Supplemental Digital Content is available in the text
Keywords: biochemical recurrence, CYP1B1, prostate cancer, radical prostatectomy, single nucleotide polymorphism
Abstract
Clinically localized prostate cancer is curative. Nevertheless many patients suffered from biochemical recurrence (BCR) after radical prostatectomy (RP). Mounting evidence suggest that estrogen and xenobiotic carcinogens play an essential role in progression of prostate cancervia oxidative estrogen metabolism. CYP1B1 is an enzyme involved in the hydroxylation of estrogens, a reaction of key relevance in estrogen metabolism. Given the role of CYP1B1 in the oxidative metabolism of endogenous/exogenous estrogen and compounds, CYP1B1 polymorphisms have the potential to modify its expression and subsequently lead to progression. We hypothesize that genetic variants of the CYP1B1 gene may influence clinical outcome in clinically localized prostate cancer patients. In this cohort study, we genotyped 9 tagging single nucleotide polymorphisms (SNPs) from the CYP1B1 gene in 312 patients treated with RP. For replication, these SNPs were genotyped in an independent cohort of 426 patients. The expression level of CYP1B1 in the adjacent normal prostate tissues was quantified by reverse transcription and real-time polymerase chain reaction. Kaplan–Meier analysis and Cox proportional hazard models were utilized to identify SNPs that correlated with BCR. CYP1B1 rs1056836 was significantly associated with BCR (hazard ratio [HR]: 0.69; 95% confidence interval [CI]: 0.40–0.89, P = 0.002) and relative CYP1B1 mRNA expression. Our findings suggest inherited genetic variation in the CYP1B1 gene may contribute to variable clinical outcomes for patients with clinically localized prostate cancer.
1. Introduction
Radical prostatectomy (RP) is the gold standard treatment modality for clinically localized prostate cancer. Prostate cancer patients treated with RP have an overall risk of biochemical recurrence (BCR) of approximately 30% at 10 years after surgery.[1] Due to the limitations of prostate-specific antigen (PSA) and other clinicopathological parameters, great efforts have been made to identify novel prognostic molecular markers to predict BCR after surgical treatment.[2] Such prognostic markers for use in localized prostate cancer would considerably improve disease management.
Previous studies suggest that estrogen and xenobiotic carcinogens play an essential role in progression of prostate cancer via oxidative estrogen metabolism.[3] CYP1B1 is an enzyme involved in the hydroxylation of estrogens, a reaction of key relevance in estrogen metabolism.[3] The overproduction of estrogen like E2 or the bioconversion of E2 into genotoxic metabolites like estradiol-3,4-quinone(E2-3,4-Q) or 4-hydroxyestradiol (4-OH E2) by CYP1B1 lead to generation of reactive oxygen species, which cause DNA adducts and enhance prostate progression.[4] Substantial evidence for a potential role of CYP1B1 in prostate cancer comes from the observations that CYP1B1 protein is overexpressed in prostate carcinoma whereas no CYP1B1 protein expression is detected in normal prostate tissue.[5]
Based on these observations, single nucleotide polymorphisms (SNPs) of CYP1B1 have been proposed as candidates for association studies on prostate cancer risk.[6–15]
CYP1B1 is polymorphous and there were 304 SNPs that have been identified.[3]
Several CYP1B1 SNPs altering the estrogen metabolism may modify an individual's prostate cancer risk. The SNP rs1056836, leading to a substitution of an amino acid leucine for valine, is associated with increased CYP1B1 messenger ribonucleic acid (mRNA) expression,[3,16] with a subsequent elevation in 4-OH E2 formation resulting in increased estrogen-mediated carcinogenicity.[3,17] By contrast, the SNP rs10012, which is responsible for Arg48Gly transition, has no effect on protein folding or stability.[3,16], Another SNPrs1800440 that results in Asn453Ser transition leads to a decrease in protein expression caused by degradation of the protein.[3,18] Given the role of CYP1B1 in the oxidative metabolism of endogenous/exogenous estrogen and compounds, CYP1B1 polymorphisms have the potential to modify its expression and subsequently lead to progression. CYP1B1 may also be involved in recurrence after RP by altering the tissue response to hormones and other genes that participate in CYP1B1-mediated pathways (e.g., estrogen receptor and aryl hydrocarbon receptor signaling pathway).[3,4] However, no studies have looked for associations between CYP1B1 polymorphisms and prognosis in prostate cancer cases. Herein, we evaluated whether genetic variations in CYP1B1 were correlated with BCR-free survival in localized prostate cancer patients treated with RP.
