 Unreacted groups on the sensor surface were deactivated through blocking with BSA, which contains primary amines and is large enough to physically occupy any spaces to minimize nonspecific absorption.42,43 Effectiveness of BSA blocking is demonstrated by the low residual response from the zero-protein controls (Figure 4). The antibody-coated sensors were suitable to use for up to 7 days (stored at 4 °C) without loss of signal (Figure S3; see SI). The presumably random orientation of antibodies is not an important factor since we were able achieve excellent detection limits due to very efficient protein capture by the magnetic detection nanoparticles (MNP) decorated with 1800 ± 350 secondary antibodies for signal amplification.
Magnetic nanoparticles allow easy manipulation with a magnet allowing easy bead attachment to labels and Ab2, and efficient washing to minimize nonspecific binding. A relatively small number of enzyme labels per MNP reduces the rate of saturation leading to five decades of linear dynamic range vs log C (Figure 5) and clinically relevant detection limits (DL, 0.05 to 2 pg mL−1). In contrast, similar assays using larger 1 μm magnetic particles (MP) with 400 000 HRP labels gave three orders of linear dynamic range vs log C, but better DLs of 5–10 fg mL−1. The ultralow DLs of assays using larger particles can be attributed to many more HRP labels on the MP conjugates. Here, the DLs for the assays employing smaller particles were specifically tailored for clinical measurements, so the ultralow DLs were sacrificed for a much wider dynamic range eliminating the need for high dilution that might cause errors. The 5-decade-wide dynamic range achieved with the 300 nm capture beads may be attributed to the larger numbers of smaller beads able to be captured on the sensors. Since sensor surface area is limited, larger magnetic beads that we have used earlier can saturate the sensor at higher protein concentration.27,29,30 A lower number of HRPs may also play a significant role in avoiding signal saturation by limiting the concentration of HRP on the sensor. While larger magnetic beads are loaded with more antibodies, only a limited number of antibodies on the beads, as defined by the contact area, can actually interact with the surface, i.e., those involved in closest contact and adhesion.44 With ~200–650 interactions per bead estimated from the contact area, our beads still can enhance binding affinity.45–47 The new microfluidic system with a miniaturized 8-port manifold increased throughput to 256 sensors compared to earlier 8-sensor systems, while achieving excellent assay speed by measuring 8 samples in less than 60 min while minimizing the sample volume (5 μL). Relative standard deviations of protein measurement in serum ranged from ±3% to ±10% for all 4 proteins. These standard deviations are acceptable for accurate assays, as shown by the good correlation between the results obtained by the new immunoarrays and single-protein ELISAs.
CONCLUSION
In summary, results above demonstrate low-cost fabrication of a high-throughput electrochemical sensor array system utilizing inexpensive rapid prototyping. The high-throughput screening system developed is capable of detecting multiple cancer biomarker proteins in serum. Protein capture using magnetic nanoparticles from 5 μL samples provides a viable strategy for multiplexed detection by minimizing nonspecific binding. Syringe-actuated fluidics enables a wide dynamic range and the possibility of adjustable detection limits. In addition, using approaches described here, prototype sensor arrays and microfluidic devices can be produced rapidly allowing laboratories without access to specialized facilities to quickly design, produce, and test prototype devices.
Supplementary Material
Acknowledgments
This work was supported financially by grant nos. EB016707 and EB014586 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.6b00256.
