ABSTRACT
Most therapeutic antibodies (Abs) target cell surface proteins on tumor and immune cells. Cloning of Ab gene libraries in E. coli and their display on bacteriophages is commonly used to select novel therapeutic Abs binding target antigens, either purified or expressed on cells. However, the sticky nature of bacteriophages renders phage display selections on cells challenging. We previously reported an E. coli display system for expression of VHHs (i.e., nanobodies, Nbs) on the surface of bacteria and selection of high-affinity clones by magnetic cell sorting (MACS). Here, we demonstrate that E. coli display is also an attractive method for isolation of Nbs against cell surface antigens, such as the epidermal growth factor receptor (EGFR), upon direct selection and screening of Ab libraries on live cells. We employ a whole cell-based strategy using a VHH library obtained by immunization with human tumor cells over-expressing EGFR (i.e., A431), and selection of bacterial clones bound to murine fibroblast NIH-3T3 cells transfected with human EGFR, after depletion of non-specific clones on untransfected cells. This strategy resulted in the isolation of high-affinity Nbs binding distinct epitopes of EGFR, including Nbs competing with the ligand, EGF, as characterized by flow cytometry of bacteria displaying the Nbs and binding assays with purified Nbs using surface plasmon resonance. Hence, our study demonstrates that E. coli display of VHH libraries and selection on cells enables efficient isolation and characterization of high-affinity Nbs against cell surface antigens.
KEYWORDS: Antibody display, bacterial display, cell surface antigen, E. coli, EGFR, nanobodies, on-cell selection, single-domain antibodies, tumor-associated antigen
Introduction
Current clinical practice for human cancers, both solid tumors and hematologic malignancies (e.g., leukemias, lymphomas), involve treatment with monoclonal antibodies (mAbs) that are mostly based on immunoglobulin G (IgG) molecules targeted to proteins expressed on the surface of malignant cells.1,2 Common targets for these mAbs include cell surface receptors of growth factors, such as members of the epidermal growth factor receptor family, erbB (e.g., EGFR, HER2, HER3), as well as surface proteins expressed by B-lymphocytes (e.g., CD20) and myeloid cells (e.g., CD33, CD38). Several immune checkpoint-blocking mAbs bind cell surface proteins on tumor cells and T-cells, such as the cytotoxic T lymphocyte-associated antigen 4 (CTLA4), and the programmed cell death protein 1 (PD-1) and its ligand (PD-L1).3,4 Furthermore, a large number of the newly identified tumor markers are associated to the cell surface.5,6 Efficient strategies for the selection of Abs recognizing cell surface antigens are thus of great interest.
Phage display, which involves the cloning of combinatorial libraries comprising the variable (V) genes from heavy (H) and light (L) chains of IgGs in E. coli and their display on filamentous bacteriophages, has been extensively used for the selection and engineering of therapeutic mAbs7,8 and smaller recombinant Ab formats with distinct functional properties (e.g., enhanced tumor penetration, multivalent and multi-specific antigen binding, customized half-life).9,10 The technique often includes incubation of bacteriophages displaying the Abs (phage antibodies or “Phabs”) with the purified antigen, either immobilized on a surface or on an affinity matrix (e.g., biotinylated antigens on streptavidin-beads), followed by the recovery of antigen-bound Phab clones.11,12 Due to the sticky nature of filamentous bacteriophages, several extensive washing steps with stringent conditions (e.g., buffers with detergents) are usually required to remove non-specific phages, a process called “biopanning.” Although biopanning with purified proteins is a robust process that has allowed the selection of high-affinity Abs against many different antigens, it has a number of limitations when used with cell surface antigens. Firstly, the purification of native membrane proteins from cells is not always practical or feasible due to low yields, poor solubility or the requirement of protein reconstitution into lipid vesicles to preserve the original conformation, all of which limit biopannings, as well as immunizations, for construction of immune Ab libraries. Secondly, purification of recombinant antigen fragments containing soluble protein domains increases yields, but may alter antigenicity due to misfolding or altered post-translational modifications (e.g., glycosylation), leading to the selection of Abs that may not recognize the native protein. Lastly, immobilization of purified antigens on solid supports and stringent washing conditions may alter conformational epitopes that could be relevant in vivo. Thus, in these cases, it is clearly advantageous to screen Ab gene libraries directly on live intact cells expressing the cell surface antigen, either endogenously or upon transfection. Screening of phage display Ab libraries on live cells requires more complex selection strategies to avoid enrichment of Phabs binding other antigens found on cells. These procedures routinely involve at least a single depletion step on cells lacking expression of the target antigen, to remove binders against non-relevant antigens (negative selection or depletion), followed by incubation of the unbound Phabs with cells expressing the antigen of interest (positive selection).13-15 However, additional steps are usually needed to improve the efficiency of phage selections on cells, such as competitive elution with a ligand or existing mAbs that bind the target antigen,16-19 washing of cells by centrifugation through an organic phase,20 removal of dead cells,21 or masking dominant epitopes with soluble Ab fragments from non-specific Phabs.22
We previously reported an Ab selection system in E. coli that does not utilize bacteriophages, but instead is based on the direct display of Ab fragments on the cell surface of bacteria, which facilitates the use of flow cytometry for rapid characterization of the selected clones.23 The E. coli display system employs fusions of Ab fragments to a N-terminal polypeptide from Intimin (called Neae), which comprises the β-barrel domain that anchors the protein in the bacterial outer membrane.24,25 We demonstrated that single VHH domains derived from camelid heavy-chain-only Abs, also referred to as nanobodies (Nbs),26 were efficiently displayed on the surface of E. coli bacteria fused to the C-terminus of Neae.23 Nbs have a number of properties that make them attractive Ab molecules, such as their small size (ca. 14 kDa), simple structure, high-affinity and specificity, distinct epitope recognition, high stability, and similarity to human VH sequences.26 We showed that E. coli bacteria displaying an immune library of VHHs against a soluble bacterial protein could be enriched for high-affinity binding clones using purified biotinylated antigen, anti-biotin magnetic beads, and magnetic cell sorting (MACS) columns for selection. Interestingly, E. coli bacteria displaying non-specific VHHs were easily washed from MACS columns using mild conditions (e.g., phosphate-buffered saline (PBS)).23
The advantageous properties of E. coli display, along with the biomedical potential of Nbs for tumor therapy and in vivo tumor imaging,27-29 prompted us to evaluate E. coli display for the selection of Nbs against tumor-associated cell surface antigens. In particular, we wanted to test its potential for the direct selection of VHH libraries on live cells. Here, we demonstrated the utility of this approach using a well-known tumor-associated cell surface antigen, the human epidermal growth factor receptor (EGFR or ErbB1).30,31 EGFR is a transmembrane tyrosine kinase (TK) receptor whose expression or activity is frequently upregulated in many human cancers of epidermal origin, including carcinomas of the head and neck, colon, breast, urinary bladder, lung, and ovary.32-34 EGFR is activated by various ligands, including EGF, transforming growth factor-α (TGF-α), heparin-binding EGF (HB-EGF), and amphiregulin. Ligand binding to the extracellular ectodomains of EGFR (eEGFR) activates the cytoplasmic TK, leading to auto-phosphorylation of the C-terminal region and initiation of downstream cell-signaling cascades promoting cell proliferation, survival, and angiogenesis.35-37 We used an experimental set up that evaluates E. coli display in a whole cell-based strategy, from tumor cell immunization to on-cell selection. We compared the efficiency of E. coli display selections on cells versus the use of MACS with purified antigen. In addition, we characterized the binding of the selected Nbs to cells expressing EGFR, determined their binding affinity and competition with the ligand, EGF. Taken together, our results demonstrate that E. coli display enables the effective isolation of high-affinity Nbs targeting different epitopes of EGFR, including competitors for EGF-binding.
Results
Construction of the E. coli display VHH library, and selections with eEGFR-Fc and on cells
We constructed an E. coli display library with VHHs obtained after immunization of 2 llamas (Llama glama) with human A431 cells, which overexpress EGFR (ca. 106 EGFR molecules/cell).38 The pool of amplified VHH gene segments were originally cloned in a phagemid vector with a library size of ca. 1 × 107 independent clones. The VHH gene segments were excised from the pool of phagemid DNA by SfiI and NotI digestion and cloned into the same sites of the E. coli display vector pNeae2, which fuses the VHH sequences to the C-end of intimin polypeptide Neae.23 The fusion polypeptides, referred to as NVHH, incorporate 2 epitope tags flanking the VHH domain for immunodetection with specific mAbs (i.e., E-tag before the VHH and myc-tag at the C-terminus).23 A library of ca. 1.3 × 107 independent clones was obtained after transformation of E. coli strain EcM1 (Table S1).
The NVHH E. coli display library was induced with isopropylthio-β-D-galactoside (IPTG) prior to its selection with eEGFR-Fc fusion protein or on live cells. A scheme of the E. coli display library construction and selections is shown in Fig. 1. Selection with purified recombinant eEGFR-Fc, a fusion of the ectodomain of human EGFR (residues 1–645) to the Fc region of human IgG1, was carried out by MACS under conditions similar to those previously reported.23 Approximately 2 × 108 bacteria were incubated with biotinylated eEGFR-Fc (100 nM) and magnetic micro-beads coated with anti-biotin mAb. The mixture was loaded onto a MACS column held on a magnet, washed with PBS containing bovine serum albumin (BSA), and the column was removed from the magnet to elute bound bacteria in Luria–Bertani (LB) medium for plating. Selection on live cells (CellS) was carried out with an initial negative selection step incubating the E. coli display library (ca. 6 × 107 bacteria) for 1 hr at 37°C with a monolayer of the murine fibroblast cell line NIH-3T3 2.2 (ca. 6 × 105 cells), which lacks expression of endogenous EGFR.39 The unbound bacteria were recovered and incubated for 15 min at 37°C with a monolayer of the murine tumor cell line Her14 (∼6 × 105 cells), which is a stably transfected NIH-3T3 2.2 clone expressing human EGFR.39 Her14 cells were washed 3 times with buffer, the bound bacteria were recovered, plated and the colony-forming units (CFU) grown on plates were determined in both cases (MACS and CellS). Further, bacteria were harvested as a pool; their plasmids isolated and transformed to fresh EcM1 bacteria for a second round of MACS or CellS under identical conditions.
