ABSTRACT
Recognition and response to prospective competitors are crucial variables that must be considered in resource distribution and utilization in plant communities. Associated behaviors are largely mediated through the exchange of low-molecular weight exudates. These cues can significantly alter the root system architecture (RSA) between neighboring plants and are routinely sensitive enough to distinguish between plants of the same or different accessions, a phenomenon known as kin recognition (KR). Such refined discrimination of identity, based on the composition and detection of patterns of exudate signals is remarkable and provides insight into the chemical ecology of plant-plant interactions. The discovery that KR occurs in Arabidopsis thaliana provides a model system to resolve many of the mechanistic questions associated with this process. We hypothesized that the low-molecular weight cues which direct changes to the RSA during KR was driven by nutrient availability. Here we present evidence in support of a nutrient-inducible model for KR. Our findings underscore how exudate production and detection are influenced by nutrient availability as well as how this information is integrated into ‘decisions’ about competition and root system architecture which may have broader impacts on community composition.
KEYWORDS: Arabidopsis thaliana, kin recognition, nutrient availability, plant growth and development, plant-plant interactions, resource utilization, root exudates, root system architecture, sociobiology
Abbreviations
- A. thaliana
Arabidopsis thaliana
- DMSO
Dimethylsulfoxide
- MES
2-(N-morpholino)ethanesulphonic acid
- MS
Murashige & Skoog Media
- RSA
Root System Architecture
The evolution, maintenance, and benefits of social interactions are the subject of considerable study in insects, birds, and other animals.1-4 Many of the ‘social’ behaviors displayed by these organisms are based on discriminating between related and unrelated conspecifics (same species) to limit both resource competition as well as inbreeding.5-7 Through careful observation the existence of analogous social behaviors in plants has been confirmed. In particular, the ability of plants to distinguish between members of the same or different accessions, the phenomenon of kin recognition (KR), has been well characterized.8-16
Plants are generally thought to be incapable of processing complex visual or auditory cues, such as plumage or song, suggesting these sensory input channels are unavailable for KR. However cryptochrome-mediated KR has been observed in model organisms such as A. thaliana.13 In this study, accession identity impacted leaf-orientation and shade responses; ultimately, impacting plant growth and resource allocation.
While photo-mediated KR is intriguing, low-molecular weight root exudates diffusing through soil have been more routinely implicated as regulatory elements of this phenomenon (Fig. 1).10,16-18 In Arabidopsis thaliana and many other plants KR manifests by a significant reduction in the number of observed lateral roots when samples are grown with plants of the same accession (Kin) versus different ones (Stranger).11-15 Sodium orthovanadate (Na3VO4), a generic ABC-transport inhibitor, which blocks the release and/or uptake of root exudates, eliminated the increased lateral root number observed during KR studies with different accessions of A. thaliana.19,20 These findings underscore the importance of such low-molecular weight secondary metabolites in mediating KR (Fig. 1). Detection of these signals is akin to the senses of smell and taste, also based on low-molecular weight molecules.
Figure 1.
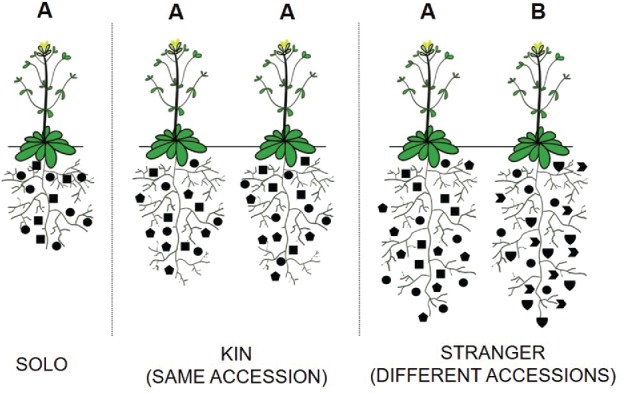
Model of exudate-mediated conspecific recognition. The root exudates (depicted by the symbols) produced by plants of 2 different accessions (A or B) regulate their interactions with other biotic elements in the soil community. When previously isolated plants (Solo) detect members of the same accession (Accession recognition, ER) exudate profiles adjust slightly, and triggers changes in the root system architecture. During encounters between 2 different accessions (A&B) recognition is limited to conspecifics. Common elements of the exudate profile are retained between but sufficient variations in exudate composition (concentration and/or identity) are sufficient to trigger more substantial changes in the RSA between the prospective competitors.