2. Methods
2.1. Study population
We retrospectively analyzed 738 men who had undergone RP from 2 independent cohorts in China as described previously.[19] The 1st cohort was composed of 312 patients from the Affiliated Hospital of Qingdao University and Fudan University Shanghai Cancer Center, and the 2nd was composed of 426 patients from Fudan University Shanghai Cancer Center. The median follow-up times were 36.3 and 37.7 months. The clinical information were abstracted from the archival medical records. The patients who received adjuvant hormone therapy or radiotherapy were excluded. BCR was defined as 2 consecutive PSA measurements >0.2 ng/mL at an interval of >3 months, and the date of this event was set to the 1st of these 2 test occasions. All participants provided written informed consent. The protocol and consent documents were approved by the Institutional Review Board of Fudan University Shanghai Cancer Center and the Affiliated Hospital of Qingdao University.
2.2. SNP selection and genotyping
Tagging SNPs were identified in the CYP1B1 gene with r2 ≥ 0.8 and a minor allele frequency ≥5% in Chinese using HapMap database. A total of 9 tagging SNPs were chosen. Genomic DNA was extracted from peripheral blood using QIAmp DNA extraction kit (Qiagen) and genotyped by TaqMan real-time PCR method according to manufacturer's instruction. All SNPs were consistent with Hardy–Weinberg equilibrium (HWE), except for rs9341250 (P < 0.01), which was excluded from further analysis. Each SNP had greater than 99% completion and the concordance was 100% for duplicated specimens.
2.3. CYP1B1 expression analysis
This analysis included 218 patients whose frozen adjacent normal prostate tissue was available for RNA extraction. Tissue specimens were collected during surgery and stored at −80 °C. The TRIzol reagent (Invitrogen, CA) was used to isolate total RNA. cDNA was synthesized using PrimeScriptRT Master Mix system (TAKARA, Osaka, Japan). Quantitative PCR was carried out using ABI 7900 Real-Time PCR System (Applied Biosystems, CA). The sequences of the CYP1B1 specific primers were forward, GCTGCAGTGGCTGCTCCT, and reverse, CCCACGACCTGATCCA AT TCT. The negative controls for each primer consisted of a reaction with no cDNA added. Quantification was achieved by the use of a standard curve for CYP1B1 and normalization of the corresponding transcripts by the ΔΔCt method and the use of the housekeeping gene GAPDH.
2.4. Statistical analysis
Categorical variables were compared by the χ2 test or Fisher exact test. HWE for the evaluation of genotype frequencies was performed by the goodness-of fit χ2 test. The association between SNPs, clinical characteristics, and BCR was assessed with hazard ratios (HRs) and 95% confidence interval (CI) estimated by Cox proportional hazards regression analysis under different genetic models. All analyses were adjusted for known prognostic factors including age, disease stage, Gleason score, lymph node involvement, and PSA at diagnosis. The BCR-free survival interval was estimated using the Kaplan–Meier method, and the significance was determined using the log-rank test. Differences in distribution of expression levels by genotype were analyzed by Student t test. All reported P-values are 2-sided. SAS version 9.1 (SAS institute Inc., NC) was used for all analyses.
