Supplemental Digital Content is available in the text
Abstract
Olfactory dysfunction is common among older adults and affects their safety, nutrition, quality of life, and mortality. More importantly, the decreased sense of smell is an early symptom of neurodegenerative diseases such as Parkinson disease (PD) and Alzheimer disease. However, the genetic determinants for the sense of smell have been poorly investigated. We here performed the first genome-wide meta-analysis on the sense of smell among 6252 US older adults of European descent from the Atherosclerosis Risk in Communities (ARIC) study, the Health, Aging, and Body Composition (Health ABC) study, and the Religious Orders Study and the Rush Memory and Aging Project (ROS/MAP). Genome-wide association study analysis was performed first by individual cohorts and then meta-analyzed using fixed-effect models with inverse variance weights. Although no SNPs reached genome-wide statistical significance, we identified 13 loci with suggestive evidence for an association with the sense of smell (Pmeta < 1 × 10−5). Of these, 2 SNPs at chromosome 17q21.31 (rs199443 in NSF, P = 3.02 × 10−6; and rs2732614 in KIAA1267–LRRC37A, P = 6.65 × 10−6) exhibited cis effects on the expression of microtubule-associated protein tau (MAPT, 17q21.31) in 447 frontal-cortex samples obtained postmortem and profiled by RNA-seq (P < 1 × 10−15). Gene-based and pathway-enrichment analyses further implicated MAPT in regulating the sense of smell in older adults. Similar results were obtained after excluding participants who reported a physician-diagnosed PD or use of PD medications. In conclusion, we provide preliminary evidence that the MAPT locus may play a role in regulating the sense of smell in older adults and therefore offer a potential genetic link between poor sense of smell and major neurodegenerative diseases.
INTRODUCTION
The human olfactory system provides us a remarkable ability to detect and discriminate odorants, and warns us of spoiled foods or dangerous environments. Olfactory dysfunction may not only result in poorer nutritional status and adverse effects on safety and quality of life, but is associated with higher mortality.1,2
The sense of smell decreases with age. Poor sense of smell affects over 20% of US older adults.3 Previous research suggests that the loss of sense of smell is among the earliest symptoms of neurodegenerative diseases such as Parkinson disease (PD)4,5 and Alzheimer disease (AD).6 For PD, this observation is consistent with the Braak hypothesis that Lewy body deposition first affects the olfactory bulb and lower brain stem before spreading into substantia nigra and cortex.7 Moreover, as the nasal cavity directly interacts with the environment, a dual-hit hypothesis was proposed that environmental neuro-toxicants may initiate PD development in the olfactory structures decades before PD clinical diagnosis.8 Therefore, research on the sense of smell in older adults may lead to a better understanding of the natural history and etiology of PD and potentially for other neurodegenerative diseases.
However, the genetic basis for the loss of the sense of smell is largely unknown. Limited studies have suggested that genetic variations at human olfactory receptors or the ApoE ε4 allele may contribute to variable olfactory sensitivity and perception.9,10 Two earlier studies also suggest potential links to regions on chromosome 4q.11,12
Here, we report the first ever genome-wide meta-analysis on the sense of smell among 6252 US older adults of European descent from 3 cohorts: the Atherosclerosis Risk in Communities (ARIC) study, the Health, Aging, and Body Composition (Health ABC) study, and the Religious Orders Study and the Rush Memory and Aging Project (ROS/MAP). We further examined suggestive loci with gene-based analyses, pathway-enrichment analyses, as well as expression quantitative trait loci (eQTL) analysis using 447 postmortem prefrontal-cortex tissues. Overall, our results suggest that microtubule-associated protein tau (MAPT), a major susceptibility gene for PD13,14 and possibly for AD,15 may also play a role in regulating the sense of smell in older adults.
METHODS
Participating Studies
Details of participating cohorts were published elsewhere.16–19 Briefly, the ARIC study is an ongoing longitudinal study that was established in 1987 to 1989 to investigate risk factors for cardiovascular diseases.16 The sense of smell was measured as part of the ARIC Neuro-Cognitive Study (NCS) in 2011 to 2013. Of the 6523 participants of ARIC NCS, 4066 were eligible after excluding non-Whites (n = 1563), and participants without valid data on the smell identification test (n = 279) or genome-wide association study (GWAS) genotyping (n = 615). The Health ABC is a prospective study established in 1997 to 1998 to investigate risk factors for disability and functional decline among older adults.17 The sense of smell was tested among 2537 participants in 1999 to 2000. After excluding self-reported non-Whites (n = 976), participants without valid data on the smell identification test (n = 106) or GWAS (n = 111), 1344 were eligible for analysis. The ROS and MAP studies are longitudinal cohorts established in mid-1990s to investigate aging and AD incidence and progression among adults 65 years or older.18,19 The sense of smell was measured as part of its clinical evaluation. Our analysis included 1065 Whites after excluding participants without data on genotyping (n = 661) or the sense of smell (n = 644).