While the effects on the root system architecture (RSA), the pattern of root growth in plants, is the most commonly observed KR phenotype other noteworthy changes also occur. For example, differential gene expression in response to the presence of Kin or Stranger plants has previously been observed in A. thaliana.16 In particular, genes associated with pathogen resistance and secondary metabolite production were influenced by this phenomenon. KR can also impact the formation of symbiotic associations as observed in studies with Ambrosia artemisiifolia which establish more robust networks with sibling plants than strangers.21 Given its effects on RSA, symbioses, and gene expression KR may play a crucial role in the formation and maintenance of plant communities.
In addition to neighbor identity, as determined by KR and other cues, nutrient availability plays a critical role in establishing the RSA in many plants.22-24 Nitrogen and phosphorus deficiencies, for example, are well established modulators of lateral root development.24,25 Nutrient availability also alters exudate production and composition with the potential to impact both interorganismal interactions as well as nutrient uptake.26-29 For example, in Sorghum bicolor reduced nitrogen and phosphorus levels promote the release of 5-deoxystrigol which encourages the recruitment of mycorrhizal fungi.26
Clearly, plants must integrate information regarding both neighbor identity as well as nutrient availability if they are to successfully compete in the environment. Work by Cahill et al. with Abutilon theophrasti (Velvetleaf) suggested that when grown in isolation these plants preferred to adopt a broad rooting strategy regardless of nutrient avaialbility.30 However, the presence of a neighboring kin plant resulted in a more restricted architecture, suggesting an effort to reduce competitive interactions between the 2 plants. Furthermore, changes in the RSA architecture could be ‘tuned’ by the placement of nutrients. These findings suggest that nutrient sensitivity and its subsequent impact on the RSA is largely driven by the presence/absence of prospective competitors.
In contrast to A. theophrasti, the RSA of A. thaliana as well as exudate production appears to be very sensitive to nutrient availability, in particular nitrogen and phosphorus, whether grown in isolation or in groups.22-29 Based on these prior observations we hypothesized that A. thaliana may utilize information regarding nutrient availability to influence decisions regarding competition based on neighbor identity. In other words, nutrient availability may influence the production of specific exudates associated with KR which could alter the RSA accordingly.
Assuming nutrient availability regulates the production of exudates associated with KR then these phenomena should be ‘tunable’ by altering nutrient concentration and observable by the subsequent changes to the RSA. As lateral root number has already been established as an indicator for both in A. thaliana we monitored this phenotype in the present study.12 Specifically, we evaluated the effects of decreasing nutrient availability, using Murashigie-Skoog (MS) media, on lateral root number in samples for KIN (same accession) or STRANGER (different accession) recognition. Using a user-defined nutrient media permits us to expand on the nutrient studies by Cahill et al. and others by varying the availability of specific nutrients to determine their effects on KR. Columbia-0 (Col-0) and Landsberg erecta (LA-1), 2 of the most common laboratory accessions of A. thaliana, were utilized in the present study. MS concentrations ranged from 1x to 0.1xMS in our initial study (Fig. 2A & B).
Figure 2.
