Abstract
Many genes determining cell identity are regulated by clusters of mediator-bound enhancer elements collectively referred to as super-enhancers. These have been proposed to manifest higher-order properties important in development and disease. Here, we report a comprehensive functional dissection of one of the strongest putative super-enhancers in erythroid cells. By generating a series of mouse models, deleting each of the five regulatory elements of the α-globin super-enhancer singly and in informative combinations, we demonstrate that each constituent enhancer appears to act independently and in an additive fashion with respect to hematologic phenotype, gene expression, chromatin structure and chromosome conformation, without clear evidence of synergistic or higher-order effects. Our study highlights the importance of functional genetic analyses for the identification of new concepts in transcriptional regulation.
Recent reports describe a new class of regulatory elements called super-enhancers1,2,3. These are defined as enhancer-like elements, bound by master regulators, particularly the Mediator complex, and bearing active chromatin marks (e.g. H3K4me1 and H3K27ac). Super-enhancers typically comprise a cluster of regulatory elements, spanning up to 12.5 kb, and are frequently flanked by CTCF binding sites, suggesting that their activity may be constrained by boundary elements4. Although originally identified in embryonic stem cells, super-enhancers have been described in many cell types5,6,7. Together these studies propose that a relatively small set of super-enhancers act as key switches to determine cell fate. However, it is unclear whether super-enhancers genuinely represent a new paradigm, describing a functional unit that is more than the sum of its parts, or whether they are simply an assembly of conventional enhancers of varying strengths8. Therefore, it is important to determine whether there are emergent functional properties uniquely associated with super-enhancers.
Here, using an unbiased approach1, we identified all super-enhancers in erythroid cells and found that the two clusters of regulatory elements controlling expression of the α- and β-globin genes are classified as super-enhancers in this cell type. The study of mammalian enhancers is hampered by the observation that these elements are defined by criteria only partially or indirectly related to their role in vivo9,10. We therefore used homologous recombination to make seven mouse models in which each constituent of the proposed α-globin super-enhancer is deleted, singly and in informative pairs, to dissect the its function.
We find that no single element is critical for globin gene expression and, although each element scores as an erythroid enhancer based on conventional chromatin signatures and enhancer assays, only two of the five elements behave as strong enhancers in vivo during embryonic, fetal, and adult erythropoiesis. These two strong enhancers fall into a subgroup of individual erythroid enhancers which are bound by the greatest amount of Mediator and the highest numbers of erythroid master regulators. Such regions have been referred to as “hotspots”6. Importantly, we find no evidence of emergent functional properties from the extended enhancer cluster; each element appears to contribute to gene expression as individual enhancers in an additive rather than synergistic manner. Thus the super-enhancer associated with the globin genes may be more simply described as a group of conventional enhancers including at least one strong enhancer, rather than as a new discrete entity with properties greater than the sum of its parts.
Results
Five α-globin regulatory elements form an erythroid super-enhancer
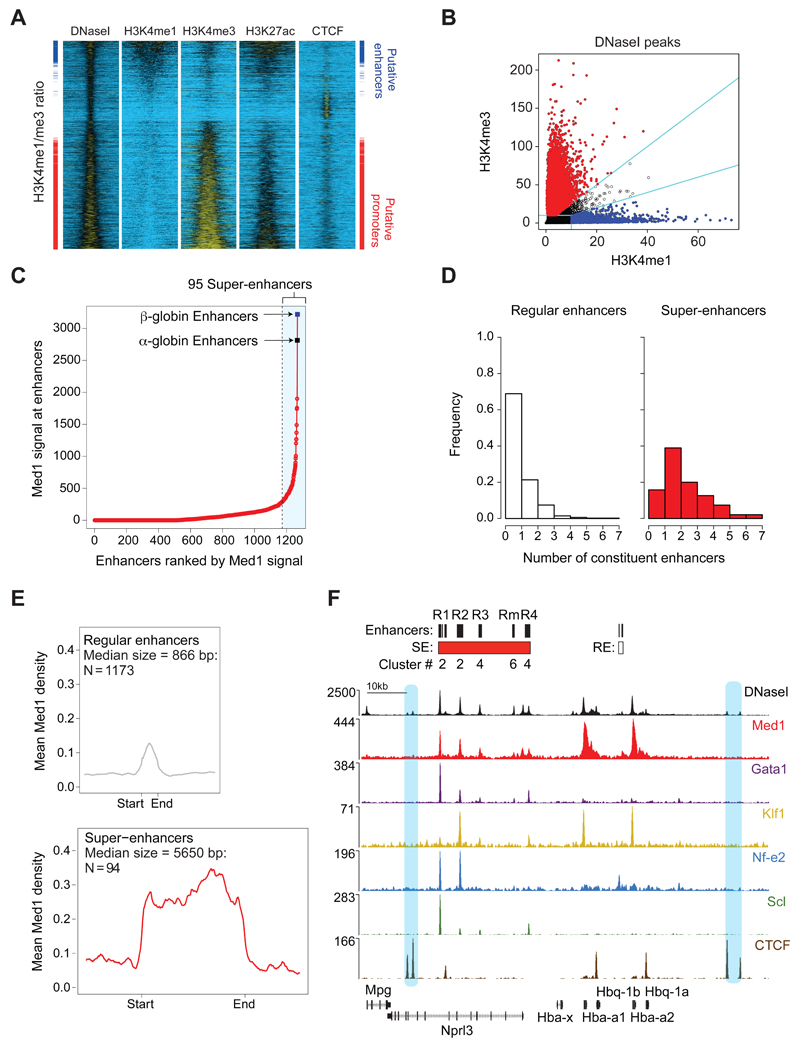
Primary mouse erythroid cells were analysed to identify and characterize super-enhancers in this cell type. All cis-regulatory elements in primary mouse erythroid cells were initially identified by characterizing DNase I hypersensitive sites (DHS)11. In total, 15,849 DHS were identified in purified erythroid (Ter119+) cells from C57BL/6 mice. After characterizing chromatin signatures at all DHS (Fig. 1A, Supplementary Fig. 1a and b), we excluded elements with low levels of H3K4me1 and H3K4me3 (<10 reads/million). A significant proportion of DHS correspond to CTCF binding sites (Supplementary Fig. 1a and b). In the remaining 10,542 DHS, enhancers were distinguished from promoters by their ratio of H3K4me3 to H3K4me1 (Fig. 1B, Supplementary Fig. 1a). Finally, this enhancer group was further refined by removing 29 DHS located within 250bp of annotated transcription start sites. The resulting set of 1963 putative enhancers has the chromatin signature previously shown to be enriched at enhancers (Fig. 1a and 1b)12,13,14.
Figure 1. The α-globin regulatory region typifies a super-enhancer in erythroid cells.
(a). Heatmap representation of DNase-seq and ChIP-seq signal +/-2kb around the DNase I peak call regions (15,849 peaks), sorted by the ratio of H3K4me1 to H3K4me3. In the side panes the annotated “putative enhancers” are marked blue, and “putative promoters” marked red. (b). Annotation of all DNaseI peak regions (black) as putative enhancers (blue) and putative promoters (red). Annotation category cut-offs are marked by cyan lines. Promoters and enhancers are identified as described. (c). All identified enhancers (n = 1,963) within 12.5kb were ‘stitched’ together, resulting in 1,268 regions that were ranked for Med1 ChIP-seq signal (input-subtracted total reads). In total, 95 stitched enhancer regions were classified as super-enhancers, including the α- and β-globin regulatory regions. (d). The number of constituent enhancers present among the 1,173 stitched regular enhancers and the 95 stitched super-enhancers. Although super-enhancers are enriched for composite enhancers with a high number of constituents, both classes contain single and composite enhancer regions. (e). Med1 binding (input-subtracted reads per million per basepair) across stitched, regular (n = 1,173) and super-enhancers (n = 95) and a region of 3kb up- and downstream. The median size of regular and super-enhancers is used to scale the region between start and end. (f). DNase-seq and ChIP-seq profiles for Med1, Gata1, Klf1, Nf-e2, Scl, and CTCF across the α-globin locus (mm9 in primary Ter119+ erythroid cells. The paired CTCF sites flanking the α-globin locus are highlighted in blue. (Coordinates, mm9: chr11:32,125,268–32,229,368.)