3. Results
3.1. Patient characteristics and treatment outcome
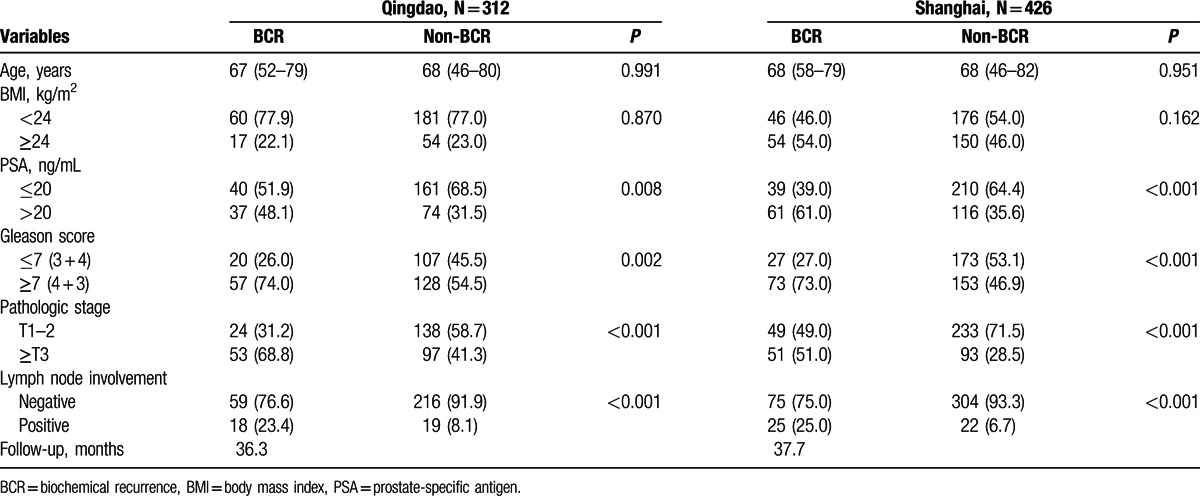
Patient characteristics of the study 1 and study 2 are described in Table 1. A total of 79 (25.3%) and 100 (23.5%) patients experienced BCR in the study 1 and study 2, respectively. PSA levels, pathologic stage, lymph node involvement, and Gleason score were significantly associated with BCR in both cohorts (P < 0.01).
Table 1.
Clinicopathologic characteristics of the study populations.
3.2. Genetic analyses of localized prostate cancer
The relative frequencies of the SNPs in patients and their corresponding HRs (95% CI) are shown in Table 2 and Supplementary Table S1. After making adjustments for known clinicopathologic variables, CYP1B1rs1056836 were found to be significantly associated with time to BCR in both study 1 and study 2 (P = 0.028 and 0.035, respectively), and upon combined analysis (HR: 0.69, 95% CI: 0.40–0.89, P = 0.002; Table 2 and Fig. 1).
Table 2.
Associations between CYP1B1 rs1056836 and biochemical recurrence.
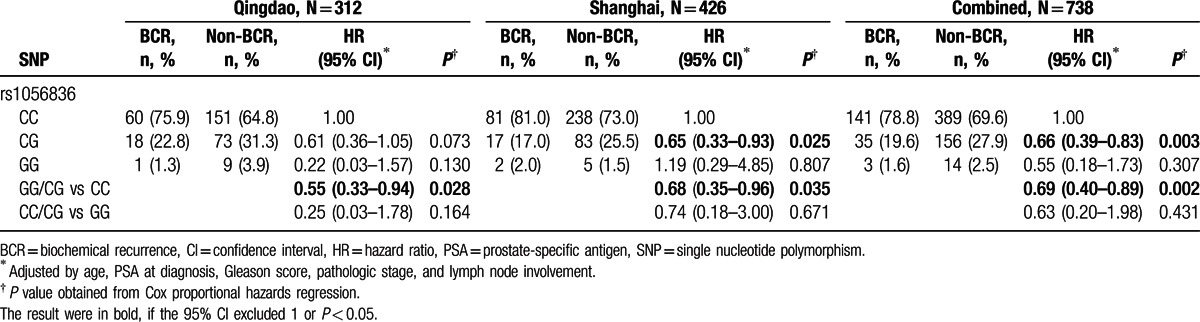
Figure 1.