Detailed experimental methods, optimizations of the array, array design, stability of arrays, and patient sample correlations (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Giljohann DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 3.Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers—blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- 4.Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discovery. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rusling JF, Bishop GW, Doan N, Papadimitrakopoulos F. Nanomaterials and biomaterials in electrochemical arrays for protein detection. J Mater Chem B. 2014;2:12–30. doi: 10.1039/C3TB21323D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rusling JF. Multiplexed electrochemical protein detection and translation to personalized cancer diagnostics. Anal Chem. 2013;85:5304–5310. doi: 10.1021/ac401058v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rusling JF, Kumar CV, Gutkind JS, Patel V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein S, van de Wijgert J, Sahabo R, Justman J, El-Sadr W, Sia SK. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 9.Chin CD, Linder V, Sia SK. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012;12:2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 10.Yu ZT, Guan H, Cheung MK, McHugh WM, Cornell TT, Shanley TP, Kurabayashi K, Fu J. Rapid, automated, parallel quantitative immunoassays using highly integrated microfluidics and AlphaLISA. Sci Rep. 2015;5:11339. doi: 10.1038/srep11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci Transl Med. 2015;7:273re1–273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 12.Kadimisetty K, Malla S, Sardesai NP, Joshi AA, Faria RC, Lee NH, Rusling JF. Automated multiplexed ECL Immunoarrays for cancer biomarker proteins. Anal Chem. 2015;87:4472–4478. doi: 10.1021/acs.analchem.5b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadimisetty K, Mosa IM, Malla S, Satterwhite-Warden JE, Kuhns T, Faria RC, Lee NH, Rusling JF. 3D-printed supercapacitor-powered electrochemiluminescent protein immunoarray. Biosens Bioelectron. 2016;77:188–193. doi: 10.1016/j.bios.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J. Electrochemical biosensors: towards point-of-care cancer diagnostics. Biosens Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 15.Beveridge Js, Stephens JR, Williams ME. The use of magnetic nanoparticles in analytical chemistry. Annu Rev Anal Chem. 2011;4:251–273. doi: 10.1146/annurev-anchem-061010-114041. [DOI] [PubMed] [Google Scholar]
- 16.Roche Diagnostics. http://www.roche.com. Accessed December 2015.
- 17.Meso Scale Diagnostics. http://www.mesoscale.com. Accessed December 2015.
- 18.Perkin Elmer. http://www.perkinelmer.com/catalog/cate-gory/id/alphatech. Accessed December 2015.
- 19.Meissner EG, Decalf J, Casrouge A, Masur H, Kottilil S, Albert ML, Duffy D. Dynamic changes of post-translationally modified forms of CXCL10 and soluble DPP4 in HCV subjects receiving interferon-free therapy. PLoS One. 2015;10(7):e0133236. doi: 10.1371/journal.pone.0133236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Guo Y, Xianyu Y, Chen W, Zhao Y, Jiang X. Nanomaterials for ultrasensitive protein detection. Adv Mater. 2013;25:3802–3819. doi: 10.1002/adma.201301334. [DOI] [PubMed] [Google Scholar]
- 21.Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat Nanotechnol. 2014;9:969–980. doi: 10.1038/nnano.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das J, Kelley SO. Protein detection using arrayed microsensor chips: tuning sensor footprint to achieve ultrasensitive readout of CA-125 in serum and whole blood. Anal Chem. 2011;83:1167–1172. doi: 10.1021/ac102917f. [DOI] [PubMed] [Google Scholar]
- 23.Lam B, Holmes RD, Live L, Sage A, Sargent EH, Kelley SO, Das J. Solution-based circuits enable rapid and multiplexed pathogen detection. Nat Commun. 2013;4 doi: 10.1038/ncomms3001. [DOI] [PubMed] [Google Scholar]
- 24.Dong H, Li CM, Zhang YF, Cao XD, Gan Y. Screen-printed microfluidic device for electrochemical immunoassay. Lab Chip. 2007;7:1752–1758. doi: 10.1039/b712394a. [DOI] [PubMed] [Google Scholar]
- 25.Fragoso A, Latta D, Laboria N, von Germar F, Hansen-Hagge TE, Kemmner W, Gartner C, Klemm R, Drese KS, O’Sullivan CK. Integrated microfluidic platform for the electrochemical detection of breast cancer markers in patient serum samples. Lab Chip. 2011;11:625–631. doi: 10.1039/c0lc00398k. [DOI] [PubMed] [Google Scholar]
