Abstract
An increased abundance of runs of homozygosity (ROH) has been associated with risk for various diseases, including schizophrenia. Here we investigate the characteristics of ROH in Palau, an Oceanic population, evaluating whether these characteristics are related to risk for psychotic disorders and the nature of this association. To accomplish these aims we evaluate a sample of 203 cases with schizophrenia and related psychotic disorders – representing almost complete ascertainment of affected individuals in the population – and contrast their ROH to that of 125 subjects chosen to function as controls.
While Palauan diagnosed with psychotic disorders tend to have slightly more ROH regions than controls, the distinguishing features are that they have longer ROH regions, greater total length of ROH, and their ROH tends to co-occur more often at the same locus. The nature of the sample allows us to investigate whether rare, highly-penetrant recessive variants generate such case-control differences in ROH. Neither rare, highly penetrant recessive variants nor individual common variants of large effect account for a substantial proportion of risk for psychosis in Palau. These results suggest a more nuanced model for risk is required to explain patterns of ROH for this population.
Keywords: Homozygosity, ROH, Schizophrenia, Psychosis, Palau
Introduction
Founder effects and population bottlenecks can distort allele frequencies, sometimes diminishing or elevating them substantially. When some of the alleles at increased frequency also affect risk for disease, the prevalence of the disease in the population can also rise. Examples of this abound in human populations, especially for Mendelian diseases of recessive inheritance, such as lysosomal storage diseases (Risch et al. 2003), cystic fibrosis (Fares et al. 2008), and glycogen storage disease (Ekstein et al. 2004). For complex diseases, the impact of founder effects and population bottlenecks is less clear regarding prevalence, but the distortion of risk allele frequencies is still an expected outcome and can be useful for gene mapping (Kallio et al. 2009).
The possibility of such population genetic effects has been a major motivation for our study of schizophrenia and psychotic disorders in Oceanic Palau. The peoples of Palau, inhabiting an archipelago of islands in the South Pacific, originated from founders from East Asia some 2,000 years ago. It is reasonable to assume the founding population was small and genetic and historical evidence suggests the population developed in relative isolation to a size of 20,000 about two centuries before present. Then, after contact with sailors from the United States and Europe during the 18th century, the population experienced a dramatic bottleneck due to the introduction of Western diseases. Since World War II and its negative impact on the Palauan population, the population has rebounded to its current size, again approximately 20,000.
None of the pedigrees that we have studied in Palau showed evidence for inbreeding. Nonetheless the limited founder population, bottlenecks, and limited population size must have the impact of elevating the degree of effective inbreeding. Indeed, in an earlier study (Devlin et al. 2001), we presented evidence for higher linkage disequilibrium and a reduction in allelic diversity in Palau relative to a European sample.
Recent studies have shown that inbreeding, through consanguineous marriage, is associated with increased risk for schizophrenia (Mansour et al. 2010; Nafissi et al. 2011). Even in the absence of frank consanguineous mating, however, long homozygous segments of chromosomes, presumably from a common distant ancestor, increase risk. The Psychiatric Genomics Consortium (Keller et al. 2012), for example, finds risk for schizophrenia rises directly with increasing fraction of the genome covered by long runs of homozygosity (ROH), namely continuous stretches of DNA sequence containing loci polymorphic at the population level but all homozygous for the individual evaluated. Recent studies of ROHs have shown their abundance in the human genome even in outbred populations, and thus they are potentially important in identifying recessive loci for complex phenotypes (Gibson et al. 2006; Li et al. 2006; Simon-Sanchez et al. 2007; McQuillan et al. 2008; Kirin et al. 2010; Wang et al. 2010).
Intriguingly risk for schizophrenia and other psychotic disorders (henceforth, psychosis or psychotic disorders) in Palau is slightly higher than that in most populations. In Palau, the rate of psychotic disorders in adults is roughly 2% whereas it is about 1% or less in most populations (Jablensky et al. 1992; Jablensky 2000; Myles-Worsley et al. 2011). We wondered if Palauan population history, which must increase effective inbreeding even in the absence of direct consanguineous mating, is directly related to the elevated risk for psychotic disorder. It is conceivable, for example, that highly-penetrant recessive alleles account for the increase in risk for Palau, something that Lencz et al. (Lencz et al. 2007) and Knight et al. (Knight et al. 2008) have explored in other population samples.
Here we investigate the nature of ROH in Palau and evaluate whether ROH is an important contributor to risk for psychotic disorders in Palau. To do so we build on our previous work, including the nearly complete ascertainment of individuals affected with psychotic disorders and their first-degree relatives in Palau (Myles-Worsley et al. 1999; Klei et al. 2005; Devlin et al. 2001; Melhem et al. 2011; Myles-Worsley et al. 2011), for whom we have pedigree information tracing beyond maternal and paternal grandparents. We thoroughly interrogate ROH regions to evaluate whether there is evidence they harbor highly-penetrant recessive alleles.
Materials and Methods
Sample and genotyping
We genotyped DNA from blood samples of all known affected subjects (n=203) and a set of control subjects (n=125). Of the 203 affected individuals, 113 were diagnosed with schizophrenia and 6 with probable schizophrenia; 33 were schizoaffective, depressed or bipolar type; 28 were diagnosed with psychosis NOS; 18 were diagnosed with bipolar disorder, 9 of whom had psychotic symptoms; and 5 had diagnoses aligned with schizophrenia or bipolar disorder (e.g., schizophreniform, schizotypal). A modified version of the Schedule for Affective Disorders and Schizophrenia-Lifetime Version (Endicott and Spitzer 1978) was administered to potentially affected individuals as identified by medical records and/or referral by a family member. An experienced clinician working with a Palauan mental health professional (JT) conducted the interviews. The SADS-L interviews were supplemented by a detailed review of psychiatric medical records. Final Best Estimate diagnoses were based on DSM-IV criteria (WB, MMW) (Spitzer et al. 1978). In this isolated population, individuals are often related to some degree. Thus, our sample of controls was a convenience sample that had no known history of psychosis, no history of psychiatric treatment, and consisted of subjects who were no closer than third-degree relatives to an affected individual and were willing to participate in the study.
