Supplemental Digital Content is available in the text
Keywords: biomarker, breast cancer, molecular subtypes, prognosis, sineoculis homeobox homolog family members, tumor development
Abstract
Sineoculis homeobox homolog (SIX) family proteins, including SIX1, SIX2, SIX3, SIX4, SIX5, and SIX6, have been implicated in the initiation and progression of breast cancer, but the role of each member in breast tumor is not fully understood. We conducted a systematic review and meta-analysis to evaluate the association between the mRNA levels of all 6 members and clinic-pathological characteristics and clinical outcome of breast cancer patients based on the PRISMA statement criteria.
ArrayExpress and Oncomine were searched for eligible databases published up to December 10, 2015. The association between the mRNA expression of SIX family members and clinic-pathological features and prognosis was measured by the odds ratio (OR), hazard ratio (HR), and the corresponding 95% confidence interval (CI), respectively. All statistical analyses were performed using STATA software.
In total, 20 published Gene Expression Omnibus (GEO) databases with 3555 patients were analyzed. Our analysis revealed that patients with SIX1 overexpression had worse overall survival (OS) (HR: 1.28, 95% CI: 1.03–1.58) and shorter relapse-free survival (RFS) (HR: 1.28, 95% CI: 1.05–1.56), and much worse prognosis for luminal breast cancer patients with SIX1 overexpression (OS: HR: 1.64, 95% CI: 1.13–2.39; RFS: HR: 1.43, 95% CI: 1.06–1.93). We found that patients with higher SIX2 level had shorter time to both relapse and metastasis. However, high SIX3 mRNA level was a protective factor for OS and RFS of basal-like breast cancer patients.
Our study suggested that members of SIX family played distinct roles in breast cancer. Detailed analysis of the expression of the SIX family members might provide useful information to predict breast cancer progression and prognosis.
1. Introduction
Breast cancer is one of the most common neoplasms and the second leading cause of cancer-related mortality in women worldwide.[1] Over the last several years, molecular signature proves the heterogeneity of breast cancer. Molecular classification provides better prediction of tumor behavior and is widely used to guide therapeutic strategies.[2] However, the current identified molecular subtypes are still not sufficient to provide information in terms of application in cancer treatment. Therefore, identifying novel biomarkers that can predict the progression and prognosis of breast cancer is becoming increasingly urgent.[3]
Sineoculis homeobox homolog (SIX) family proteins are a group of evolutionarily conserved transcription factors that play important roles in cell proliferation, differentiation, apoptosis, adhesion, and migration. This family has 6 members, including SIX1, SIX2, SIX3, SIX4, SIX5, and SIX6.[4] Each member plays a distinct role in the regulation of cell functions. For example, SIX1 is required for the development of murine kidney, muscle, and inner ear.[5] Combinational activation of SIX1, SIX2, and SIX4 was confirmed to be essential to brain development[6]; absence or inactivation of these three genes partly accounted for various brain defects.[6] It has been shown that loss of SIX3/6 expression can lead to pinhole-eye evolution in Nautilus.[7]
Aberrant expression of SIX class has been linked to cancer formation and progression.[8,9] SIX1, the most studied SIX family member, was reported to play a role in the development of tumors, including pancreatic cancer,[10] colorectal cancer,[11] gastric cancer,[12] and especially breast cancer.[13–16] It promoted cell proliferation via reactivating the cell cycle-related proteins cyclin A[17] and cyclin D,[10] and stimulated malignant transformation of nontumorigenic cells.[18] Ectopic expression of SIX1 led to tumor invasion and metastasis partly by modulating epithelial–mesenchymal transition (EMT).[19,20] In addition, high SIX1 level is associated with paclitaxel resistance in breast cancer cells.[15] More importantly, it was found to be closely linked to poor clinical prognosis of cancer patients.[14,21] In patients with Wilms tumors, mutations of SIX1 and SIX2 may contribute to a higher rate of relapse and death.[22] Further, SIX2 promoted breast cancer metastasis by downregulation of E-cadherin.[23] However, high expression of SIX3 contributed to the improved clinical outcome of lung adenocarcinoma patients, and restoration of SIX3 in lung cancer cells led to the suppression of cell proliferation and migration.[24] High protein abundance of SIX4 was closely correlated with poor differentiation and increased depth of invasion in esophageal squamous cell carcinoma.[25]
Although a variety of studies have been conducted to explore the association between SIX and breast cancer, the SIX family member expression signatures in breast cancer and their relation to molecular features remain unclear. Therefore, we conducted a meta-analysis to assess mRNA expression profile of SIX family in breast cancer and analyzed their correlation with molecular subtypes and clinical significance.
2. Methods
Ethical committee or institutional review board approvals were not necessary for this study because it was a meta-analysis based on existing literature.
2.1. Search strategy
The electronic databases including ArrayExpress and Oncomine were searched for relevant Gene Expression Omnibus (GEO) datasets of human breast cancer with the mRNA expression of SIX family members up to December 10, 2015, by using the search term “breast cancer.” Only the datasets which met the inclusion criteria were included in this meta-analysis.
2.2. Inclusion criteria
Databases we used fulfilled the following inclusion criteria: samples in the datasets were human breast cancer tissues or normal breast tissues; the mRNA expression of SIX family members was measured in these databases; the datasets were about mRNA, rather than DNA or microRNA; the sample capacity was more than 45; required clinic-pathological and prognosis information of breast cancer patients was available in these databases, such as grade, T stage, N stage, TNM stage, molecular subtypes, and clinical outcome. We only chose the most complete datasets, when several datasets had some patient population in common.
2.3. Data extraction
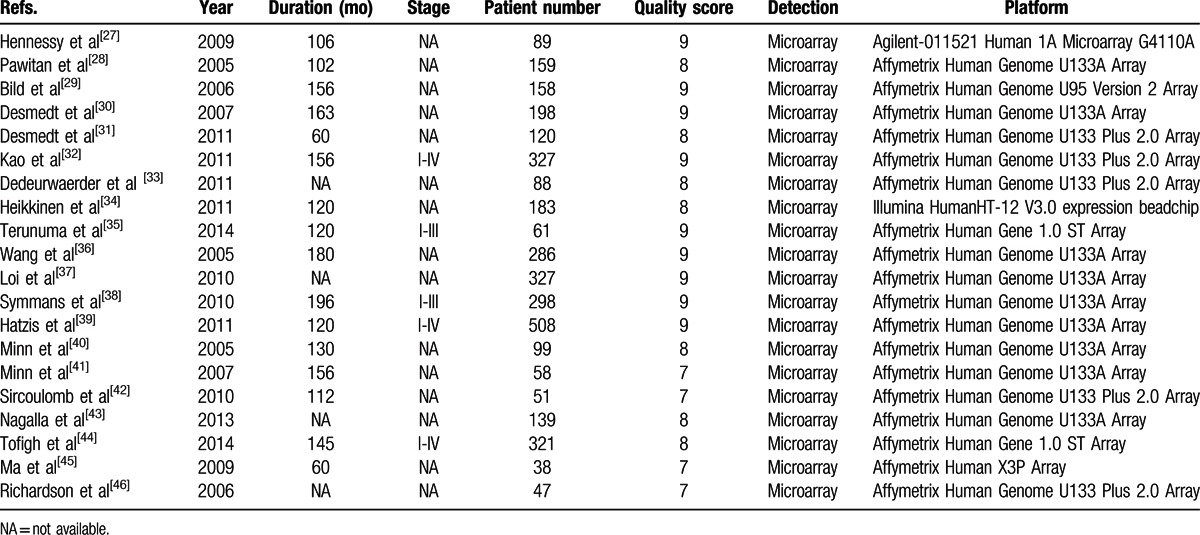
Data analysis was performed independently by 2 individuals. All data were extracted in a predefined table by using a standardized data collection form: first author's name, publication year, follow-up duration, tumor stage, patient number, detection methods, and platform. Cutoff values for SIX1–6 were median expression. We reviewed ArrayExpress and Oncomine, and found 20 human breast cancer microarray datasets with mRNA expression of SIX family members and clinical data. For genes with more than 1 probe, the probe with maximum expression value was selected in our analysis. Overall survival (OS), relapse-free survival (RFS), and metastasis-free survival (MFS) were evaluated by Cox proportional hazard ratio (HR) and 95% confidence interval (CI).