To identify erythroid super-enhancers, we used the ROSE algorithm3, ‘stitching’ together individual enhancers within 12.5 kb of each other to define a single entity spanning a contiguous genomic region. The stitched enhancers and the remaining individual enhancers (those without a neighbouring enhancer within 12.5 kb) were then ranked by the level of Med1 signal within the extended genomic region. A small number (95) of these enhancer regions (<7%) bound very high levels of Med1. By definition1,3, elements with a Med1-value above a cut-off where the slope of the distribution plot of Med1 ChIP-seq intensity is 1 were designated super-enhancers (Fig. 1c). The remaining enhancers were considered to be regular enhancers. As in other cell types1, and by definition, Med1 levels were most informative in distinguishing between super-enhancers and regular enhancers (Supplementary fig. 1c).
As in previous studies1,15, super-enhancers often spanned large genomic regions, and contained multiple constituent enhancer elements (Fig. 1d). In erythroid cells, their median size is an order of magnitude larger than that of regular erythroid enhancers (5650 bp versus 866 bp). As well as the defining enrichment of Med1 (Fig. 1e), we also found significant enrichment of key erythroid-specific transcription factors (Supplementary Fig.1d and e) and their binding motifs (Supplementary Fig. 1f). The previously reported α- and β-globin gene regulatory regions were the two highest scoring super-enhancers in erythroid cells using Mediator-binding as the defining parameter (Fig. 1c and Supplementary Table 1). This was also true using any of the other reported diagnostic parameters (Supplementary Table 1). Like other super-enhancers15, the globin super-enhancer lies close to the cell-specific gene it regulates (Fig. 1f). .
The mouse α-globin super-enhancer identified using these methods spans 24 kb and contains five enhancer-like elements, four of which lie in the introns of an adjacent, widely-expressed gene (Nprl3) (Fig. 1f). The entire α-globin cluster is flanked by two pairs of CTCF binding sites (Fig. 1f), a common observation for super-enhancers15.
Given that the α-globin super-enhancer typifies this newly proposed class of element, we used it as a model to examine in detail the structure and function of super-enhancers.
Transcription factor binding varies between super-enhancer components
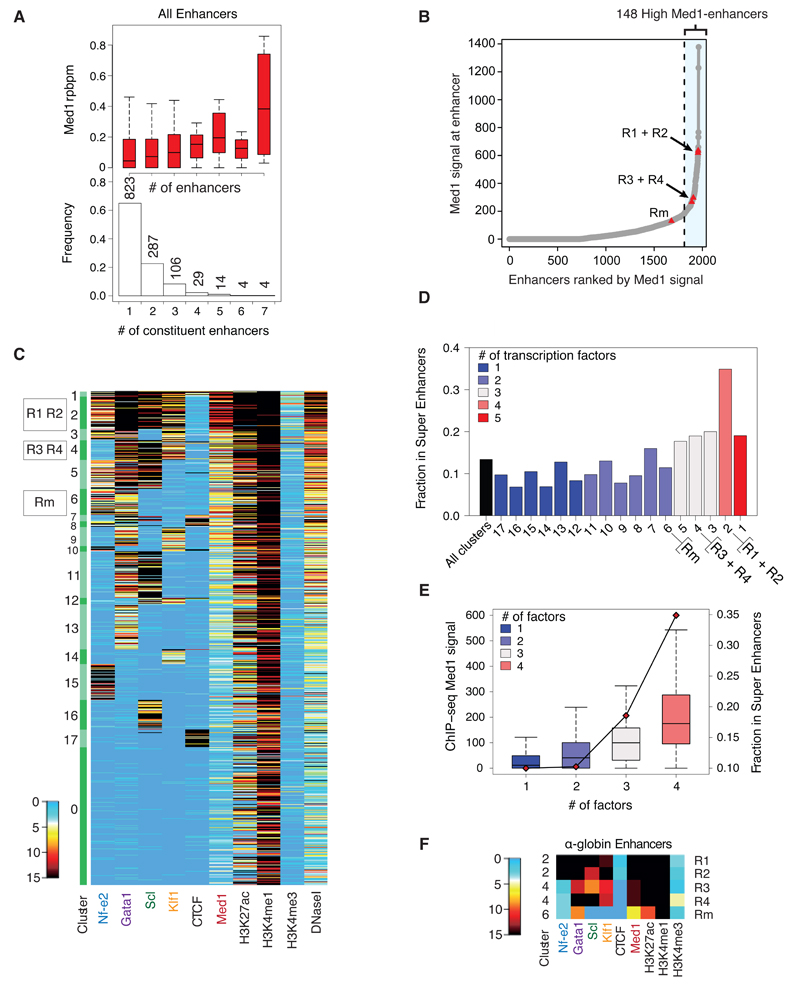
Each of the constituents of the α-globin super-enhancer is bound by different combinations of erythroid transcription factors and varying levels of Med1 (Fig. 1f). Analysis of the composition of regular enhancers and super-enhancers showed that, as expected, super-enhancers are enriched for composite enhancers (Fig. 1d); the greater the number of constituent elements, the more likely the region will be classified as a super-enhancer (Supplementary Fig. 1g). An increase in the number of constituent elements correlated to higher levels of Med1 (Fig. 2a). When individual enhancers were ranked for Med1 occupancy without stitching, Med1 binding varied considerably between the five constituent enhancers of the α-globin super-enhancer (Fig. 2b). Using the same methodology as described above to determine a cut-off, the four evolutionarily conserved α-globin regulatory elements (R1-4)16 still fell into the class of highly Med1 bound regions when analysed as single elements, whereas Rm (a species-specific cis-element) fell below the proposed criterion (Fig. 2b).
Figure 2. Erythroid super-enhancer constituents vary in transcription factor binding and chromatin signature.
(a). Boxplot showing normalized Med1 ChIP-seq density (input-subtracted reads/basepair/million) to constituent enhancer regions as a function of the number of constituents present in the stitched enhancer region. (Box plot shows median and interquartile range; whiskers define 1.5 × IQR) The histogram displays the relative distribution of composite enhancers. (b). All identified enhancers (n = 1,963) were ranked for Med1 ChIP-seq signal (input-subtracted total reads), and 148 individual enhancers were classified as High-Med1 enhancers. The α-globin enhancers have been highlighted as red triangles. (c). PAM-clustering results for the “putative enhancers”. Clusters 2–17 are ranked by mean Med1 signal (cluster 2 highest, cluster 17 lowest signal). Raw read counts, downscaled, input-corrected, background-subtracted and normalized to Klf1 ChIP-seq data within the peak regions. Regions having >15 reads are shown in black. (d). The fraction of individual enhancers that are constituents of super-enhancers (as defined in Fig. 1C) in each cluster. Clusters are ranked by mean Med1 signal of individual enhancers within the cluster. The colour of the bars indicates the number of master erythroid transcription factors bound to the individual enhancers of the cluster. (e). Med1 ChIP-seq signal (input-subtracted total reads) at individual enhancers and the fraction of individual enhancers that are constituents of super-enhancers as a function of the number of bound transcription factors. In total 570 enhancers are bound by one factor; 459 by two factors; 237 by three factors; and 129 by four factors.
We applied an unbiased clustering approach to further characterize the individual components of all erythroid super-enhancers. A Partitioning Around Medoids (PAM) algorithm was used to group similar regulatory elements based on the binding levels of the four master erythroid transcription factors Nf-e2, Gata1, Scl and Klf1, and the ubiquitously expressed factor CTCF. In addition to groups of elements that were bound by none (cluster 0) or all (cluster 1) of the transcription factors, 16 further clusters each binding distinct combinations of transcription factors were identified. PAM clusters were ranked for average Med1 signal per cluster and visualized in a heatmap (Fig. 2c). Motif analysis supported the clustering results, confirming enrichment of binding motifs that matched the different combinations of the four investigated transcription factors found in each cluster by ChIP-seq (Supplementary Table 2). Despite not showing enrichment for transcription factor binding by ChIP-seq, DNase I hypersensitive elements in cluster 0 (containing approximately 25% of all enhancers) were enriched for Gata1, Gata-Scl and Klf1 binding motifs indicating that low levels of these erythroid transcription factors may bind to these elements.