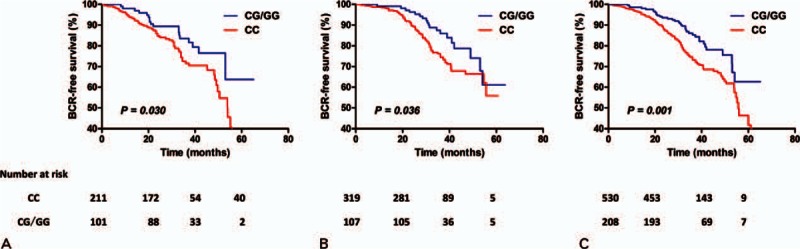
Kaplan–Meier survival curves for biochemical recurrence (BCR)-free survival by rs1056836 dominant model in (A) study 1, (B) study 2, and (C) combined analysis. P value obtained from log-rank t test.
3.3. Relationship between rs1056836 and expression of the CYP1B1
The distributions of CYP1B1 expression in the prostate tissue of patients are shown in Fig. 2. Compared with CG/GG genotype, rs1056836CC genotype was significantly associated with higher expression levels of CYP1B1 mRNA (P = 0.025).
Figure 2.
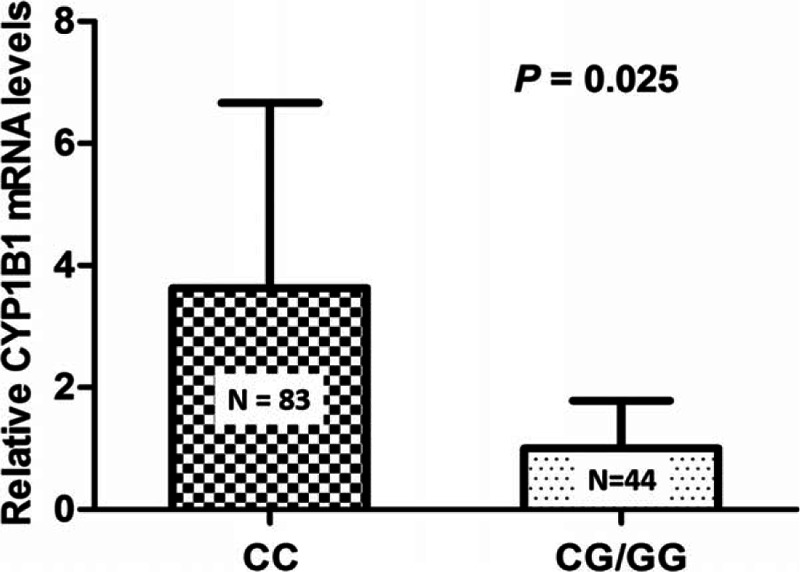
The relative CYP1B1 messenger ribonucleic acid (mRNA) expression levels in prostate tissue by rs1056836 dominant model.
4. Discussion
In this hypothesis-driven study, we observed significant associations between CYP1B1 rs1056836 and BCR in patients with localized prostate cancer. Our results are biologically plausible considering that elevated CYP1B1 gene expression was associated with more aggressive cancers.[20] These results support the hypothesis of a link between the genetic variants of the CYP1B1 gene and prostate cancer progression.
The SNP rs1056836 is a nonsynonymous polymorphism in exon 3, which encodes the heme-binding domain, and results in a G-to-C and subsequent amino acid substitutions of valine-to-leucine. A number of studies have suggested that the rs1056836 may increase prostate cancer risk, but the evidence has not been entirely consistent.[6–15] When stratifying by ethnicity, a meta-analysis 12 independent studies revealed rs1056836 was significantly associated with an increased risk of prostate cancer in Asians.[21] In contrast, no association was found between the polymorphism and prostate cancer risk in either the Caucasian or mixed subject subgroup. The apparent inconsistency of these results may underlie differences in distribution of allele frequencies among ethnicities, lifestyle, and disease prevalence. In the current study of Chinese origin, the minor allele frequency of rs1056836 was lower than Caucasian or mixed population, which is in accordance with Asian studies.[13,15]
Five previous studies have investigated the relationship between the CYP1B1 rs1056836 and prostate cancer aggressiveness, but the definitions of low and high aggressiveness vary among these research reports.[8,10,12,13,15] In spite of these limitations, there is some evidence that the CYP1B1 rs1056836 showed a significant decrease in the risk for disease aggressiveness (OR: 0.45, 95% CI: 0.24–0.84, P = 0.01).[8] Overall, our data combined with previous observations reinforce the importance of this critical gene in cancer progression at numerous disease stages.