Research protocols and procedures were approved by institutional review boards (IRBs) at each of the sites in the US and the Republic of Palau. All subjects provided written informed consent to participate after receiving a full explanation of the study in both English and Palauan.
DNA samples were genotyped using Affymetrix Genomewide Human SNP Array 5.0. Quality control (QC) was conducted at the individual and then SNP levels. For individuals, samples were excluded for one of three reasons: called genotypes fell below 98%; discrepant nominal and genetically-determined sex; or a large number of Mendelian errors (> 20,000). SNPs, based on genotypes called by Birdseed (Korn et al. 2008), were excluded if they were unmapped or monomorphic, had minor allele frequency (MAF) < 0.025, had a call rate below 99% or more than 10 Mendelian errors. After this step, all SNPs passed a test for Hardy-Weinberg Equilibrium and there were 267,754 SNPs for analysis.
Identifying ROH regions
We used Plink (Purcell et al. 2007) to evaluate ROH on autosomes, based on the following settings: a sliding window of 2Mb; an ROH must consist of at least 75 consecutive homozygous SNPs, with no heterozygotes allowed, but up to 2 missing genotypes were allowed; the density specified is a minimum of 1 SNP per 50 kb; the default proportion of overlapping windows that must be called homozygous to define a SNP to be in an ROH region (0.05); and the maximum allowed gap between SNPs in an ROH region was 2 Mb. The last criterion is chosen to prevent ROH spanning the centromere. (PLINK command: [--homozyg-window-kb 2000] [--homozyg-snp 75] [--homozyg-window-het 0] [--homozyg-window-missing 2] [--homozyg-density 50] [--homozyg-gap 2000])
Contrasting cases and controls on ROH
We contrasted cases and controls for the occurrence and length of ROH regions using linear mixed effects models taking into account the genetic-based relationship matrix (lmekin function in R). If some or all of risk for psychosis traces to recessive inheritance, we would expect clustering of homozygous haplotypes (ROH) in specific regions of the genome.
We next sought to determine whether ROH tended to cluster in specific regions and more often in cases than in controls. To do so we needed to account for the different number of cases and controls and the fact that SNPs were dependent within an ROH. To account for the former, we express counts of ROH in terms of fraction of cases and fraction of controls. To account for the latter, we sampled locations in the genome sparsely and implemented a resampling procedure as follows: to circumvent the dependence of SNPs, we sampled 1000 SNPs independently and without replacement; for each SNP, the fraction of cases and fraction of controls who were ROH for the SNP were calculated; taking the difference of this pair of fractions and evaluating the distribution of differences over all 1000 SNPs produced a z-statistic for the sample; finally, the experiment was performed a 1000 times to get a distribution of z-statistics and a proper test for clustering as the difference of the mean and its expectation (zero). For this experiment, we randomly included only one subject in any nuclear family, thereby eliminating concerns about dependence induced by close genetic relationships.
Searching for penetrant recessive risk alleles
A motivation for this study was to search for regions of the genome in which highly-penetrant recessive alleles reside. The simplest approach is single-SNP recessive analyses comparing cases and controls on homozygosity for each SNP. For these analyses, we used the Modified Quasi-Likelihood Score test (Thornton and McPeek 2007) (MQLS). To control for relatedness, we used a genetic-based relationship matrix instead of the pedigree information, which is a slight modification of the original MQLS method. We conducted the MQLS analyses two ways using: 1) the major allele as the recessive allele; and 2) the minor allele as the recessive allele. The genotypes for SNPs with small p-values were visually inspected for valid genotype clustering. Any SNP showing poor genotype clustering was excluded.
Although single SNP-analysis is simple, it does not capture the hallmark feature of a highly penetrant recessive locus. If such loci existed, and if they accounted for the affection status of multiple affected subjects, then we reasoned we should be able to identify such loci by evaluating haplotype sharing among affected subjects. There are various ways to identify regions where one would investigate haplotype sharing. In one approach we identified regions where there is a clustering of 5 or more consecutive SNPs with −log10(P) ≥ 2.5 from the MQLS recessive analyses that fall within larger ROH regions. We determined, for each cluster of consecutive SNPs, the SNP with the minimum p-value from the MQLS single-SNP recessive analyses. Let the minimum p-value be “minP”. Then, for each of these clusters of consecutive SNPs, we used MQLS to compare cases and controls on the frequency of the full homozygous haplotype: to perform the MQLS test for recessive inheritance, we coded a particular (homozygous) haplotype as an allele and all other haplotypes as the other allele. There could be more than one homozygous haplotype in each of these regions, thus we iterated through these haplotypes. For a region where the MQLS p-value for any haplotype was less than or equal to minP, i.e., the homozygous haplotype was more significantly associated with affection status than any of its constituent SNPs, the region was interrogated for haplotype sharing among affected subjects. To do so, we evaluated extended haplotype sharing across subjects and looked for regions in which cases were more likely to share the same stretch of the homozygous haplotype compared to controls.
There is more than one approach to investigate haplotype-sharing. Another approach that we used is to identify overlapping ROH regions from PLINK where 10 or more cases and/or controls overlapped in these regions and identified shared homozygous haplotypes within each region. We then used MQLS to compare cases and controls on homozygous haplotype sharing in these overlapping regions as determined by PLINK to control for relatedness. Similar to the above-described analyses, haplotypes were coded as alleles with the homozygous haplotype as one allele and all other haplotypes as the other allele. We also evaluated extended homozygous haplotype sharing across subjects in regions of interest.
Common variants in ROH regions
It is possible that the homozygous regions contain variants of modest additive impact on risk, which are joined by chance as a haplotype; and homozygosity of that haplotype increases the risk for schizophrenia and related psychotic disorders. To address this question, we conducted single-SNP analyses using the additive model using MQLS and assigned a score of 1 for each SNP across the genome when the p-value from the MQLS additive is lower than the minP from the recessive model (major and minor) and -1 when minP from recessive model was lower than the p-value from the additive model. Then, for each case, we computed their total SNP score across ROH regions throughout their genome and divided it by the total number of SNPs in these ROH regions, which we will refer to as “average SNP score”. We normed the average SNP score across ROH regions by subtracting the average SNP score across the whole genome (−0.39).
