Abstract
To investigate the clinicopathologic characteristics and the prognostic impact of PIK3CA gene amplification in curatively resected esophageal squamous cell carcinoma (ESCC). Using 534 curatively resected ESCCs, the PIK3CA gene copy number was evaluated with fluorescent in situ hybridization. PIK3CA amplification was defined as PIK3CA/centromere 3 ratio is ≥ 2.0 or average number of PIK3CA signals/tumor cell nucleus ≥ 5.0. PIK3CA mutations in exon 9 and 20, encoding the highly conserved helical and kinase domains were assessed by direct sequencing in 388 cases. PIK3CA amplification was detected in 56 (10.5%) cases. PIK3CA amplification was significantly associated with higher T-stage (P=0.026) and pathologic stage (P=0.053). PIK3CA amplification showed a significantly shorter disease free survival (DFS) compared with that of non-amplified group (33.4 vs 63.1 months, P=0.019). After adjusting for gender, tumor location, pathologic stage, histologic grade and adjuvant treatment, PIK3CA amplification was significantly associated with a shorter DFS (adjusted hazard ratio [AHR] 1.53; 95% CI, 1.10-2.17; P=0.02). Though the statistical insignificance, PIK3CA amplification showed tendency of shorter OS (52.1 vs 96.5 moths, P=0.116). PIK3CA mutations were detected in 6 (1.5%) of 388 cases; 5 cases with exon 9 mutations in E545K while one exon 20 mutation in H1047L. PIK3CA amplification is a frequent oncogenic alteration and associated with shorter survival, suggesting its role as a prognostic biomarker in resected ESCC. PIK3CA amplification may represent a promising therapeutic target for ESCC.
Keywords: PIK3CA, esophageal squamous cell carcinoma, amplification, mutation, fluorescent in situ hybridization
INTRODUCTION
Esophageal cancer (EC) is the sixth most common cause of cancer death and the eighth most common cancer worldwide [1]. Despite the improvement in diagnosis and multidisciplinary treatment, the prognosis remains poor, even for patients who undergo complete resection [2]. Overall, though the conventional chemotherapy incorporating 5-fluorouracil, cisplatin, irinotecan and taxane, median overall survival remains 10-12 months with frequent toxicities [3]. The limited improvement with conventional therapies prompts us to explore the molecular biology and identify of prognostic and druggable biomarkers.
There are two main histologic types of esophageal cancer, esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). EAC is associated with gastroesophageal reflux disease, Barrett's esophagus, and obesity [4–6], predominant in the United States and most other Western countries. However, ESCC dominates with 80% of all cases worldwide and predominant in Asia. In contrast to EAC, smoking and alcohol abuse contribute to the development of ESCC [7]. With those distinct biologic and epidemiologic differences, EAC and ESCC may need different therapeutic approaches. Over the past decade, molecular targeted therapy blocking important oncogenic pathway including human epidermal growth factor receptor-2 (HER2; also known as ERBB2) have led to remarkable progress in EAC including human epidermal growth factor receptor-2 (HER2; also known as ERBB2) [8]. Despite the improved outcome of EAC, ESCC still lacks therapeutically relevant predictive or prognostic target. A comparative genomic study revealed different genetic alterations between EAC and ESCC [9, 10]. Copy number gains of SOX2, PIK3CA, CCND1, and FGFR1 were more frequent in ESCC than in EAC, implicating these genes as therapeutic targets for ESCC. Very recently, we reported that FGFR1 amplification is frequently observed and an independent prognostic factor in resected ESCC [11]. Taken together, PIK3CA may also represent an attractive molecular target for ESCC.
Phosphoinisitide 3-kinase (PI3K)/Akt signaling pathway regulates cell proliferation, growth, survival, apoptosis, and glucose metabolism [12]. Activation of the PI3K pathway occurs upon engagement with mutation or amplification of PIK3CA, the p110α catalytic subunit of PI3K. Amplification and mutation of PIK3CA is generally associated with increased PIK3CA expression, PI3K activation, and phosphorylation of downstream Akt, supporting the oncogenic role of PI3K aberration. PIK3CA gene amplification was found in 10-30% of non-small cell lung cancer, breast cancer, colon cancer and head/neck cancer [13–16]. Activating somatic mutations (codons 542 and 545 in exon 9 and codon 1047 in exon 20) were also identified in various solid tumors [17]. Despite accumulating evidence of biologic role, only a few studies have reported the frequency of PIK3CA aberration in ESCC and its prognostic role is still controversial [18–22].
In this study, we evaluated the frequency of PIK3CA amplification and mutation in surgically resected ESCC. Furthermore, we also determined the prognostic impact of genetic aberration of PIK3CA in ESCC.
RESULTS
Patient characteristics
A total of 534 patients with curative esophagectomy were analyzed and their clinicopathologic features are presented in Table 1. The majority of patients were male (93.4%) with a median age of 65 years (range 31-90). Median tumor size was 4 cm and approximately half of the tumors were stage pT3 or pN0. Approximately two-thirds of cases (54.9%) were located in the lower esophagus and one-third in the middle esophagus. All patients received radical surgery, with evidence of pathologic stage I in 19.9%, stage II in 44.8%, and stage III in 35.4%. Two-thirds (63.9%) were moderately differentiated carcinoma, and more than half of patients located in the lower esophagus. The majority of patients were current (39.3%) or former (36.7%) smokers, and the median smoking dosage was 25 pack-years (range 0-150). Adjuvant treatment was given to 138 patients (25.8%), and 62 of these (44.9%) were treated with concurrent chemoradiotherapy. Adjuvant therapy was introduced in 4.7% for stage I, 21.3% for stage II, and 42.9% for stage III patients.