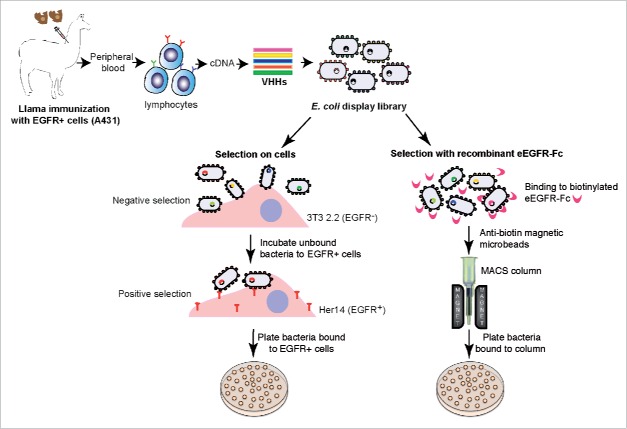
Figure 1.
Scheme of the generation and display of the anti-EGFR VHH immune library on E. coli and the methodology adopted for selection of anti-EGFR Nbs on live cells (CellS) and with recombinant eEGFR-Fc. Two llamas (Llama glama) were immunized with human A431 cells overexpressing EGFR. After immunization, the VHH gene segments amplified from peripheral blood lymphocytes by RT-PCR, were cloned into the E. coli display vector pNeae2. Next, selection of the E. coli display library was carried out on mouse tumor cell line transfected with human EGFR (CellS) or by using magnetic cell sorting (MACS) with recombinant eEGFR-Fc. In MACS, E. coli bacteria binding the biotinylated antigen are incubated with anti-biotin magnetic beads and captured in an iron column held in a magnet. Elution of bound bacteria is done with fresh LB media upon column removal from the magnet and grown by plating. In CellS, E. coli bacteria are initially incubated with the parental mouse cell line NIH-3T3 2.2 lacking expression of EGFR (negative selection). Bacteria that do not bind NIH-3T3 2.2 are recovered and immediately incubated with the isogenic mouse cell line transfected with human EGFR (Her14) for positive selection. Bacteria bound to Her14 cells after washing are recovered by lysis of the mammalian cells and grown by plating.
In MACS selections, CFU of column-bound bacteria were ∼0.6% of the total bacteria used in the first round, and ∼4.8% in the second round (Table S2). In CellS, CFU of Her14-bound bacteria were ∼0.3% of the total bacteria used in the first round, and ∼1.2% in the second round (Table S3). The increase in the percentage of bound bacteria in the second round of MACS and CellS suggested an enrichment of specific binders in both cases. The higher percentage of bound bacteria in MACS selections also suggested the better selection efficiency of MACS vs. CellS.
To evaluate the enrichment of EGFR-binding clones after MACS or CellS, we analyzed the binding of bacteria to eEGFR-Fc by flow cytometry (Fig. 2). Induced bacteria from the E. coli display library and from each selection round of MACS and CellS were incubated with biotinylated eEGFR-Fc (50 nM) and secondary streptavidin-phycoerythrin (PE) conjugate. Flow cytometry analysis revealed a clear increase in the fluorescence intensity of bacteria after selection rounds in both MACS and CellS. The percentage of bacteria binding eEGFR-Fc increased from background levels found in the library (∼0.2% positives) to ca. 28% and 74% positives in rounds 1 and 2 of MACS (Fig. 2A, upper panels), and to ca. Five% and 35% positives in rounds 1 and 2 of CellS (Fig. 2B, upper panels). In addition, flow cytometry analysis of bacteria with anti-myc mAb showed a similar level of surface display of the NVHH fusions in the library and the enriched bacterial pools after selections (Fig. 2A and 2B, bottom panels).
Figure 2.
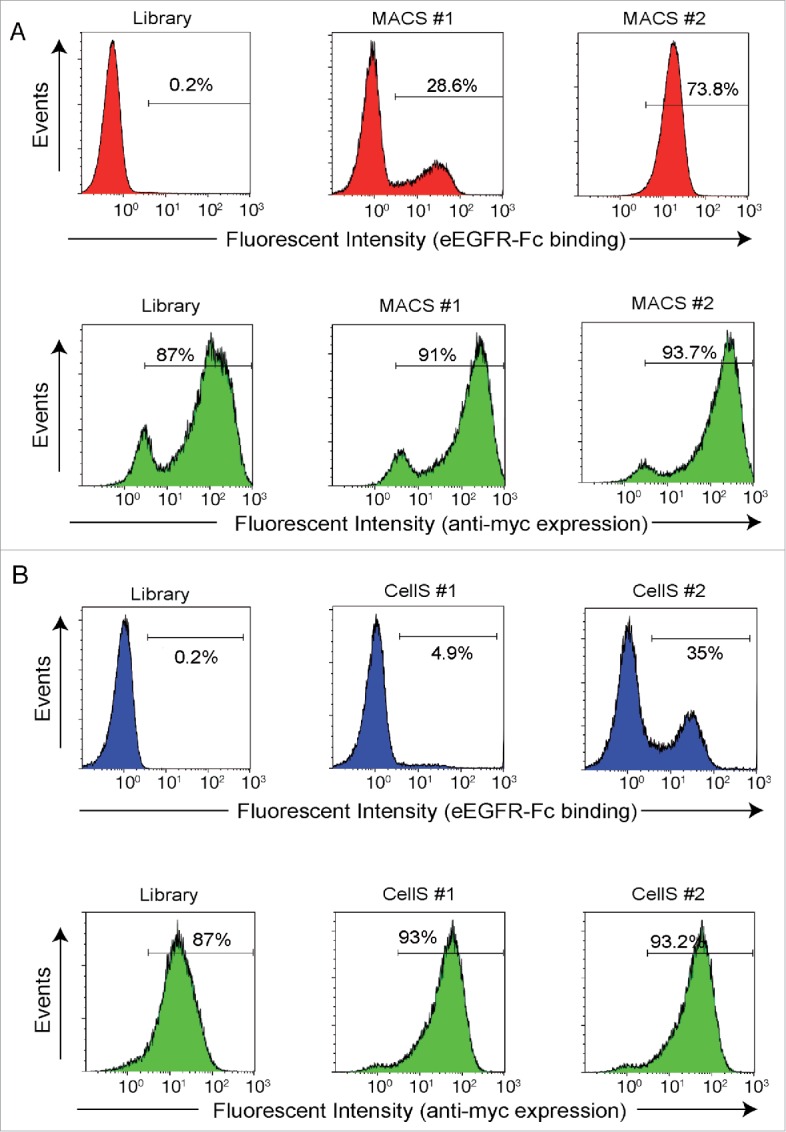
Binding of E. coli display VHH library to biotinylated eEGFR-Fc along selection cycles by MACS and CellS. Flow cytometry analysis of induced E. coli EcM1 bacteria displaying anti-EGFR VHH immune library, or their respective sub-libraries enriched after the indicated round (#1, #2) of magnetic cell sorting (MACS) with biotinylated eEGFR-Fc (A) or cell selection (CellS) with Her14 cell line (B). Histograms show the fluorescence intensity of bacteria incubated with biotinylated eEGFR-Fc and secondary Streptavidin-PE (top panels in A and B) to determine eEGFR-Fc binding, or anti-myc tag mAb and anti-mouse mAb conjugated to Alexa 488 fluorophore (bottom panels in A and B) to determine display levels of NVHH fusions.
Screening of bacterial clones binding EGFR
Given the good enrichment of eEGFR-Fc binding clones observed after the second round of MACS (Fig. 2), 96 bacterial clones were randomly picked from this selection round and the sequence of their cloned VHHs was determined. Bacteria displaying different VHH sequences were individually assessed for their binding to eEGFR-Fc and BSA (negative control) by flow cytometry (Fig. 3). Five different VHHs binding specifically to eEGFR-Fc were identified from these clones: VEGFR1 (72 clones), VEGFR2 (10 clones), VEGFR3 (a single clone), VEGFR4 (3 clones) and VEGFR5 (a single clone) (Table 1), while 9 clones did not bind eEGFR-Fc. To screen clones from CellS, 96 clones were picked from the second round of CellS and their specific binding to Her14 (EGFR+), but not to NIH-3T3 2.2 (EGFR-), cells was examined by light microscopy. Induced bacteria were added at a multiplicity of infection (MOI) of 100 to either Her14 cells or NIH-3T3 2.2 cells and incubated for 20 min. Clones that did not bind either cell line were classified as “non-binders,” those that bound both NIH-3T3 2.2 cells and Her14 cells were classified as “non-specific binders” and those that bound only Her14 cells were called “specific Her14-binders.” Out of the 96 clones screened, 45 were non-binders, 26 were non-specific binders and 25 were specific Her14-binders. VHH DNA sequences from these specific Her14-binders were determined, with VEGFR1 being identified in 18 clones, VEGFR2 in 3 clones, VEGFR4 in 2 clones, and a new VHH sequence, VEGFR6, was identified in 2 clones (Table 1). Like the other 5 EGFR-binding VHHs, VEGFR6 also showed specific binding to eEGFR-Fc in flow cytometry (Fig. 3). VEGFR3 and VEGFR5 were not found among the 25 clones analyzed from CellS, likely due to their low frequency (both single clones in MACS screening). We confirmed that bacteria displaying these VHHs bound specifically to EGFR-positive Her14 cells, but not NIH-3T3 2.2 cells (Fig. 4). Negative control E. coli bacteria displaying a VHH against an unrelated antigen, fibrinogen (Fib), did not bind either of these cell lines. This indicates that MACS and CellS are complementary methods of selection that can be used in parallel to generate a larger panel of specific Abs.
Figure 3.
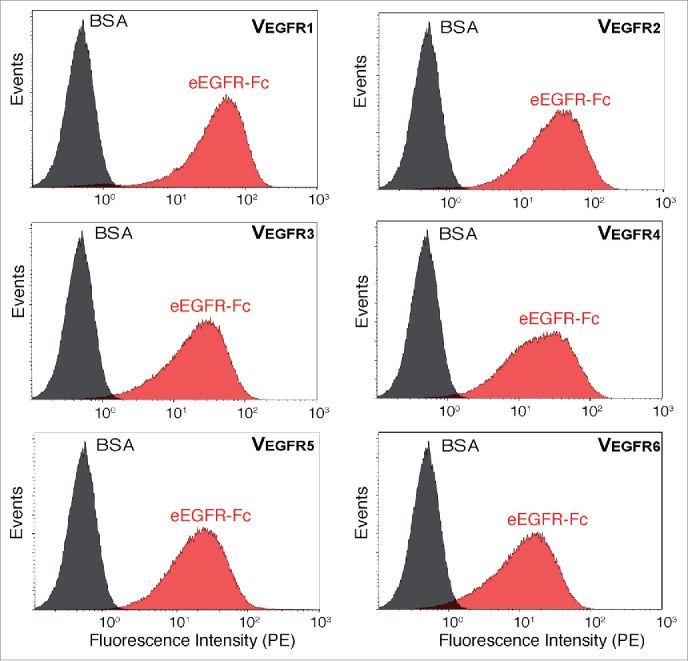
Binding of E. coli bacteria displaying selected VHH clones to eEGFR-Fc. Fluorescent flow cytometry analysis of induced E. coli EcM1 cells displaying the indicated Nb (VEGFR1-6) incubated with biotinylated eEGFR-Fc (red) or BSA (black) and secondary Streptavidin-PE.