We speculated that rs1056836 in the coding region of CYP1B1 may lead to increases in consequent expression levels, thus increasing the risk of cancer by elevating the generation of bioactivated carcinogenic metabolites. Subsequent RT-PCR assays consistently showed CYP1B1 rs1056836 was associated with mRNA level. Transcription level of CYP1B1 in prostate cancer was readily increased more than in normal prostate cells.[22] In prostate cancer, CYP1B1 activity is regulated via several factors including aryl hydrocarbon receptor (AhR), endogenous estrogen levels, and ERs.[4,23] AhR is constitutively active in advanced prostate cancer cell lines.[20] The overproduction of estrogen such as E2 via ERs into carcinogenic metabolites such as estradiol-3,4-quinone or 4-hydroxyestradiol by CYP1B1 can induce proliferation and malignant transformation of prostate cells.[4] Taken together, our data suggest that rs1056836 alters expression of CYP1B1, which in turn influence AhR and estrogen-mediated carcinogenic activity, consequently contributing to the more aggressive phenotype and poorer clinical outcome in prostate cancer. Further biological and functional studies should be accompanied to determine the role of this SNP/gene during prostate cancer progression.
A limitation of our study is that RP case number and the relatively short follow-up period may have limited the power to detect existing associations between the other SNPs studied and prostate cancer outcomes. Due to the multiple comparisons in our data analysis, we could not completely rule out the possibility of false positive findings. However, the replication of data in an independent population reduced the possibility of chance findings. Furthermore, because our analysis was limited to homogeneous Chinese Han population, the generalize ability to other ethnic groups is uncertain.
In conclusion, we found that an SNP located in the coding region of the CYP1B1 gene was associated with prostate cancer clinical outcome. Men with CYP1B1 rs1056836 CC genotype may be at higher risk of recurrence and thus genotyping CYP1B1 has the potential of improving risk stratification and adjuvant therapeutic decision making in localized prostate cancer. Future epidemiological and functional studies are warranted to confirm these findings for translational application and to uncover the mechanistic basis of the CYP1B1 genes in affecting cancer progression.
Supplementary Material
Footnotes
Abbreviations: BCR = biochemical recurrence, CI = confidence interval, HR = hazard ratio, PSA = prostate-specific antigen, RP = radical prostatectomy, SNP = single nucleotide polymorphism.
Cheng-yuan Gu and Xiao-jian Qin contributed equally to this work.
Funding: This study was supported by Fudan University Shanghai Cancer Center Foundation (YJ201506), Shanghai Municipal Commission of Health and Family Planning (No. 20134237), National Natural Science Foundation of China (Grant No.81272837), and Shanghai municipal hospital emerging advanced technology joint research project (Grant No.SHDC12013122)
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- 1.Boorjian SA, Thompson RH, Tollefson MK, et al. Long-term risk of clinical progression after biochemical recurrence following radical prostatectomy: the impact of time from surgery to recurrence. Eur Urol 2011; 59:893–899. [DOI] [PubMed] [Google Scholar]
- 2.Klotz L. Active surveillance for prostate cancer: overview and update. Curr Treat Options Oncol 2013; 14:97–108. [DOI] [PubMed] [Google Scholar]
- 3.Gajjar K, Martin-Hirsch PL, Martin FL. CYP1B1 and hormone-induced cancer. Cancer Lett 2012; 324:13–30. [DOI] [PubMed] [Google Scholar]
- 4.Go RE, Hwang KA, Choi KC. Cytochrome P450 1 family and cancers. J Steroid Biochem Mol Biol 2015; 147:24–30. [DOI] [PubMed] [Google Scholar]
- 5.Carnell DM, Smith RE, Daley FM, et al. Target validation of cytochrome P450 CYP1B1 in prostate carcinoma with protein expression in associated hyperplastic and premalignant tissue. Int J Radiat Oncol Biol Phys 2004; 58:500–509. [DOI] [PubMed] [Google Scholar]