Table 1. Patient characteristics according to PIK3CA amplification.
| Characteristics | All patients | Amplification* | No amplification | P† | |||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | Ratio | Number | Total | (%) | No. | % | ||
| No of patients | 534 | 100 | 56 | 10.5 | 478 | 89.5 | |||
| Age, years | 0.183 | ||||||||
| Median | 65 | 66 | 65 | 66 | 65 | (0.128) | |||
| Range | 31-90 | 51-80 | 44-72 | 44-80 | 31-90 | ||||
| Sex | 0.851 | ||||||||
| Male | 499 | 93.4 | 38 | 14 | 52 | 92.9 | 447 | 93.5 | (0.979) |
| Female | 35 | 6.6 | 3 | 1 | 4 | 7.1 | 31 | 6.5 | |
| Tumor size, cm | 0.148 | ||||||||
| Median | 4.0 | 4.0 | 4.0 | 4.0 | 3.0 | (0.093) | |||
| Range | 0.2-14.5 | 1-9 | 1-8 | 1.0-9.0 | 0.2-14.5 | ||||
| pT stage‡ | 0.026 | ||||||||
| T1 | 153 | 28.7 | 7 | 1 | 8 | 14.3 | 145 | 30.3 | (0.020) |
| T2 | 113 | 21.2 | 7 | 7 | 14 | 25.0 | 99 | 20.7 | |
| T3 | 252 | 47.2 | 27 | 7 | 34 | 60.7 | 218 | 45.6 | |
| T4 | 16 | 3.0 | 0 | 0 | 0 | 0 | 16 | 3.3 | |
| pN stage‡ | 0.426 | ||||||||
| N0 | 261 | 48.9 | 16 | 6 | 22 | 39.3 | 239 | 50.0 | (0.540) |
| N1 | 244 | 45.7 | 21 | 9 | 30 | 53.6 | 214 | 44.8 | |
| N2 | 18 | 3.4 | 3 | 0 | 3 | 5.4 | 15 | 3.1 | |
| N3 | 11 | 2.1 | 1 | 0 | 1 | 1.8 | 10 | 2.1 | |
| pTMN stage‡ | 0.053 | ||||||||
| I | 106 | 19.9 | 7 | 1 | 8 | 14.3 | 98 | 20.5 | (0.075) |
| II | 239 | 44.8 | 12 | 8 | 20 | 35.7 | 219 | 45.8 | |
| III | 189 | 35.4 | 22 | 6 | 28 | 50.0 | 161 | 33.7 | |
| Location | 0.707 | ||||||||
| Cervical | 13 | 2.4 | 2 | 0 | 2 | 3.6 | 11 | 2.3 | (0.806) |
| Upper | 70 | 13.1 | 7 | 2 | 9 | 16.1 | 61 | 12.8 | |
| Middle | 158 | 29.6 | 12 | 6 | 18 | 32.1 | 140 | 29.3 | |
| Lower | 293 | 54.9 | 20 | 7 | 27 | 48.2 | 266 | 55.6 | |
| Histologic grade | 0.158 | ||||||||
| Well | 106 | 19.9 | 9 | 7 | 16 | 28.6 | 90 | 18.8 | (0.080) |
| Moderate | 341 | 63.9 | 28 | 6 | 34 | 60.7 | 307 | 64.2 | |
| Poorly | 87 | 16.3 | 4 | 2 | 6 | 10.7 | 81 | 16.9 | |
| Smoking status¶ | 0.391 | ||||||||
| Never-smoker | 128 | 24.0 | 11 | 5 | 16 | 28.6 | 112 | 23.4 | (0.634) |
| Former smoker | 196 | 36.7 | 13 | 3 | 16 | 28.6 | 180 | 37.7 | |
| Current smoker | 210 | 39.3 | 17 | 7 | 24 | 42.9 | 186 | 38.9 | |
| Smoking dosage (pack-years) | |||||||||
| Median | 25 | 30 | 15 | 26 | 25 | 0.924 | |||
| Range | 0-150 | 0-150 | 0-68 | 0-150 | 0-150 | (0.645) | |||
| Adjuvant therapy | 0.077 | ||||||||
| Yes | 138 | 25.8 | 6 | 3 | 9 | 6.5 | 129 | 27.0 | (0.099) |
| No | 396 | 74.2 | 38 | 9 | 47 | 83.9 | 349 | 73.0 | |
| PIK3CA FISH§ | |||||||||
| Number (median, range) | 2.2 (0-16.0) | 4.6 (4.0-16.0) | 5.6 (5.0-6.0) | 5.4 (4.0-16.0) | 2.1 (0-3.9) | <0.001 | |||
| Ratio (mean, range) | 1.1 (0-6.0) | 2.5 (2.0-6.0) | 1.5 (0.9-1.9) | 2.3 (0.9-6.0) | 1.1 (0-1.9) | <0.001 | |||
Abbreviations:
PIK3CA amplification was defined as if one of the following criteria is fulfilled: (1) Ratio: PIK3CA/CEP3 ratio is ≥ 2.0, (2) Number: average number of PIK3CA signal per nucleus ≥ 5.0
χ2 test, Fisher's exact test, or Mann-Whitney U test. Parenthesis indicates comparisons among ratio, numbers, and non-amplified groups
Pathologic stage at the time of surgical resection was determined according to the American Joint Committee on Cancer (seventh edition) guidelines.