Interestingly, the ranking of clusters for Med1 binding revealed higher levels of Med1 at enhancers containing a larger number of transcription factor binding sites (Fig. 2c and 2d). These elements were also more likely to be incorporated in super-enhancers (Fig. 2e). Although this was the case for elements contained within both regular enhancers and super-enhancers, constituents of the latter had higher levels of Med1 binding than enhancer regions outside super-enhancers.
We investigated whether any of the clusters were overrepresented in super-enhancers (Fig. 2d). Cluster 2, containing enhancers binding all four erythroid transcription factors, was significantly enriched, being present in over 30% of erythroid super-enhancers. Consistent with the recent identification of transcription factor “hotspots” as key components of adipogenic super-enhancers 6, enhancer elements bound by a high number of transcription factors are enriched in erythroid super-enhancers.
We next applied this analysis to the constituent elements of the α-globin super-enhancer (Fig. 2f). Of its five constituent elements, R1 and R2 were both present in cluster 2, and had the highest levels of Med1 (Fig. 2c). The elements R3 and R4 were found in cluster 4, binding lower levels of Med1 and only three of the transcription factors (Gata1, Scl and Klf1). Finally, the mouse-specific regulatory element, Rm, to which only two of the key transcription factors were bound (Nf-e2 and Gata1), was part of PAM cluster 6. Thus, the constituents of the α-globin super-enhancer show considerable variation in Med1 and transcription factor binding, raising the possibility that they may vary in their contribution to α-globin gene regulation.
Enhancer assays do not reliably describe super-enhancer elements
Mammalian enhancers are defined as distal genetic elements that positively regulate expression in an orientation-independent manner in heterologous gain of function expression experiments. Using a transient, non-integrated, in-vivo Citrine reporter assay in developing chicken embryos, we tested the enhancer activity of the five elements.
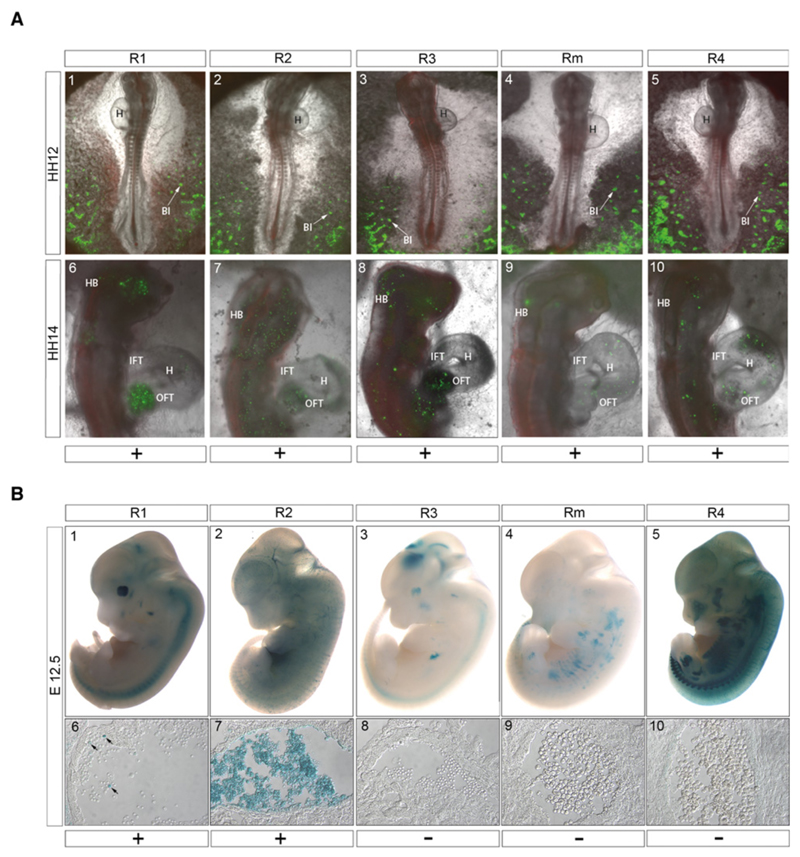
Enhancer activity was initially detected in Hamilton-Hamburger stage 9 embryos in the developing blood islands, where it persisted throughout development (Fig. 3a), consistent with the expression of globin here. Later during development, enhancer activity was also detected in the circulating blood (Fig. 3a and Supplementary Video 1), and was most prominent in the heart and head where the greatest density of red blood cells is found (Fig. 3a, panels 6-10). Expression was most prominent for the R1 enhancer, while the activity of the R2 enhancer is detected more broadly across the head and trunk. R3 and R4 had comparable patterns of expression to R2. Rm had the lowest level of late activity in regions other than blood islands, with only low levels of Citrine reporter expression detected in the head. No activity was observed with the negative control (Supplementary Fig. 2a). Consistent with our findings in the PAM analysis the effects of these non-integrated constructs in transient assays reflect the numbers and types of transcription factor binding sites present in each element.
Figure 3. Enhancer assays of individual elements.
(a) Reporter assays for activity of 5 α–globin enhancers during chick embryonic development. Panels A1-5: Activity of all five enhancers was detected in the developing blood islands, indicated by arrows, in the posterior part of the embryo, at Hamilton Hamburger stage 12 (HH12). Anterior is oriented to the top. Panels A6-10. HH14 reporter activity, driven by all enhancers, was also detected in circulating blood, most notably in the head and heart. Ubiquitous expression of Histone 2B-tethered RFP was used as electroporation control. BI, blood islands; OFT, outflow tract; IFT, inflow tract; H, heart; HB, hindbrain. Scale bar represents 1 mm. Further details are given in Supplementary Table 3.
(b) Functional analysis of enhancer activity in E12.5 mouse embryos. The upper panels (b1-5) show a representative LacZ stained embryo for each of the enhancer constructs indicated. Lower panels (b6-10) show a section through the heart (R1) or dorsal aorta (R2-4 and Rm), showing a population of hematopoietic cells. R1 shows a low level of activity in a subset of cells (arrowed), R2 shows robust activity in the majority of hematopoietic cells, but there is no detectable activity in these cells from R3, R4 or the Rm element. Scale bar represents 1 mm in panels 1–5 and 50μm in panels 6–10. Further details are given in Supplementary Table 3.
Each element was also tested using the well-established mouse transgenic system17,18, where a vector containing a candidate enhancer, a minimal promoter and the LacZ gene is stably integrated into chromatin at random positions in the mouse genome. Examination of whole embryos at E12.5 following LacZ staining suggested that only R2 exhibited positive enhancer activity in hematopoietic cells at this time point, with 5 of 7 LacZ-stained embryos exhibiting an expression pattern consistent with erythroid enhancer activity (Fig. 3b and Supplementary Table 3). We prepared tissue sections from three LacZ positive mice for each enhancer construct tested; these confirmed strong enhancer activity for R2 and also demonstrated weaker activity for R1, while no activity was detected for the remaining three elements (R3, R4 and Rm) (Fig. 3b). Thus, despite their chromatin signatures, these three elements would not be classified as enhancers by this standard assay; only those two elements with the most extensive transcription factor binding profile as determined by PAM analysis scored positively in the assay.