We also examined whether risk loci identified in the latest GWAS analysis for schizophrenia (Ripke et al. 2013) were more likely to fall in ROH regions in cases compared to controls. If one of the 27 associated SNPs reported in Ripke et al. (2013) was not genotyped, we selected the nearest SNP that was genotyped in our sample. Similar to analyses contrasting the clustering of ROH in cases versus controls described previously, for each of the 27 SNPs we determined the difference between the fraction of cases and fraction of controls who were ROH for SNPs. This resulted in a distribution for the differences, the mean of which was contrasted against the expected mean (zero) and by using the standard deviation derived from the earlier experiment to test for association.
Results
Identifying ROH segments
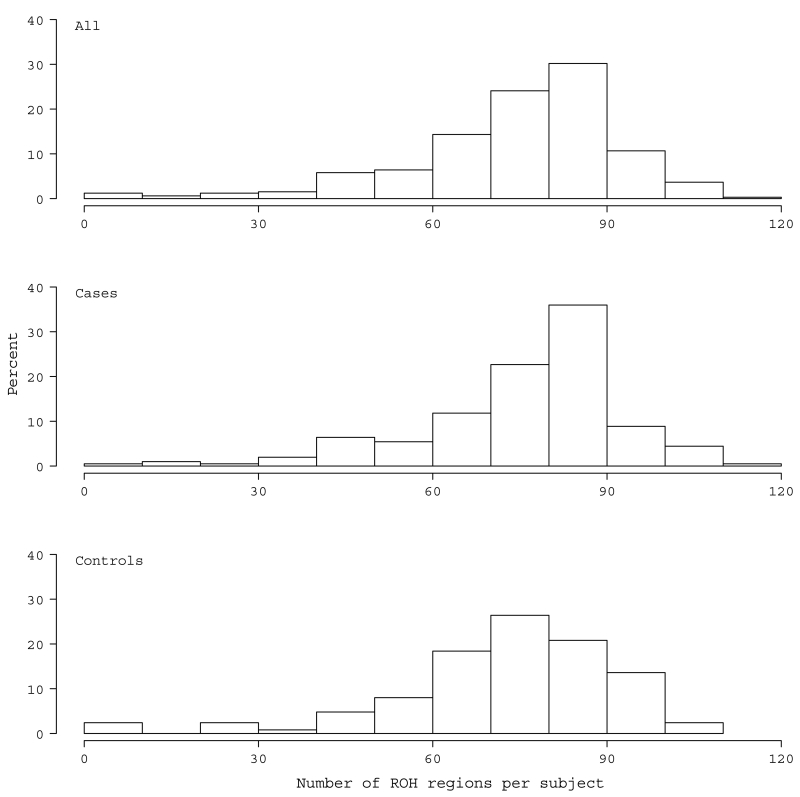
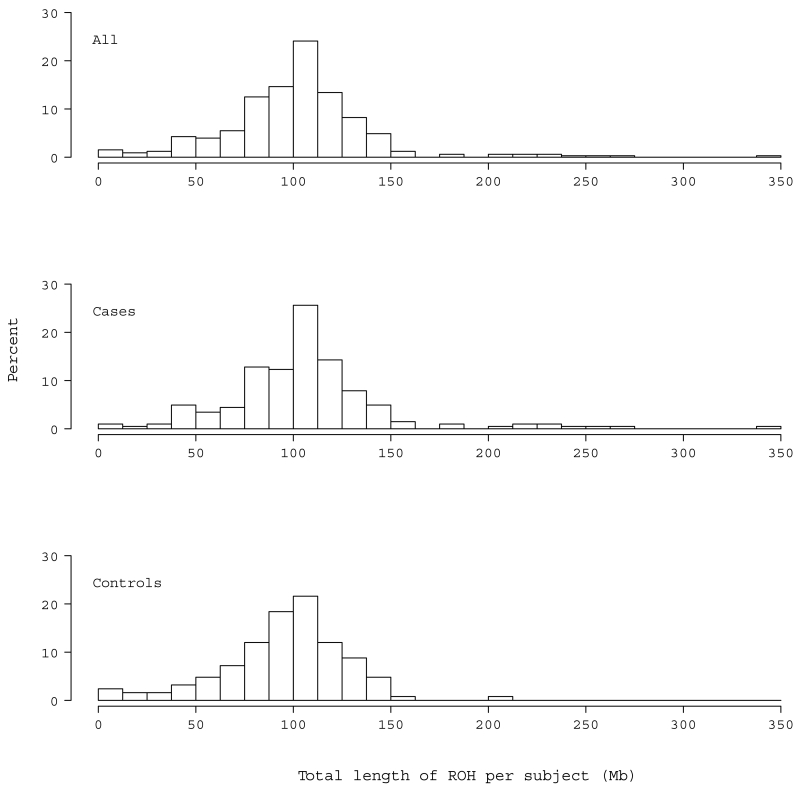
There were many regions identified as ROH in Palauan subjects, which were distributed across the genome roughly in proportion to chromosome size. The correlation between the number of ROH regions per chromosome and its size was 0.943 (p < 0.001). All subjects were found to carry at least one ROH. The average number of ROH per subject was 74.8 (Standard Deviation (SD)= 18.2, min=5, max=119) (Fig. 1) and the average length of an ROH region was 1.33 Mb (SD= 0.27, min=0.71, max=2.89 Mb). The average total length of ROH per subject was 102.1 Mb (SD= 39, min=5.2, max=343.4), representing roughly 3.6% (SD= 1.4%) of the genome (Fig. 2).
Fig. 1.
Distribution of the number of ROH regions per subject in the total sample (a) and in cases (b) and controls (c)
Fig. 2.