Never-smokers; a lifetime smoking dose of fewer than 100 cigarettes; former smokers, those who have stopped smoking for more than 1 year; current smokers, those who currently smoke or have quit for less than 1 year.
PIK3CA numbers are average numbers of PIK3CA signals per nucleus, and ratios are PIK3CA/CEN3 ratios.
Association of PIK3CA amplification status and clinicopathologic features
Among 534 patients, 41 (7.7%) of cases satisfied both criteria of PIK3CA: CEN3 ratio of ≥ 2.0 and an average number of PIK3CA signal per nucleus ≥5.0, whereas 15 (2.8%) cases only satisfied the criterion of PIK3CA ≥ 5.0 (Supplementary Figure S1). With our criteria, PIK3CA amplification was detected in 56 (10.5%) cases (Table 1, Figure 1). The median PIK3CA gene copy number was 5.4 (range, 4.0-16.0) and 2.1 (range, 0-3.9) in PIK3CA amplified and non-amplified groups, respectively. The mean PIK3CA/CEN3 ratio was 2.3 (0.9-6.0) for the amplification group and 1.08 (0-1.9) for the no amplification group.
Figure 1.
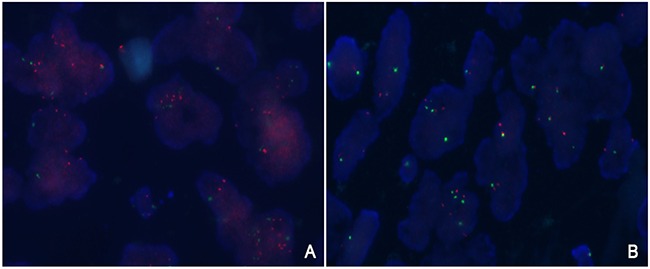
Representative fluorescent in situ hybridization of tumors with A. and without B. PIK3CA amplification. FISH analysis demonstrated an increase in the signals of PIK3CA (red signals) compared to reference CEN3 (green signals).
There was no significant difference in PIK3CA amplification according to age, sex, tumor size, location, histologic grade, smoking and adjuvant therapy as shown in Table 1. However, PIK3CA amplification was significantly associated with a higher T-stage (P=0.026). Although only marginal statistical significance was achieved, there was a positive association between PIK3CA amplification and higher pathologic stage (P=0.053). Based on the pattern of amplification (PIK3CA: CEN3 ratio of ≥ 2.0 and signal per nucleus ≥5.0 vs. PIK3CA signal per nucleus ≥5.0), there was similar statistical significance compared to no amplification group.
Prognosis according to PIK3CA amplification
The 5 year DFS and OS rates for all patients were 46.3% and 59.8% with a median follow-up time of 56.4 months. The 5-year DFS rate according to pTNM stages was 64.6% in stage I, 55.4% in stage II, and 28.9% in stage III. The 5-year OS rate according to pTNM stages were 76.7% for stage I, 61.3% for stage II, and 32.4% for stage III.
PIK3CA amplification showed a significantly shorter DFS compared with that of non-amplified group (33.4 vs 63.1 months, P=0.019, Figure 2A). Though the statistical insignificance probably due to small sample size, PIK3CA amplification showed a tendency of shorter OS than no amplification (52.1 vs 96.5 months, P=0.116, Figure 2B). In the Cox proportional hazard model adjusted for gender, tumor location, pathologic stage, histologic grade and adjuvant treatment, PIK3CA amplification was significantly associated with a shorter DFS (adjusted hazard ration [AHR] 1.53; 95% CI, 1.10-2.17; P=0.02, Table 2). There was trend toward worse DFS for the pattern of amplification (AHR 1.53; 95% CI 1.1-2.3 for PIK3CA: CEN3 ratio of ≥ 2.0 vs AHR 1.32; 95% CI, 0.97-2.56; PIK3CA signal per nucleus ≥5.0; P=0.05) compared to non-amplified group. There was no significant difference in OS according to PIK3CA amplification, gender and adjuvant treatment in multivariate analysis. Regarding the prognostic role of PIK3CA amplification according to pathologic stage, there were no significant difference with PIK3CA amplification in DFS (5-year DFS 38.1% vs 57.3% in stage II [P=0.074]; 16.1% vs 28.8% in stage III [P=0.356], Supplementary Figure S2).
Figure 2. Survival analysis according to PIK3CA amplification.
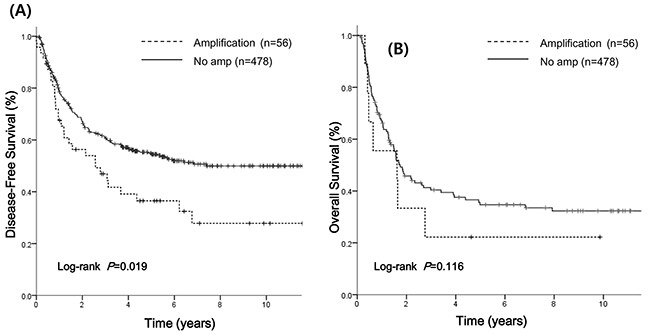
A. Median disease-free survival (DFS) was 33.4 months in the PIK3CA amplification group and 63.1 months in the no amplification group. B. Median overall survival (OS) was 52.1 months in the amplification group and 96.5 months in the no amplification group.