Table 1.
Nbs against EGFR selected by E. coli display with MACS and CellS.
| Number of clones | |||
|---|---|---|---|
| Nb name | Amino acid sequence of CDR-H3 | MACS | CellS |
| VEGFR1 | DKWSSSRRSVDYDY | 72/96 | 18/96 |
| VEGFR2 | TYNPYSRDHYFPRMTTEYDY | 10/96 | 03/96 |
| VEGFR3 | RYSDVIFTLPERYAY | 01/96 | — |
| VEGFR4 | STYSRDSVFTKWANYNY | 03/96 | 02/96 |
| VEGFR5 | GPILGSSESYRSSRRYAY | 01/96 | — |
| VEGFR6 | DLNFIGIVTTTSEKYDY | — | 02/96 |
Figure 4.
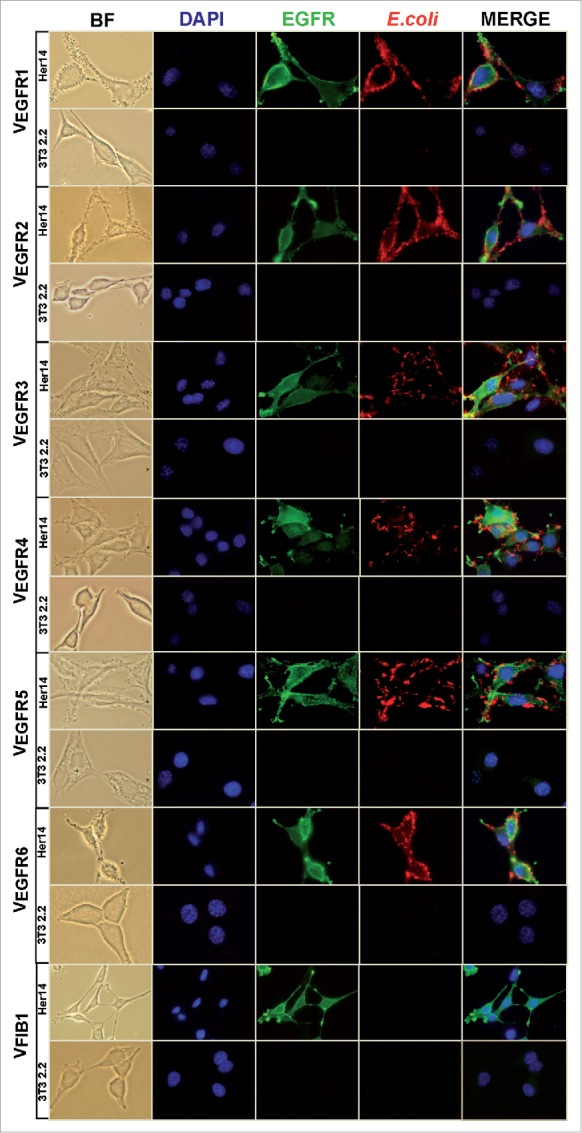
Adhesion to tumor cells of E. coli bacteria displaying the selected anti-EGFR Nbs. Bright field (BF) and fluorescence microscopy images of Her14 (EGFR+) and NIH-3T3 2.2 (EGFR-) cells grown in culture and infected with E. coli bacteria displaying the indicated Nb clone. Microscopy images showing the specific adhesion of bacteria with selected anti-EGFR Nbs to Her14 cells, but not to NIH-3T3 2.2 cells. Also included is an E. coli bacterial clone displaying a non-relevant Nb (VFIB1). Bacteria were labeled with anti-E. coli polyclonal Ab (red), EGFR was labeled with anti-EGFR mAb (green), DNA and cell nuclei were labeled with DAPI (blue).
Characterization of anti-EGFR nanobodies selected by E. coli display
Overlay of flow cytometry histograms and mean fluorescence intensities of E. coli bacteria suggested that the selected VHHs had different binding affinities to eEGFR-Fc (Fig. S1). To determine the actual binding properties and kinetic data of the selected VHHs, the soluble Nbs from the 6 clones were expressed with C-terminal 6xHis- and myc-tags in the periplasm of E. coli strain WK6 (Table S1). The Nbs were purified by a 2-step process using metal affinity chromatography followed by gel filtration to collect the monomeric Nbs of ca. 20 kDa. A control Nb binding GFP (VGFP) was purified in the same manner. Enzyme-linked immunosorbent assay (ELISA) performed by titration of the purified Nbs confirmed their specific binding to eEGFR-Fc (Fig. 5) and revealed that Nbs VEGFR1, VEGFR2 and VEGFR3 have a higher affinity than VEGFR4 and VEGFR5, with VEGFR6 being the clone with the lowest binding signals (Fig. 5). Binding of a control Nb (VGFP) to eEGFR-Fc, and of anti-EGFR Nbs to BSA, were in all cases at background levels (OD490 ≤ 0.05; signals subtracted from data in Fig. 5).
Figure 5.
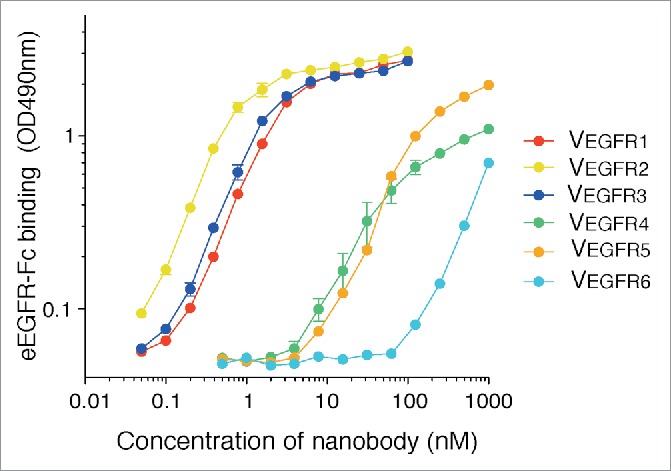
Binding activity of the selected Nbs to eEGFR-Fc determined by ELISA. Binding curves determined by ELISA using the purified anti-EGFR Nbs with C-terminal 6xHis and myc tags. The ELISA was developed with anti-myc-tag mAb and anti-mouse-POD. ELISA signals against a control antigen (BSA) were subtracted from the represented values. The plot shows the OD values at 490 nm with standard error from 2 independent experiments with duplicates using the purified Nbs at the indicated concentrations (0.05–1000 nM).
The purified Nbs were also tested for their ability to specifically bind EGFR-expressing cells by immunofluorescence microscopy (Fig. 6). In parallel, cells were also stained with a commercial mAb against EGFR (positive control) and with VGFP (negative control). This experiment revealed that all 6 different Nbs stained specifically Her14 cells, but not 3T3 2.2 cells, with clones VEGFR1, VEGFR2 and VEGFR3 having the highest fluorescence signals (Fig. 6). Her14 cells labeled with Nbs VEGFR4 and VEGFR5 showed intermediate levels of fluorescence, whereas VEGFR6 only produced a faint staining (Fig. 6). Taken together, these data confirm the specific binding of the 6 selected Nbs to EGFR, albeit with different binding affinities.
Figure 6.
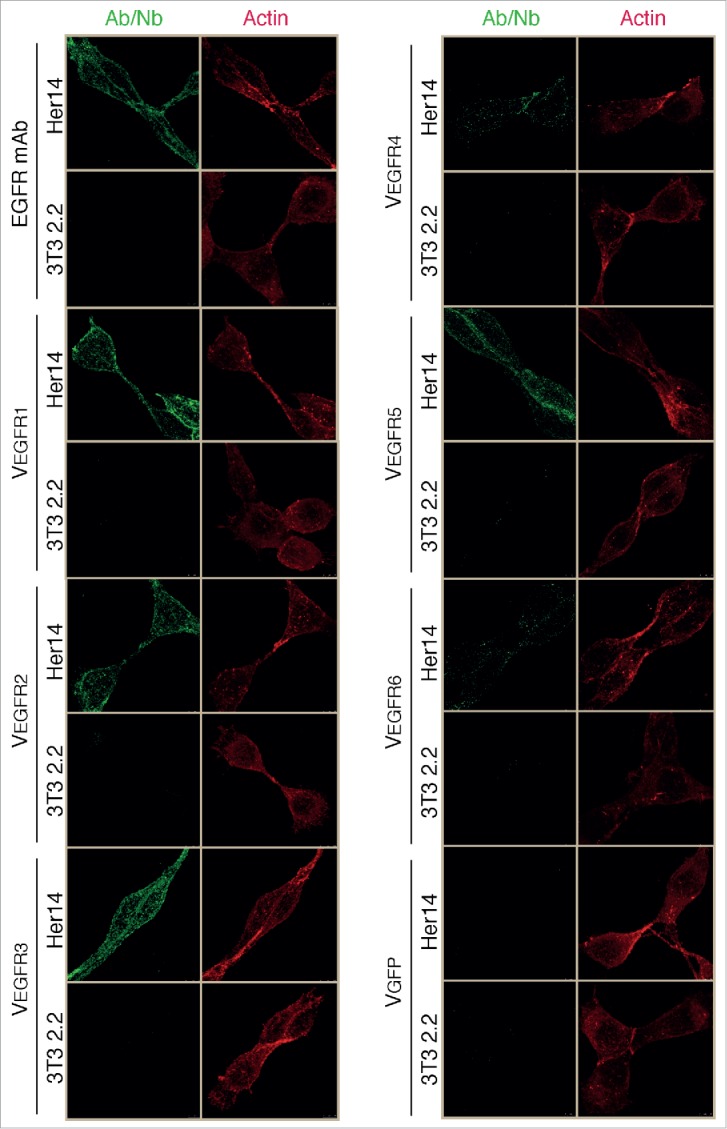
Specific staining of mammalian cells expressing human EGFR with the selected Nbs. Confocal microscopy images of Her14 (EGFR+) and NIH-3T3 2.2 (EGFR-) cells stained (as indicated) with a commercial anti-human EGFR mAb, the purified Nbs selected against EGFR, and a negative control Nb (VGFP). Binding of anti-EGFR mAb or Nbs were detected using anti-myc mAb and/or anti-mouse IgG (green). Cells were stained with phalloidin (red) to reveal F-actin cytoskeleton.