The Newcastle-Ottawa Quality Assessment Scale (NOS) was employed to assess the quality of the studies. Based on the criteria, 8 sources of potential study bias estimating patient selection, study comparability, and outcomes were required to be identified.
2.4. Statistical analysis
The method we used to perform the statistical analysis was as described in our previous meta-analysis on CD44.[26] The association between SIX mRNA expression and clinic-pathological parameters of breast cancer was assessed by the odds ratio (OR) and its corresponding 95% CI. HR was utilized to evaluate the effects of high expression of SIX family members on the clinical outcome of breast cancer patients and HR > 1 indicated that patients with higher mRNA expression of SIX1–6 were more likely to have worse survival. Heterogeneity of publication across studies was assessed by a Chi-square-based-Q statistic and inconsistency index (I2) statistic. We employed the random-effect model if I2 value was more than 50% which indicated that heterogeneity could not be ignored. The fixed-effect model was considered when I2 value was less than 50% which suggested there was no heterogeneity or only moderate heterogeneity. Publication bias was measured by Begg test and Egger test. All statistical analyses were carried out using STATA software package (version 12.0) (Stata Corp LP, College Station, TX).
3. Results
3.1. Search result
The flow diagram for the screening and identification of relevant studies is shown in Fig. 1. One thousand six hundred ninety-five datasets were initially identified, including 1577 records from ArrayExpress and 118 from Oncomine. A total of 1207 datasets were excluded because of duplicates, small sample capacity (n < 45) and data on DNA or microRNA level. We eliminated a total of 385 records after title and abstract screening because of irrelevant topics. After full-text review, a total of 83 datasets were excluded. Among these, 5 datasets were excluded because other datasets included in our meta-analysis contained the patient population from these 5 databases and we only chose the latest and most complete datasets, and other 78 databases were excluded due to no required clinical information. After the complicated screening, 20 studies with 3555 patients met the standard. Table 1 shows the characteristics of all 20 studies.[27–46] These studies mainly assessed the association between the mRNA expression of SIX1, SIX2, SIX3, SIX4, SIX5, and SIX6 with clinical parameters of breast cancer. Tumor size (T stage) 1 and 2 were identified as early T stage, and 3 and 4 were identified as late T stage. No lymph node metastasis (N0) was identified to be N-negative stage, while N1, N2, and N3 were classified into N-positive group. Tumor-node-metastasis (TNM) stages I and II were grouped as early-staged disease whereas III and IV were grouped as late-staged disease. Histological grade I and II were pooled as low-grade disease, while grade III was identified as high-grade disease.
Figure 1.
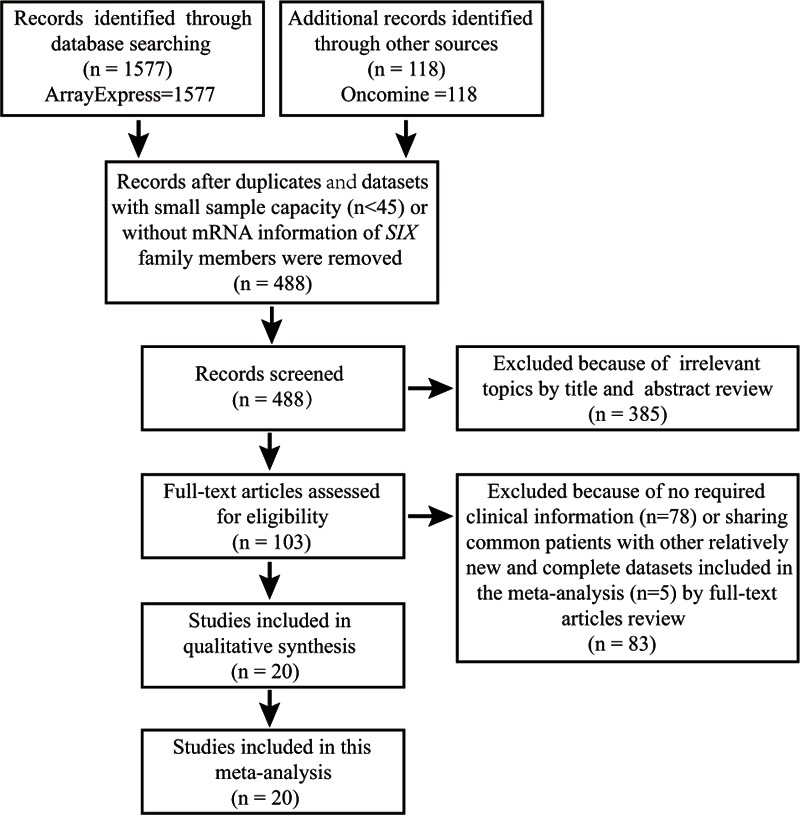
Flow diagram of article selection.
Table 1.
Characteristics of the included studies in the meta-analysis.
3.2. The mRNA levels of SIX family members are correlated with breast cancer risk
There are a total of 6 studies that assessed the association between the mRNA level of SIX family members and breast cancer risk. Our analysis indicated that the mRNA expression of SIX1 (OR: 2.13, 95% CI: 1.28–3.54; P = 0.040 and I2 = 57.0%; Fig. 2A), SIX2 (OR: 1.79, 95% CI: 1.06–2.99; P = 0.444 and I2 = 0.0%; Fig. 2B), SIX3 (OR: 2.04, 95% CI: 1.17–3.56; P = 0.362 and I2 = 6.3%; Fig. 2C), and SIX4 (OR: 5.37, 95% CI: 3.01–9.57; P = 0.776 and I2 = 0.0%; Fig. 2D) was increased in breast cancer tissues when compared with normal breast tissues.
Figure 2.