Distribution of the total length of ROH across the genome per subject in the total sample (a) and in cases (b) and controls (c)
Contrasting cases and controls ROH
The average number of ROH in cases, 76.1 (SD= 17.4, min=7, max=119), was larger than that in controls, 72.8 (SD= 19.3, min=5, max=106), but not significantly so (p=0.11) (Fig. 1). The average length of an ROH was stochastically longer in cases, 1.36 Mb (SD= 0.31, min=0.74, max=2.89), whereas in controls it was 1.29 Mb (SD= 0.19, min=0.71, max=2.2) (β=67.3, Standard Error (SE)=30.8, z=2.2, p=0.029). Cases were also more likely to have an increased number of large ROH regions that were greater than 2 Mb (β=0.044, SE=0.02, z=2.02, p=0.043) and 5 Mb (β=0.127, SE=0.065, z=1.96, p=0.05) compared to controls. Cases, on average, showed greater total length of ROH, 105.9 Mb (SD= 42.4), as compared to the average control, who carried a total ROH of 95.9 Mb (SD= 32.0) (β=9.96, SE=4.4, z=2.3, p=0.023) (Fig. 2). Cases also had the most extreme value for total ROH, with one case having ~343 Mb of their genome in ROH. For the latter, the longest stretch was 21.7 Mb spanning 89,952,531 to 111,628,901 on chromosome 5. For controls, the longest total ROH was 204.9 Mb with the longest stretch ~18.7Mb.
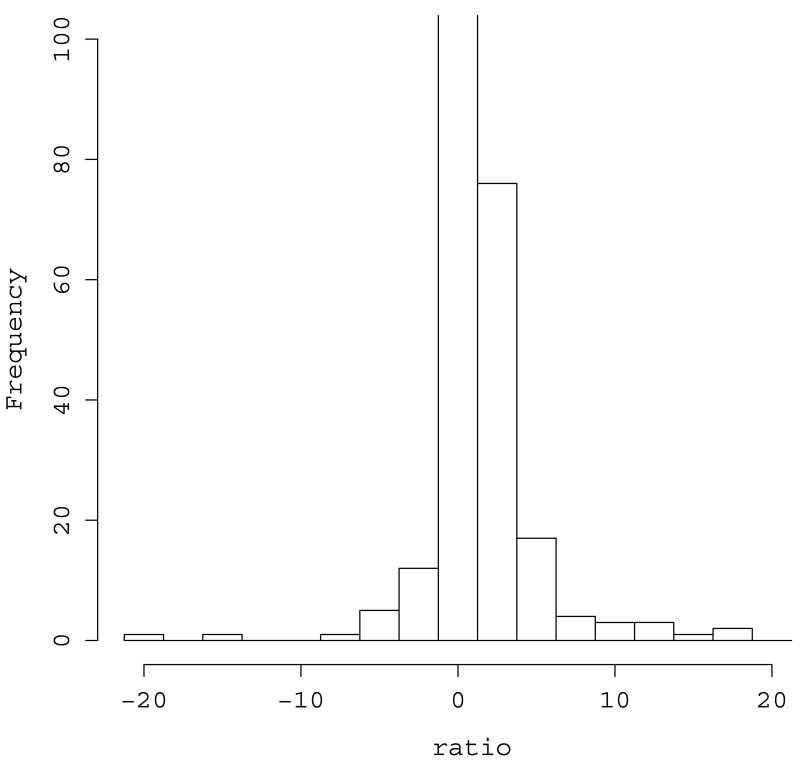
To determine whether the ROH found in cases tended to co-occur in the same region more than would be expected by chance, we randomly sampled SNPs throughout the genome and counted co-occurrence of ROH in cases and controls (see Methods). We found that ROH tended to clump in the same region for cases significantly more often than in controls (Z=3.87, p=5.47.10−5). The clumping is pronounced even after controlling for the count of cases and controls (Fig. 3) and there are some extreme clusters (Supplementary Fig. 1).
Fig. 3.
Co-occurrence of ROH regions in cases and controls: Distribution of Aij/Aji normalized ratio where Aij is the count of number of i controls and j cases for which a SNP was in an ROH region. A continuity correction was used and we obtained 1−[(Aij/Aji)/(171/121)] where 171/121 is the ratio of the number of cases to the number of controls. For values < 0, we used the inverted value, i.e., − Aji/Aij. Cells in which there were 0 cases and 0 controls received 0 as their normalized value. The y-axis is truncated at 100.
Searching for penetrant recessive risk alleles
We first conducted single-SNP analyses in which realized homozygosity of each SNP was tested for significant difference between cases and controls using MQLS and recessive mode of inheritance (Supplementary Fig. 2). There were 25 SNPs with notably small p-values (< 10−4) from the analyses where the major allele was the recessive allele and 13 SNPs from minor recessive analyses. Although none reached genomewide significance (i.e., 5 × 10−8), one SNP on chromosome 6, rs1923415, approached it with p-value=8.8.10−7 (MQLS statistic=24.2, df=1) from minor recessive analyses. The smallest p-value from the major recessive analyses was 2.5·10−6 (MQLS statistic=22.2, df=1) for rs1002074 on chromosome 10. Indeed it would require a SNP with large recessive impact on risk (see Discussion) to generate a significant signal for this population and sample.
We next examined regions where there was a cluster of 5 or more SNPs with −log10(P-value) ≥ 2.5 (Supplementary Fig. 2, Supplementary Table 1). Of these regions, only five fell within loci also meeting the criterion of containing ROH. Of these five, one showed a p-value for one of its haplotypes less than or equal to the minimum p-value of individual SNPs in the cluster, suggesting that one or more haplotypes in this region carried more information for association than individual SNPs. This region falls on chromosome 3 (chr3: 141,659,241-141,818,850) and intersects 10 out of 11 exons of TFDP2. When we inspected homozygous haplotype sharing, we found one haplotype shared by 11 cases and 2 controls (Table 1). We next examined extended haplotype sharing from each side of this haplotype, only two unrelated cases were homozygous for a 174-SNP haplotype (chr3: 140,563,185-142,819,277) and they shared the same haplotype (Table 1). Homozygous haplotypes in these regions were not accounted for by deletions from copy number variants (Melhem et al. 2011). When we evaluated regions intermediate in size between the 174 and 5 SNP regions there were a few notable sub-haplotypes. One was homozygous in four cases, including the two unrelated cases homozygous for the full haplotype, as well as one control; however, the difference between the frequency in cases versus that in controls was not significant (OR=2.49; 95% CI, 0.28-22.6; Fisher’s exact test (FET), p=0.653). Two of the cases showed a genetically-based estimated relationship consistent with second-degree relatives and both showed relationship estimates consistent with third-degree relatedness to the control. All other cases were not related to each other or to the control subject. The second sub-haplotype was homozygous in five cases and no controls (OR=3.8; 95% CI, 0.45-31.9; FET, p=0.258), including three cases who were carriers of the first sub-haplotype. Of the 10 possible pairs of these five subjects, only two cases were estimated to be related as second-degree relatives and they were the same two related cases carrying the first sub-haplotype. The third sub-haplotype was carried by five cases and one control (OR=3.1; 95% CI, 0.36-27.1; FET, p=0.413). Of these subjects there was an affected sibling pair and all others were essentially unrelated. Of note, the parents of the affected sibling pair and another affected sibling were also genotyped; however, only the unaffected father was homozygous for the same sub-haplotype. Thus, we were unable to detect a pattern of haplotype-sharing that was consistent with inheritance of a highly penetrant recessive variant at this locus.