No single element is critical for globin gene expression
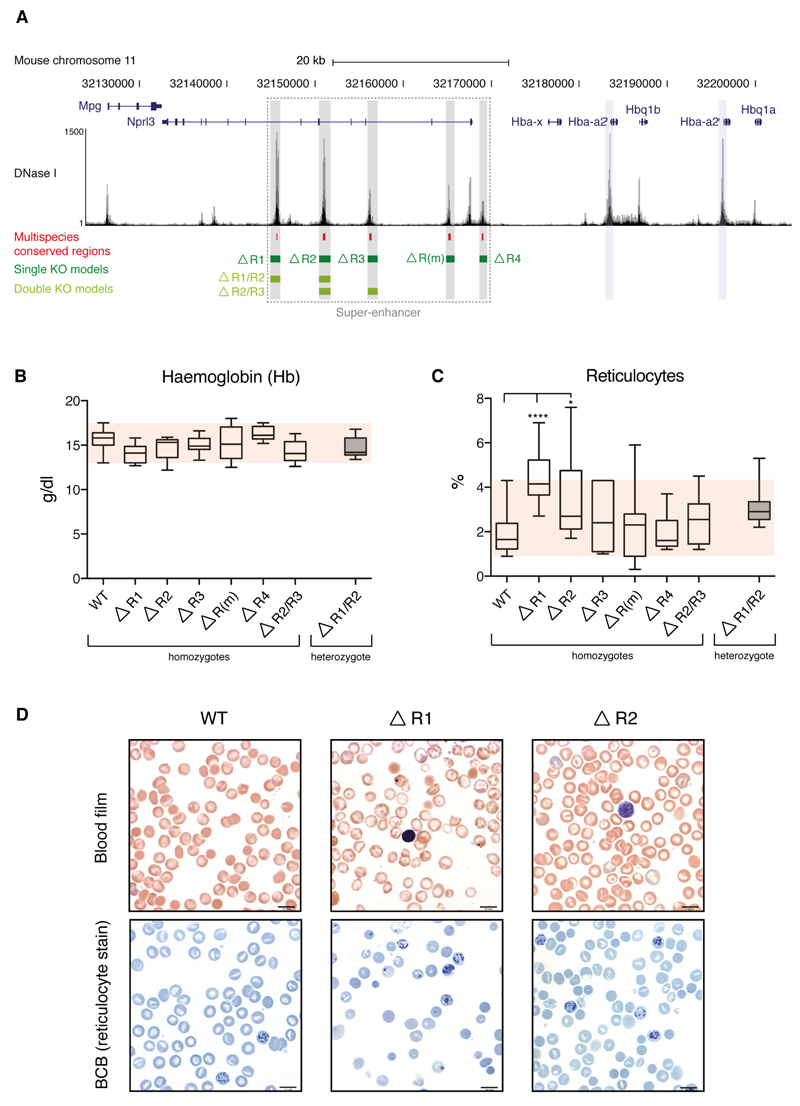
To determine the contribution of each enhancer to α-globin transcription within the context of the super-enhancer (Fig. 4a) we generated knockout mouse models for each constituent element. Knockouts were generated by homologous recombination for all five components of the α-globin super-enhancer (plus two informative combinations of knockouts, see below) Two of these deletions (R2-/- and R3-/-) have been previously described19,14.
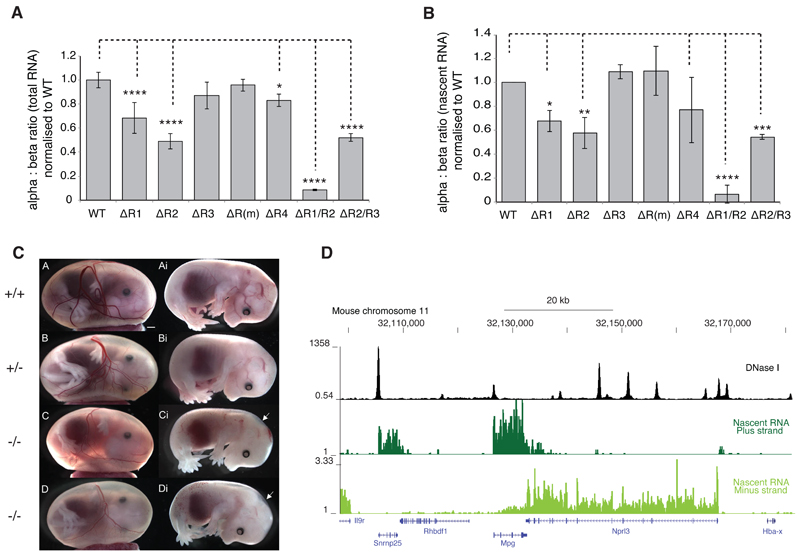
Figure 4. Hematologic impact of single and double enhancer knockouts.
(a). Mouse α-globin locus (chromosome 11), illustrating the α-globin genes (Hba-a1 and Hba-a2, blue highlight); the five α-globin enhancers (R1, R2, R3, Rm and R4, grey) and the regions deleted in each enhancer knockout models (green) in relation to multispecies conserved regions (red). (b). Hemoglobin was measured in adult blood from mice homozygous for each individual enhancer knockout, and for homozygotes for the R2/R3-/- double deletion. None of the enhancer knockout models exhibits hemoglobin levels outside of the normal range (red shaded box). Hematologic parameters for the R1/R2-/- double knockout could not be analysed due to its embryonic lethality and hematologic data shown for this model are from adult heterozygotes only. (c). Total reticulocyte count for each model. A significantly elevated reticulocyte count is observed in the R1-/- and R2-/- knockout models. All other models fall within the normal range (red shaded box). (d). Blood films (top panel) and brilliant cresyl blue (BCB) stained blood (lower panel) from the blood of wild type (WT), R1-/- and R2-/- mice. Whilst the blood films from the three genotypes are essentially identical, increased reticulocytes can be observed in BCB films from the R1-/- and (to a lesser extent) R2-/- mice. Data shown represent means and standard deviation. All data are from independent biological replicates, with numbers per group given in Supplementary Table 5. Statistical analysis was performed using a one-way ANOVA with Dunnett’s correction. No randomization was required for any mouse analysis.
We analysed the viability of mice homozygous for each individual deletion. (Supplementary Table 3). In all cases, offspring were seen in expected Mendelian ratios, and homozygotes survived to adulthood and bred normally.
To assess whether the loss of individual enhancer elements influenced the hematologic phenotype, blood counts and smears were examined. No single knockout reduced the hemoglobin concentration in adult mice below the normal range (Fig. 4b) nor did they result in a significant reduction in either mean red cell volume or mean red cell hemoglobin beyond the normal range (Supplementary Fig. 3), although we noted a non-significant trend towards a lower MCV and MCH in the R2-/- mice (as previously reported19). All hematologic data are summarised in Supplementary Table 4.
We noticed an increased variation in staining (polychromasia) in R1-/- and R2-/- mice (Fig. 4d), suggesting that stress erythropoiesis is required to maintain normal hematologic parameters in these models. We evaluated this using the reticulocyte count and noted significantly increased counts in R1-/- and R2-/- mice (Fig. 4c and d). suggesting these mice maintain normal red cell parameters only through stressed erythropoiesis. Knockouts of R3, R4 and Rm appear to have little, if any, effect on α-globin expression or erythropoiesis.
Quantification of the impact of each enhancer on α-globin transcription was initially performed using an RNA protection assay and qPCR. These assays showed no consistent evidence of altered α-globin RNA expression in homozygotes for knockouts of individual sites, although we noted a trend towards reduced α-globin RNA expression in R2-/- (as previously described19). However, these techniques evaluate accumulated RNA and, at best, identify fold changes in expression, precluding the identification of fractional changes. We therefore developed a modified method20 to quantify total and metabolically-labeled nascent RNA in populations of synchronously maturing primary erythroid cells from each mouse model. Fetal liver cells were isolated at E12.5 to permit assessment of definitive erythropoiesis in homozygotes of all models, including the embryonically lethal R1/R2-/- model. RNA from equivalent populations of synchronously maturing erythroid cells (Supplementary Fig. 4a) was quantified using Nanostring21, enabling simultaneous measurement of expression of all globin genes plus five control genes expressed throughout erythroid maturation. The approach was validated using nascent RNA-seq (Fig. 5d and supplementary Fig. 4b).
Figure 5. α-globin transcription in single and double enhancer knockouts.
(a) NanoString quantification of the ratio between α- and β-globin transcripts in steady state RNA isolated from primary mouse fetal liver cells from homozygote mice at E12.5. Samples were taken 12 hours after exposure to high levels of Epo (intermediate erythroblasts). A variable but significant reduction in the α:β-transcript ratio is observed in the R1-/-, R3-/-, R4-/- and R2-/-/R3-/- knockouts. A 90% reduction is seen in the R1-/-/R2-/- double knockout. (b). NanoString quantification of the α:β-globin transcript ratio in nascent RNA isolated from primary mouse fetal liver cells at the same stage as Figure 5a. Modest effects are observed in the R1-/-, R2-/- R4-/- and R2-/-/R3-/- models, with the greatest reduction in α:β-transcript ratio observed in the R1-/-/R2-/- double knockout. All data are from a minimum of 3 independent biological replicates, and are shown as mean with standard deviation. Statistical analysis is by one-way ANOVA with Dunnett’s correction (**** reflects p<0.0001). (c). Embryos from WT (++), heterozygote (+/-) and homozygotes (-/-) for the R1/R2 knockout taken at E14.5 Homozygotes are smaller and paler with hemorrhagic areas and evidence of nuchal edema (arrowed). (d). RNA sequencing of nascent RNA obtained from primary fetal liver cultures using metabolic labeling. Unspliced directional transcripts are shown across the α–globin cluster
Fig. 5a and 5b demonstrate that individual elements of the super-enhancer are not functionally equivalent. Deletions of R1 and R2 have a significant and reproducible impact on the synthesis of α-globin RNA. Deletion of R4 has a minor impact. Deletions of R3 and Rm, however, have no discernible impact on α-globin RNA synthesis (Fig. 5b).