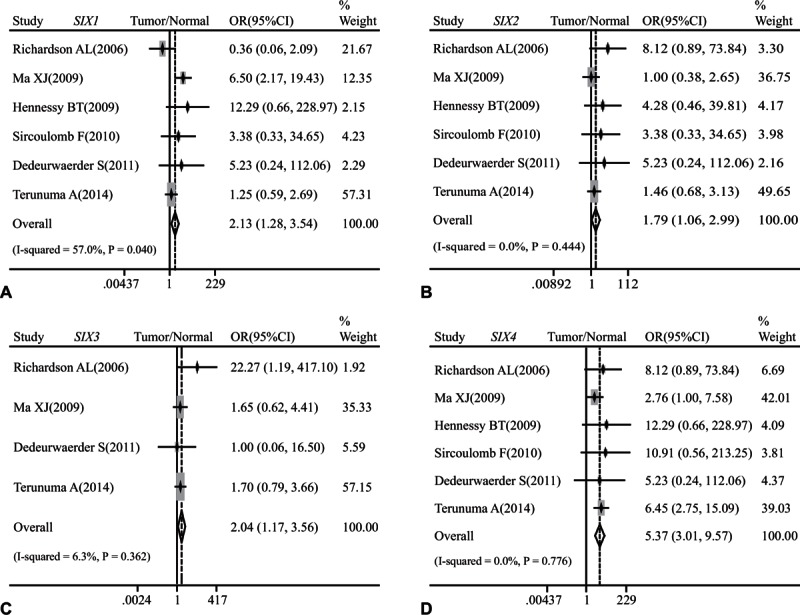
Forest plot of odds ratio (OR). CI = confidence interval. (A). Association between the mRNA expression of SIX1 and breast cancer risks in comparison to normal breast tissues. (B). Association between the mRNA expression of SIX2 and breast cancer risks in comparison to normal breast tissues. (C). Association between the mRNA expression of SIX3 and breast cancer risks in comparison to normal breast tissues. (D). Association between the mRNA expression of SIX4 and breast cancer risks in comparison to normal breast tissues.
3.3. The mRNA levels of SIX family members are correlated with clinic-pathological features in breast cancer
Our results suggested that breast cancer patients with higher histological grade were likely to have a larger amount of SIX1 (OR: 1.50, 95% CI: 1.23–1.82; P = 0.177 and I2 = 28.1%; Fig. 3A), SIX2 (OR: 1.50, 95% CI: 1.23–1.83; P = 0.844 and I2 = 0.0%; Fig. 3B), or SIX3 (OR: 1.31, 95% CI: 1.07–1.60; P = 0.174 and I2 = 30.5%; Fig. 3C) at mRNA level. But, we failed to find any association between the mRNA expression of SIX1–6 and T stage (Supplementary Figure 1), N status (Supplementary Figure 2), or TNM stage (Supplementary Figure 3).
Figure 3.
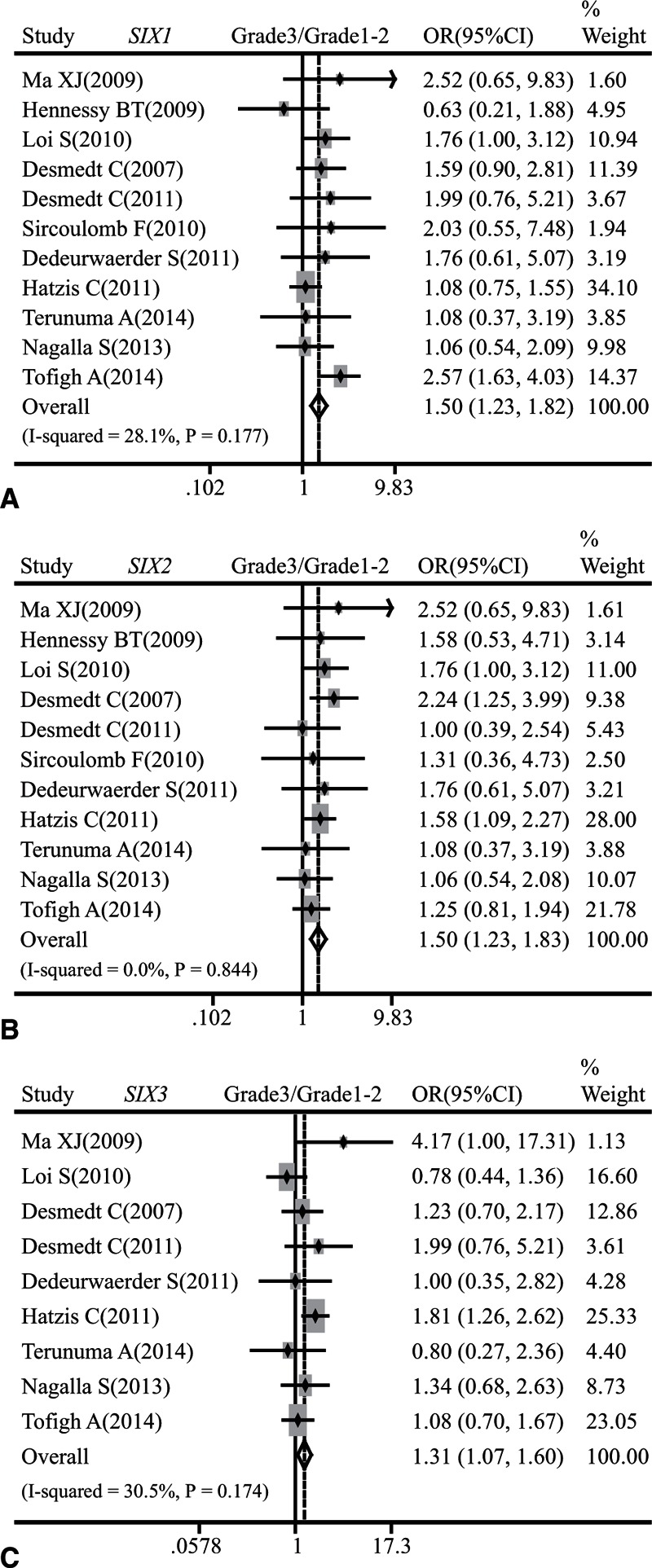
Forest plot of odds ratio (OR). CI = confidence interval. (A). Association between the mRNA expression of SIX1 and histological grade of breast cancer. (B). Association between the mRNA expression of SIX2 and histological grade of breast cancer. (C). Association between the mRNA expression of SIX3 and histological grade of breast cancer.