Table 1.
Haplotype sharing analyses for the extended 105 SNP haplotype on chr3: 140,563,185-142,819,277
| ID | Diagnosis | Haplotype |
|---|---|---|
| 60041 | Probable schizophrneia |
******CTGACCCGTGACAGTTCGTTGACCTGGCGGTACTGGACGTGCACAGGATAGTACCAATAGTACAGTCCCTTGGGTGTTACCCGCCCCAGGGGT*** |
| 22445 | Psychosis NOS | ******CTGACCCGTGACAGTTCGTTGACCTGGCGGTACTGGACGTGCACAGGATAGTACCAATAGTACAGTCCCTTGGGTGTTACCCGCCCCAGGGGT*** |
| 3915 | Schizophrneia | CTGACCCGTGACAGTTCGTTGACCTGGCGGTACTGGACGTGCACAGGATAGTACCAATAGTACAGTCCCTTGGGTGTTACCCGCCCCAGGGGT |
| 22361 | Bipolar disorder with psychosis |
CTGACCCGTGACAGTTCGTTGACCTGGCGGTACTGGAC |
| 26070 | control | CTGACCCGTGACAGTTCGTTGACCTGGCGGTACTGGAC |
| 60006 | Schizophrneia | TGGACGTGCACAGGATAGTACCAATAGTACAGTCCCTTGGGTGTTACCCGCCCCAGGGGT |
| 22587 | Schizophreniform | TGGACGTGCACAGGATAGTACCAATAGTACAGTCCCTTGGGTGTTACCCGCCCCAGGGGT |
| 60026 | Probable schizophrenia |
ACAGACTTGTCAACTCTGGAC |
| 22397 | Schizophrneia | ACAGACTTGTCAACTCTGGAC |
| 22522 | Schizophrneia | ACAGACTTGTCAACTCTGGAC |
| 25012 | Schizophrneia | ACAGACTTGTCAACTCTGGAC |
| 22534 | Schizoaffective | ACAGACTTGTCAACTCTGGAC |
| 60080 | control | ACAGACTTGTCAACTCTGGAC |
Only homozygous stretches are represented; The original 5-SNP haplotype is represented in larger font, Bold, and Underline; 60041 and 22445 are homozygous for 174-SNP haplotype; Three sub-haplotypes are represented; 22397 and 22522 are siblings.
We also sought evidence for highly-penetrant recessive alleles by comparing counts of locus-specific ROH for cases versus controls. For this comparison we used the MQLS to account for the relatedness of subjects (Table 2). None of the results conclusively document a recessive risk locus, but a region on chromosome 10 (86,893,742-87,102,314) showed the most promising evidence for recessive inheritance, with seven cases and one control carrying the same homozygous haplotype. When the regions given in Table 2 were extended by adding 50 SNPs on each side, only one of the seven cases was homozygous for the extended haplotype (Chromosome 10: 86,561,322-87,553,409 bp) and no subject was homozygous for the extended haplotypes on chromosomes 2 (99,496,928-101,464,524) and 12 (59,298,290-62,286,036). For the chromosome 6 region, the extended haplotype (chr6: 28,033,174-29,675,676) resulted in three haplotypes that were homozygous in several subjects; however, none was associated with risk for psychosis.
Table 2.
Regions with p<0.01 from homozygous haplotype sharing analysis in overlapping ROH regions identified by PLINK
| Chr | Start | End | Number of SNPs |
MQLS P-value for homozygous haplotype |
Gene(s) or closest gene(s) |
|---|---|---|---|---|---|
| 10 | 86,900,441 | 87,102,314 | 37 | 7.8·10−5 | GRID1 |
| 6 | 28,512,017 | 29,225,795 | 38 | 8.7·10−3 |
ZNF311, TRIM27,
SCAND3, LOC401242, OR2W1, OR2J3, OR2B3, LOC100129636, OR2J2 |
| 12 | 59,917,023 | 61,612,551 | 114 | 9.6·10−3 | SLC16A7 |
| 2 | 100,246,927 | 101,139,604 | 108 | 9.9·10−3 |
AFF3, BC105019,
LONRF2, CHST10, NMS |
Common variants in ROH regions
Supplementary Figure 3 presents the distribution of the average SNP scores in ROH regions in cases, which shows more SNPs acting additively than recessively in these ROH regions (Shapiro-Wilk test p<0.001). We also examined ROH regions where there were cases and no controls overlapping and found 31.7% of SNPs in these regions (271 SNPs) to act additively rather than recessively, despite the obvious bias due to selection of ROH pile-up in the region. Finally, SNPs identified as risk loci for schizophrenia from the most recent GWAS analysis (Ripke et al. 2013) (or their closest SNP) showed a higher fraction of cases than controls to have these SNPs fall in ROH regions, which did not reach statistical significance (z=0.81, p=0.20).