To determine the dissociation constant at equilibrium (KD), the kinetic association (ka) and dissociation constants (kd) of the isolated Nbs, we conducted surface plasmon resonance (SPR) experiments in which eEGFR-Fc was covalently immobilized to a SPR-sensor chip at ca. 1000 response units (RUs). Dilutions of the purified Nbs were injected at different concentrations, from 0.14 nM to 100 nM depending of the clone (Fig. 7). All clones were analyzed by SPR, except VEGFR6 given its weak binding in ELISA to eEGFR-Fc at concentrations below 100 nM (see above, Fig. 5). Association and dissociation of each Nb were recorded at the different concentrations (in duplicates) and the resulting sensorgrams were overlaid, aligned and analyzed using a 1:1 Langmuir binding model (Fig. 7). According to this analysis, VEGFR1 showed the highest affinity for eEGFR-Fc (KD= 0.47 nM), followed by clones VEGFR2 and VEGFR3, with KDs of ca. 2 nM in both cases. Clones VEGFR4 (KD = 15.9 nM) and VEGFR5 (KD= 24.2 nM) showed lower affinity for eEGFR-Fc. No binding signals were observed when the negative control Nb was injected over the sensor chip, or when the different Nbs were injected over a chip channel lacking eEGFR-Fc (data not shown). The KD and kinetic constants (ka, kd) of these Nbs are summarized in Table 2.
Figure 7.
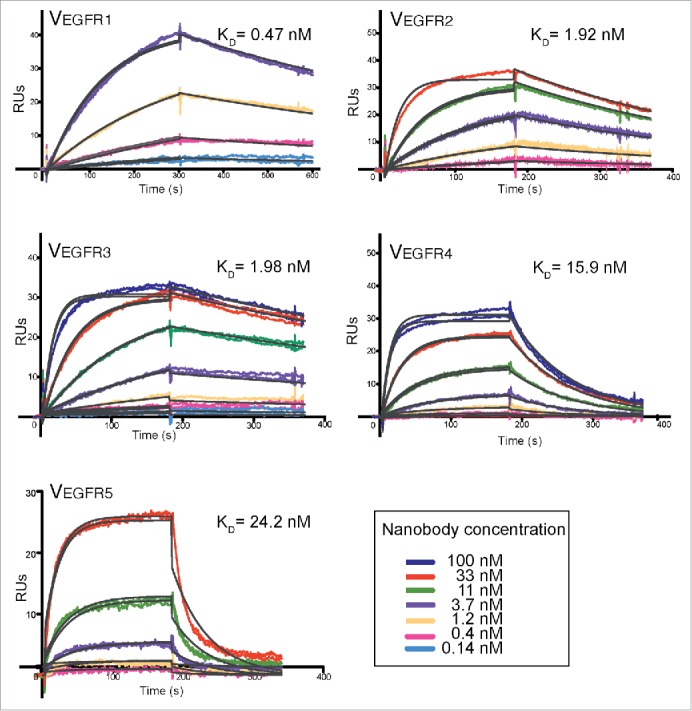
Determination of the equilibrium dissociation constant (KD) of the selected anti-EGFR Nbs using surface plasmon resonance. SPR sensorgrams monitoring real-time association and dissociation of purified Nbs VEGFR1-5 (at the concentrations indicated in the color code box) to eEGFR-Fc immobilized onto a CM5 sensor chip. The increase in resonance units (RU) was recorded along time (in seconds) and dissociation of the Nbs was evaluated after injection of buffer. Sensorgrams from 2 independent injections are shown. The curve was fitted by non-linear least squares regression using the 1:1 Langmuir binding model (theoretical sensorgrams in black).
Table 2.
Kinetic and affinity constants (ka, kd and KD) to eEGFR-Fc determined by SPR.
| Nb | Association (ka) M−1 s−1 | Dissociation (kd) s−1 | KD (kd/ka) M |
|---|---|---|---|
| VEGFR1 | 2.22 × 106 | 1.06 × 10−3 | 0.47 × 10−9 |
| VEGFR2 | 1.49 × 106 | 2.87 × 10−3 | 1.92 × 10−9 |
| VEGFR3 | 6.86 × 105 | 1.36 × 10−3 | 1.98 × 10−9 |
| VEGFR4 | 7.25 × 105 | 1.15 × 10−2 | 15.9 × 10−9 |
| VEGFR5 | 8.33 × 105 | 2.00 × 10−2 | 24.2 × 10−9 |
Next, we investigated whether these Nbs recognize different epitopes of EGFR by analyzing their competition for binding to eEGFR-Fc. In these assays, the Nbs were sequentially injected over the SPR sensor chip in order to record their binding in the presence of a different pre-bound Nb. These experiments showed that the binding of VEGFR1 had no effect on the binding of all of the other Nbs (Fig. S2), indicating the recognition of a unique epitope. On the contrary, VEGFR2 blocked completely the binding of Nb clones VEGFR3 and VEGFR4, and vice versa (Fig. S2 and data not shown). Binding of Nb VEGFR5 was found to be independent of any other clone (Fig. S2 and data not shown). Therefore, these data suggest that at least 3 different epitopes groups could be identified within the panel of selected anti-EGFR Nbs, one formed by VEGFR1, a second one by clones VEGFR2, VEGFR3 and VEGFR4, and a third one by VEGFR5. Results of the competitive binding assays using the Nbs are summarized in Table 3.
Table 3.
Summary of Nb and EGF binding competition to eEGFR-Fc by SPR.
| VEGFR1 | VEGFR2 | VEGFR3 | VEGFR4 | VEGFR5 | |
|---|---|---|---|---|---|
| VEGFR1 | + | − | − | − | − |
| VEGFR2 | − | + | + | + | − |
| VEGFR3 | − | + | + | + | − |
| VEGFR4 | − | + | + | + | − |
| VEGFR5 | − | − | − | − | + |
| EGF | − | + | + | + | − |
Competition of EGF and nanobodies for EGFR binding
We investigated whether the selected Nbs could compete with EGF ligand for binding to EGFR. We first tested EGF-binding competition by SPR analysis. Purified Nbs VEGFR1 to VEGFR5 (100 nM) were injected onto an eEGFR-Fc sensor chip in the presence or absence of EGF (300 nM). The sensorgrams obtained were superimposed at the time of Nb injection (Fig. 8), and showed that EGF does not affect the binding of Nbs VEGFR1 and VEGFR5, whereas it reduces the binding of Nbs VEGFR2, VEGFR3 and VEGFR4 to roughly half. Since clones VEGFR2, VEGFR3 and VEGFR4 compete with each other and with EGF (Table 3; Figs. 8 and S2), these Nbs should likely bind to an epitope partially overlapping the EGF-binding site of EGFR or inhibit the conformational change necessary for the receptor to bind EGF.40 On the contrary, Nbs VEGFR1 and VEGFR5 bind to 2 distinct epitopes of EGFR that are independent of EGF binding.
Figure 8.
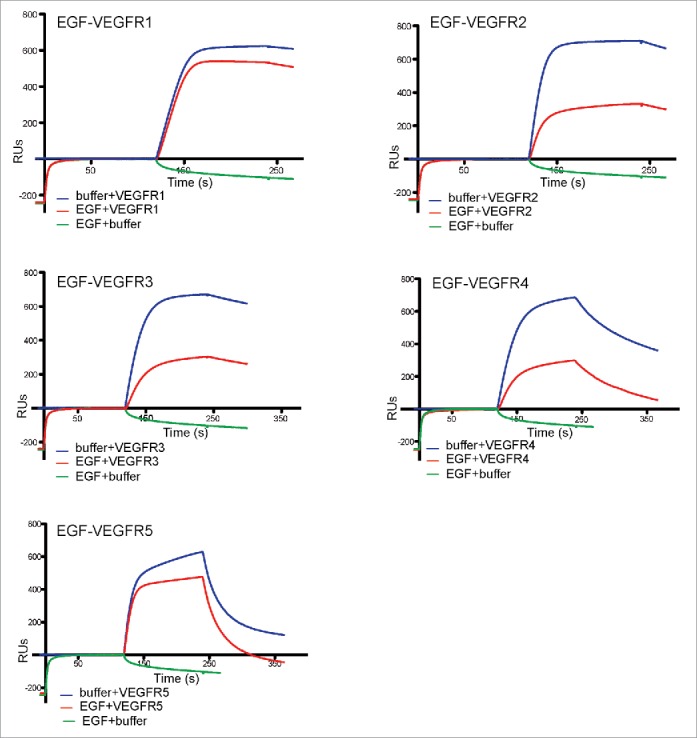
Competition of EGF and the selected Nbs for binding to eEGFR-Fc determined by SPR. SPR sensorgrams monitoring real-time association and dissociation of the indicated purified Nb (VEGFR1-5) at 100 nM to eEGFR-Fc immobilized onto a CM5 sensor chip in the presence or absence of pre-bound EGF (300 nM). Sensorgrams are aligned at the point of Nb injection. Since EGF starts to dissociate from eEGFR-Fc at the time of Nb injection (see sensorgrams EGF+buffer), this reduction in RUs of the sensorgrams with Nbs when EGF was pre-bound does not indicate competition. This is the case of Nbs VEGFR1 and VEGFR5. Higher reductions in RUs when EGF was pre-bound are indicative of EGF competition, as is the case for Nbs VEGFR2, VEGFR3 and VEGFR4, whose binding is partially abrogated in the presence of pre-bound EGF.