- 6.Holt SK, Kwon EM, Fu R, et al. Association of variants in estrogen-related pathway genes with prostate cancer risk. Prostate 2013; 73:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catsburg C, Joshi AD, Corral R, et al. Polymorphisms in carcinogen metabolism enzymes, fish intake, and risk of prostate cancer. Carcinogenesis 2012; 33:1352–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beuten J, Gelfond JA, Byrne JJ, et al. CYP1B1 variants are associated with prostate cancer in non-Hispanic and Hispanic Caucasians. Carcinogenesis 2008; 29:1751–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndt SI, Chatterjee N, Huang WY, et al. Variant in sex hormone-binding globulin gene and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev 2007; 16:165–168. [DOI] [PubMed] [Google Scholar]
- 10.Cussenot O, Azzouzi AR, Nicolaiew N, et al. Combination of polymorphisms from genes related to estrogen metabolism and risk of prostate cancers: the hidden face of estrogens. J Clin Oncol 2007; 25:3596–3602. [DOI] [PubMed] [Google Scholar]
- 11.Sobti RC, Onsory K, Al-Badran AI, et al. CYP17, SRD5A2, CYP1B1, and CYP2D6 gene polymorphisms with prostate cancer risk in North Indian population. DNA Cell Biol 2006; 25:287–294. [DOI] [PubMed] [Google Scholar]
- 12.Cicek MS, Liu X, Casey G, et al. Role of androgen metabolism genes CYP1B1, PSA/KLK3, and CYP11alpha in prostate cancer risk and aggressiveness. Cancer Epidemiol Biomarkers Prev 2005; 14:2173–2177. [DOI] [PubMed] [Google Scholar]
- 13.Fukatsu T, Hirokawa Y, Araki T, et al. Genetic polymorphisms of hormone-related genes and prostate cancer risk in the Japanese population. Anticancer Res 2004; 24:2431–2437. [PubMed] [Google Scholar]
- 14.Chang BL, Zheng SL, Isaacs SD, et al. Polymorphisms in the CYP1B1 gene are associated with increased risk of prostate cancer. Br J Can 2003; 89:1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka Y, Sasaki M, Kaneuchi M, et al. Polymorphisms of the CYP1B1 gene have higher risk for prostate cancer. Biochem Biophys Res Commun 2002; 296:820–826. [DOI] [PubMed] [Google Scholar]
- 16.Landi MT, Bergen AW, Baccarelli A, et al. CYP1A1 and CYP1B1 genotypes, haplotypes, and TCDD-induced gene expression in subjects from Seveso, Italy. Toxicology 2005; 207:191–202. [DOI] [PubMed] [Google Scholar]
- 17.Hanna IH, Dawling S, Roodi N, et al. Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res 2000; 60:3440–3444. [PubMed] [Google Scholar]
- 18.Bandiera S, Weidlich S, Harth V, et al. Proteasomal degradation of human CYP1B1: effect of the Asn453Ser polymorphism on the post-translational regulation of CYP1B1 expression. Mol Pharmacol 2005; 67:435–443. [DOI] [PubMed] [Google Scholar]
- 19.Gu C, Qu Y, Zhang G, et al. A single nucleotide polymorphism in ADIPOQ predicts biochemical recurrence after radical prostatectomy in localized prostate cancer. Oncotarget 2015; 6:32205–32211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richmond O, Ghotbaddini M, Allen C, et al. The aryl hydrocarbon receptor is constitutively active in advanced prostate cancer cells. PloS One 2014; 9:e95058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Xu DL, Lu Q, et al. Prostate cancer risk and aggressiveness associated with the CYP1B1 4326C/G (Leu432Val) polymorphism: a meta-analysis of 2788 cases and 2968 controls. Asian J Androl 2012; 14:560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokizane T, Shiina H, Igawa M, et al. Cytochrome P450 1B1 is overexpressed and regulated by hypomethylation in prostate cancer. Clin Cancer Res 2005; 11:5793–5801. [DOI] [PubMed] [Google Scholar]
- 23.Chang JT, Chang H, Chen PH, et al. Requirement of aryl hydrocarbon receptor overexpression for CYP1B1 up-regulation and cell growth in human lung adenocarcinomas. Clin Cancer Res 2007; 13:38–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