Table 2. Survival outcome in multivariate analysis.
| Variable | Category | DFS | OS | |||||
|---|---|---|---|---|---|---|---|---|
| Univariate P value | HR | 95% CI | P | HR | 95% CI | P | ||
| Sex | Female vs Male (ref) | 0.32 | 0.80 | 0.48-1.33 | 0.39 | 0.82 | 0.46-1.41 | 0.47 |
| Location | Lower vs upper/middle | 0.07 | 1.63 | 1.13-2.35 | 0.008 | 1.53 | 1.04-2.24 | 0.03 |
| Pathologic stage* | II/III vs I (ref) | <0.001 | 2.32 | 1.80-2.99 | <0.001 | 2.79 | 2.14-3.66 | <0.001 |
| Histology | Poor vs well/moderate (ref) | 0.06 | 1.34 | 0.99-1.80 | 0.06 | 1.32 | 0.96-1.80 | 0.08 |
| Adjuvant treatment | Yes vs no (ref) | 0.05 | 1.22 | 0.93-1.61 | 0.16 | 1.05 | 0.79-1.39 | 0.75 |
| PIK3CA amplification | Amplification vs no amplification (ref) | 0.01 | 1.53 | 1.10-2.17 | 0.02 | 1.21 | 0.83-1.77 | 0.30 |
Abbreviations: DFS, disease-free survival; OS, overall survival; ref, reference; amp, amplification;
Clinical stage at the time of initial diagnosis was determined according to the American Joint Committee on Cancer (seventh edition) guidelines
We next explored the role of adjuvant treatment according to PIK3CA amplification. We additionally analyzed survival outcome for two subgroups: one group without treatment (n=396) and another group with adjuvant chemotherapy and/or radiotherapy (n=138). In the adjuvant group, PIK3CA amplification showed an inferior DFS but it was statistically insignificant (median DFS 21.0 vs 22.8 months, P=0.39, Supplementary Figure 3A). Among the patients without adjuvant treatment, the PIK3CA amplification group had a significantly shorter DFS compared with non-amplified group (median DFS 33.5 vs 96.5 months, P=0.011, Supplementary Figure S3B).
PIK3CA mutations
Among the 526 patients, we examined PIK3CA exon 9 and 20 mutations using direct sequencing in 388 cases with FFPE tissues available. PIK3CA mutations were detected in 6 (1.5%) of 388 cases; 5 cases in exon 9 only, and 1 case in exon 20 only. The clinicopathologic characteristics of the 6 patients with PIK3CA mutations are listed in Table 3. PIK3CA exon 9 mutations were identified in 5 tumors of E545K (GAG to AAG), while exon 20 mutation was identified in H1047L (CAT to CTT) (Figure 3). No co-occurrence of exon 9 and exon 20 mutations was identified. Of 6 PIK3CA mutants, the single case with exon 20 mutation satisfied the criteria of PIK3CA amplification with PIK3CA/CEN3 ratio of 2.5. The patient was diagnosed as stage IIB, upper ESCC with well-differentiated histology. He had experienced recurrence within 6 months after esophagectomy and died of disease. Though the statistical insignificance, all 6 cases were high T stage (5 with T3 and 1 with T4) and 4 out of 6 cases (66.7%) were lymph node negative. Furthermore, all tumors with PIK3CA mutations originated from the mid-to-lower esophageal region. No prognostic difference in DFS (P=0.876) and OS (P=0.695) was detected according to the presence of PIK3CA mutation.
Table 3. Clinicopathologic characteristics of 6 patients with PIK3CA mutation.
| No. | Mutation | Domain | Age/Sex | Stage | Histology | Recur | Death | PIK3CA amplification |
|---|---|---|---|---|---|---|---|---|
| 1 | E545K | Helical | Male/56 | IIA (T3N0) | Well | No | No | No |
| 2 | E545K | Helical | Male/54 | IIB (T3N0) | Moderate | Yes | Yes | No |
| 3 | E545K | Helical | Male/65 | IIIA (T3N1) | Well | No | No | No |
| 4 | E545K | Helical | Male/61 | IIIA (T3N1) | Moderate | Yes | Yes | No |
| 5 | E545K | Helical | Male/59 | IIIA (T4N0) | Moderate | No | No | No |
| 6 | H1047L | Kinase | Male/66 | IIB (T3N0) | Well | Yes | Yes | Amplified |
Figure 3.
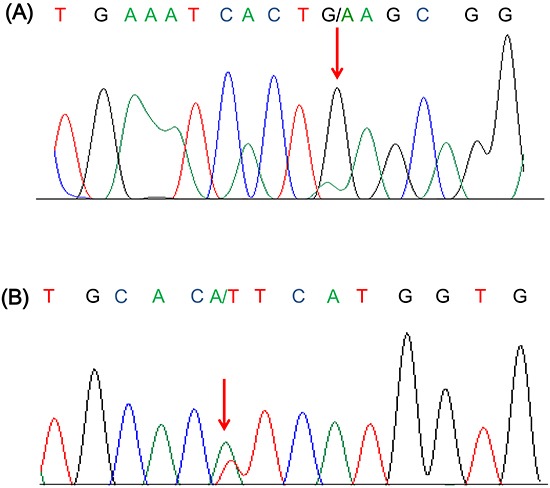
Representative sequence chromatogram of A. E545K (G1634A) and B. H1047L (A3140T) mutation.