While the impact of homozygosity for the R1 and R2 deletions on the α:β globin mRNA ratio is significant, there appears to be compensation for this imbalance in the peripheral blood. The hemolysis and ineffective erythropoiesis induced by the skewed α:β ratio result in an increased production of erythroid cells (Fig. 4c and 4d); those cells with the greatest imbalance in globin synthesis are likely to be destroyed prior to leaving the bone marrow, leaving little change in the peripheral red blood cells or hematologic indices.
Thus, the regulatory effect of the α-globin super-enhancer depends mainly on two elements, R1 and R2; the other three elements contribute very little, if at all, to the activity of the super-enhancer in vivo. Importantly, it can be seen that the super-enhancer does not act as a multi-component structure; removal of any single element cannot abolish its function.
The enhancer elements act in an additive manner
To determine if the apparently inactive enhancer-like elements play a compensatory role when a major element is removed, or if the removal of two elements rather than one might collapse the super-enhancer structure, we generated mice in which both an active (R2) and inactive enhancer-like element (we arbitrarily chose R3) were deleted in cis. We found that homozygotes for this double deletion were born in the normal Mendelian ratios and were viable into adulthood. Adults are indistinguishable from R2-/- mice both hematologically (Fig. 4b) and transcriptionally (Fig. 5).
Next we made mice in which both active elements R1 and R2 are removed in cis. Heterozygotes for the R1/R2 knockout were viable and were able to breed normally. However cross breeding of heterozygotes showed significantly reduced litter sizes and no homozygotes at term. The sacrifice of pregnant females following timed matings showed that R1/R2-/- mice were smaller and paler than WT or heterozygote littermates, with nuchal edema (arrowed, Figure 5c; Supplementary Table 4). These mice die in utero at ~E14.5
The embryonic lethality of R1/R2-/- mice means that hematologic indices could be assessed only in heterozygotes. These have reduced MCH and MCV and raised reticulocyte counts typical of thalassemia (Fig. 4). Analysis of α:β globin RNA ratios showed that R1/R2-/- fetal liver cells produced very little α-globin RNA (Fig. 5). Further, within the limits of the experimental method, the contributions of R1 and R2 to transcription appear to be additive rather than clearly synergistic. The remaining <10% of normal α-globin RNA levels, is likely to be transcribed under the influence of the intact cis-elements. Our data suggest that R4 may contribute to this.
The chromatin environment does not depend on the intact super-enhancer
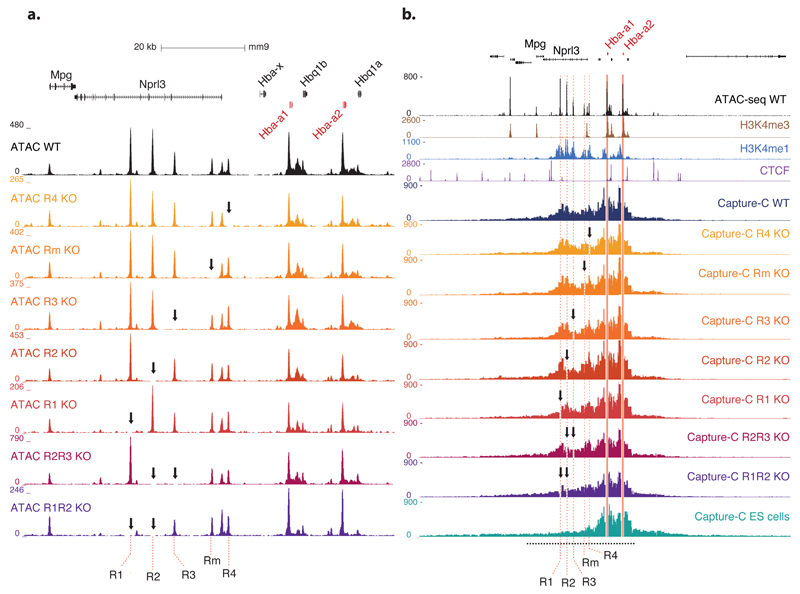
Each cis element in the α-globin regulatory domain is associated with a nuclease-sensitive site, demonstrated by DNase I sensitivity and ATAC-seq11. To determine if the hypersensitive sites within the α–globin super-enhancer are interdependent, we analysed the ATAC-Seq profile in fetal liver culture erythroid cells from homozygotes for each regulatory element knockout (Fig. 6a). We show that the presence of no individual hypersensitive site critically depends on any other: they appear to form independently. Even in the most phenotypically severe R1/R2-/- model, hypersensitive sites at R3, R4 and Rm still form.
Figure 6. Analysis of chromatin structure.
(a). Open chromatin landscape (ATAC-seq) at the α-globin cluster in wild type, five individual single enhancer knockout and two double enhancer knockout mice. Formation of the elements in the cluster is not impaired by deletion of any individual enhancers, nor by the double deletions. The position of each individual element of the predicted α globin super-enhancer is named and highlighted by red dashed lines. (Coordinates, mm9: chr11:32,123,000–32,209,000.)
(b). Comparison of the interaction profiles from the Hba-a1&2 promoters in primary erythroid cells from WT mice, each of the single and double enhancer knockout models and ES cells (E14). NG Capture-C was performed using the Hba-a1&2 promoters as a viewpoint (since the genes are virtually identical this represents a composite interaction profile from both promoters). The X-axis displays the number of unique interactions from the promoter fragments with each DpnII fragment genome-wide, normalised for total number of interactions. DpnII fragments overlapping the deleted regions removed for visual clarity. The region displayed in panel a is indicated by a black dashed line below the interaction profiles. (Coordinates, mm9: chr11:32,032,001–32,332,000.)
In the absence of R1 and R2, the peaks of hypersensitivity at the α-globin promoters are reduced when compared to wild type (Fig. 6a, Supplementary Fig. 5a). However, on genome-wide analysis, of R1/R2-/- cells, we found no changes to global hypersensitive site formation beyond those commensurate with inter-individual variation (Supplementary Fig. 5b).
Using Next Generation Capture-C22, we have previously shown that a region of approximately 70 kb, including the α-globin genes and the super-enhancer, are contained within a tissue-specific “compartment” in which the individual regulatory elements interact with each other, the promoters of the globin genes, and the surrounding chromatin. Assessing this in fetal liver culture erythroid cells from the knockout models demonstrates very little change in the compartment between the wild type and any single knockout (Fig. 6b). The intact super-enhancer is therefore not required for the formation or maintenance of the chromatin compartment.
Although the interactions between the α-globin promoters and the regions around the R1 and R2 enhancers are markedly reduced in R1/R2-/- cells, they remain above the baseline levels found in ES cells (Supplementary Fig. 6. The interaction between the promoters and the CTCF site located between the enhancers is also reduced, though still present in R1/R2-/-, as is the interaction between the promoters and the weaker enhancer R4. In R2/R3-/-,cells, the promoters’ interactions with R1 are diminished but those with Rm and R4 are virtually unchanged compared to wild type. These data suggest that distal strong enhancers R1 and R2 potentiate the interactions between the elements (R3, Rm and R4) and the α-globin promoters.
When we extended the assessment of chromosomal conformation genome-wide using next-generation Capture-C, the only statistically significant changes seen in even the most phenotypically severe double knockouts were those at the α-globin promoters (Supplementary Fig. 7).