We also wanted to investigate whether E. coli display and flow cytometry could be used to determine EGF competition to these Nbs. Bacteria displaying VEGFR1-6 were analyzed by flow cytometry after incubation with biotinylated eEGFR-Fc (50 nM) in the presence or absence of EGF (25 µM). We found that clones VEGFR2, VEGFR3, VEGFR4, and VEGFR6 had clearly reduced binding signals to eEGFR-Fc in the presence of EGF (Fig. 9). On the other hand, binding of VEGFR1 and VEGFR5 was either not affected, or only marginally affected in the presence of EGF (Fig. 9). Therefore, flow cytometry data with E. coli bacteria displaying the Nbs and SPR data with the purified Nbs were in perfect agreement and indicated that both EGF-competitive and non-competitive VHHs binding EGFR were selected by E. coli display using MACS and CellS.
Figure 9.
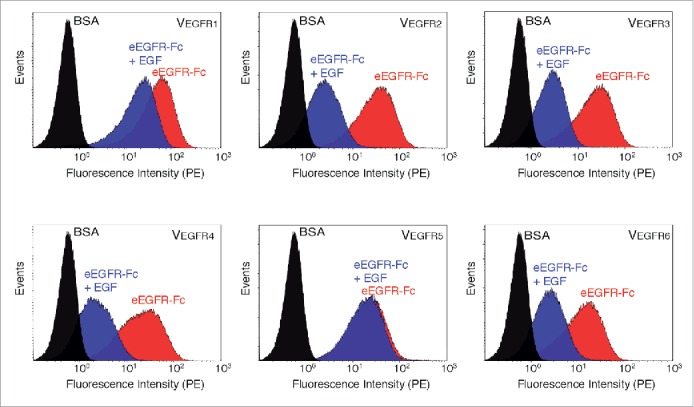
Competition of EGF and the selected Nbs for binding to eEGFR-Fc determined by flow cytometry analysis of E. coli bacteria. Flow cytometry analysis of induced E. coli EcM1 bacteria displaying the indicated Nb (VEGFR1-6) and incubated with 50 nM of biotinylated BSA (black) or eEGFR-Fc in the absence (red) or presence (blue) of 25 µM EGF. Overlay of histograms showing the fluorescence intensity of bacteria after staining with secondary Streptavidin-PE.
Discussion
Selection of Nbs against cell surface proteins is of great interest for therapeutic and diagnostic applications,27,28 and direct selection on live cells (CellS) is advantageous for identification of Nbs binding epitopes in vivo or when antigens are difficult to produce.22 In this work, we demonstrated that E. coli display enables the efficient isolation of high-affinity Nbs against a cell surface receptor protein expressed on human tumor cells (i.e., EGFR) by screening an immune VHH library directly on live intact cells. We used a whole cell-based strategy employing a VHH library generated by llama immunization with a human tumor cell line overexpressing EGFR (i.e., A431) and performed selections on a mouse cell line transfected to express human EGFR (i.e., Her14) after an initial subtractive selection step with the non-transfected parental mouse cell line (i.e., 3T32.2). A simple visual screening of E. coli bacteria bound to transfected Her14 cells, and not to control 3T32.2 cells, in light microscopy was sufficient to identify clones binding the target cell-surface antigen, EGFR. One clear advantage of this approach is that it can be easily extended to other cell surface antigens that may be transfected and expressed on mammalian cell lines. In parallel, we performed selections with purified eEGFR-Fc to compare the relative efficiency of using purified antigen and MACS versus CellS. Both approaches successfully isolated high-affinity Nbs against EGFR, although the efficiency of selection with purified eEGFR-Fc and MACS was higher than that of CellS. The percentage of bacteria binding eEGFR-Fc in flow cytometry after 2 rounds of MACS was ca. 74%, whereas that of CellS was ca. 35%. Upon screening of 96 random clones after MACS2, 87 bound eEGFR-Fc in flow cytometry. In contrast, of 96 randomly picked clones from CellS, 25 clones bound specifically to Her14 cells and eEGFR-Fc. Despite the reduced efficiency, the percentage of specific binders obtained by E. coli display and subtractive CellS method is higher than that reported by phage display of VHH libraries with subtractive CellS, and similar to that obtained with optimized masked-CellS on transfected cells.22 In addition, E. coli display has some interesting practical advantages, in particular with regard to the direct screening of bacteria binding target cells for selection and the use of flow cytometry for rapid characterization of the antigen-binding properties of the selected Nbs even before protein purification. In this context, it is worth mentioning the good correlation found between SPR data (using purified Nbs) and flow cytometry data (with E. coli bacteria) when the competition between EGF and Nbs for binding to EGFR-Fc was analyzed.
Among the 6 different anti-EGFR Nbs selected, both CellS and MACS screenings identified high affinity binders against different epitopes of EGFR, some of which compete with the EGF ligand. Four different Nbs were identified among the 25 positive clones selected by CellS that bound specifically to Her14 cells (VEGFR1, VEGFR2, VEGFR4, VEGFR6). Among them, Nbs VEGFR2, VEGFR4 (and VEGFR6 likely) binds to an epitope of EGFR that at least partially overlaps with the binding site of EGF or prevents the extracellular region of EGFR from adopting the extended conformation needed for binding to the ligand, EGF. On the other hand, Nb VEGFR1 binds to a unique epitope and has no effect on the EGF binding to EGFR. It is probable that screening a higher number of clones from CellS could have identified additional Nb sequences binding EGFR. Nonetheless, the screening of 96 clones from each selection method was sufficient to compare the efficiencies of MACS and CellS and identify a good variety of Nbs. The more frequent Nbs (VEGFR1, VEGFR2 and VEGFR4) were found in both MACS and CellS, whereas Nbs from single clones (VEGFR3, VEGFR5) were identified only in MACS, likely due the higher number of EGFR-positive binders analyzed (87/96). The Nb VEGFR3 competes with EGF and with the binding of Nbs VEGFR2 and VEGFR4, which shows that they bind overlapping epitopes of EGFR. On the contrary, Nb VEGFR5 binds a distinct epitope, not recognized by any of the identified Nb clones and does not compete with EGF for binding.
The Nb clone (VEGFR6) with lowest affinity was isolated in 2 of the 25 positive clones from CellS, but not in MACS. Although serendipity cannot be totally ruled out during the random sampling of bacterial colonies, this finding could suggest that E. coli display and selection on cells might not only favor the isolation of high affinity binders, but also lead to the identification of low affinity clones due to avidity effects caused by the display of multiple molecules of antigen and Nbs on the surface of cells and bacteria, respectively. It has been estimated that ca. 105 EGFR molecules are expressed on Her14 cells38,41 and 6000 – 8000 Nbs are displayed on E. coli surface with the intimin vector pNeae2.23 Further, in addition to the known dimerization of EGFR (and eEGFR-Fc),42 the intimin β-barrel is reported to form dimers on the bacterial surface.43 Thus, an avidity effect caused by the expression of high levels of EGFR on the tumor cell plasma membrane and that of Nbs on the bacterial surface, along with their dimerization capacity, could explain the significant binding of bacteria displaying VEGFR6 to Her14 cells and eEGFR-Fc, whereas the monomeric Nb VEGFR6 only stained weakly Her14 cells and bound eEGFR-Fc at high concentrations (> 100 nM). However, this Nb could target a unique epitope, thus enlarging the panel of specific Abs selected by E. coli display. Increased avidity caused by dimerization or multimerization of Ab molecules has also been reported in phage display and yeast display,44-46 and is a commonly engineered property in Ab fragments to enhance binding.9 Despite the possibility that avidity caused by multimeric display of Nbs on E. coli surface and of target antigen on the plasma cell membrane could have a positive effect on the selection of low affinity binders, it is clear that this effect is not significant enough to interfere with the selection of high-affinity binders, since clones of high affinity were equally predominant in both CellS and MACS with soluble eEGFR-Fc.
In conclusion, our results demonstrate that E. coli display and selection on live cells is an effective method for the isolation of specific and high affinity Nbs capable of binding different epitopes on a cell-surface antigen, such as EGFR. The immune library can be obtained from animals immunized with tumor cells overexpressing the cell surface antigen (e.g., A431) and selections can be performed by simply incubating bacteria with tumor cells of different origin (e.g., mouse) expressing the target antigen (e.g., EGFR) by transfection. Also, a library generated by cell-based immunization can be used to select Abs against many target cell surface antigens in parallel. Finally, E. coli display also enables the rapid characterization of the selected Nbs using flow cytometry for analysis of antigen binding or competition with ligand prior to Nb purification and further in vitro analysis.
Materials and methods
Bacterial strains, growth and induction conditions
The E. coli strains used in this work are listed in Table S1. Bacteria carrying any plasmid vector with a VHH gene were grown at 30°C in LB liquid medium or on agar plates with the appropriate antibiotic [ampicillin (Ap) or chloramphenicol (Cm)] for plasmid selection. Otherwise bacteria were grown at 37°C in LB. LB plates and pre-inoculum media prior to induction contained 2% (w/v) glucose for repression of the lac promoter. The preinocula cultures were started from individual colonies (for single clones) or from a mixture of clones (in case of libraries), freshly grown and harvested from plates, diluted to an initial OD600 of 0.5, and grown overnight (o/n) under static conditions. For induction of pNeae2-derivatives, bacteria (corresponding to an OD600 of 0.5) were harvested by centrifugation (4000 × g, 5 min), and grown in the same media with 0.05 mM IPTG, but without glucose for 3 h with agitation (160 rpm), unless indicated otherwise. Upon plating on LB agar, an OD600 of 1.0 of these cultures resulted in about ∼4 × 108 CFU/ml. For over-expression of soluble VHH in the periplasm, E. coli WK6 cells with the corresponding pHEN6-derivative (ApR) were grown to an OD600 of 0.6–0.8 and induced overnight at 30°C with 1 mM IPTG.
Growth of mammalian cell cultures
The mouse fibroblast cell lines NIH 3T3 2.2 and Her14 were grown as monolayers in Dulbecco's modified Eagle's medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS, Sigma) and 2 mM L-glutamine (complete DMEM), at 37°C with 5% CO2. In experiments of selection of E. coli display libraries on cells, growing cells in culture were trypsinized and seeded in 6-well plates (Falcon) containing growth media to a confluency of ∼20% the day before the experiment. For immunofluorescence microscopy (IFM) experiments, cells were seeded and grown on circular coverslips (previously irradiated with UV for 30 mins) placed in 24-well plates (Falcon), prior to infection with bacteria, staining and analysis under the microscope.