DISCUSSION
We conducted this study to examine the frequency and prognostic impact of PIK3CA amplification among curatively resected ESCCs. To our knowledge, this is the first study of PIK3CA amplification in a large cohort of East Asian ESCC patients. We found that PIK3CA amplification is a common genetic event and an independent poor prognostic factor in ESCC.
Though the known therapeutic options in EAC such as HER2 inhibitor, ESCC still lacks therapeutically relevant genetic alterations. In the era of personalized medicine, EAC and ESCC show different genetic aberrations for therapeutic strategies. A recent comparative genome study reported focal regions of DNA amplification or loss including SOX2, PIK3CA, CCND1, and FGFR1 are more frequent in ESCC than EAC [9]. A study of DNA copy number profiles also revealed frequent alterations of the 3q22-26 regions containing PIK3CA in ESCC patients [10]. Therefore, PIK3CA may be a putative drive gene for ESCC with the available therapeutic agents and examining PIK3CA aberrations in a large cohort of ESCC is worthwhile.
Activation of the PI3K pathway, generally as a result of PIK3CA amplification, has been reported in upper aerodigestive tract cancers including 12% of lung squamous cell carcinoma (SCC) [23], 18.2% of nasopharyngeal carcinoma [24], 20% of oropharyngeal SCC [25], and 32.2% of HNSCC [16]. For esophageal carcinoma, one study demonstrated 26.7% of PIK3CA gene amplification in ESCC [18]. With FISH analysis, we found an overall 10.5% frequency of PIK3CA amplification.
Regarding the prognostic significance of PIK3CA amplification, previous reports yielded controversial results. In nasopharyngeal carcinoma, PIK3CA amplification was strongly associated with distant metastasis, lymph node involvement, advanced tumor stage, and ultimately with reduced overall survival [24]. For non-lymph node metastatic HNSCC, patients with PIK3CA amplification showed earlier recurrence than those without (10% vs 31% disease free at 2 years) [16]. Angulo et al reported that PIK3CA amplification was significantly more frequent in lung SCC compared with adenocarcinoma (42% vs 3%, P<0.001), however, no association was found with other clinicopathologic characteristics [15]. Clinicopathologic heterogeneity including primary tumor, pathologic stage, and adjuvant treatment, may contribute to the controversial results. Furthermore, conclusions based on small sample sizes may lead to inconsistent results. In our study, by carefully assessing a large cohort of ESCC cases, we were able to clarify the prognostic value of PIK3CA amplification in a homogenous ESCC patients. Therefore, PIK3CA amplification was significantly associated with shorter DFS regardless of sex, histologic grade, and adjuvant therapy, implying a potential role as an independent negative prognostic factor in curatively resected ESCC.
To investigate PIK3CA gene copy number gain, most commonly used methods are real-time quantitative polymerase chain reaction (PCR) and in situ hybridization [15, 16, 18, 24, 26, 27]. With PCR studies, it is difficult to compare results among studies because of different cut-off value; some studies used copy number more than 4 [16, 28] whereas other group considered 2 or 3 copies as amplification [18, 23]. In contrast, relatively consistent criteria are used for FISH studies. The majority of studies defined PIK3CA amplification as PIK3CA/CEP3 ≥ 2 [25–27] and another group defined it as copy number more than 5 [15]. In our large cohort study, we used a combination of these criteria and demonstrated a significant difference in prognostic value. In our study, 26.7% of cases with copy number ≥5 might not have been considered amplified if only the ratio criterion of ratio ≥ 2.0 was used. FISH is also more powerful than PCR by allowing visualization of individual cancer cells in a routine clinical environment. Identification PIK3CA amplification by FISH analysis may contribute to easy subgroups selection. Our criteria should be further validated in other solid tumors and in clinical trials with PI3K inhibitors.
Hotspot mutations of PIK3CA in exon 9 and exon 20 have been shown to increase lipid kinase activity, leading to downstream activation [29]. The frequency of PIK3CA mutation has been variously reported as 2.2 to 21% of ESCC cases, with controversial prognostic value [19–22]. Mori et al identified a frequency of 2.2% using PCR-based direct sequencing [30] and Hou et al reported a frequency of 12.5 % with mutant-enriched liquid chip technology [19]. More recently, Shingaki et al identified PIK3CA mutations in 21% of cases by employing a pyrosequencing approach [22]. Here, we report PIK3CA mutations in 1.6% of ESCCs using standard, bidirectional Sanger sequencing. Different sequencing methodologies may have an important influence on the reported frequencies. The limited sensitivity of Sanger sequencing may result in an apparent the low frequency and further studies to compare these multiple methods are warranted.