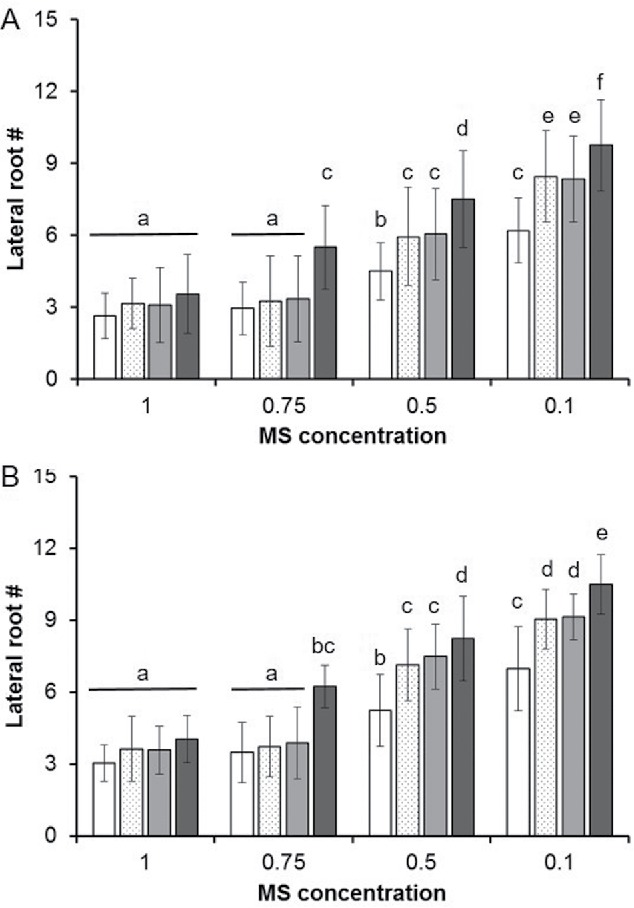
Inducing CR and ER in Arabidopsis thaliana. Seedlings of the A. thaliana (A) Col-0 and (B) LA-1 accessions were grown on plates of the indicated concentration of MS. Lateral root number for all samples (n = 50) were scored after 18 d of growth (See Methods). White Bars: SOLO (individual seedlings), Matted dots: SIBLINGS (2 seedlings from the same seed pod), Light Gray Bars: Accession-resolution (ER, 2 seedlings of the same accession), and Dark gray: Conspecific-resolution (CR, 2 seedlings of different accessions). Results are expressed as the mean of the lateral root number +/− standard deviation. Different letters indicate statistically significant differences samples based on Tukey's post-hoc test (p-value < 0.05).
Lateral root number was inversely correlated with nutrient concentration in all samples but the magnitude of this effect varied. KIN samples displayed increased lateral root number compared to SOLO samples (single plant) at concentrations ≤0.5xMS (Fig. 2A & B). This is in sharp contrast to STRANGER samples (Col-0 and LA-1) which were significantly different from SOLO controls even at 1xMS (Fig. 2A & B). Lateral root numbers in STRANGER populations were also significantly greater than those in KIN populations at concentrations ≤0.75xMS in these studies. These trends were consistent for both the Col-0 (Fig. 2A) and LA-1 (Fig. 2B) accessions. Similar trends were observed for KR studies between the Col-0 and Bensheim-0 (Be-0) accessions (Data not shown).
In plants like Cakile edentula (American searocket) neighbor recognition can even distinguish between ‘kin’ and ‘sibling’ plants, the latter derived from the same parent(s).15 In these samples, lateral root development was reduced in sibling plants relative to Kin controls. The limits of A. thaliana to distinguish siblings has yet to be established and is important for determining the ultimate utility of this model system for understanding such recognition events. In order to determine this recognition boundary, Seedlings of either Col-0 or LA-1 derived from the same self-fertilized seed pod (SIBLINGS) were also evaluated in our nutrient reduction growth assays. At all concentrations, SIBLINGS (Col-0/Col-0 or LA-1/LA-1) were indistinguishable from their corresponding KIN samples (Fig. 2A & B). This suggests that unlike C. edentula and several other plants, the sensitivity of neighbor recognition in A. thaliana may be limited to accession-level resolution. Further studies to resolve ‘sibling’ effects in A. thaliana are currently underway but these results may underscore a limit to the viability of A. thaliana as a model system for the study of the KR phenomenon.
Having established conditions for ‘tuning’ recognition, specific nutrients were evaluated for their ability to affect KR. SOLO, KIN, and STRANGER samples were grown on plates containing 0.10xMS media supplemented with an excess of the basal salts for either: Nitrogen (N), Phosphorus (P), Sulfur (S), Iron (Fe), Calcium (Ca), or Potassium (K) equivalent to 1xMS for that nutrient (Fig. 3). Only N or P treated samples were different from 0.1xMS controls. Samples supplemented with nitrogen (N) or phosphorus (P) showed significant reductions in the total lateral root number in both STRANGER and KIN samples consistent with 1xMS controls. KIN and STRANGER samples supplemented with both N+P were statistically indistinguishable from one another (Fig. 3). Modified 1xMS in which N and P were both reduced to 0.10xMS concentrations, showed increased lateral root allocation in KIN and STRANGER samples (Fig. 3).
Figure 3.