Plasmid and library construction and oligonucleotides
Plasmids used in this study are summarized in Table S1. DNA manipulation, ligation, transformation and plasmid preparation were performed following standard molecular cloning techniques. The E. coli DH10B-T1R strain was used for plasmid propagation, isolation and cloning experiments. DNA constructs were sequenced by Secugen SL. PCR reactions for cloning were performed with proof-reading Vent DNA polymerase (New England Biolabs) or Taq DNA polymerase (Roche). Oligonucleotides used for DNA sequencing and amplification are listed in Table S4. Oligonucleotides were synthesized by Sigma Genosys, except those used for VHH amplification (VHH-Sfi2, VHH-Not2), which were from Scandinavian Gene Synthesis (SGS). The plasmid pNeae2 (CmR) carries the lacI and lac promoter region controlling a gene fusion between intimin residues 1–654 (from EHEC O157:H7 strain EDL933stx-) followed by the E-tag (GAPVPYPDLEPA), the hexahistidine (6xHis) epitope (flanked by SfiI and NotI sites), and a C-terminal myc-tag (EQKLISEED).23 The pool of amplified VHH cloned in the phagemid vector pUR5068,47 which is identical to pHEN6,48 were excised from the pool of phagemid DNA by SfiI and NotI digestion and cloned into the same sites of E. coli display vector pNeae2, replacing the 6xHis tag. Ligations were electroporated into E. coli DH10B-T1R for library construction with CFU determined in serial dilutions to determine library size. Plasmids from this library were prepared from bacteria harvested from LB-Cm-glucose plates and electroporated into E. coli EcM1 strain for selections. The pHEN6 (ApR) vector is a phagemid derivative of pHEN149 carrying the lac promoter controlling a gene fusion encoding a signal peptide (PelBss), SfiI and NotI sites, and C-terminal 6xHis and myc-tag epitopes. Plasmid derivatives of pHEN6 with cloned VHH in SfiI-NotI sites were employed for periplasmic expression of soluble Nbs into E. coli WK6 strain.50
Protein extracts and SDS-PAGE
Whole cell protein extracts were prepared by harvesting bacteria after induction (1 ml of OD600 1.5), resuspended in 50 μl of 10 mM Tris HCl pH 8.0, mixed with the same volume of SDS-sample buffer (2X) or urea-SDS sample buffer (2X) -for samples with intimin fusions, as reported previously.24 Samples were boiled 10 min or 30 min (for samples with intimin fusions), sonicated briefly (5 sec; Labsonic B Braun), centrifuged (14,000 × g, 5 min) to pellet insoluble material, loaded onto 10% SDS-PAGE gels and run using a Miniprotean III electrophoresis system (Bio-Rad). Proteins were either stained with Coomassie or transferred to a membrane for Western blot for specific detection of myc-tagged proteins with anti-myc mAb clone 9B11 (1:2000; Cell Signaling Technology 2276S) and anti-mouse IgG-peroxidase (POD) conjugate (1:5000; Sigma A-2554), as previously described.23
Protein biotinylation
Biotinamidocaproate N-hydroxysuccinimide ester (Biotin-NHS; Sigma) was re-constituted at 25 mg/ml in dimethylsulfoxide (DMSO, Fluka) and immediately used. Purified protein (0.05–0.5 mg) [human eEGFR-Fc (R&D Biosystems), GFP (Upstate, Merck Millipore) and BSA (Sigma)] was mixed with biotin-NHS (20-fold molar excess) in 1 ml of PBS and incubated for 2 h at room temperature (RT). The reaction was stopped by addition of Tris-HCl (pH 7.5) at a final concentration of 50 mM and the samples were placed on ice for 1 h. The reaction mix was loaded onto a pre-packed column for gel filtration chromatography (Sephadex G25 PD-10; GE Healthcare) and the biotinylated protein was eluted in 500-μl fractions with PBS. Protein concentration was estimated using the Bicinchoninic acid (BCA) Pierce protein assay kit (Thermo Scientific).
Magnetic cell sorting (MACS)
Induced E. coli bacteria (equivalent to a final OD600 of 5.0) were harvested by centrifugation (4000 × g, 3 min), washed 3 times with 2 ml PBS (sterile filtered and degassed), and resuspended in a final volume of 1 ml of PBS. Biotinylated EGFR-Fc (100 nM) was added to 100 μl of bacteria, the final volume was adjusted to 200 μl with PBS-BSA (PBS supplemented with 0.5% w/v BSA, sterile filtered and degassed), and incubation was carried out at RT for 1 h. After incubation, bacteria were washed 3 times with 1 ml of PBS-BSA, resuspended in 100 μl of the same buffer containing 20 μl of anti-biotin paramagnetic beads (Miltenyi Biotec) and incubated at 4°C for 20 min. Next, bacteria were washed 3 times with 1 ml of PBS-BSA, resuspended in 500 μl of the same buffer, of which 10 μl was kept aside to calculate the input bacteria before the procedure, while the rest (490 μl) was applied onto a MACS MS column (Miltenyi Biotec), previously equilibrated with 500 μl of PBS-BSA and placed on the OctoMACS Separator (Miltenyi Biotec). The flow through of unbound cells was collected and the column was washed 3 times with 500 μl of PBS-BSA. The wash was combined with the flow-through as “unbound fraction.” Next, the column was removed from the OctoMACS Separator and placed onto a new collection tube, 2 ml of LB was added and the cells were eluted. This fraction was labeled as the “Bound fraction.” Serial dilutions of Unbound and Bound fractions were plated to determine CFU and to harvest Bound bacteria. All experiments were carried out in duplicates.
Selection of E. coli display VHH libraries on cells
Mouse fibroblast cell lines NIH-3T3 2.2 (EGFR-) and Her14 (EGFR+)39,51 were grown as monolayers in 6-well culture plates (BD Falcon) containing culture media to a confluency of ∼40%, i.e., ∼6 × 105 cells, and washed with Hank's balanced salt solution (HBSS, Sigma). Induced E. coli bacteria (equivalent to a final OD600 of 1) were harvested by centrifugation (4000 × g, 3 min) and washed with 2 ml HBSS. Washed bacteria (300 μl; ca. ∼6 × 107 bacteria) were added to wells containing NIH-3T3 2.2 cells and incubated for 1 h at 37°C. After incubation, the unbound bacteria were recovered, added to wells containing Her14 cells, and incubated for 15 mins at 37°C. Next, the Her14 cells were washed 3 times with 1 ml HBSS, to remove any non-specifically bound bacteria and subsequently lysed with HBSS supplemented with 0.2% SDS and 0.1% DNAse. Serial dilutions of the cell lysate containing bacteria were plated to determine CFU of bacteria recovered after the procedure. All experiments were carried out in duplicates.
Flow cytometry analysis of bacteria
Induced bacterial cells (equivalent to a final OD600 of 1.0; ∼4 × 108 CFU) were harvested by centrifugation (4,000 × g, 3 min), washed twice with 500 μl of PBS (filter-sterilized) and resuspended in a final volume of 400 μl of PBS. Next, 190 μl of this cell suspension (∼2 × 108 CFU) was incubated with the primary antibody or antigen (50 nM eEGFR-Fc, in the presence or absence of its ligand; 25 μM EGF as indicated) and PBS was added to adjust the total volume to 200 μl. The primary antibody (for assay of expression level) was anti-myc mAb clone 9B11 (1:200; Cell Signaling Technology 2276S), while biotinylated antigens (EGFR-Fc, BSA) were used at 50 nM for assay of antigen binding, unless otherwise indicated. The samples were incubated at RT for 1h. After incubation, the cells were washed once with 500 μl of PBS, and resuspended either in 500 μl of PBS containing 1 μl of goat anti-mouse-IgG1 conjugated to Alexa 488 Fluor (2 mg/ml, Invitrogen A-21121) or in 200 μl of PBS containing 30 μl of 1:200 dilution of streptavidin-phycoerythrin (PE) (0.5 mg/ml, Beckman Coulter). The mixture was incubated for 30 min at 4°C in the dark. The cells were washed once with 500 μl of PBS and resuspended in a final volume of 1 ml in PBS. For each experiment, at least 100,000 cells were analyzed in a cytometer (Gallios, Beckman Coulter).
Purification of Nbs from the periplasm of E. coli
Soluble Nbs with 6xHisHis and myc tags in their C-termini were induced in the periplasm of E. coli WK6 cells carrying pHEN6-derivatives. Bacteria were harvested by centrifugation (4000 × g, 15 min, 4°C) from 2 L cultures, resuspended in 30 ml Periplasmic Extraction buffer [50 mM sodium phosphate pH 7.4, 200 mM NaCl, 5 mM EDTA and 1 mg/ml polymyxin B sulfate (Sigma)] and stirred at 4°C for 3 h using an orbital shaker. The periplasmic extract was obtained by centrifugation (20000 × g, 30 min, 4°C) and dialyzed o/n at 4°C against 5 L of column buffer (CB: 50 mM sodium phosphate pH 7.4, 200 mM NaCl). Dialyzed extract was loaded onto a Cobalt-containing affinity resin (Talon, Clontech), washed, and bound protein eluted in CB containing 150 mM imidazole. Eluted Nb was dialyzed against HEPES-buffer (20 mM HEPES pH 7.4, 200 mM NaCl, sterile filtered and degassed), concentrated to 1 ml using 3-kDa centrifugal filter unit (Amicon Ultra-15), and loaded onto a gel filtration column (HiLoad 16/600 Superdex 75 preparative grade, GE Healthcare). The gel filtration column was calibrated with protein markers (Bio-Rad) and equilibrated with 3 column volumes of HEPES-buffer prior to use. The fractions corresponding to the monomeric Nbs (apparent molecular mass of ca. 17–25 kDa) were collected and concentrated in a 3-kDa centrifugal filter unit (Amicon Ultra-15). Protein concentration was estimated using the bicinchoninic acid (BCA) Pierce protein assay kit (Thermo Scientific).
Enzyme-linked immunosorbent assay
Human eEGFR-Fc (R&D Biosystems) or BSA (Sigma) was adsorbed at 4°C o/n onto 96-well immunoplates (Maxisorb; Nunc) at a concentration of 5 μg/ml in PBS. All steps were subsequently done at room temperature. Immunoplates were washed in PBS and blocked by incubation with 200 μl of 3% (w/v) Milk-PBS for 2 h. The purified Nbs were diluted in 3% (w/v) Milk-PBS, added at the indicated concentrations (0.05–1000 nM) in duplicates and incubated for 1 h. After incubation, the wells were washed 3 times with PBS (Immunowash 1575, Bio-Rad) and the bound Nbs were detected by the addition of anti-myc mAb (1:1000; Cell Signaling Technology 2276S) followed by anti-mouse IgG-peroxidase (POD) conjugate (1:1000; Sigma A-2554), and incubation of the plates for 1 h. The plates were washed 3 times with PBS and developed with H2O2 and o-phenylenediamine (OPD; Sigma), as described previously.52 The plates were read at 490 nm using the iMark ELISA plate reader (Bio-Rad).