Discussion
Based on genome-wide studies pioneered by the ENCODE project (www.encodeproject.org), a surprisingly large number of regulatory elements have been identified (2.9 million DHS across various human cell types23 or ~300,000 cis-regulatory elements in various murine tissues24). This equates to 15 -150 elements for every structural gene. Yet assays of their functional roles remain sub-optimal, typically examining elements outside their natural chromosomal environment, which can give, as we describe, conflicting results.
Further classification of regulatory elements has been driven by the observation that some elements bearing the signatures of enhancers cluster around particular genes in specific tissues. Such clusters of enhancers were first referred to as locus control regions25, then stretch enhancers26 and super-enhancers1. Clearly, it is important to distinguish between models which propose an emergent property from this grouping (and thus a new type of element, e.g. super-enhancers), from a more simple clustering of regular enhancers.
We have systematically addressed this at the mammalian α-globin locus. Here we have shown that the linked α-globin regulatory elements fulfil the criteria for being among the highest-ranking super-enhancers in erythroid cells. Each component is marked by a DHS, binds key cell-specific transcription factors and the Mediator complex, is modified by H3K4me1 and H3K27ac, and produces eRNAs and lncRNAs14. Each element scores positively in a transient enhancer assay. Although we have not assessed possible changes in Med1 binding, activating chromatin marks, or eRNA production in the context of each enhancer knockout27, we have found that each element within the cluster appears to act independently rather than co-operatively, with removal of any one element having little or no effect on the function of the others. Similarly, removal of two elements does not appear to abrogate the formation of other enhancer elements showing that the cluster does not act as an interdependent holo-complex. Previous less extensive studies have also shown that the individual regulatory elements of the β-globin LCR, which we show here also corresponds to an erythroid super-enhancer, behave in a similar independent and additive manner28. While we cannot extrapolate from these findings to all super-enhancers, previous principles of gene regulation first established for the globin genes have, without exception, been shown to be generally applicable.
Importantly we have shown that although all components of the cluster would be classified as enhancers they are not functionally equivalent. Whereas R1 and R2 are strong enhancers in their natural chromosomal environment, R3, Rm and R4 are either very weak enhancers or are not enhancers at all, despite their chromatin marks. Of interest only R1 and R2 score positively in an enhancer assay in which the sequences are incorporated into chromatin. Further sub-classification of all erythroid enhancers shows that R1 and R2 are distinguished by binding the greatest number of cell-specific transcription factors and the greatest amount of Mediator, and they are also associated with the most extensively modified chromatin. These sites correspond to elements that others have referred to as “hotspots”. However, they could more simply be referred to as strong individual enhancers.
From the data presented here and from others29, 30, it seems likely that multiple enhancers may provide robustness in gene expression. Removal of either strong enhancer (R1 or R2) at the α-globin locus produces stressed erythropoiesis to ensure normal hemoglobin production. Under conditions of environmental or physiological compromise it is possible that having two enhancers rather than one would provide a selective advantage. Such enhancers have previously been referred to as “shadow enhancers”31.
One puzzle emerging from this study is why enhancer-like elements which clearly score positively on indirect assays have no discernible function in their natural chromosomal environment. One possibility is that they do have a weak enhancer function which is not easily revealed by current physiological assays. Alternatively, they play a more significant role at different stages of development or may define the polarity of the enhancer: this has not been tested in this study.
We would suggest, however, that these elements reflect evolutionary turnover in cis-acting elements as seen in other studies32. It is unlikely that cis-acting elements emerge fully formed during evolution or that they disappear entirely when they lose function; the apparently functionless elements we identify could therefore represent evolving enhancers. Indeed, it has been proposed that transcription factor hotspots, such as R1 and R2, create a permissive chromatin environment for the emergence of new transcription factor binding regions6.
Our data do not support the suggestion that super-enhancers have a singular composite function, but rather that strong enhancers simply augment the interactions between their chromatin environment and cognate promoters. This generates a compartment of interaction between the chromatin surrounding the enhancer and the promoter, which results in an environment that might favor the evolution of other enhancers.
Our study, together with previous work at the β-globin LCR, does not support further sub-classification of enhancers beyond the simple designation of strong to weak elements. These may be usefully ranked by the number of lineage-specific transcription factors that they bind, in turn reflected by the degree of binding of Mediator and by the associated activated chromatin modifications. When tested at the α-globin locus, clustering of enhancers in super-enhancers does not appear to lead to any unexplained emergent properties. However, genetic dissection of other super-enhancers in other cell types is warranted to provide a more comprehensive understanding of super-enhancer function.
URLs
http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf (Stark, R. & Brown, G. (2011). DiffBind: differential binding analysis of ChIP-Seq peak data.)
Online Methods
Analysis of cis-elements
DNase I hypersensitive sites, ATAC accessibility and ChIP sequencing to characterize cis-acting elements were performed and analysed as previously described14, 33. For ChIP-seq analysis, all data were aligned using Bowtie (version 1.0.0)34 to the NCBI37/MM9 build of the mouse genome. Alignments were performed with the following parameters: -m 2, -k 1,–best. Enhancers were identified from a peak call of C57BL/6 mouse DNaseI hypersensitive regions.
SeqMonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) was used for an independent peak call of two technical replicates of biological duplicate experiments11. The ploidy regions, as well as peaks higher or equal in signal in sonication input were excluded. Each of the files was manually curated, to exclude remaining background peaks in MIG35. The peak calls were merged to yield a single high-confidence DNaseI hypersensitive regions file containing 15,849 peaks. To identify putative enhancer elements, and to avoid a bias towards a limited set of transcription factors, DNaseI hypersensitive peaks were classified based on the ratio between the coverage of H3K4me1 and H3K4me3 signal within each peak call region. H3K4me1 and H3K4me3 coverage was normalised to million mapped reads before determining the ratio (in house script: Quantbam.pl). A total of 5,307 peaks were annotated to be low in both markers, as both H3K4me1 and H3K4me3 marks had <10 rpm coverage. The rest of the peaks were further divided into putative enhancers and promoters. Regions were annotated as enhancers if the ratio between H3K4me3:H3K4me1 was <1. Annotated enhancers that were within 250bp of refGene annotated transcription start sites were removed from the set.
Super-enhancers were identified using the ROSE tool1. Briefly, individual enhancers within 12.5kb were stitched together to form a single, larger enhancer domain. Stitched enhancer domains were then ranked for input-normalized ChIP-seq occupancy of Med1. The x-axis point at which the tangent to a scaled graph with X and Y axes ranging from 0-1 had a slope of 1 was used as a cut-off, above which stitched enhancers were classified as super-enhancers. This same methodology was followed to analyse the erythroid enhancers for the occupancy of DNaseI, H3K4me1, H3K27ac and a combination of four erythroid transcription factors (Gata1, Scl, Klf1, Nf-e2). The signal for each of the factors was normalized to a maximum value of 1.0 and visualized after sorting the enhancers for the relative signal. To identify individual high-Med1 enhancers we used the ROSE tool in a similar manner as described above, only specifying a stitching distance (option -s) of 0 and thereby preventing the stitching of individual enhancers elements before downstream analysis.
Factor Enrichment Analysis
For quantification of ChIP-seq signal we calculated the normalized read density after removal of PCR duplicates. Reads were extended by 200 bp and we calculated the number of reads per basepair (bp). The calculated read densities were then normalized to the total number of million mapped reads to produce the reads per million mapped per basepair (rpm/bp). Finally, ChIP-seq rpm/bp values were normalized to input by subtraction.
PAM-clustering
PAM-clustering analysis was done with library “cluster” in R.36 Before clustering, the coverages of all TFs of interest and CTCF (and the sonication input) were counted, and down-sampled to the Klf1 read counts (the dataset with the lowest total number of mapped reads). The sonication input value was subtracted from the coverages, and 13 reads (approximate max height of Klf1 baseline read depth) was subtracted from all coverages. All regions which had at least 4 reads were set to have 4 reads as the coverage. This essentially led to close to binary clustering, as most regions now had either 0 or 4 reads. After that the regions were subjected to PAM-clustering of 2 to 24 clusters. The clustering results were visually inspected, and validated in silhouette plots, and the best clustering (fewest outliers, high average silhouette value for all clusters) was seen when our annotated enhancers were divided to 16 clusters. These are clusters 2 to 17, clusters 0-1 being excluded from the clustering (as invariable regions cannot be clustered). Cluster 1 has high signal in all the TFs and CTCF, and cluster 0 has no signal in any of the TFs or CTCF. To visualize the PAM-clusters, they were plotted along mean Mediator coordinate, so that cluster 2 has the highest mean Mediator signal, and cluster 17 the lowest. In the visualization all read counts higher than 15 reads within the region are shown as signal “15 reads”. All the reads are down-sampled to the KLF signal, input-subtracted, KLF-background 13 reads subtracted. The histone marker coverages are counted by adding +/-500 bases overhang to the peaks before counting the coverage. Sonication INPUT is subtracted using the same +/- 500 bases overhang, otherwise histone coverages normalized as above. Data are not row-normalized, but show the raw signal.