3.4. The mRNA expression of SIX family members is correlated with molecular subtypes of breast cancer
The association between SIX mRNA expression with the status of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor-2 (HER2), and basal-like breast cancer was also analyzed. The mRNA levels of SIX1 (OR: 1.56, 95% CI: 1.30–1.88; P < 0.001 and I2 = 91.5%; Fig. 4A), SIX2 (OR: 1.72, 95% CI: 1.52–1.96; P = 0.038 and I2 = 47.8%; Fig. 4B), and SIX3 (OR: 1.44, 95% CI: 1.26–1.64; P = 0.038 and I2 = 50.9%; Fig. 4C) were negatively correlated with the status of ER. As for PR status, the mRNA expression of SIX2 (OR: 1.63, 95% CI: 1.24–2.14; P = 0.649 and I2 = 0.0%; Fig. 4E) and SIX3 (OR: 2.06, 95% CI: 1.54–2.76; P = 0.222 and I2 = 31.7%; Fig. 4F) was inversely correlated with PR status. No significant association was found between PR status and SIX1 (OR: 0.90, 95% CI: 0.69–1.18; P = 0.393 and I2 = 3.7%; Fig. 4D). Furthermore, the mRNA levels of SIX1 (OR: 0.66, 95% CI: 0.48–0.92; P = 0.030 and I2 = 54.9%; Supplementary Figure 4A) and SIX2 (OR: 0.61, 95% CI: 0.45–0.84; P = 0.196 and I2 = 29.1%; Supplementary Figure 4B) were positively correlated with HER2 status, but we failed to find significant association between HER2 status and the mRNA expression of SIX3 (OR: 1.16, 95% CI: 0.84–1.61; P = 0.164 and I2 = 36.4%; Supplementary Figure 4C), SIX4 (OR: 1.02, 95% CI: 0.93–1.12; P = 0.594 and I2 = 0.0%; Supplementary Figure 4D), SIX5 (OR: 1.01, 95% CI: 0.96–1.06; P = 0.839 and I2 = 0.0%; Supplementary Figure 4E), and SIX6 (OR: 1.01, 95% CI: 0.96–1.05; P = 0.787 and I2 = 0.0%; Supplementary Figure 4F).
Figure 4.
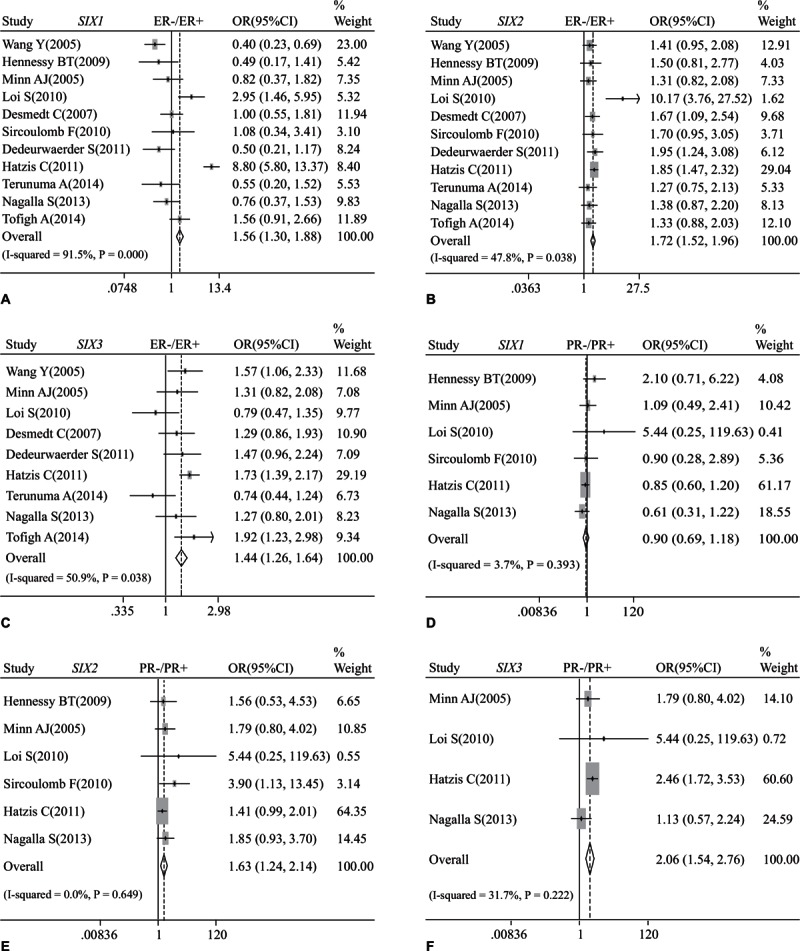
Forest plot of odds ratio (OR). CI = confidence interval. (A). Association between the mRNA expression of SIX1 and ER status of breast cancer. (B). Association between the mRNA expression of SIX2 and ER status of breast cancer. (C). Association between the mRNA expression of SIX3 and ER status of breast cancer. (D). Association between the mRNA expression of SIX1 and PR status of breast cancer. (E). Association between the mRNA expression of SIX2 and PR status of breast cancer. (F). Association between the mRNA expression of SIX3 and PR status of breast cancer.
Furthermore, the mRNA expression of SIX2 (OR: 1.70, 95% CI: 1.31–2.21; P = 0.669 and I2 = 0.0%; Fig. 5B) and SIX3 (OR: 2.53, 95% CI: 1.91–3.36; P = 0.879 and I2 = 0.0%; Fig. 5C) was statistically higher in basal-like tumors than in the luminal subtype of breast cancer, while that of SIX1 (OR: 0.56, 95% CI: 0.43–0.73; P = 0.949 and I2 = 0.0%; Fig. 5A) was obviously lower in basal-like breast cancer in comparison with luminal subtype.
Figure 5.
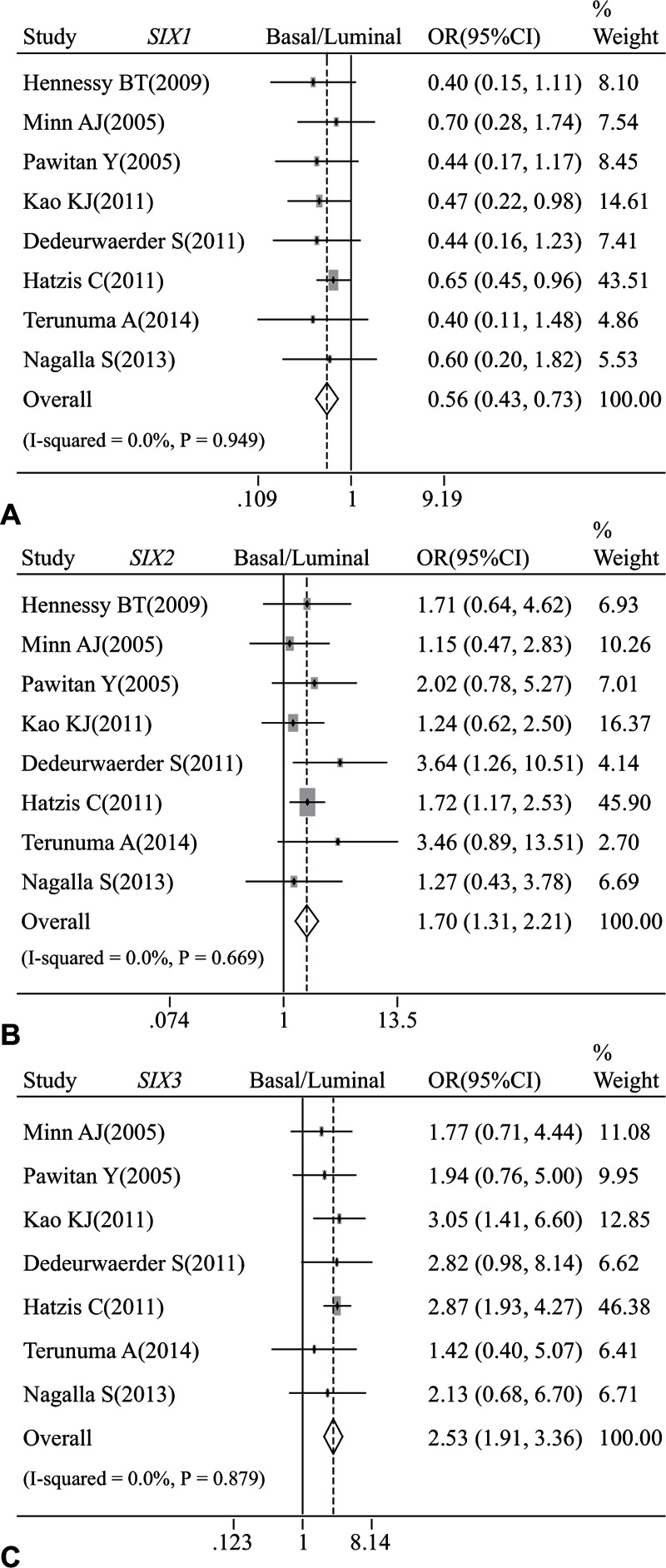
Forest plot of odds ratio (OR). CI = confidence interval. (A). Association between the mRNA expression of SIX1 and basal-like breast cancer in comparison to luminal subtype. (B). Association between the mRNA expression of SIX2 and basal-like breast cancer in comparison to luminal subtype. (C). Association between the mRNA expression of SIX3 and basal-like breast cancer in comparison to luminal subtype.