Assay of bacterial adhesion to in vitro cultured cells for immunofluorescence microscopy
Induced E. coli bacteria (1OD600, ∼4 × 108 bacteria) were harvested by centrifugation (4000 × g, 3 min), washed in HBSS and resuspended at 4 × 107 CFU/ml in the same buffer. For an infection at MOI 100:1, a 1 ml sample of this bacterial suspension was added per well to a 24-well cell culture plate having the indicated cell line (NIH-3T3 2.2 or Her14, ∼5 × 104 cells/well). The cell lines were grown on sterile coverslips (13 mm diameter, VWR International) placed at the bottom of the well. After 20 min infection at 37°C, the wells were aspirated and washed 3 times with 1 ml HBSS at RT. The coverslips were fixed in a 4% (w/v) paraformaldehyde solution for 20 min at RT and washed 3 times with 1 ml HBSS. Coverslips were blocked and stained for 45 mins in a wet chamber at RT with 50 μl of PBS-10% goat serum solution containing primary Abs, i.e., anti-E. coli all antigens rabbit polyclonal serum (1:1500, Amsbio B65003R-1) and anti-human EGFR mouse mAb clone 528 (1:750, EMD Millipore GR01). The coverslips were washed by immersion 15 times in a large volume of HBSS (200 ml), placed again in the wet chamber and incubated with 50 μl of PBS-10% goat serum solution containing secondary Abs, i.e., goat anti-rabbit IgG conjugated to Alexa 594 Fluor (1:500, Life Technologies A-11012), in order to stain E. coli red, rabbit anti-mouse IgG conjugated to Alexa 488 Fluor (1:500, Life Technologies A-11059), to stain EGFR as green and 4′,6-diamidino-2-phenilindole (DAPI, Life Technologies) to stain nuclear material. Next, the coverslips were washed in PBS as above, the excess liquid was removed by touching a kimwipe to the edge of the coverslip, and the coverslips were mounted with 2 μl ProLong Gold anti-fade reagent (Life Technologies) on glass slides. The samples were visualized using an epifluorescence microscopy (Zeiss Axioimager microscope).
Staining of in vitro cultured cells with Nbs for immunofluorescence microscopy
The mouse cell lines NIH-3T3 2.2 or Her14 were seeded (∼2 × 104 cells/well) and grown on sterile coverslips (13 mm diameter, VWR International) placed at the bottom of the wells in a 24-well tissue culture plate for 36 hrs. The wells were aspirated and washed once with 1 ml HBSS at RT. Coverslips were blocked and stained for 45 mins in a wet chamber at RT with 50 μl of PBS-10% goat serum solution containing primary Ab, i.e., anti-human EGFR mouse mAb clone 528 (1:750, EMD Millipore GR01) or the indicated Nb in HBSS (500 nM). The coverslips were washed by immersion 15 times in a large volume of HBSS (200 ml), fixed in a 4% (w/v) paraformaldehyde solution for 20 min at RT and washed 3 times with 1 ml HBSS. The coverslips were washed by immersion 15 times in a large volume of HBS (200 ml), placed in the wet chamber and incubated for 45 min with 50 μl of PBS-10% goat serum solution containing anti-myc mAb clone 9B11 (1:500; Cell Signaling Technology 2276S) and Phalloidin-Alexa Fluor 594 conjugate (1:500; Life Technologies) for F-actin staining. Next, the coverslips were washed by immersion 10 times in a large volume of HBSS (200 ml), placed in the wet chamber and incubated with 50 μl of PBS/10% goat serum solution containing secondary Ab, i.e., Goat anti-mouse IgG1 conjugated to Alexa 488 Fluor (1:500, Life Technologies A-21121). Coverslips were washed in PBS and mounted with 2 μl ProLong Gold anti-fade reagent (Life Technologies) on glass slides, as described above. The samples were visualized using confocal microscopy (Leica TCS SP5 multispectral confocal system).
Affinity determination and competition assays using surface plasmon resonance
SPR measurements were performed using a Biacore 3000 instrument (GE Healthcare). All proteins solutions were dialyzed against HEPES-buffer [20 mM HEPES, 200 mM NaCl (pH 7.4), sterile filtered and degassed] at 4°C o/n. For affinity determination assays, human eEGFR-Fc (R&D Biosystems) (1000 response units, RUs) was immobilized on a CM5 chip (GE Healthcare) using the amino-coupling method by injection of EDC/NHS at 5 μl/min for 6 min. Then, eEGFR-Fc at 5 μg/ml in 10 mM Sodium Acetate (NaAc) pH 5.0 was injected for 2 min. Finally, the immobilization process was stopped by injection of 1 M ethanolamine-HCl, pH 8.5 for 6 min. For competition assays, eEGFR-Fc was immobilized at 6000 RUs. For this immobilization, EDC/NHS at 5 μl/min was injected for 7 min, followed by injection of eEGFR-Fc at 10 μg/ml in 10 mM NaAc (pH 5.0) for 7 min, and the blocking of the immobilization process by injection of 1 M ethanolamine-HCl, pH 8.5 for 7 min. For the determination of binding kinetics, dilutions of purified Nb (analyte) at the indicated concentrations were injected at a flow of 30 μl/min. Then, HEPES-buffer was injected at the same flow rate. For competition assays, VEGFR1-5 were diluted to 100 nM and EGF at 300 nM. The two competitor molecules were injected sequentially at a flow of 30 μl/min. Sensorgrams were generated and the chip was regenerated after every cycle using 3 injections (10 μl) of 10 mM glycine-HCl pH 2.5. For binding kinetics, sensorgrams with the different concentrations of analyte were overlaid, aligned and analyzed with BIAevaluation 4.1 software (GE Healthcare) compared to predicted 1:1 binding model. All data were processed using a double-referencing method.53
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of Protein Tools unit of CNB-CSIC for their excellent technical support on SPR experiments. Work in the laboratory of LAF is supported by research grants from the Spanish Ministerio de Economía y Competitividad BIO2014-60305R, BACFITERed SAF2014-56716-REDT, Comunidad Autónoma de Madrid (S2010-BMD-2312), and the European Research Council (ERC-2012-ADG_20120314). VS and CM obtained PhD contracts of La Caixa Foundation. CP was supported by FPI contract BES-2009-024051 from MINECO.
Supplemental Material
Supplemental Material may be downloaded here: publisher's website
References
- 1.Redman JM, Hill EM, AlDeghaither D, Weiner LM. Mechanisms of action of therapeutic antibodies for cancer. Mol Immunol 2015; 67:28-45; PMID:25911943; http://dx.doi.org/ 10.1016/j.molimm.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer 2012; 12:278-87; PMID:22437872; http://dx.doi.org/ 10.1038/nrc3236 [DOI] [PubMed] [Google Scholar]
- 3.Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol 2015; 33:1974-82; PMID:25605845; http://dx.doi.org/ 10.1200/JCO.2014.59.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al.. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014; 515:568-71; PMID:25428505; http://dx.doi.org/ 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salanti A, Clausen TM, Agerbaek MO, Al Nakouzi N, Dahlback M, Oo HZ, Lee S, Gustavsson T, Rich JR, Hedberg BJ, et al.. Targeting Human Cancer by a Glycosaminoglycan Binding Malaria Protein. Cancer Cell 2015; 28:500-14; PMID:26461094; http://dx.doi.org/ 10.1016/j.ccell.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SY, Theunissen JW, Balibalos J, Liao-Chan S, Babcock MC, Wong T, Cairns B, Gonzalez D, van der Horst EH, Perez M, et al.. A novel antibody-drug conjugate targeting SAIL for the treatment of hematologic malignancies. Blood Cancer J 2015; 5:e316; PMID:26024286; http://dx.doi.org/ 10.1038/bcj.2015.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol 2010; 10:345-52; PMID:20414207; http://dx.doi.org/ 10.1038/nri2747 [DOI] [PubMed] [Google Scholar]
- 8.Frenzel A, Schirrmann T, Hust M. Phage display-derived human antibodies in clinical development and therapy. mAbs 2016; in press; PMID:27416017; http://dx.doi.org/ 10.1080/19420862.2016.1212149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuesta ÁM, Sainz-Pastor N, Bonet J, Oliva B, Álvarez-Vallina L. Multivalent antibodies: when design surpasses evolution. Trends Biotechnol 2010; 28:355-62; PMID:20447706; http://dx.doi.org/ 10.1016/j.tibtech.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 10.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol 2005; 23:1126-36; PMID:16151406; http://dx.doi.org/ 10.1038/nbt1142 [DOI] [PubMed] [Google Scholar]
- 11.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23:1105-16; PMID:16151404; http://dx.doi.org/ 10.1038/nbt1126 [DOI] [PubMed] [Google Scholar]
- 12.Hoogenboom HR. Overview of antibody phage-display technology and its applications. Methods Mol Biol 2002; 178:1-37; PMID:11968478; http://dx.doi.org/ 10.1385/1-59259-240-6:001 [DOI] [PubMed] [Google Scholar]
- 13.Hoogenboom HR, Lutgerink JT, Pelsers MM, Rousch MJ, Coote J, Van Neer N, De Bruine A, Van Nieuwenhoven FA, Glatz JF, Arends JW. Selection-dominant and nonaccessible epitopes on cell-surface receptors revealed by cell-panning with a large phage antibody library. Eur J Biochem 1999; 260:774-84; PMID:10103007; http://dx.doi.org/ 10.1046/j.1432-1327.1999.00214.x [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Conrad F, Cooperberg MR, Kirpotin DB, Marks JD. Mapping tumor epitope space by direct selection of single-chain Fv antibody libraries on prostate cancer cells. Cancer Res 2004; 64:704-10; PMID:14744788; http://dx.doi.org/ 10.1158/0008-5472.CAN-03-2732 [DOI] [PubMed] [Google Scholar]
- 15.Siva AC, Kirkland RE, Lin B, Maruyama T, McWhirter J, Yantiri-Wernimont F, Bowdish KS, Xin H. Selection of anti-cancer antibodies from combinatorial libraries by whole-cell panning and stringent subtraction with human blood cells. J Immunol Methods 2008; 330:109-19; PMID:18096183; http://dx.doi.org/ 10.1016/j.jim.2007.11.008 [DOI] [PubMed] [Google Scholar]
- 16.Figini M, Obici L, Mezzanzanica D, Griffiths A, Colnaghi MI, Winter G, Canevari S. Panning phage antibody libraries on cells: isolation of human Fab fragments against ovarian carcinoma using guided selection. Cancer Res 1998; 58:991-6; PMID:9500461 [PubMed] [Google Scholar]