Motif Analysis and Heatmaps
Transcription factor binding sites in enhancer and promoter elements were identified using HOMER software37, and were further analysed by PAM-clustering using library “cluster” in R36.
Motif analysis was done with HOMER code annotate Peaks.pl, by using 1) peak widths as they were given in the peak call file, 2) +/400 bases 3) +/- 200 bases, and 4) +/- 100 bases from the peak center. The results were inspected, and used to validate the PAM clustering analysis. To validate the enrichment of erythroid transcription factor motifs in super-enhancers, the findMotifs.pl tool from the HOMER suite was used with a peak width of +/-200bp from the center of constituent peaks within super-enhancers. P-values correspond to corrected p in the HOMER output. Heatmaps were produced using HOMER, version 4.7. The heatmaps were visualized in R, using library heatmap.2 and RcolorBrewer.
Analysis of Enhancer Elements in Transient Assays
Chick reporter construct cloning: Enhancer regions were amplified from mouse genomic DNA using KAPA HiFi polymerase and primers described in the table below. Gel purified amplicons were cloned using a Golden gate assembly approach37 into a novel Citrine (YFP variant) reporter vector containing BsmBI-flanked LacZ expression cassette upstream of the mouse minimal alpha globin promoter (HB2) and Citrine fluorescent protein ORF. Negative control constructs were made by inserting a short negative region instead of an enhancer. The negative region was assembled by annealing two oligonucleotides. Endotoxin-free maxi preps were prepared for in vivo electroporation using Qiagen’s kit (12362).
Primers for construct generation: R1Fwd, TTTTTTCGTCTCgccagggcatcgagtggagagaaggg; R1Rev, TTTTTTCGTCTCcaacagtcgagtttatgctgcgtcct; R2Fwd, TTTTTTCGTCTCgccaggtttgctaaacatctgtcaggga; R2Rev, TTTTTTCGTCTCcaacagcgagaagtctgcccaggttt; R3Fwd, TTTTTTCGTCTCgccagggcccttcccctgaacactta; R3Rev, TTTTTTCGTCTCcaacagtagcctgtctcccttccctc; R4Fwd, TTTTTTCGTCTCgccagggccataccttccgactctga; R4Rev TTTTTTCGTCTCcaacagtcaacttccgacccagtgtg; R(m)Fwd, TTTTTTCGTCTCgccagggacacagtaaatcccaagcca; R(m)Rev, TTTTTTCGTCTCcaacagccacatggtaagatcctgtc; NegFwd, ccaggAGCTGGATCGATgatatcCGATCGATCGTAGCAC; NegRev, aacagGTGCTACGATCGATCGgatatcATCGATCCAGCT.
Individual reporter constructs together with control construct, ubiquitously expressing histone2B-tethered RFP, were co-electroporated into the entire epiblast of the early chick gastrula embryos (Hamburger-Hamilton stage 4, HH4), as previously described39. Following electroporation, embryos were allowed to develop to desired stages using ex-ovo culture in thin albumin, supplemented with 1% penicillin/streptomycin. Reporter reporter activity was monitored up to 3 days post electroporation using fluorescence microscopy.
Enhancer activity was also assayed using an established mouse transgenic system where a vector containing a candidate enhancer, a minimal promoter, and the LacZ gene is stably integrated into the mouse genome via standard pronuclear injection17,18. The coordinates of enhancer elements assayed in this work are the same as those used for the chick enhancer assays. To visualize LacZ staining on histological sections, embryos were fixed in 4% paraformaldehyde at 4°C, washed three times in phosphate-buffered saline, embedded in OCT-compound (Sakura Finetek), and cut on a cryostat (Leica, Deerfiled, IL) at 10 µM. Specimens were viewed and photographed using a Nikon Eclipse E600 microscope and DXM1200C (Nikon) camera.
Generation of knockout mouse models
All mouse work was performed in accordance with UK Home Office regulations under approved project licences. Targeting and mouse model generation was performed as previously described14,19. A region of ~1kb encompassing the conserved sequence for each regulatory element (R1, R2, R3, Rm and R4, as previously described16) was identified for deletion. The region was chosen to include all binding sites for erythroid specific transcription factor binding sites, without impinging on other active elements such as CTCF sites. The coding sequence of Nprl3 was preserved. Regions of homology totalling ~7kb were identified on either side of the planned deletion and were amplified from a mouse 129/Ola specific α-globin bacterial artificial chromosome using a high fidelity Taq polymerase. These were cloned into the vector pGemT-easy for sequencing before excision with EcoR1, blunting with T4 polymerase, and ligation into the unique Srf1 and Bst1107I sites in the pNTFlox targeting vector. In this vector, the homology arms flank a floxed Neomycin resistance gene, driven by a PGK promoter. Outwith the homology arms, a herpes simplex thymidine kinase cassette, again driven by a PGK promoter, is included. Sequenced and purified vector was linearized at its unique XhoI site, and purified prior to electroporation into 129/Ola ES cells, which were then cultured in GMEM. Positive selection with G418 and negative selection with FIAU were used to isolate cells incorporating the construct. Correctly targeted clones were identified by Southern blotting, and were karyotyped prior to blastocyst injection and chimera generation.
Following germline transmission, mice bearing the floxed neomycin resistance gene were crossed with GATA1-Cre expressing mice to excise the neomycin resistance cassette in all tissues40 as this has previously been shown to affect local gene expression41. This recombination leaves only a single residual loxP site and multiple cloning site footprint from the targeting vector. Southern blotting and sequencing were used to confirm the correct deletion. Heterozygotes were then crossed to yield a line of mice homozygous for the single enhancer deletion.
For double knockouts, targeted ES cell clones heterozygous for the floxed neomycin resistance cassette at the site of the first targeted enhancer were subject to in vitro cre recombination with pCAGGS-Cre. After loss of neomycin resistance and southern blotting to confirm the expected sequence, a further round of targeting was performed with pNTFLox bearing homology arms for the second enhancer to be deleted. Newly neomycin-resistant clones were then subject to a second recombination stage with a weaker cre-recombinase (MC-Cre), with the aim of achieving a discrete second enhancer deletion (avoiding a maximal deletion product which would employ the residual loxP from the preceding cre recombination involved in the deletion of the first enhancer). Southern blotting was used to identify clones in which the second enhancer had been targeted in cis, and deleted in isolation, maintaining the coding sequence of Nprl3. Correctly targeted clones were then karyotyped and injected into blastocysts to yield chimeras, with subsequent germline transmission. Coordinates of the five single knockouts im mm9 are: R1, chr11:32145028-32146147; R2, chr11:32150550-32151858; R3. chr11:32156048-32157191; R(m), chr11:32165011-32165922; R4, chr11:32168783-32169689.
Hematologic analysis of mouse models
Hematologic phenotypes of a balanced mix of male and female mice for all models were examined at more than 7 weeks of age to avoid inter-individual variation42. All mice were generated on the same complex background. To compare the mouse models we have pooled wild type samples from different pedigrees. Reticulocyte preparations were made using Brilliant Cresyl Blue staining, and counts were made by two independent assessors blinded to genotype. Sample sized were chosen with a view to detecting a 25% reduction in haemoglobin; smaller confirmatory datasets were obtained where the analysis of phenotype had already been reported (as for R2 and R3 deletions14, 19. Data were subject to Levene’s test to confirm homogeneity of variance, followed by one-way ANOVA with Dunnett’s correction.