Elevated PI3K signaling correlates with PIK3CA mutation and/or amplification. In addition it is associated with increased activity of PI3K effector protein kinase B (PKB) [26] and pAkt [31], suggesting that these may be susceptible to PI3K inhibitors. In lung cancer cell lines, 37% of squamous cell harbored PIK3CA amplification and they were sensitive to PI3K inhibitor GDC-0941 with less than 1μmol/L of IC50 [31]. In a preclinical platform from Cancer Cell Line Encyclopedia, PIK3CA-amplified tumors were sensitive to BYL719, a PI3K α-selective inhibitor [32]. Cell lines with PIK3CA amplification was positively associated with BYL719 sensitivity (P=0.0037) and tumor-bearing mice with PIK3CA amplification responded to BYL719, leading to a response rate of −18% (lung cancer) and −80% (gastric cancer). Despite the preliminary data, recent phase I trials have explored the potential predictive role of the PIK3CA gene. Pan-PI3K (BKM120 and GDC-0941) and α-selective inhibitors (BYL719 and GDC-0032) demonstrated responses and prolonged stable disease in patients with PIK3CA mutation [33–36]. In addition to PIK3CA mutation, PIK3CA amplification was positively associated with sensitivity to BYL719 in PIK3CA wild type cell lines [32]. In a phase I trial of GDC-0941, a heavily treated ovarian cancer patient with PIK3CA amplification achieved disease stabilization for 4 months with significant pharmacodynamic changes [37]. To date, clinical trials with PI3K inhibitors have been reported in un-selected patients, and current trials with PI3K inhibitors are for patients with PIK3CA gene alterations are ongoing (ClinicalTrials.gov number NCT01928459 and NCT01608022). In addition, as shown in our study, frequency of PIK3CA amplification increases from 14.3% (stage I) to 50% (stage III) indicating more advanced patients may have benefit from PI3K inhibitors. Our results require further validation with PI3K inhibitors in clinical trials.
The main limitations of our study include its retrospective nature and patient selection. Therefore, our findings should be validated in an independent cohort and response data to PIK3CA-targeted therapies in the future clinical trials.
In conclusion, in our large cohort study, we demonstrated that PIK3CA amplification is an independent poor prognostic factor in resected ESCC. Our findings also provide strong implication that PIK3CA amplification and mutation is a promising therapeutic target in ESCC.
MATERIALS AND METHODS
Patients and tissue samples
A total of 534 patients with ESCC who underwent radical esophagectomy at Severance Hospital and Samsung Medical Center, Seoul, Korea between 2002 and 2010 were enrolled in this study. The criteria used for patient selection included (1) surgically resected SCC of the thoracic esophagus (R0 resection), (2) availability of primary tumor tissue, (3) no distant metastasis, and (4) no preoperative treatment. Tumor samples were available for 664 patients, of which we excluded 107 cases (16.1%) who received neoadjuvant treatment. Twenty-three patients (3.5%) were excluded because of incomplete survival follow-up. All diagnosis were reviewed by two experienced pathologists (Y.L.C. and H.K.) and confirmed by hematoxylin and eosin staining. Paraffin-embedded tumor specimens were used to construct a tissue microarray with three representative cores of 2-mm-diameter.
Clinicopathologic characteristics and survival outcome was collected by reviewing the medical records. Staging was determined using the 7th edition of the American Joint Committee on Cancer guideline of tumor, node, and metastasis (TNM) classification. Smoking status of never-smoker, former smoker, and current smoker were defined as in previous studies (23). The study was approved by the institutional review board of Severance Hospital and Samsung Medical Center.
PIK3CA fluorescence in situ hybridization
To assess the presence of PIK3CA amplification, we performed fluorescence in situ hybridization (FISH) on tissue microarrays. PIK3CA (Spectrum Green) and CEP3 (Spectrum Orange) FISH was performed as per manufacturer's recommendation. (Abbott Molecular, Abbott Park, IL, USA). Evaluation was performed independently by two experienced pathologists (Y.L.C. and H.K.K) blinded to clinical information, and at least 100 nuclei per case were evaluated. Based on the previous studies(24-27), PIK3CA amplification was defined as fulfillment of one of the following criteria: (1) PIK3CA/CEP3 ratio ≥ 2.0 and (2) average number of PIK3CA signals per nucleus ≥ 5.0
Mutation analysis
Genomic DNA was extracted from 388 formalin-fixed paraffin-embedded (FFPE) tissue specimens using a QIAamp DNA Micro kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instruction. The extracted DNA was used in a PCR amplification reaction with primers were designed to amplify the following regions at codon E542, E545 and H1047; exon 9; 5′;- AGAGACAATGAATTAAGGGAAAATGAC-3′;; 5′;- TTTAGCACTTACCTGTGACTCCA-3′;, exon 20; 5′;-TATTCGACAGCATGCCAATC-3′;; 5′;- TGTGTGGAAGATCCAATCCA-3′;,.
PCR was carried out with the following conditions: initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 30 s, 54°C for 60 s, 72°C for 45 s and a final polymerization step of 72°C for 5 min in a GeneAmp PCR system 2720 (Life technologies, CA, USA). The amplified DNA product was visualized by gel electrophoresis and PCR products were sequenced using the Big Dye terminator sequencing kit (Life technologies, CA, USA) according to the manufactures’ instruction. Sequence reactions were the subjected to electrophoresis on an Applied Biosystems 3130XL DNA Analyzer (Life Technologies, CA, USA).
Statistical analysis
We analyzed the association between PIK3CA amplification status and clinical significance using the χ2 test or Fisher's exact test. We also assessed the prognostic value of PIK3CA amplification on survival outcome using Kaplan-Meier curves with a log-rank test. Disease free survival (DFS) was defined from the time of surgery to initial relapse or death. Overall survival (OS) was measured from the time of surgery to death or the last follow-up date, and 95% confidence intervals (CIs) were evaluated by survival analysis using the Kaplan-Meier method. Statistical significance was defined as P < 0.05 for all analyses. Multivariate analysis was done using Cox regression analysis for following variables: gender, location, pathologic stage, histology, adjuvant treatment, and PIK3CA amplification status. All statistical analysis was performed using SPSS version 18.0 (SPSS, Chicago, IL, USA).
SUPPLEMENTARY FIGURES
Footnotes
CONFLICTS OF INTEREST
The authors have no potential conflicts of interest to disclose.