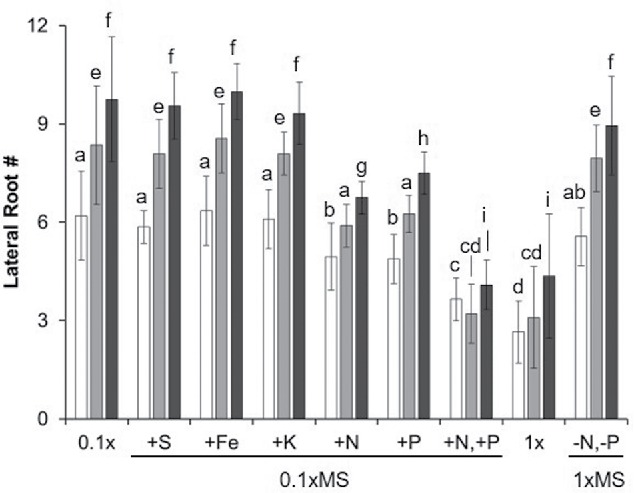
Supplementing nutrients limits CR and ER. Seedlings of A. thaliana Col-0 and LA-1 are grown on plates containing either: (i) 0.10xMS with the indicated nutrient added (+) using MS basal salts to yield 1XMS or (ii) 1xMS with nitrogen and phosphorus content reduced to 0.1xMS equivalent (-N,-P). Lateral root number was scored after 18 d and the results expressed as the mean +/− one standard deviation (n = 30). White SOLO, Light Gray: Accession-resolution (ER, 2 seedlings of the same accession), and Dark gray: Conspecific-resolution (CR, 2 seedlings of different accessions). Different letters indicate statistically significant differences samples based on Tukey's post-hoc test (p-value < 0.05).
Finally we sought to determine if nutrient availability altered exudate composition and/or sensitivity thereby inducing the observed phenotypes (Fig. 4). Exudates were extracted from seedlings of LA-1 grown at ‘nutrient limiting’ (0.1xMS, white bars) or ‘nutrient rich’ (1xMS, gray bars) conditions and applied by agar overlays to individual seedlings (Fig. 4). LA-1 exudates from nutrient-limited plants significantly increased lateral root number in Col-0 seedlings grown under the same conditions, but had little effect on Col-0 seedlings grown under nutrient rich conditions (Fig. 4). Exudates from LA-1 seedlings grown under nutrient rich conditions failed to significantly increase lateral root number in Col-0 seedlings under either nutrient limiting or nutrient rich conditions (Fig. 4).
Figure 4.
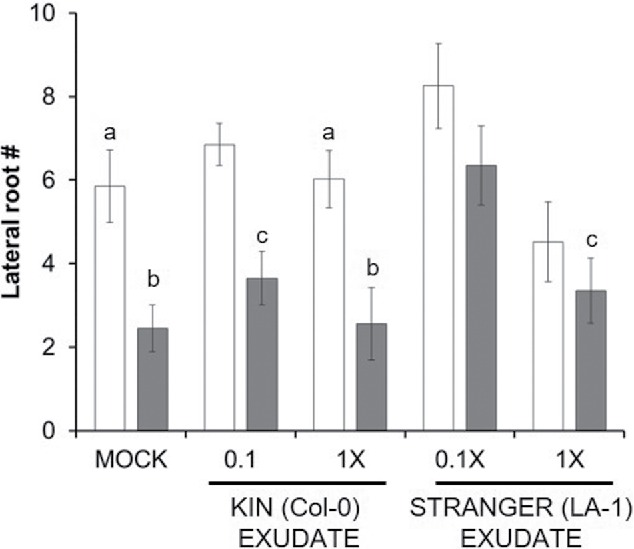
Low nutrient conditions promote recognition. Seven day-old Col-0 seedlings grown at 0.10x (white) or 1x (gray) MS were treated with exudates from Col-0 (KIN) or LA-1 (STRANGER) plants also grown at 0.10x or 1xMS (indicated on x-axis). MOCK seedlings were treated with the same agar overlay without the exudate. Seedlings were scored for lateral root number after an additional 11 d of growth. Results are expressed as the mean lateral root number of each sample (n = 30) +/− one standard deviation. Different letters indicate statistically significant differences samples based on Tukey's post-hoc test (p-value < 0.05).
In order to determine if the observed trends in exudate production and sensitivity were consistent with KIN samples these experiments were repeated with Col-0 seedlings as the source of the exudates. Col-0 exudates derived from nutrient limited plants were also able to stimulate lateral root production in Col-0 seedlings grown under nutrient limiting but not nutrient rich conditions (Fig. 4). Finally, nutrient rich exudates from Col-0 seedlings failed to significantly induce lateral root formation in either Col-0 samples (Fig. 4).