The Smell Identification Tests
All 3 cohorts used validated smell identification tests to examine the sense of smell: the 12-item Sniffin’ Sticks screening test (Burghart, Wedel, Germany)20 in ARIC and the 12-item Brief Smell Identification Test (B-SIT, Sensonics, Haddon Heights, NJ)21 in the Health ABC and ROS/MAP. Both tests assess participant's sense of smell to correctly identify 12 daily odorants, although the exact odorants were somewhat different. Briefly, participants were instructed to smell each odorant and choose the correct odorant from 4 possible answers in a multiple-choice format. One point was given for each correct answer with a total score from 0 to 12. Both tests have been validated and widely used in clinical and epidemiological studies.20,21
All cohorts assessed global cognitive function during the same study visit in which the sense of smell was evaluated. The ARIC and ROS/MAP studies used the Mini-Mental State Examination (MMSE) that has a maximal score of 30, whereas the Health ABC study used the Modified Mini-Mental Status Examination (MMMSE) with a maximal score of 100. Each cohort also determined ApoE genotypes by genotyping 2 ApoE variants at codons 130 and 176 (formerly 112 and 158) separately.
Genotyping, Imputation, and Statistical Analysis
GWAS genotyping was performed using the Affymetrix Genome-wide Human SNP array 6.0 in ARIC and ROS/MAP, and Illumina Human1M-Duo BeadChip in Health ABC. Imputation was performed in each cohort using MACH based on HapMap Phase II CEU build 36 reference panel. Prespecified criteria were used for quality control of each cohort.
Each cohort performed its own GWAS analysis following a standardized procedure with ProbABEL22 or PLINK,23 using a linear regression model with analysis of allele dosages. The outcome was the natural log of the sense of smell score plus 1 to account for the few participants who had a score of 0. Potential confounders were adjusted for in 2 separate models: first for age, gender, study sites, cognitive function, and the first 2 principal components (PCAs) in Model 1, and then additionally for ApoE ε4 allele in Model 2. We further adjusted for ApoE ε4 because it is the most important genetic risk factor for AD and predicts poor sense of smell in all participating cohorts.
We performed meta-analysis using a fixed-effect model with inverse variance weights in METAL,24 after filtering out SNPs with MAF < 1% 25–27 or low imputation quality (r2 < 0.3). We applied genomic control for each dataset before meta-analysis (e-Methods). We considered P value < 5 × 10−8 as genome-wide significant and P value between 1 × 10−5 and 5 × 10−8 as suggestive evidence for an association.
We used VEGAS28 (Versatile Gene-based Association Study) for gene-based analysis and calculated individual gene-wise P values for each gene in relation to the sense of smell, and used binomial test to identity enriched ingenuity canonical pathways (http://www.ingenuity.com; IPA, Ingenuity® Systems) based on the results of meta-analysis in Model 2, the enrichment test P values were derived from 10,000 rounds of permutation tests (e-Methods).
To explore the functional relevance of top SNPs from the GWAS analysis, we applied the publicly available data from the RegulomeDB (http://regulomedb.org/)29 (e-Methods). We then generated RNA expression data using frozen postmortem dorsolateral prefrontal cortex (DLPFC) brain tissue from 447 ROS/MAP participants to define the biological consequence of the risk variants of identified SNPs (e-Methods).
Ethical Aspects
Individual study protocols were approved by relevant institutional review boards and all study participants provided written consent.
RESULTS
Table 1 presents characteristics of study participants. As expected, the sense of smell decreased with age and was lower in men than women. Further, higher cognitive score was associated with better sense of smell, whereas the ApoE ε4 allele was related to poorer sense of smell. These population characteristics were similarly associated with the sense of smell across cohorts (Table S1, http://links.lww.com/MD/A526). Although these studies used different screening tests of smell identification, the distributions of the sense of smell scores were comparable across cohorts (Figure S1, http://links.lww.com/MD/A526).
TABLE 1.