GRANT SUPPORT
This study was supported by a grant from the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI12C1440, B. C. Cho), by faculty research grant of Yonsei University College of Medicine for 2012 (6-2012-0044, H. Kim) and by the National Research Foundation of Korea(NRF) grant funded by the Korea government (MSIP) (No. 2015R1C1A2A01055617, H. S. Kim).
REFERENCES
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. Journal of Clinical Oncology. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Medical progress - Esophageal cancer. New England Journal of Medicine. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 3.Ilson DH. Oesophageal cancer: new developments in systemic therapy. Cancer Treat Rev. 2003;29:525–532. doi: 10.1016/s0305-7372(03)00104-x. [DOI] [PubMed] [Google Scholar]
- 4.Wong A, Fitzgerald RC. Epidemiologic risk factors for Barrett's esophagus and associated adenocarcinoma. Clin Gastroenterol Hepatol. 2005;3:1–10. doi: 10.1016/s1542-3565(04)00602-0. [DOI] [PubMed] [Google Scholar]
- 5.Morita M, Saeki H, Mori M, Kuwano H, Sugimachi K. Risk factors for esophageal cancer and the multiple occurrence of carcinoma in the upper aerodigestive tract. Surgery. 2002;131:S1–6. doi: 10.1067/msy.2002.119287. [DOI] [PubMed] [Google Scholar]
- 6.Freeman HJ. Risk of gastrointestinal malignancies and mechanisms of cancer development with obesity and its treatment. Best Pract Res Clin Gastroenterol. 2004;18:1167–1175. doi: 10.1016/j.bpg.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Brown LM, Hoover R, Silverman D, Baris D, Hayes R, Swanson GM, Schoenberg J, Greenberg R, Liff J, Schwartz A, Dosemeci M, Pottern L, Fraumeni JF. Excess incidence of squamous cell esophageal cancer among US black men: Role of social class and other risk factors. American Journal of Epidemiology. 2001;153:114–122. doi: 10.1093/aje/153.2.114. [DOI] [PubMed] [Google Scholar]
- 8.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 9.Bandla S, Pennathur A, Luketich JD, Beer DG, Lin L, Bass AJ, Godfrey TE, Little VR. Comparative Genomics of Esophageal Adenocarcinoma and Squamous Cell Carcinoma. Annals of Thoracic Surgery. 2012;93:1101–1106. doi: 10.1016/j.athoracsur.2012.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rumiato E, Pasello G, Montagna M, Scaini MC, De Salvo GL, Parenti A, Cagol M, Ruol A, Ancona E, Amadori A, Saggioro D. DNA copy number profile discriminates between esophageal adenocarcinoma and squamous cell carcinoma and represents an independent prognostic parameter in esophageal adenocarcinoma. Cancer Letters. 2011;310:84–93. doi: 10.1016/j.canlet.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Kim HS, Lee SE, Bae YS, Kim DJ, Lee CG, Hur J, Chung H, Park JC, Jung da H, Shin SK, Lee SK, Lee YC, Kim HR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival in patients with resected esophageal squamous cell carcinoma. Oncotarget. 2015;6:2562–2572. doi: 10.18632/oncotarget.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, Trink B, Sidransky D. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005;7:R609–616. doi: 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jehan Z, Bavi P, Sultana M, Abubaker J, Bu R, Hussain A, Alsbeih G, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Uddin S, et al. Frequent PIK3CA gene amplification and its clinical significance in colorectal cancer. J Pathol. 2009;219:337–346. doi: 10.1002/path.2601. [DOI] [PubMed] [Google Scholar]
- 15.Angulo B, Suarez-Gauthier A, Lopez-Rios F, Medina PP, Conde E, Tang M, Soler G, Lopez-Encuentra A, Cigudosa JC, Sanchez-Cespedes M. Expression signatures in lung cancer reveal a profile for EGFR-mutant tumours and identify selective PIK3CA overexpression by gene amplification. J Pathol. 2008;214:347–356. doi: 10.1002/path.2267. [DOI] [PubMed] [Google Scholar]
- 16.Suda T, Hama T, Kondo S, Yuza Y, Yoshikawa M, Urashima M, Kato T, Moriyama H. Copy number amplification of the PIK3CA gene is associated with poor prognosis in non-lymph node metastatic head and neck squamous cell carcinoma. BMC Cancer. 2012;12:416. doi: 10.1186/1471-2407-12-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 18.Akagi I, Miyashita M, Makino H, Nomura T, Hagiwara N, Takahashi K, Cho K, Mishima T, Ishibashi O, Ushijima T, Takizawa T, Tajiri T. Overexpression of PIK3CA is associated with lymph node metastasis in esophageal squamous cell carcinoma. Int J Oncol. 2009;34:767–775. doi: 10.3892/ijo_00000202. [DOI] [PubMed] [Google Scholar]