Plants compete with each other for available light, pollinators, nutrients, water, and mutualistic symbionts all of which may provide selective advantages.31 Resource scarcity is also a major driving force in competition and can alter root exudate composition, the source of the signals responsible for KR. 26,30,32,33 Our findings support our hypothesis that nutrient availability modulates changes in the RSA associated with KR. Nutrient reduction resulted in a general increase in lateral root number, our metric for ‘competition’ in all samples. This effect was significantly greater than SOLO samples in both the KIN and STRANGER samples, supporting KR as a nutrient dependent phenomenon (Fig. 2). This effect was eliminated by supplementing the 0.1xMS samples with excess nitrogen and/or phosphorus, 2 nutrients which play crucial roles in regulating lateral root density/number (Fig. 3).24,25 Furthermore, exudates collected from seedlings grown at 0.10xMS, rather than 1xMS, are more effective at inducing lateral root formation (Fig. 4). Sensitivity to these exudates also appears nutrient dependent as seedlings grown at 0.10xMS are more sensitive to 0.10xMS exudates than seedlings grown at 1xMS (Fig. 4). Taken together these results support the hypothesis that such recognition events are nutrient-dependent (nitrogen and phosphorus, specifically), inducible phenomena in plants.
The increased lateral root number observed in KIN and STRANGER samples relative to SOLO plants may simply reflect increased nutrient scarcity due to the introduction of an additional plant, independent of any specific recognition. However, our extraction method separates organic low-molecular weight root exudates from the macro and micronutrients which are retained in the aqueous layer. The constituents of the organic phase are then dissolved into the agar without any additional nutrients and applied to individual test plants. While this method results in a single dose being applied to our samples rather than continuously over the growth period, they are still able to influence the RSA of test plants. Such experimental limitations are a reasonable explanation for the reduced activity of the exudates (Fig. 4) relative to the direct growth studies (Fig. 2).
While compelling, the studies presented here have several limitations. First, gel-based media provides a uniform distribution of water and nutrients across the surface, unlike the heterogeneous soil environment and may accentuate these effects. Future studies will explore these phenotypes in soil analogs to better mimic this environment. Second, while lateral root number is easy to document, it may not be the most effective measure of ‘competition’ between root systems. Root system volume and other metrics may ultimately prove more useful indicators of competition. Third, as the petri dish is a closed, sterile system it is difficult to determine what responses arise from an exaggerated concentration of exudates or volatile organic compounds that have accumulated or might normally have been eliminated by microorganisms within the rhizosphere. Finally, given that Nitrogen and Phosphorus appear to be crucial regulators of lateral root formation during KR conditions, it is difficult to distinguish between pure nutrient-mediated effects and those arising from the perception of perspective competitors. Indeed, it may be impossible to completely uncouple these 2 processes.
Future studies will focus on exploiting our ability to ‘dial-in’ competition to identify molecular elements associated with KR, including changes in gene expression, protein synthesis, secondary metabolite production, and exudate composition. The direct comparison of plants engaged in STRANGER or KIN with SOLO plants may also permit us to distinguish elements associated with nutrient detection and acquisition as well as conspecific recognition. This is particularly important as root exudates are not only crucial to mediating biotic interactions with other soil organisms but can also facilitate nutrient acquisition (See 29 for a review). Long-term growth and physiological assays are currently underway in A. thaliana to determine what effects KR may have on fitness and secession under nutrient limiting conditions.
Resolving the molecular elements associated with these phenomena can provide insights into improving soil nutrient utilization and optimization of plant-plant interactions. As genes associated with pathogen resistance are differentially regulated during such interactions it further suggests that research into KR may impact our understanding of how host-pathogen interactions are regulated under nutrient limiting conditions in the presence of multiple plants, approximating real world conditions more accurately than the isolated inoculations of lone plants.16 The study of the molecular elements underpinning these processes also refines our understanding of the forces at work under the limited resources which drive succession and maintenance in plant communities.
Finally, the study of plant-plant recognition strategies broadens our appreciation for the social behaviors present in a Kingdom (Plantae) which appears to communicate primarily through chemical signals. In some respects these chemical exchanges are not entirely unlike the vocalizations utilized by songbirds to coordinate social behaviors.4 The continued exploration of the convergence of the prokaryotic and eukaryotic world on these solutions for resource utilization and competition has substantial value in the natural world as well as across multiple disciplines.