- 26.Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Microfluidic Electrochemical Immunoarray for Ultrasensitive Detection of Two Cancer Biomarker Proteins in Serum. Biosens Bioelectron. 2011;26:4477–4483. doi: 10.1016/j.bios.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otieno BA, Krause CE, Latus A, Chikkaveeraiah BB, Faria RC, Rusling JF. On-line protein capture on magnetic beads for ultrasensitive microfluidic immunoassays of cancer biomarkers. Biosens Bioelectron. 2014;53:268–274. doi: 10.1016/j.bios.2013.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malhotra R, Patel V, Chikkaveeraiah BV, Munge BS, Cheong SC, Zain RB, Abraham MT, Dey DK, Gutkind JS, Rusling JF. Ultrasensitive detection of cancer biomarkers in the clinic by use of a nanostructured microfluidic array. Anal Chem. 2012;84:6249–6255. doi: 10.1021/ac301392g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause CE, Otieno BA, Bishop GW, Choquette PG, Lalla RV, Peterson DE, Rusling JF, Phadke G. Ultrasensitive microfluidic array for serum pro-inflammatory cytokines and C-reactive protein to assess oral mucositis risk in cancer patients. Anal Bioanal Chem. 2015;407:7239–7243. doi: 10.1007/s00216-015-8873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause CE, Otieno BA, Latus A, Faria RC, Patel V, Gutkind JS, Rusling JF. Rapid microfluidic immunoassays of cancer biomarker proteins using disposable inkjet-printed gold nanoparticle arrays. ChemistryOpen. 2013;2:141–145. doi: 10.1002/open.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang CK, Vaze A, Rusling JF. Fabrication of immunosensor microwell arrays from gold compact discs for detection of cancer biomarker proteins. Lab Chip. 2012;12:281–286. doi: 10.1039/c1lc20833k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang CK, Vaze A, Rusling JF. Paper-based electrochemical immunoassay for rapid, inexpensive cancer biomarker protein detection. Anal Methods. 2014;6:8878–8881. doi: 10.1039/C4AY01962H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mani V, Wasalathanthri DP, Joshi AA, Kumar CV, Rusling JF. Highly efficient binding of paramagnetic beads bioconjugated with 100 000 or more antibodies to protein-coated surfaces. Anal Chem. 2012;84:10485–10491. doi: 10.1021/ac3028257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 35.Chikkaveeraiah BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Single-wall carbon nanotube forest arrays for immunoelectrochemical measurement of 4 protein biomarkers for prostate cancer. Anal Chem. 2009;81:9129–9134. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y, Zhao X, Kim E, Whitesides GM. A Selective Etching Solution for Use with Patterned Self-Assembled Monolayers of Alkanethiolates on Gold, Silver and Copper. Chem Mater. 1995;7:2332–2337. [Google Scholar]
- 37.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang S, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. The human serum proteome: Display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3:1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 38.Coltro WKT, de Jesus DP, da Silva JAF, do Lago CL, Carrilho E. Toner and paper-based fabrication techniques for microfluidic applications. Electrophoresis. 2010;31:2487–2498. doi: 10.1002/elps.201000063. [DOI] [PubMed] [Google Scholar]
- 39.Liddell E. Immunoassay Components. In: Wild D, editor. The Immuunoassay handbook. 4th. Elsevier; Great Britain: 2013. pp. 245–265. [Google Scholar]
- 40.Khoshtariya DE, Dolidze TD, Shushanyan M, Davis KL, Waldeck DH, van Eldik R. Fundamental signatures of short- and long-range electron transfer for the blue copper protein azurin at Au/SAM junctions. Proc Natl Acad Sci U S A. 2010;107:2757–2762. doi: 10.1073/pnas.0910837107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B, Bard AJ, Mirkin MV, Creager SE. Electron transfer at self-assembled monolayers measured by scanning electrochemical microscopy. J Am Chem Soc. 2004;126:1485–1492. doi: 10.1021/ja038611p. [DOI] [PubMed] [Google Scholar]
- 42.Jeyachandran YL, Mielczarski JA, Mielczarski E, Rai B. Efficiency of blocking of non-specific interaction of different proteins by BSA adsorbed on hydrophobic and hydrophilic surfaces. J Colloid Interface Sci. 2010;341:136–142. doi: 10.1016/j.jcis.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Reimhult K, Petersson K, Krozer A. QCM-D analysis of the performance of blocking agents on gold and polystyrene surfaces. Langmuir. 2008;24:8695–8700. doi: 10.1021/la800224s. [DOI] [PubMed] [Google Scholar]
- 44.Cooper K, Gupta A, Beaudoin S. Simulation of the adhesion of particles to surfaces. J Colloid Interface Sci. 2001;234:284–292. doi: 10.1006/jcis.2000.7276. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Veracoechea FJ, Frenkel D. Designing super selectivity in multivalent nano-particle binding. Proc Natl Acad Sci U S A. 2011;108:10963–10968. doi: 10.1073/pnas.1105351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soukka T, Harma H, Paukkunen J, Lovgren T. Utilization of kinetically enhanced monovalent binding affinity by immunoassays based on multivalent nanoparticle-antibody aioconjugates. Anal Chem. 2001;73:2254–2260. doi: 10.1021/ac001287l. [DOI] [PubMed] [Google Scholar]
- 47.Soukka T, Paukkunen J, Harma H, Lonnberg S, Lindroos H, Lovgren T. Supersensitive Time-resolved Immunofluorometric assay of free prostate-specific antigen with nanoparticle label technology. Clin Chem. 2001;47:1269–1278. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