Discussion
The nature of homozygous segments in genomes from a population reflects its genetic history. Here we describe features of homozygous segments found in the Palauan population. Consistent with other Oceanic populations, we find extensive regions of homozygosity from the Palauan population and, on average, 3.5% of the genome is covered by runs of homozygosity (ROH). For populations of European ancestry, ROH were estimated in one study to cover roughly 2.6% of the genome (McQuillan et al. 2008), whereas the Human Genome Diversity Project (HGDP) reports Oceanic populations have almost three times more of their genome in ROH, compared to European and African populations (Kirin et al. 2010). Populations with high prevalence of consanguineous marriages have even larger coverage of the genome by ROH (Hunter-Zinck et al. 2010). It is important to note that differences in the definition of ROH used in these studies and in genotyping platforms limit the comparison of results from these studies. However, results in Palau are consistent with its history of limited mating amongst relatives and with multiple population bottlenecks.
The rate of schizophrenia and psychotic disorders in Palau is roughly twofold higher than the worldwide average rate (2% versus 1%). In the prequel we asked if the elevated ROH resulting from Palauan population history could be an important contributor to risk for psychotic disorders in Palau. We do find that cases tend to have more ROH than controls, but the significant difference between the two groups is that ROH segments are significantly longer in cases, a greater fraction of their genomes is covered by ROH, and their ROH tend to cluster more in the same locus. Therefore some form of recessive inheritance could play a role in risk for psychotic disorders in Palau.
Interpreting the impact of increased ROH on risk depends on population genetic structure, which for Palauans is quite different from that of most populations of European ancestry. For example, Keller et al. (2012) find that the risk of schizophrenia, as measured by the odds, increase by 17% for every 1% increase in genome-wide ROH in subjects of European ancestry. In those subjects roughly 0.15% of the genome exists in long ROH – technically long stretches of autozygosity – translating to about 4.5 Mb of DNA in ROH out of a 3,000 Mb genome. For Palauan controls, we find 95.9 Mb of the genome involves ROH, or 3.1%, which is twentyfold higher than subjects of European ancestry, although here we ignore the difference between long ROH and autozygosity. For context, if ROH were equivalent to autozygosity, a European offspring of parents who are fourth-degree relatives would be expected to have 3.1% of their genome ROH. In subjects diagnosed with psychotic disorders in Palau, there is a 10.4% increase in total ROH (105.9 Mb) relative to Palauan control subjects. In other words, there is a 1.1 fold increase in ROH; within the context of first cousin marriage of randomly-chosen Palauans, the 10 Mb increase in ROH is only 5.1% of that expected due to such a mating. For subjects of European ancestry, Keller et al. (2012) estimate the risk of schizophrenia is increased 2.74 fold for first cousin marriages; such a mating results in expected ROH that is fortyfold greater than that for an offspring of random mating. From our results, and using the approximations described above, first-cousin marriage in Palau would result in roughly 3.8 fold increased risk. Still, in terms of ROH and autozygosity, most risk for schizophrenia for Europeans traces to subtle inbreeding and it appears to be the case for Palauans too.
When we searched regions in which more cases than controls shared homozygous segments and conducted haplotype sharing analyses in these regions using techniques from fine-mapping of recessive Mendelian loci, however, we could not identify any locus with strong evidence that it likely harbors a recessive, highly penetrant risk variant. Given the size of the population of Palau, power to detect minor deviations from statistical expectation in a sample of subjects with psychosis is small. For this reason we cannot eliminate the possibility that very rare variants affect risk in a recessive manner, such as those identified by a recent investigation of the genetic architecture of autism (Lim et al. 2013; Yu et al. 2013; Gamsiz et al. 2013). Nonetheless, if a variant or haplotype were to occur as a homozygote in 12 or more cases and no controls, mimicking a rare highly penetrant variant found in less than 6% of cases, we would expect a notable result even by an exact test (exact test p-value < 0.005). Keeping in mind the twofold-increased rate of psychotic disorders in Palau (i.e., 100 more cases), the complete ascertainment of cases, and the pedigrees and inter-relatedness amongst cases, this scenario is not extreme. Yet nothing so extreme is observed for a specific haplotype involved in ROH regions.
We also examined common recessive variants and found several SNPs with small p-values (< 104) with one SNP approaching genomewide significance. For a common variant to achieve genomewide significance in this sample would require a substantial effect on risk under a recessive model; still, treating cases and controls as independent and for a recessive risk allele frequency of 0.5, 0.4 or 0.2, a genotype relative risk on the order of 3.5, 4 or 8 would achieve good power in this sample (approximately 80% power for a significance level of 5 × 10−8 (Purcell et al. 2003). No such signals were observed, consistent with our expectations and the expected small effect sizes of common variants that confer risk to schizophrenia.
Thus the nature of the population and sample allow us to exclude two possible mechanisms of recessive risk, namely highly penetrant recessive variants accounting for more than a few cases or common variants of large effect on risk. Previous studies have reported increased risk of schizophrenia with inbreeding from consanguineous marriages (Mansour et al. 2010; Nafissi et al. 2011; Keller et al. 2012) and with increase in ROH in outbred populations (Keller et al. 2012). Our findings are consistent with these studies in many features. Several mechanisms for the association between homozygous segments and risk for psychotic disorders in Palau seem plausible. Our results are compatible with the possibility of partially recessive risk variants. It is also possible that homozygous regions contain variants of modest additive impact on risk. We assess this possibility and find some evidence that homozygous regions in cases contain variants with additive impact on risk, which are joined by chance as a haplotype; and homozygosity of that haplotype increases the risk. Finally, it is possible that increased homozygosity is in general associated with reduced vigor and fitness, called inbreeding depression (Charlesworth and Willis 2009), which is thought to accrue by modest effects at many loci. Inbreeding depression itself could elevate risk for psychotic disorders. In addition to this classical genetic effect, a recent study shows evidence for the involvement of epigenetic processes in inbreeding depression (Vergeer et al. 2012), which would not be detected by classical genetic analysis.
While we find no evidence for penetrant recessive risk variants in Palau, examples of successful mapping of recessive risk loci do exist, even in the context of complex psychiatric phenotypes (Gamsiz et al. 2013; Yu et al. 2013; Lim et al. 2013). For example, Yu et al. (Yu et al. 2013) recently identified three genes affecting risk for autism by homozygosity mapping. Complementing these findings, Lim et al. (Lim et al. 2013) estimate that 3% of risk for autism traces to complete knockout of autosomal genes. Insofar as we are aware, no such estimate is available for schizophrenia or psychotic disorders more generally, but it would not be surprising to find that a similar portion of risk for psychosis traces to recessive inheritance.