Materials and methods
Reagents & materials
Columbia-0, Landsberg, and Bensheim accessions were purchased from Lehle Seeds and propagated in the Florida Institute of Technology greenhouse. All reagents were purchased from Sigma-Aldrich, Fisher Scientific, or Caisson Labs.
Seed sterilization and germination
Seeds of A. thaliana are surface sterilized by a 5 minute wash with a 2% sodium hypochlorite solution supplemented with 0.5% Tween-20, followed by a 3 minute treatment with 90% EtOH. Seeds were rinsed in triplicate with sterile dH2O then transferred to germination plates (0.25xMS, 8 g/L agar, pH: 6.0) and stored at 4°C for 3 d. Seeds were germinated by removing them from the refrigerator and placing them under cool white lights for ≈3 d.
Growth studies
Following germination, all plants were transferred to agar plates containing the indicated concentrations of Murashigie-Skoog (MS) media (10 g/L agar, 5 g/L sucrose, pH: 5.8).34 Nutrient supplementation studies were performed using the same basal salts utilized for the preparation of MS. Three day-old seedlings were transferred from germination plates to growth plates to initiate experiments. Each plate contained: (i) a single seedling (SOLO), (ii) 2 seedlings of the same accession (KIN), or (iii) 2 seedlings of distinct accessions (STRANGER). All seedling pairs were placed approximately 2 cm apart. Plates were wrapped in Parafilm and grown vertically for 18 d at 23°C with a 16:8 h day:night cycle. Total lateral root number was recorded at the end of the growth period. The results of the nutrient limited or supplementation growth studies were analyzed by an ANOVA followed by a Tukey's post-hoc evaluation using the PAST v3.0 software package (http://folk.uio.no/ohammer/past/).35
Extraction experiments
Col-0 or LA-1 seedlings were grown individually in 6-well plates with 5 ml of the indicated nutrient concentration on an orbital shaker (90 rpm) for 18 d at 23°C with a 16:8 h day:night cycle.36 At the end of the growth period exudates from 24 seedlings were extracted from the growth media with ethyl acetate (EtOAc), evaporated to dryness, and dissolved in 1 ml of DMSO. Extracts were added by agar overlays to isolated test seedlings. Overlays were prepared by adding 50 μl aliquots of the extracts to 9.95 ml of warm 10 mM 2-(N-morpholino)ethanesulphonic acid (MES), a biologically inert buffer, supplemented with 8 g/L agar. 37
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. Arijit Mukherkee of the University of Southern Arkansas, Dr. Anil Mehta of Emory University, and Dr. Glenn Miller of the Florida Institute of Technology for their insight and thoughtful comments.
Reference
- 1.Krause ET, Kruger O, Kohlmeier P, Caspers BA. Olfactory kin recognition in a songbird. Biol Lett 2012; 8:327-9; PMID:22219391; http://dx.doi.org/15858573 10.1098/rsbl.2011.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamboa GJ. Kin recognition in eusocial wasps. Ann Zool Fennici [Internet] 2016; 41:789-808; http://www.annzool.net/PDF/anzf41/anzf41-789.pdf [Google Scholar]
- 3.Lihoreau M, Rivault C. Kin recognition via cuticular hydrocarbons shapes cockroach social life. Behav Ecol 2009; 20:46-53; http://dx.doi.org/ 10.1093/beheco/arn113 [DOI] [Google Scholar]
- 4.Sharp SP, McGowan A, Wood MJ, Hatchwell BJ. Learned kin recognition cues in a social bird. Nature 2005; 434:1127-30; PMID:15858573; http://dx.doi.org/ 10.1038/nature03522 [DOI] [PubMed] [Google Scholar]
- 5.Kurland JA. Kin selection theory: A review and selective bibliography. Ethol Sociobiol [Internet] 1980; 1:255-74; http://www.sciencedirect.com/science/article/B6X2B-45XSNN1-1T/1/6b78fd18975a5954a7c01b6fe463917b [Google Scholar]