Analysis of Steady State and Nascent RNA in Mouse Models
Fetal livers were dissected from E12.5 mouse embryos and the dissociated cell suspension was expanded for 5-7 days in Stempro (Invitrogen) supplemented with Epo (1U/ml), SCF (50ng/ml) and dexamethasone (1μM). Following expansion of the cultures, mature red cells were removed by negative selection for Ter119 using anti-Ter-119 magnetic beads (Miltenyi), to obtain a population of erythroid precursors. Cultures were then switched to medium containing high Epo (5U/ml) to induce differentiation to mature erythroid cells. Samples for total and nascent RNA were collected after 12 hours of differentiation. Cells were lysed in TriReagent (Sigma) and RNA was extracted according to the manufacturer’s instructions. Samples were DNase I treated using Turbo DNA-free (Ambion). To obtain nascent RNA, fetal liver cultures were pulsed with 50μM 4-thiouridine (Sigma) for 1 hour followed by immediate lysis in TriReagent. Nascent RNA was then extracted according to the method described by Dölken et al20. Quantification of total and nascent RNA transcripts was performed using NanoString nCounter technology, using a customised probe set. Data were subject to Levene’s test to confirm homogeneity of variance, followed by one-way ANOVA with Dunnett’s correction.
Analysis of Chromatin Landscape Using ATAC-seq
ATAC-Seq was performed on each mouse model as previously published33, using either cultured or uncultured fetal liver cells. 8-10x105 cells were used per biological replicate. For the double knockout models, fetal liver cells were cultured as described above. For the single knockout models, E14.5 fetal livers were disaggregated, filtered, stained with PE anti-mouse Ter119+ antibody (Ly-76, BD Pharmingen) and purified using MACS beads (Miltenyi Biotec) on a magnetic column. Cells were lysed and nuclei were isolated prior to transposition with Tn5 transposase (Nextera, Illumina) for 30 minutes at 37°C. DNA was purified using a MinElute kit (Qiagen). Libraries were amplified and barcoded using the NEBNext 2x Mastermix (NEB) and the custom primers as previously described32. ATAC-Seq libraries profiles were visualized using D1000 tape on the Tapestation (Agilent) and quantified using a universal library quantification kit (KAPA Biosystems). Samples were sequenced using 75 cycles paired end kit on the NextSeq Illumina platform. Paired-end reads for ATAC-Seq were processed using our in-house customised DNase and CHIP pipeline: samples were aligned to the appropriate genome build (mm9) using bowtie (version 1.0.0)34. To prevent the exclusion of the duplicated globin genes bowtie was run with the –m reporting option set to 2 to allow reads to map twice to the genome. To exclude over- amplified products from these data sets, reads that map to the exact same genomic position were collapsed into a single representative read. Ploidy regions which represent regions in the genome which strongly overreact in high-throughput sequencing experiments due to large copy number differences between the real genome and the genome build and normalize poorly were excluded at this stage from downstream analysis. For the mouse data we generated this set of regions in-house. Genome-wide tracks were produced using the in-house perl tool sam2bigwig.pl, which produces a track of read density over a set window size and increment of movement across the genome. Bowtie alignments were thus converted to genome wide density tracks (BigWig) and the output was displayed in UCSC Genome Browser43. Peak detection for ATAC-Seq was performed with the MACS2 algorithm44. Differential analysis between the double knockout R1-/-/R2-/- and WT was performed using the R package DiffBind.
Analysis of Chromatin Landscape by NG Capture-C
NG Capture-C was performed as described22. Cells were obtained using the fetal liver culture system outlined above. 3C libraries were made using standard methods similar to the protocol for in situ Hi-C. Prior to oligonucleotide capture, 3C libraries were sonicated to 200 bp and Illumina paired-end sequencing adaptors (NEB E6040 / E7335 / E7500; Agilent Herculase II) were added. Samples were indexed, allowing multiple samples to be pooled prior to oligonucleotide capture using biotinylated DNA oligonucleotides designed for the Hba-a1&2, Hbb-b1&2 and Slc25A37 promoters (Sigma). The first hybridisation reaction was scaled up relative to the number of samples included in the reaction to maintain library complexity (Nimblegen Roch SeqCap EZ). Following a 72h hybridisation step a streptavidin bead pull down (Invitrogen M270) was performed followed by multiple bead washes (Nimblegen SeqCap EZ) and PCR amplification of the captured material (Kappa / Nimblegen SeqCap EZ accessory kit v2). A single volume, double capture step was performed. The material was sequenced using the Illumina MiSeq with 150bp PE reads (300bp V2 chemistry). Data were analysed using analysis scripts (available via Github) and R was used to normalize data and generate differential tracks.
Supplementary Material
Acknowledgements
The work was supported by the Wellcome Trust (D.H.), the UK Medical Research Council (MRC), and the National Institute for Health Research Biomedical Research Council, Oxford. L.A.P. was supported by NIDCR FaceBase grant U01DE020060NIH and by NHGRI grants R01HG003988, and U54HG006997, and research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California. This manuscript is dedicated to the memory of Professor Bill Wood who initiated the project and died in 2014 during the course of this work.
Footnotes
Accession codes
The Geo submission accession code is GSE78835 (reference series for all datasets).
Author contributions:
D.R.H and W.G.W conceived the project; D.R.H., J.R.H., D.H., J.O.J.D., L.H., M.S., and J.T. designed experiments; J.R.H., C.B, D.H., J.O.J.D., B.J.G., L.H., M.T.K, J.A.S, M.C.S. J.T, R.W., P-S.L, J.A.S-S., H.A. and S.B performed experiments; D.R.H., J.R.H., C.B., D.H., J.O.J.D., B.J.G., L.H., M.T.K, M.C.S. J.T, R.W. J.A.S., and P-S.L analysed data; D.R.H., J.R.H., C.B., D.H., J.O.J.D., B.J.G., L.H., M.T.K, M.C.S. J.T, R.W. and J.A.S. wrote the manuscript; L.A.P., A.J.H. and T.S-S. provided reagents and expertise; D.R.H., J.R.H., R.J.G, and A.J.H.S provided supervision.
Statement of financial interest:
The authors declare that they have no competing financial interests.
References
- 1.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ing-Simmons E, et al. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015;25:504–513. doi: 10.1101/gr.184986.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siersbæk R, et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014;7:1434–42. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 7.Vahedi G, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 9.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 10.Kvon EZ. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 2015;106:185–192. doi: 10.1016/j.ygeno.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Hosseini M, et al. Causes and Consequences of Chromatin Variation between Inbred Mice. PLoS Genet. 2013;9(6):e1003570. doi: 10.1371/journal.pgen.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada-Iglesias A, Baipai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalczyk MS, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2010;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes JR, et al. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci USA. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 18.Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 19.Anguita E, Sharpe JA, Sloane-Stanley JA, Tufarelli C, Higgs DR, Wood WG. Deletion of the mouse alpha-globin regulatory element (HS -26) has an unexpectedly mild phenotype. Blood. 2002;100:3450–3456. doi: 10.1182/blood-2002-05-1409. [DOI] [PubMed] [Google Scholar]
- 20.Dolken L, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 22.Davies JOJ, et al. Multiplexed analysis of chromosome conformation at vastly increased sensitivity. Nat Methods. 2016;13:74–80. doi: 10.1038/nmeth.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 26.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA. 2013;2110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev Cell. 2016;36:9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bender MA, et al. The hypersensitive sites of the murine β-globin locus control region act independently to affect nuclear localization and transcriptional elongation. Blood. 2012;119:3820–3827. doi: 10.1182/blood-2011-09-380485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villar D, et al. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGowan SJ, Hughes JR, Han ZP, Taylor S. MIG: Multi-Image Genome viewer. Bioinformatics. 2013;29:2477–2478. doi: 10.1093/bioinformatics/btt406. [DOI] [PubMed] [Google Scholar]
- 36.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. Cluster Analysis Basics and Extensions. R package version 2.0.3. 2015 [Google Scholar]
- 37.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engler C, Gruetzner R, Kandzia R, Mariollonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauka-Spengler T, Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- 40.Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase mediated DNA excisions in mice. Proc Natl Acad Sci USA. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiering S, et al. Targeted deletion of 5’HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 1995;15:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 42.Russell ES, Berstein SE. In: Biology of the Laboratory Mouse. 2nd edn. Green EL, editor. Chapter 17. Dover Publications Inc; New York: 1966. [Google Scholar]
- 43.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.