- 17.Klimka A, Matthey B, Roovers RC, Barth S, Arends JW, Engert A, Hoogenboom HR. Human anti-CD30 recombinant antibodies by guided phage antibody selection using cell panning. Br J Cancer 2000; 83:252-60; PMID:10901379; http://dx.doi.org/ 10.1054/bjoc.2000.1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beiboer SH, Reurs A, Roovers RC, Arends JW, Whitelegg NR, Rees AR, Hoogenboom HR. Guided selection of a pan carcinoma specific antibody reveals similar binding characteristics yet structural divergence between the original murine antibody and its human equivalent. J Mol Biol 2000; 296:833-49; PMID:10677285; http://dx.doi.org/ 10.1006/jmbi.2000.3512 [DOI] [PubMed] [Google Scholar]
- 19.Veggiani G, Ossolengo G, Aliprandi M, Cavallaro U, de Marco A. Single-domain antibodies that compete with the natural ligand fibroblast growth factor block the internalization of the fibroblast growth factor receptor 1. Biochem Biophys Res Commun 2011; 408:692-6; PMID:21539817; http://dx.doi.org/ 10.1016/j.bbrc.2011.04.090 [DOI] [PubMed] [Google Scholar]
- 20.Lipes BD, Chen YH, Ma H, Staats HF, Kenan DJ, Gunn MD. An entirely cell-based system to generate single-chain antibodies against cell surface receptors. J Mol Biol 2008; 379:261-72; PMID:18455737; http://dx.doi.org/ 10.1016/j.jmb.2008.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavoni E, Vaccaro P, Anastasi AM, Minenkova O. Optimized selection of anti-tumor recombinant antibodies from phage libraries on intact cells. Mol Immunol 2014; 57:317-22; PMID:24240062; http://dx.doi.org/ 10.1016/j.molimm.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 22.Even-Desrumeaux K, Nevoltris D, Lavaut MN, Alim K, Borg JP, Audebert S, Kerfelec B, Baty D, Chames P. Masked selection: a straightforward and flexible approach for the selection of binders against specific epitopes and differentially expressed proteins by phage display. Mol Cell Proteomics 2014; 13:653-65; PMID:24361863; http://dx.doi.org/ 10.1074/mcp.O112.025486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salema V, Marín E, Martínez-Arteaga R, Ruano-Gallego D, Fraile S, Margolles Y, Teira X, Gutierrez C, Bodelón G, Fernández LÁ. Selection of single domain antibodies from immune libraries displayed on the surface of E. coli cells with two β-domains of opposite topologies. PLoS ONE 2013; 8:e75126; PMID:24086454; http://dx.doi.org/ 10.1371/journal.pone.0075126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodelón G, Marín E, Fernández LA. Role of periplasmic chaperones and BamA (YaeT/Omp85) in folding and secretion of Intimin from enteropathogenic Escherichia coli strains. J Bacteriol 2009; 191:5169-79; PMID:Can't; http://dx.doi.org/ 10.1128/JB.00458-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairman JW, Dautin N, Wojtowicz D, Liu W, Noinaj N, Barnard TJ, Udho E, Przytycka TM, Cherezov V, Buchanan SK. Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure 2012; 20:1233-43; PMID:22658748; http://dx.doi.org/ 10.1016/j.str.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem 2013; 82:775-97; PMID:23495938; http://dx.doi.org/ 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- 27.Kijanka M, Dorresteijn B, Oliveira S, van Bergen En Henegouwen PM. Nanobody-based cancer therapy of solid tumors. Nanomedicine (London, England) 2015; 10:161-74; PMID:25597775; http://dx.doi.org/24578722 10.2217/nnm.14.178 [DOI] [PubMed] [Google Scholar]
- 28.Chakravarty R, Goel S, Cai W. Nanobody: The “Magic Bullet” for Molecular Imaging? Theranostics 2014; 4:386-98; PMID:24578722; http://dx.doi.org/ 10.7150/thno.8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira S, Heukers R, Sornkom J, Kok RJ, van Bergen En Henegouwen PM. Targeting tumors with nanobodies for cancer imaging and therapy. J Control Release 2013; 172:607-17; PMID:24035975; http://dx.doi.org/ 10.1016/j.jconrel.2013.08.298 [DOI] [PubMed] [Google Scholar]
- 30.Capdevila J, Elez E, Macarulla T, Ramos FJ, Ruiz-Echarri M, Tabernero J. Anti-epidermal growth factor receptor monoclonal antibodies in cancer treatment. Cancer Treat Rev 2009; 35:354-63; PMID:19269105; http://dx.doi.org/ 10.1016/j.ctrv.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 31.Martinelli E, De Palma R, Orditura M, De Vita F, Ciardiello F. Anti-epidermal growth factor receptor monoclonal antibodies in cancer therapy. Clin Exp Immunol 2009; 158:1-9; PMID:19737224; http://dx.doi.org/ 10.1111/j.1365-2249.2009.03992.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira S, van Bergen en Henegouwen PM, Storm G, Schiffelers RM. Molecular biology of epidermal growth factor receptor inhibition for cancer therapy. Expert Opin Biol Ther 2006; 6:605-17; PMID:16706607; http://dx.doi.org/ 10.1517/14712598.6.6.605 [DOI] [PubMed] [Google Scholar]
- 33.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol 2005; 23:2445-59; PMID:15753456; http://dx.doi.org/ 10.1200/JCO.2005.11.890 [DOI] [PubMed] [Google Scholar]
- 34.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001; 2:127-37; PMID:11252954; http://dx.doi.org/ 10.1038/35052073 [DOI] [PubMed] [Google Scholar]
- 35.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and membrane interactions of the EGF receptor. Cell 2013; 152:557-69; PMID:23374350; http://dx.doi.org/ 10.1016/j.cell.2012.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birtwistle MR, Hatakeyama M, Yumoto N, Ogunnaike BA, Hoek JB, Kholodenko BN. Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol Syst Biol 2007; 3:144; PMID:18004277; http://dx.doi.org/ 10.1038/msb4100188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 2000; 19:3159-67; PMID:10880430; http://dx.doi.org/ 10.1093/emboj/19.13.3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roovers RC, Laeremans T, Huang L, De Taeye S, Verkleij AJ, Revets H, de Haard HJ, van Bergen en Henegouwen PM. Efficient inhibition of EGFR signaling and of tumour growth by antagonistic anti-EFGR Nanobodies. Cancer Immunol Immunother 2007; 56:303-17; PMID:16738850; http://dx.doi.org/ 10.1007/s00262-006-0180-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honegger AM, Kris RM, Ullrich A, Schlessinger J. Evidence that autophosphorylation of solubilized receptors for epidermal growth factor is mediated by intermolecular cross-phosphorylation. Proc Natl Acad Sci U S A 1989; 86:925-9; PMID:2915986; http://dx.doi.org/ 10.1073/pnas.86.3.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz KR, Bagchi A, Roovers RC, Van Bergen en Henegouwen PM, Ferguson KM. Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure 2013; 21:1214-24; PMID:23791944; http://dx.doi.org/ 10.1016/j.str.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honegger AM, Dull TJ, Felder S, Van Obberghen E, Bellot F, Szapary D, Schmidt A, Ullrich A, Schlessinger J. Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell 1987; 51:199-209; PMID:3499230; http://dx.doi.org/ 10.1016/0092-8674(87)90147-4 [DOI] [PubMed] [Google Scholar]
- 42.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, et al.. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 2013; 152:543-56; PMID:23374349; http://dx.doi.org/ 10.1016/j.cell.2012.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leo JC, Oberhettinger P, Chaubey M, Schutz M, Kuhner D, Bertsche U, Schwarz H, Gotz F, Autenrieth IB, Coles M, et al.. The Intimin periplasmic domain mediates dimerisation and binding to peptidoglycan. Mol Microbiol 2015; 95:80-100; PMID:25353290; http://dx.doi.org/ 10.1111/mmi.12840 [DOI] [PubMed] [Google Scholar]
- 44.Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: Where are we now? Combinatorial chemistry & high throughput screening 2008; 11:127-34; PMID:18336206; http://dx.doi.org/ 10.2174/138620708783744516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee CV, Sidhu SS, Fuh G. Bivalent antibody phage display mimics natural immunoglobulin. J Immunol Methods 2004; 284:119-32; PMID:14736422; http://dx.doi.org/ 10.1016/j.jim.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 46.Rondot S, Koch J, Breitling F, Dubel S. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol 2001; 19:75-8; PMID:11135557; http://dx.doi.org/ 10.1038/83567 [DOI] [PubMed] [Google Scholar]
- 47.De Haard HJ, Bezemer S, Ledeboer AM, Muller WH, Boender PJ, Moineau S, Coppelmans MC, Verkleij AJ, Frenken LG, Verrips CT. Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection. J Bacteriol 2005; 187:4531-41; PMID:15968064; http://dx.doi.org/ 10.1128/JB.187.13.4531-4541.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother 2001; 45:2807-12; PMID:11557473; http://dx.doi.org/ 10.1128/AAC.45.10.2807-2812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res 1991; 19:4133-7; PMID:1908075; http://dx.doi.org/ 10.1093/nar/19.15.4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zell R, Fritz HJ. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J 1987; 6:1809-15; PMID:3038536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felder S, LaVin J, Ullrich A, Schlessinger J. Kinetics of binding, endocytosis, and recycling of EGF receptor mutants. J Cell Biol 1992; 117:203-12; PMID:1556153; http://dx.doi.org/ 10.1083/jcb.117.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jurado P, Ritz D, Beckwith J, de Lorenzo V, Fernández LA. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J Mol Biol 2002; 320:1-10; PMID:12079330; http://dx.doi.org/ 10.1016/S0022-2836(02)00405-9 [DOI] [PubMed] [Google Scholar]
- 53.Myszka DG. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol 2000; 323:325-40; PMID:10944758; http://dx.doi.org/ 10.1016/S0076-6879(00)23372-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.