- 6.Foster KR, Wenseleers T, Ratnieks FLW. Kin selection is the key to altruism. Trends Ecol. Evol. 2006; 21:57-60; PMID:16701471; http://dx.doi.org/6487228 10.1016/j.tree.2005.11.020 [DOI] [PubMed] [Google Scholar]
- 7.Rushton JP, Russell RJ, Wells PA. Genetic similarity theory: beyond kin selection. Behav Genet 1984; 14:179-93; PMID:6487228; http://dx.doi.org/ 10.1007/BF01065540 [DOI] [PubMed] [Google Scholar]
- 8.Lepik A, Abakumova M, Zobel K, Semchenko M. Kin recognition is density-dependent and uncommon among temperate grassland plants. Funct Ecol 2012; 26:1214-20; http://dx.doi.org/ 10.1111/j.1365-2435.2012.02037.x [DOI] [Google Scholar]
- 9.Marler TE. Kin recognition alters root and whole plant growth of split-root Cycas edentata seedlings. HortScience 2013; 48:1266-9; http://dx.doi.org/ 10.4161/cib.28009 [DOI] [Google Scholar]
- 10.Semchenko M, Saar S, Lepik A. Plant root exudates mediate neighbour recognition and trigger complex behavioural changes. New Phytol 2014; 204:631-7; PMID:25039372; http://dx.doi.org/ 10.1111/nph.12930 [DOI] [PubMed] [Google Scholar]
- 11.Murphy GP, Dudley SA. Kin recognition: Competition and cooperation in Impatiens (Balsaminaceae). Am J Bot 2009; 96:1990-6; PMID:21622319; http://dx.doi.org/ 10.3732/ajb.0900006 [DOI] [PubMed] [Google Scholar]
- 12.Biedrzycki ML, Jilany T. Root exudates mediate kin recognition in plants. 2010; 3:28-35; PMID:20539778; http://dx.doi.org/ 10.4161/cib.3.1.10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crepy MA, Casal JJ. Photoreceptor-mediated kin recognition in plants. New Phytol 2015; 205:329-38; PMID:25264216; http://dx.doi.org/ 10.1111/nph.13040 [DOI] [PubMed] [Google Scholar]
- 14.Bhatt M V. Khandelwal A, Dudley SA. Kin recognition, not competitive interactions, predicts root allocation in young Cakile edentula seedling pairs. New Phytol 2011; 189:1135-42; PMID:21118260; http://dx.doi.org/ 10.1111/j.1469-8137.2010.03548.x [DOI] [PubMed] [Google Scholar]
- 15.Dudley SA, File AL. Kin recognition in an annual plant. Biol Lett 2007; 3:435-8; PMID:17567552; http://dx.doi.org/ 10.1098/rsbl.2007.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biedrzycki ML, L V, Bais HP. Transcriptome analysis of Arabidopsis thaliana plants in response to kin and stranger recognition. Plant Signal. Behav. 2011; 6:1515-24; PMID:21900741; http://dx.doi.org/16469918 10.4161/psb.6.10.16525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin IT, Halitschke R, Paschold A, von Dahl CC, Preston CA. Volatile signaling in plant-plant interactions: “talking trees” in the genomics era. Science 2006; 311:812-5; PMID:16469918; http://dx.doi.org/ 10.1126/science.1118446 [DOI] [PubMed] [Google Scholar]
- 18.Tsuchiya Y, McCourt P. Strigolactones as small molecule communicators. Mol Biosyst [Internet] 2012; 8:464-9; http://www.ncbi.nlm.nih.gov/pubmed/22027812; PMID:22027812; http://dx.doi.org/ 10.1039/C1MB05195D [DOI] [PubMed] [Google Scholar]
- 19.Callaway RM. The detection of neighbors by plants. Trends Ecol. Evol. 2002; 17:104-5; http://dx.doi.org/ 10.1016/S0169-5347(01)02438-7 [DOI] [Google Scholar]
- 20.Badri D V, Loyola-Vargas VM, Broeckling CD, De-la-Peña C, Jasinski M, Santelia D, Martinoia E, Sumner LW, Banta LM, Stermitz F, et al.. Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol 2008; 146:762-71; http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2245854&tool=pmcentrez&rendertype=abstract; PMID:18065561; http://dx.doi.org/ 10.1104/pp.107.109587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.File AL, Klironomos J, Maherali H, Dudley SA. Plant Kin Recognition Enhances Abundance of Symbiotic Microbial Partner. PLoS One 2012; 7:1-10; PMID:23029158; http://dx.doi.org/ 10.1371/journal.pone.0045648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malamy JE. Intrinsic and environmental response pathways that regulate root system architecture. Plant, Cell Environ 2005; 28:67-77; PMID:16021787; http://dx.doi.org/17684758 10.1111/j.1365-3040.2005.01306.x [DOI] [PubMed] [Google Scholar]