3.5. The mRNA expression of SIX family members is correlated with breast cancer survival
Our analysis indicated that SIX1, SIX2, and SIX4 were associated with clinical prognosis of whole breast cancer population at mRNA level. High mRNA level of SIX1 was statistically associated with a poor OS (HR: 1.28, 95% CI: 1.03–1.58; P = 0.963 and I2 = 0.0%; Fig. 6A) and RFS (HR: 1.28, 95% CI: 1.05–1.56; P = 0.206 and I2 = 26.8%; Fig. 6B) of whole population of breast cancer. However, we could not find any significant association between SIX1 mRNA expression and MFS of whole breast cancer population (HR: 1.08, 95% CI: 0.84–1.39; P = 0.244 and I2 = 22.4%; Fig. 6C). Furthermore, SIX2 was statistically associated with RFS (HR: 1.22, 95% CI: 1.02–1.45; P = 0.327 and I2 = 12.9%; Fig. 6E) and MFS (HR: 1.24, 95% CI: 1.00–1.53; P = 0.478 and I2 = 0.0%; Fig. 6F), but not correlated with OS (HR: 1.08, 95% CI: 0.86–1.36; P = 0.748 and I2 = 0.0%; Fig. 6D) of whole breast cancer population. Furthermore, patients with higher SIX4 level tended to display worse OS (HR: 1.39, 95% CI: 1.04–1.86; P = 0.770 and I2 = 0.0%; Supplementary Figure 5A) of whole breast cancer population, while did not exhibit significant difference on RFS (HR: 1.24, 95% CI: 0.80–1.92; P = 0.689 and I2 = 0.0%; Supplementary Figure 5B) and MFS (HR: 0.84, 95% CI: 0.59–1.20; P = 0.266 and I2 = 24.3%; Supplementary Figure 5C).
Figure 6.
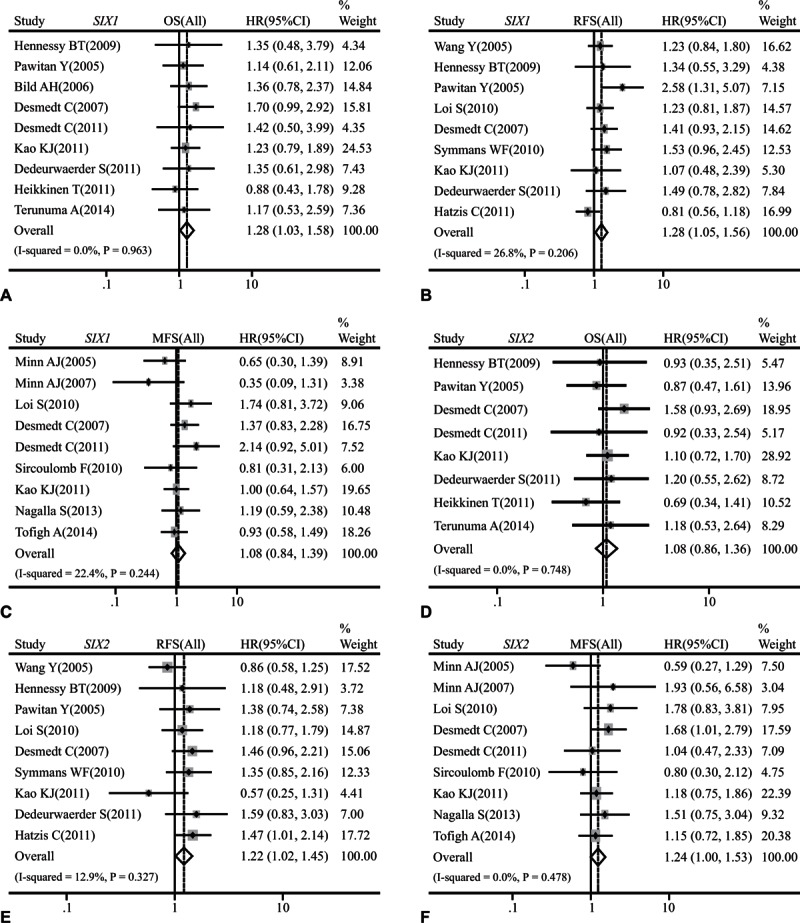
Forest plot of hazard radio (HR). CI = confidence interval. (A). Association between the mRNA expression of SIX1 and OS of breast cancer. (B). Association between the mRNA expression of SIX1 and RFS of breast cancer. (C). Association between the mRNA expression of SIX1 and MFS of breast cancer. (D). Association between the mRNA expression of SIX2 and OS of breast cancer. (E). Association between the mRNA expression of SIX2 and RFS of breast cancer. (F). Association between the mRNA expression of SIX2 and MFS of breast cancer.
Moreover, subgroup analysis showed that some SIX class members had impact on survival performance of patients with a certain molecular subtype. High SIX1 contributed to poor OS (HR: 1.64, 95% CI: 1.13–2.39; P = 0.705 and I2 = 0.0%; Fig. 7A) and RFS (HR: 1.43, 95% CI: 1.06–1.93; P = 0.112 and I2 = 38.4%; Fig. 7B) of luminal breast cancer patients. SIX6 was also found to be linked to poor OS of patients with luminal breast cancer (HR: 1.54, 95% CI: 1.06–2.25; P = 0.456 and I2 = 0.0%; Fig. 7C), but not associated with RFS (HR: 1.26, 95% CI: 0.96–1.64; P = 0.207 and I2 = 26.7%; Fig. 7D) of this subgroup. On the contrary, high SIX3 level was found to be associated with better OS (HR: 0.44, 95% CI: 0.20–0.96; P = 0.593 and I2 = 0.0%; Fig. 7E) and RFS (HR: 0.49, 95% CI: 0.32–0.76; P = 0.451 and I2 = 0.0%; Fig. 7F) of basal-like breast cancer patients.