If it were the case that about 3% of risk for psychotic disorders traced to recessive variants, on average, this quantity would vary from population to population. Given our results, it is possible that no such variants exist in the Palauan population. The history of Palau, with presumably a small founder population teamed with repeated population bottlenecks, would have led to stochastic loss of many allelic variants. Natural selection could also play a role in eliminating risk variants, although its impact on recessive risk variants is much less than on dominant variants. In contrast to recessive variants, we have successfully mapped dominant risk variants in Palau, both by CNV(Myles-Worsley et al. 2013) and by identity-by-decent (unpublished data) analyses. Whatever the explanation for our ROH results, they shed light on the complexity of homozygosity mapping for gene discovery in complex diseases.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institute of Health MH080375, MH080373, MH080299, and MH077930. We are grateful to the people of Palau for their participation in this study.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10(11):783–796. doi: 10.1038/nrg2664. doi:10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Otto C, Tiobech S, Byerley W. Genome-wide distribution of linkage disequilibrium in the population of Palau and its implications for gene flow in Remote Oceania. Hum Genet. 2001;108(6):521–528. doi: 10.1007/s004390100511. [DOI] [PubMed] [Google Scholar]
- Ekstein J, Rubin BY, Anderson SL, Weinstein DA, Bach G, Abeliovich D, Webb M, Risch N. Mutation frequencies for glycogen storage disease Ia in the Ashkenazi Jewish population. Am J Med Genet A. 2004;129A(2):162–164. doi: 10.1002/ajmg.a.30232. doi:10.1002/ajmg.a.30232. [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35(7):837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- Fares F, Badarneh K, Abosaleh M, Harari-Shaham A, Diukman R, David M. Carrier frequency of autosomal-recessive disorders in the Ashkenazi Jewish population: should the rationale for mutation choice for screening be reevaluated? Prenat Diagn. 2008;28(3):236–241. doi: 10.1002/pd.1943. doi:10.1002/pd.1943. [DOI] [PubMed] [Google Scholar]
- Gamsiz ED, Viscidi EW, Frederick AM, Nagpal S, Sanders SJ, Murtha MT, Schmidt M, Triche EW, Geschwind DH, State MW, Istrail S, Cook EH, Devlin B, Morrow EM. Intellectual disability is associated with increased runs-of-homozygosity in simplex autism. Am J Hum Genet. 2013 doi: 10.1016/j.ajhg.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15(5):789–795. doi: 10.1093/hmg/ddi493. doi:10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Hunter-Zinck H, Musharoff S, Salit J, Al-Ali KA, Chouchane L, Gohar A, Matthews R, Butler MW, Fuller J, Hackett NR, Crystal RG, Clark AG. Population genetic structure of the people of Qatar. Am J Hum Genet. 2010;87(1):17–25. doi: 10.1016/j.ajhg.2010.05.018. doi:10.1016/j.ajhg.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablensky A. Epidemiology of schizophrenia: the global burden of disease and disability. Eur Arch Psychiatry Clin Neurosci. 2000;250(6):274–285. doi: 10.1007/s004060070002. [DOI] [PubMed] [Google Scholar]
- Jablensky A, Sartorius N, Ernberg G, Anker M, Korten A, Cooper JE, Day R, Bertelsen A. Schizophrenia: manifestations, incidence and course in different cultures. A World Health Organization ten-country study. Psychol Med Monogr Suppl. 1992;20:1–97. doi: 10.1017/s0264180100000904. [DOI] [PubMed] [Google Scholar]
- Kallio SP, Jakkula E, Purcell S, Suvela M, Koivisto K, Tienari PJ, Elovaara I, Pirttila T, Reunanen M, Bronnikov D, Viander M, Meri S, Hillert J, Lundmark F, Harbo HF, Lorentzen AR, De Jager PL, Daly MJ, Hafler DA, Palotie A, Peltonen L, Saarela J. Use of a genetic isolate to identify rare disease variants: C7 on 5p associated with MS. Hum Mol Genet. 2009;18(9):1670–1683. doi: 10.1093/hmg/ddp073. doi:10.1093/hmg/ddp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC, Simonson MA, Ripke S, Neale BM, Gejman PV, Howrigan DP, Lee SH, Lencz T, Levinson DF, Sullivan PF. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 2012;8(4):e1002656. doi: 10.1371/journal.pgen.1002656. doi:10.1371/journal.pgen.1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM, Wilson JF. Genomic runs of homozygosity record population history and consanguinity. PLoS One. 2010;5(11):e13996. doi: 10.1371/journal.pone.0013996. doi:10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klei L, Bacanu SA, Myles-Worsley M, Galke B, Xie W, Tiobech J, Otto C, Roeder K, Devlin B, Byerley W. Linkage analysis of a completely ascertained sample of familial schizophrenics and bipolars from Palau, Micronesia. Hum Genet. 2005;117(4):349–356. doi: 10.1007/s00439-005-1320-1. doi:10.1007/s00439-005-1320-1. [DOI] [PubMed] [Google Scholar]
- Knight HM, Maclean A, Irfan M, Naeem F, Cass S, Pickard BS, Muir WJ, Blackwood DH, Ayub M. Homozygosity mapping in a family presenting with schizophrenia, epilepsy and hearing impairment. Eur J Hum Genet. 2008;16(6):750–758. doi: 10.1038/ejhg.2008.11. doi:10.1038/ejhg.2008.11. [DOI] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40(10):1253–1260. doi: 10.1038/ng.237. doi:10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, Kucherlapati R, Malhotra AK. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A. 2007;104(50):19942–19947. doi: 10.1073/pnas.0710021104. doi:10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LH, Ho SF, Chen CH, Wei CY, Wong WC, Li LY, Hung SI, Chung WH, Pan WH, Lee MT, Tsai FJ, Chang CF, Wu JY, Chen YT. Long contiguous stretches of homozygosity in the human genome. Hum Mutat. 