- 19.Hou J, Jiang D, Zhang J, Gavine PR, Xu S, Liu Y, Xu C, Huang J, Tan Y, Wang H, Lu Y, Zheng L, Hou Y, et al. Frequency, characterization, and prognostic analysis of PIK3CA gene mutations in Chinese esophageal squamous cell carcinoma. Hum Pathol. 2014;45:352–358. doi: 10.1016/j.humpath.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Maeng CH, Lee J, van Hummelen P, Park SH, Palescandolo E, Jang J, Park HY, Kang SY, MacConaill L, Kim KM, Shim YM. High-throughput genotyping in metastatic esophageal squamous cell carcinoma identifies phosphoinositide-3-kinase and BRAF mutations. PLoS One. 2012;7:e41655. doi: 10.1371/journal.pone.0041655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips WA, Russell SE, Ciavarella ML, Choong DY, Montgomery KG, Smith K, Pearson RB, Thomas RJ, Campbell IG. Mutation analysis of PIK3CA and PIK3CB in esophageal cancer and Barrett's esophagus. Int J Cancer. 2006;118:2644–2646. doi: 10.1002/ijc.21706. [DOI] [PubMed] [Google Scholar]
- 22.Shigaki H, Baba Y, Watanabe M, Murata A, Ishimoto T, Iwatsuki M, Iwagami S, Nosho K, Baba H. PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res. 2013;19:2451–2459. doi: 10.1158/1078-0432.CCR-12-3559. [DOI] [PubMed] [Google Scholar]
- 23.Kawano O, Sasaki H, Okuda K, Yukiue H, Yokoyama T, Yano M, Fujii Y. PIK3CA gene amplification in Japanese non-small cell lung cancer. Lung Cancer. 2007;58:159–160. doi: 10.1016/j.lungcan.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Fendri A, Khabir A, Mnejja W, Sellami-Boudawara T, Daoud J, Frikha M, Ghorbel A, Gargouri A, Mokdad-Gargouri R. PIK3CA amplification is predictive of poor prognosis in Tunisian patients with nasopharyngeal carcinoma. Cancer Sci. 2009;100:2034–2039. doi: 10.1111/j.1349-7006.2009.01292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiosea SI, Grandis JR, Lui VW, Diergaarde B, Maxwell JH, Ferris RL, Kim SW, Luvison A, Miller M, Nikiforova MN. PIK3CA, HRAS and PTEN in human papillomavirus positive oropharyngeal squamous cell carcinoma. BMC Cancer. 2013;13:602. doi: 10.1186/1471-2407-13-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massion PP, Kuo WL, Stokoe D, Olshen AB, Treseler PA, Chin K, Chen C, Polikoff D, Jain AN, Pinkel D, Albertson DG, Jablons DM, Gray JW. Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res. 2002;62:3636–3640. [PubMed] [Google Scholar]
- 27.Woenckhaus J, Steger K, Werner E, Fenic I, Gamerdinger U, Dreyer T, Stahl U. Genomic gain of PIK3CA and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol. 2002;198:335–342. doi: 10.1002/path.1207. [DOI] [PubMed] [Google Scholar]
- 28.Hou P, Liu D, Shan Y, Hu S, Studeman K, Condouris S, Wang Y, Trink A, El-Naggar AK, Tallini G, Vasko V, Xing M. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13:1161–1170. doi: 10.1158/1078-0432.CCR-06-1125. [DOI] [PubMed] [Google Scholar]
- 29.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori R, Ishiguro H, Kimura M, Mitsui A, Sasaki H, Tomoda K, Mori Y, Ogawa R, Katada T, Kawano O, Harada K, Fujii Y, Kuwabara Y. PIK3CA mutation status in Japanese esophageal squamous cell carcinoma. J Surg Res. 2008;145:320–326. doi: 10.1016/j.jss.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 31.Spoerke JM, O'Brien C, Huw L, Koeppen H, Fridlyand J, Brachmann RK, Haverty PM, Pandita A, Mohan S, Sampath D, Friedman LS, Ross L, Hampton GM, et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin Cancer Res. 2012;18:6771–6783. doi: 10.1158/1078-0432.CCR-12-2347. [DOI] [PubMed] [Google Scholar]
- 32.Fritsch C, Huang A, Chatenay-Rivauday C, Schnell C, Reddy A, Liu M, Kauffmann A, Guthy D, Erdmann D, De Pover A, Furet P, Gao H, Ferretti S, et al. Characterization of the novel and specific PI3Kalpha inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol Cancer Ther. 2014;13:1117–1129. doi: 10.1158/1535-7163.MCT-13-0865. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Angulo A JD, Argil es G, Schellens J, Burris H, Berlin J, et al. Safety, pharmacokinetics, and preliminary activity of the a-specific PI3K inhibitor BYL719: Results from the first-in-human study. J Clin Oncol. 2013 [Google Scholar]
- 34.Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, Demanse D, De Buck SS, Ru QC, Peters M, Goldbrunner M, Baselga J. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30:282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 35.D. D. Von Hoff PL, Demetri G. D, Weiss G. J, Shapiro G, Ramanathan R. K, Ware J. A, Raja R, Jin J, Levy G. G, Mazina K. E, Wagner A. J. A phase I dose-escalation study to evaluate GDC-0941, a pan-PI3K inhibitor, administered QD or BID in patients with advanced or metastatic solid tumors. J Clin Oncol. 2011;29 [Google Scholar]
- 36.Juric D KI, Ramanathan R, Xiao J, Sanabria S, Wilson T, et al. GDC-0032, a beta isoform-sparing PI3K inhibitor: results of a first-in-human phase Ia dose escalation study. Cancer Res. 2013 [Google Scholar]
- 37.Sarker D, Ang JE, Baird R, Kristeleit R, Shah K, Moreno V, Clarke PA, Raynaud FI, Levy G, Ware JA, Mazina K, Lin R, Wu J, et al. First-in-human phase I study of pictilisib (GDC-0941), a potent pan-class I phosphatidylinositol-3-kinase (PI3K) inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2015;21:77–86. doi: 10.1158/1078-0432.CCR-14-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