Figure 7.
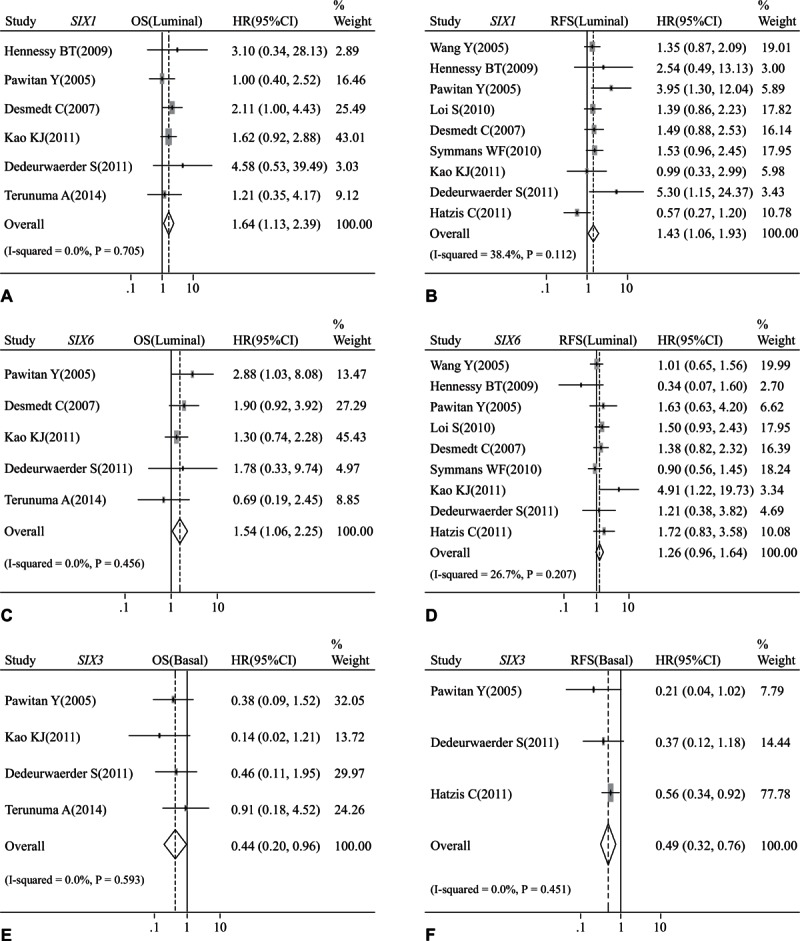
Forest plot of hazard radio (HR). CI = confidence interval. (A). Association between the mRNA expression of SIX1 and OS of luminal breast cancer. (B). Association between the mRNA expression of SIX1 and RFS of luminal breast cancer. (C). Association between the mRNA expression of SIX6 and OS of luminal breast cancer. (D). Association between the mRNA expression of SIX6 and RFS of luminal breast cancer. (E). Association between the mRNA expression of SIX3 and OS of basal-like breast cancer. (F). Association between the mRNA expression of SIX3 and RFS of basal-like breast cancer.
3.6. Publication bias
Publication bias statistics were obtained using Begg test and Egger test. There is no significant publication bias for the following analysis: mRNA expression of SIX family members: breast cancer risk: SIX1: Begg test P = 0.707, Egger test P = 0.568; SIX3: Begg test P = 0.734, Egger test P = 0.474; SIX4: Begg test P = 0.707, Egger test P = 0.381. Histologic grade: SIX1: Begg test P = 1.000, Egger test P = 0.872; SIX2: Begg test P = 0.755, Egger test P = 0.894; SIX3: Begg test P = 0.754, Egger test P = 0.996. ER status: SIX1: Begg test P = 0.276, Egger test P = 0.058; SIX2: Begg test P = 0.755, Egger test P = 0.578; PR status: SIX3: Begg test P = 1.000, Egger test P = 0.789. Basal-like breast cancer: SIX2: Begg test P = 0.266, Egger test P = 0.549; SIX3: Begg test P = 0.133, Egger test P = 0.072. OS (All): SIX1: Begg test P = 0.754, Egger test P = 0.814. RFS (All): SIX1: Begg test P = 0.466, Egger test P = 0.231; SIX2: Begg test P = 0.466, Egger test P = 0.699. MFS (All): SIX2: Begg test P = 0.602, Egger test P = 0.756. OS (luminal): SIX1: Begg test P = 0.707, Egger test P = 0.523; SIX6: Begg test P = 1.000, Egger test P = 0.931. RFS (luminal): SIX1: Begg test P = 0.348, Egger test P = 0.362; OS (basal): SIX3: Begg test P = 1.000, Egger test P = 0.450. RFS (basal): SIX3: Begg test P = 0.296, Egger test P = 0.121.
4. Discussion
Members of the SIX family are expressed at the low level in normal adult tissues but increased in human cancers.[47,48] We found that mRNA levels of SIX1, SIX2, SIX3, and SIX4 were higher in breast cancer as compared to normal counterparts, suggesting their overexpression may contribute to the development of breast cancer. Consistent with this notion, Jin et al[14] analyzed SIX1 expression by immunohistochemistry analysis in 262 breast cancer tissues and found that SIX1 protein was elevated in breast cancer. The mechanism by which SIX1 promoted breast tumor formation may be reinstating its properties normally displayed in early developmental tissues, including stimulation of proliferation and inhibition of apoptosis.[49] SIX1 transcriptionally induces the expression of growth-promoting genes, such as cyclin A1, cyclin D1, and c-Myc.[50,51] By increasing these gene expression, SIX1 promoted malignant transformation.[17,18]
Based on our results, histological grade of breast cancer tended to be positively associated with the mRNA expression of SIX1–3, which may indicate that high SIX1–3 levels were linked to poor differentiation. In agreement, immunohistochemistry analysis on breast phyllodes cancer showed that tumor grade was positively correlated with SIX1 protein level.[16] By activating proproliferative and prosurvival mechanisms, SIX family members promoted expansion of progenitor cell populations prior to differentiation.[52–54] In addition to breast cancer, higher SIX1 level was also linked to poor differentiation in gastric tumor[47] and prostate cancer.[55]
Currently, association between the SIX family members and ER status, PR status or basal-like breast cancer remains unclear. Based on our analysis, SIX1, SIX2, and SIX3 were negatively linked to ER status at mRNA level. SIX2 and SIX3 were negatively correlated with PR status. ER+/PR+ breast tumors were most likely to be low grade.[2] We also found that expressions of SIX1–3 were positively correlated with histological grade and inversely correlated with the status of ER and PR. Based on the status of ER, PR, and HER2, breast cancers are grouped into 5 distinct molecular subtypes, namely luminal A, luminal B, HER2-overexpressing, basal-like, and normal-like.[2] Among these subtypes, luminal breast cancer accounted for the majority of breast cancer and tended to be with a better outcome, while patients with basal-like subtype have a poor survival rate.[2] In this study, we found that in contrast to high expression of SIX2 and SIX3, the level of SIX1 mRNA was significantly lower in basal-like tumors as compared to luminal subtype. However, the expression of SIX1 mRNA was positively associated with HER2 status. A further study revealed that high level of SIX1 protein was significantly associated with HER2+ status.[14] About 67.2% of HER2+ breast tissues were SIX1 strongly positive, while only 49.4% of HER2− tumor tissues were with strong staining of SIX1.[14] We assumed that high SIX1 mRNA level of HER2-overexpressing compensated the low SIX1 mRNA of basal-like breast cancer, contributing to the negative correlation between SIX1 mRNA and ER status at general level. Tumors of basal-like subtype are highly heterogeneous and tend to be high grade.[2] Additionally, our results showed that elevated level of SIX2 and SIX3 was correlated with higher histological grade. Thus, it is not surprising that the mRNA levels of SIX2 and SIX3 was much higher in basal-like tumors than in luminal one.