2006;27(11):1115–1121. doi: 10.1002/humu.20399. doi:10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, Macarthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M, Lek M, Liu L, Flannick J, Ripke S, Nagaswamy U, Muzny D, Reid JG, Hawes A, Newsham I, Wu Y, Lewis L, Dinh H, Gross S, Wang LS, Lin CF, Valladares O, Gabriel SB, Depristo M, Altshuler DM, Purcell SM, State MW, Boerwinkle E, Buxbaum JD, Cook EH, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Daly MJ. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77(2):235–242. doi: 10.1016/j.neuron.2012.12.029. doi:10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour H, Fathi W, Klei L, Wood J, Chowdari K, Watson A, Eissa A, Elassy M, Ali I, Salah H, Yassin A, Tobar S, El-Boraie H, Gaafar H, Ibrahim NE, Kandil K, El-Bahaei W, El-Boraie O, Alatrouny M, El-Chennawi F, Devlin B, Nimgaonkar VL. Consanguinity and increased risk for schizophrenia in Egypt. Schizophr Res. 2010;120(1-3):108–112. doi: 10.1016/j.schres.2010.03.026. doi:10.1016/j.schres.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, Macleod AK, Farrington SM, Rudan P, Hayward C, Vitart V, Rudan I, Wild SH, Dunlop MG, Wright AF, Campbell H, Wilson JF. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83(3):359–372. doi: 10.1016/j.ajhg.2008.08.007. doi:10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhem N, Middleton F, McFadden K, Klei L, Faraone SV, Vinogradov S, Tiobech J, Yano V, Kuartei S, Roeder K, Byerley W, Devlin B, Myles-Worsley M. Copy number variants for schizophrenia and related psychotic disorders in Oceanic Palau: risk and transmission in extended pedigrees. Biol Psychiatry. 2011;70(12):1115–1121. doi: 10.1016/j.biopsych.2011.08.009. doi:10.1016/j.biopsych.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles-Worsley M, Coon H, Tiobech J, Collier J, Dale P, Wender P, Reimherr F, Polloi A, Byerley W. Genetic epidemiological study of schizophrenia in Palau, Micronesia: prevalence and familiality. Am J Med Genet. 1999;88(1):4–10. doi: 10.1002/(sici)1096-8628(19990205)88:1<4::aid-ajmg2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Myles-Worsley M, Tiobech J, Blailes F, Middleton FA, Vinogradov S, Byerley W, Faraone SV. Familial transmission of schizophrenia in Palau: A 20-year genetic epidemiological study in three generations. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(3):247–254. doi: 10.1002/ajmg.b.31171. doi:10.1002/ajmg.b.31171. [DOI] [PubMed] [Google Scholar]
- Myles-Worsley M, Tiobech J, Browning SR, Korn J, Goodman S, Gentile K, Melhem N, Byerley W, Faraone SV, Middleton FA. Deletion at the SLC1A1 glutamate transporter gene co-segregates with schizophrenia and bipolar schizoaffective disorder in a 5-generation family. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(2):87–95. doi: 10.1002/ajmg.b.32125. doi:10.1002/ajmg.b.32125. [DOI] [PubMed] [Google Scholar]
- Nafissi S, Ansari-Lari M, Saadat M. Parental consanguineous marriages and age at onset of schizophrenia. Schizophr Res. 2011;126(1-3):298–299. doi: 10.1016/j.schres.2010.11.029. doi:10.1016/j.schres.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Holmans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O’Neill FA, Owen MJ, Milovancevic MP, Posthuma D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Levinson DF, Gejman PV, Laurent C, Mowry BJ, O’Donovan MC, Pulver AE, Schwab SG, Wildenauer DB, Dudbridge F, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Van Os J, Walshe M, Weisbrod M, Wiersma D, Donnelly P, Barroso I, Blackwell JM, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, McCarthy MI, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45(10):1150–1159. doi: 10.1038/ng.2742. doi:10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Tang H, Katzenstein H, Ekstein J. Geographic distribution of disease mutations in the Ashkenazi Jewish population supports genetic drift over selection. Am J Hum Genet. 2003;72(4):812–822. doi: 10.1086/373882. doi:10.1086/373882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, Crawley A, Keen JC, Nash J, Borgaonkar D, Hardy J, Singleton A. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16(1):1–14. doi: 10.1093/hmg/ddl436. doi:10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Endicott J, Robins E. Research diagnostic criteria: rationale and reliability. Arch Gen Psychiatry. 1978;35(6):773–782. doi: 10.1001/archpsyc.1978.01770300115013. [DOI] [PubMed] [Google Scholar]
- Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81(2):321–337. doi: 10.1086/519497. doi:10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergeer P, Wagemaker NC, Ouborg NJ. Evidence for an epigenetic role in inbreeding depression. Biol Lett. 2012;8(5):798–801. doi: 10.1098/rsbl.2012.0494. doi:10.1098/rsbl.2012.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LS, Hranilovic D, Wang K, Lindquist IE, Yurcaba L, Petkovic ZB, Gidaya N, Jernej B, Hakonarson H, Bucan M. Population-based study of genetic variation in individuals with autism spectrum disorders from Croatia. BMC Med Genet. 2010;11:134. doi: 10.1186/1471-2350-11-134. doi:10.1186/1471-2350-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DA, Adli M, Malik AN, D’Gama AM, Lim ET, Sanders SJ, Mochida GH, Partlow JN, Sunu CM, Felie JM, Rodriguez J, Nasir RH, Ware J, Joseph RM, Hill RS, Kwan BY, Al-Saffar M, Mukaddes NM, Hashmi A, Balkhy S, Gascon GG, Hisama FM, Leclair E, Poduri A, Oner O, Al-Saad S, Al-Awadi SA, Bastaki L, Ben-Omran T, Teebi AS, Al-Gazali L, Eapen V, Stevens CR, Rappaport L, Gabriel SB, Markianos K, State MW, Greenberg ME, Taniguchi H, Braverman NE, Morrow EM, Walsh CA. Using Whole-Exome Sequencing to Identify Inherited Causes of Autism. Neuron. 2013;77(2):259–273. doi: 10.1016/j.neuron.2012.11.002. doi:10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.