- 23.Berntson GM. Modelling root architecture: are there tradeoffs between efficiency and potential of resource acquisition? New Phytol 1994; 127:483-93; http://onlinelibrary.wiley.com/doi/10.1111/j.1469-8137.1994.tb03966.x/abstract; http://dx.doi.org/ 10.1111/j.1469-8137.1994.tb03966.x [DOI] [Google Scholar]
- 24.López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 2002; 129:244-56; http://dx.doi.org/ 10.1104/pp.010934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Rong H, Pilbeam D. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. In: J Exp Bot 2007; 58(9):2329-38 [DOI] [PubMed] [Google Scholar]
- 26.Yoneyama K, Xie X, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K. Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 2007; 227:125-32; PMID:17684758; http://dx.doi.org/ 10.1007/s00425-007-0600-5 [DOI] [PubMed] [Google Scholar]
- 27.Aoki M, Fujii K, Kitayama K. Environmental Control of Root Exudation of Low-Molecular Weight Organic Acids in Tropical Rainforests. Ecosystems 2012; 15:1194-203; http://dx.doi.org/ 10.1007/s10021-012-9575-6 [DOI] [Google Scholar]
- 28.Wang BL, Shen JB, Zhang WH, Zhang FS, Neumann G. Citrate exudation from white lupin induced by phosphorus deficiency differs from that induced by aluminum. New Phytol 2007; 176:581-9; PMID:17725555; http://dx.doi.org/ 10.1111/j.1469-8137.2007.02206.x [DOI] [PubMed] [Google Scholar]
- 29.Dakora FD, Phillips DA. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 2002; 245:35-47; http://dx.doi.org/ 10.1023/A:1020809400075 [DOI] [Google Scholar]
- 30.Cahill JF, McNickle GG, Haag JJ, Lamb EG, Nyanumba SM, St Clair CC. Plants integrate information about nutrients and neighbors. Science 2010; 328:1657; PMID:20576883; http://dx.doi.org/ 10.1126/science.1189736 [DOI] [PubMed] [Google Scholar]
- 31.Mitchell RJ, Flanagan RJ, Brown BJ, Waser NM, Karron JD. New frontiers in competition for pollination. Ann Bot 2009; 103:1403-13; http://dx.doi.org/ 10.1093/aob/mcp062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masclaux F, Hammond RL, Meunier J, Gouhier-Darimont C, Keller L, Reymond P. Competitive ability not kinship affects growth of Arabidopsis thaliana accessions. New Phytol 2010; 185:322-31; http://dx.doi.org/10.1111/j.1469-8137.2009.03057.x\nhttp://www3.interscience.wiley.com/cgi-bin/fulltext?ID=122671343&PLACEBO=IE.pdf&mode=pdf; PMID:19886895; http://dx.doi.org/ 10.1111/j.1469-8137.2009.03057.x [DOI] [PubMed] [Google Scholar]
- 33.Lizé A, Khidr SK, Hardy ICW. Two components of kin recognition influence parasitoid aggression in resource competition. Anim Behav 2012; 83:793-9; http://dx.doi.org/ 10.1016/j.anbehav.2012.01.001 [DOI] [Google Scholar]
- 34.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant [Internet] 1962; 15:473-97; http://onlinelibrary.wiley.com/doi/10.1111/j.1399-3054.1962.tb08052.x/full [Google Scholar]
- 35.Hammer Ø, Harper DAT, Ryan PD. Paleontological statistics software package for education and data analysis. Palaeontol Electron 2001; 4:9-18; http://palaeo-electronica.org/2001_1/past/issue1_01.htm [Google Scholar]
- 36.Badri D V, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 2009; 151:2006-17; PMID:19854857; http://dx.doi.org/ 10.1104/pp.109.147462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bugbee BG, Salisbury FB. An evaluation of MES (2(N-Morpholino)ethanesulfonic acid) and Amberlite IRC-50 as pH buffers for nutrient solution studies. J Plant Nutr 1985; 8:567-83; PMID:11539688; http://dx.doi.org/ 10.1080/01904168509363369 [DOI] [PubMed] [Google Scholar]