Our results indicated that some SIX members had distinct impact on the survival of breast cancer patients. For example, high SIX1 mRNA level was significantly correlated with poor OS and RFS of breast cancer population, but not correlated with MFS. This is consistent with a study on 262 breast cancer tissues showing that breast cancer patients with higher SIX1 protein level had remarkably lower 5-year OS rate than those with low SIX1 expression.[14] Furthermore, patients with higher SIX1 mRNA level were also found to exhibit obviously worse RFS. By activating transforming growth factor-beta (TGF-β) and mitogen-activated protein kinase (MEK)/ERK signaling, SIX1 obviously enriched breast cancer stem population.[13] However, SIX1 level did not have effects on MFS. Aberrant expression of SIX1 was found not only in about half of primary breast cancer, but also even in the majority of metastatic lesions.[56] SIX1 was found to potently promote the metastatic spread of breast cancer MCF-7 cells.[19] Several molecular studies on SIX1 could explain why SIX1 has unfavorable impact on breast cancer patient metastasis. SIX1 suppressed the expression of epithelial marker E-cadherin by activating TGF-β, which promoted EMT and finally resulted in tumor metastasis.[57] In addition, SIX1 promoted lymphanogenesis by upregulating vascular endothelial growth factor (VEGF)-C to contribute to tumor metastasis.[57,58] However, tumor metastasis was regulated by a complex network. A large variety of molecules were involved in this process, such as epithelial growth factor receptor (EGFR) and TGF-β.[59] Considering this complex regulation of breast cancer metastasis process, the effects of SIX1 on MFS might be covered.
In addition, patients with high SIX2 mRNA expression tended to have shorter time to both relapse and metastasis at overall level. SIX2 was reported to be a novel regulator of human breast tumor metastasis.[23] SIX2 can promote tumor metastasis by downregulating the epithelial marker E-cadherin. The underlying mechanisms involve the upregulation of Zeb2 that is a direct suppressor of E-cadherin and direct promotion of the methylation of E-cadherin.[23]
Additionally, subcategory analysis indicated that some members play crucial roles in the survival performance of a certain molecular subtype group. For instance, SIX1 was associated with poor OS and RFS of luminal breast cancer patients. SIX6 was linked to poor OS of luminal cancer patients. SIX1's unfavorable impact on clinical outcome of luminal group was supported by Iwanaga R’ research.[13] Apart from these, higher SIX3 mRNA level was strikingly found to contribute to a better OS and RFS in basal-like breast cancer population, indicating that SIX3 is an anticancer factor for basal-like breast tumor. Although the protective role of SIX3 in the clinical outcome of basal-like breast cancer has not been reported, this role in lung adenocarcinoma has been identified.[24]
Both heterogeneity tests and publication bias are essential to a meta-analysis. In this study, evidence of minor heterogeneities was noted. The production of heterogeneity in this result might be due to the following aspects: the platforms used to assess the SIX expression were different. Different platforms mean different design of probe sets for a certain gene; the sample size is limited, indicating that multicenter prospective studies are needed; the demographic data from different datasets were diverse, such as sex, age, disease stage; patients came from different countries. The expression level of a certain gene may be different in different races. In this meta-analysis, no big significant publication bias was found, suggesting our results may be very close to reality.
5. Conclusions
Taken together, our meta-analysis provides evidence that SIX family members play distinct and crucial roles in progression and prognosis of breast cancer. SIX1, SIX2, and SIX4 are activated in breast cancer patients. Increased SIX1–3 expression is linked to high histological grade and ER status, and that SIX2 and SIX3 are upregulated in basal-like breast cancer. High levels of SIX1 and SIX2 predict poor clinical outcome. SIX1 and SIX6 could serve as an unfavorable factor for prognosis of luminal breast cancer patients, while SIX3 is capable of playing a protective role in prognosis of basal-like breast cancer patients. Our meta-analysis reveals an association between SIX family members and clinic-pathological features and prognosis. The role of SIX family as biomarkers for predicting breast cancer progression and prognosis is worthy of further validation.
Supplementary Material
Footnotes
Abbreviations: EGFR = epithelial growth factor receptor, EMT = epithelial–mesenchymal transition, ER = estrogen receptor, GEO = Gene Expression Omnibus, HER2 = human epidermal growth factor receptor-2, LNM = lymph node metastasis, MEK = mitogen-activated protein kinase, MFS = metastasis-free survival, NOS = Newcastle-Ottawa Quality Assessment Scale, OS = overall survival, PR = progesterone receptor, PRISMA = preferred reporting items for systematic reviews and meta-analyses, RFS = relapse-free survival, SIX = sineoculis homeobox homolog, TGF-β = transforming growth factor-beta, TNM = tumor-node-metastasis, VEGF = vascular endothelial growth factor.
All the authors reviewed the ICMJE criteria for authorship and agreed with manuscript results and conclusions. The funders did not participate in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors’ contributions: H-XX and K-JW searched literatures, collected and analyzed the data. Y-JT, QL, NH, X-LH, and XY collected and analyzed data. K-MW and GSW designed the study and wrote the manuscript. All authors edited and approved the final manuscript.
This review was supported by National Natural Science Foundation of China (grant no. 81572608 and 81502209) and Wuhan Yellow Crane Medical Talent Program (grant no. 2015-12).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Makki J. Diversity of breast carcinoma: histological subtypes and clinical relevance. Clin Med Insights Pathol 2015; 8:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radenkovic S, Milosevic Z, Konjevic G, et al. Lactate dehydrogenase, catalase, and superoxide dismutase in tumor tissue of breast cancer patients in respect to mammographic findings. Cell Biochem Biophys 2013; 66:287–295. [DOI] [PubMed] [Google Scholar]
- 4.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet 2005; 6:893–904. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Oghi KA, Zhang J, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 2003; 426:247–254. [DOI] [PubMed] [Google Scholar]
- 6.Garcez RC, Le Douarin NM, Creuzet SE. Combinatorial activity of Six1–2–4 genes in cephalic neural crest cells controls craniofacial and brain development. Cell Mol Life Sci 2014; 71:2149–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogura A, Yoshida MA, Moritaki T, et al. Loss of the six3/6 controlling pathways might have resulted in pinhole-eye evolution in Nautilus. Sci Rep 2013; 3:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer 2002; 2:777–785. [DOI] [PubMed] [Google Scholar]
- 9.Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer 2005; 41:2428–2437. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Tian T, Lv F, et al. Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS One 2013; 8:e59203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahlert C, Lerbs T, Pecqueux M, et al. Overexpression of SIX1 is an independent prognostic marker in stage I-III colorectal cancer. Int J Cancer 2015; 137:2104–2113. [DOI] [PubMed] [Google Scholar]
- 12.Lv H, Cui A, Sun F, et al. Sineoculis homeobox homolog 1 protein as an independent biomarker for gastric adenocarcinoma. Exp Mol Pathol 2014; 97:74–80. [DOI] [PubMed] [Google Scholar]
- 13.Iwanaga R, Wang CA, Micalizzi DS, et al. Expression of Six1 in luminal breast cancers predicts poor prognosis and promotes increases in tumor initiating cells by activation of extracellular signal-regulated kinase and transforming growth factor-beta signaling pathways. Breast Cancer Res 2012; 14:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H, Cui M, Kong J, et al. Sineoculis homeobox homolog 1 protein is associated with breast cancer progression and survival outcome. Exp Mol Pathol 2014; 97:247–252. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Tian T, Hu X, et al. Six1 mediates resistance to paclitaxel in breast cancer cells. Biochem Biophys Res Commun 2013; 441:538–543. [DOI] [PubMed] [Google Scholar]
- 16.Tan WJ, Thike AA, Bay BH, et al. Immunohistochemical expression of homeoproteins Six1 and Pax3 in breast phyllodes tumours correlates with histological grade and clinical outcome. Histopathology 2014; 64:807–817. [DOI] [PubMed] [Google Scholar]
- 17.Coletta RD, Christensen K, Reichenberger KJ, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A 2004; 101:6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coletta RD, Christensen KL, Micalizzi DS, et al. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res 2008; 68:2204–2213. [DOI] [PubMed] [Google Scholar]
- 19.Micalizzi DS, Christensen KL, Jedlicka P, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest 2009; 119:2678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radisky DC. Defining a role for the homeoprotein Six1 in EMT and mammary tumorigenesis. J Clin Invest 2009; 119:2528–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong J, Zhou X, Liu S, et al. Overexpression of sineoculis homeobox homolog 1 predicts poor prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol 2014; 7:3018–3027. [PMC free article] [PubMed] [Google Scholar]
- 22.Walz AL, Ooms A, Gadd S, et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015; 27:286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang CA, Drasin D, Pham C, et al. Homeoprotein Six2 promotes breast cancer metastasis via transcriptional and epigenetic control of E-cadherin expression. Cancer Res 2014; 74:7357–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mo ML, Okamoto J, Chen Z, et al. Down-regulation of SIX3 is associated with clinical outcome in lung adenocarcinoma. PLoS One 2013; 8:e71816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Q, Yu WW, Zhao KL, et al. Expression of Six1 and Six4 in esophageal squamous cell carcinoma and their correlation with clinical prognosis. Zhonghua Bing Li Xue Za Zhi 2013; 42:446–450. [DOI] [PubMed] [Google Scholar]
- 26.Xu H, Tian Y, Yuan X, et al. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. Onco Targets Ther 2016; 9:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 2009; 69:4116–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pawitan Y, Bjöhle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res 2005; 7:R953–R964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439:353–357. [DOI] [PubMed] [Google Scholar]
- 30.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 2007; 13:3207–3214. [DOI] [PubMed] [Google Scholar]
- 31.Desmedt C, Di Leo A, de Azambuja E, et al. Multifactorial approach to predicting resistance to anthracyclines. J Clin Oncol 2011; 29:1578–1586. [DOI] [PubMed] [Google Scholar]
- 32.Kao KJ, Chang KM, Hsu HC, et al. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer 2011; 11:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dedeurwaerder S, Desmedt C, Calonne E, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med 2011; 3:726–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heikkinen T, Greco D, Pelttari LM, et al. Variants on the promoter region of PTEN affect breast cancer progression and patient survival. Breast Cancer Res 2011; 13:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terunuma A, Putluri N, Mishra P, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 2014; 124:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005; 365:671–679. [DOI] [PubMed] [Google Scholar]
- 37.Loi S, Haibe-Kains B, Majjaj S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci U S A 2010; 107:10208–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol 2010; 28:4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 2011; 305:1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature 2005; 436:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minn AJ, Gupta GP, Padua D, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A 2007; 104:6740–6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sircoulomb F, Bekhouche I, Finetti P, et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer 2010; 10:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagalla S, Chou JW, Willingham MC, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol 2013; 14:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tofigh A, Suderman M, Paquet ER, et al. The prognostic ease and difficulty of invasive breast carcinoma. Cell Rep 2014; 9:129–142. [DOI] [PubMed] [Google Scholar]
- 45.Ma XJ, Dahiya S, Richardson E, et al. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 2009; 11:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006; 9:121–132. [DOI] [PubMed] [Google Scholar]
- 47.Emadi-Baygi M, Nikpour P, Emadi-Andani E. SIX1 overexpression in diffuse-type and grade III gastric tumors: features that are associated with poor prognosis. Adv Biomed Res 2015; 4:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorburn A, Behbakht K, Ford H. TRAIL receptor-targeted therapeutics: resistance mechanisms and strategies to avoid them. Drug Resist Updat 2008; 11:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichenberger KJ, Coletta RD, Schulte AP, et al. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res 2005; 65:2668–2675. [DOI] [PubMed] [Google Scholar]
- 50.Ford HL, Kabingu EN, Bump EA, et al. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc Natl Acad Sci U S A 1998; 95:12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang CA, Jedlicka P, Patrick AN, et al. SIX1 induces lymphangiogenesis and metastasis via upregulation of VEGF-C in mouse models of breast cancer. J Clin Invest 2012; 122:1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu PX, Zheng W, Huang L, et al. Six1 is required for the early organogenesis of mammalian kidney. Development 2003; 130:3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Perissi V, Liu F, et al. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 2002; 297:1180–1183. [DOI] [PubMed] [Google Scholar]
- 54.Zuber ME, Perron M, Philpott A, et al. Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell 1999; 98:341–352. [DOI] [PubMed] [Google Scholar]
- 55.Zeng J, Shi R, Cai CX, et al. Increased expression of Six1 correlates with progression and prognosis of prostate cancer. Cancer Cell Int 2015; 15:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ford HL, Landesman-Bollag E, Dacwag CS, et al. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J Biol Chem 2000; 275:22245–22254. [DOI] [PubMed] [Google Scholar]
- 57.Zeng J, Wei M, Shi R, et al. MiR-204-5p/Six1 feedback loop promotes epithelial-mesenchymal transition in breast cancer. Tumour Biol 2016; 37:2729–2735. [DOI] [PubMed] [Google Scholar]
- 58.Liu D, Li L, Zhang XX, et al. SIX1 promotes tumor lymphangiogenesis by coordinating TGF beta signals that increase expression of VEGF-C. Cancer Res 2014; 74:5597–5607. [DOI] [PubMed] [Google Scholar]
- 59.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13:97–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