Population Characteristics of Participating Cohorts
The Q–Q plots for Model 1 showed relatively low genomic inflation factors (overall λ = 0.996), suggesting minimal population stratification (Figure S2, http://links.lww.com/MD/A526). Although no SNP reached genome-wide significance, 35 SNPs had a suggestive association with the sense of smell (Pmeta < 1 × 10−5, Figure 1A). We examined LD among these SNPs and identified 19 SNPs that either had the lowest P value in each LD block (r2 ≥ 0.8) or were potentially functional as annotated in the RegulombDB (Tables S2 and S3, http://links.lww.com/MD/A526). Among these, 3 SNPs were predicted as eQTLs in frontal-cortex and cerebellum tissues in the RegulomeDB: rs199443 (NSF, Pmeta = 4.01 × 10−6), rs2075650 (TOMM40, Pmeta = 4.24 × 10−6), and rs2732614 (KIAA1267–LRRC37A, Pmeta = 8.34 × 10−6). The rs199443 and rs2732614 were cis-eQTLs for MAPT in both frontal-cortex and cerebellum tissues,30 while rs2075650 was cis-eQTL for TOMM40 in lymphoblastoid cells.31
FIGURE 1.
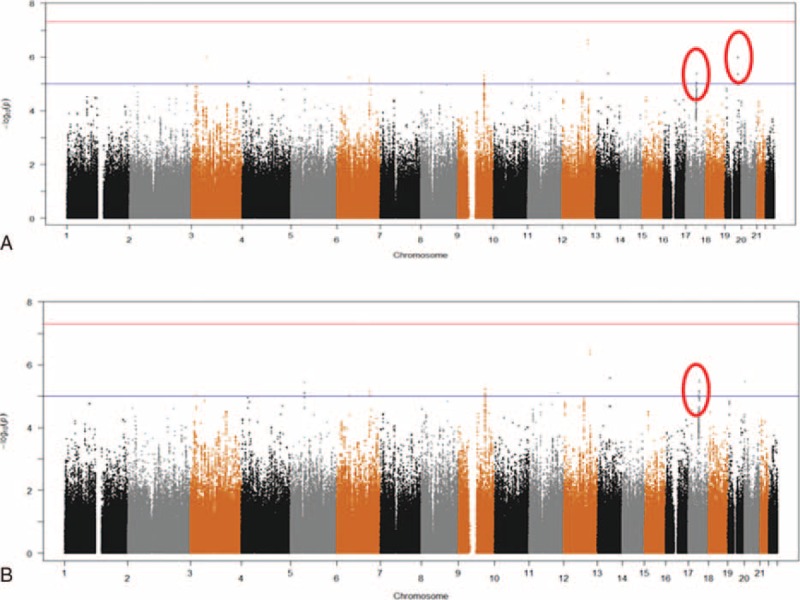
Manhattan plots of the genome-wide meta-analysis on the sense of smell. The red line indicates the genome-wide significance threshold (5 × 10−8) and the blue line indicates the suggestive threshold (1 × 10−5). (A) Model 1 adjusted for age, gender, study center, cognitive function, and the first 2 principal components; (B) Model 2 further adjusted for ApoE ε4 allele.
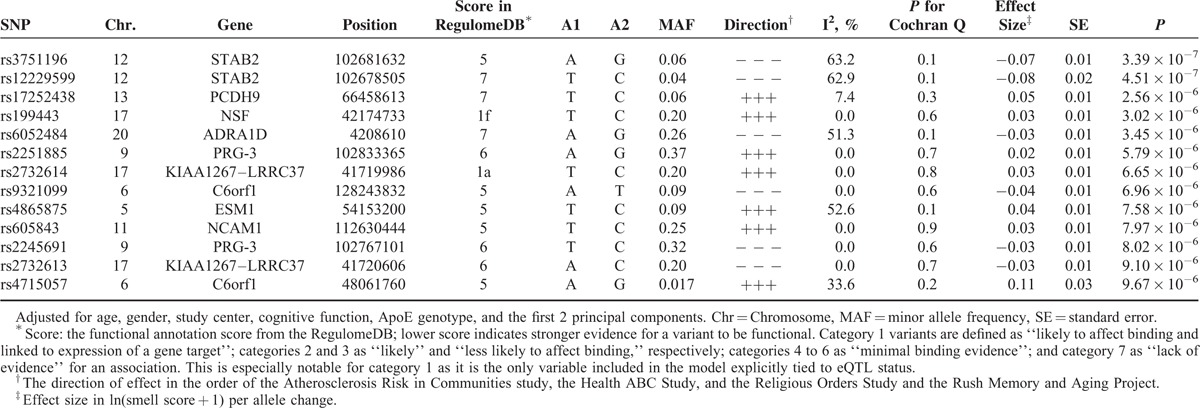
In the meta-analysis of Model 2 that further adjusted for ApoE ε4, the Q–Q plot also showed a relatively low genomic inflation factor with an overall λ of 0.996 (Figure S2, http://links.lww.com/MD/A526). As shown in Table 2 and Figure 1B, 13 SNPs had suggestive associations with the sense of smell (Pmeta < 1 × 10−5, Figure S3, http://links.lww.com/MD/A526); another 47 SNPs were in LD (r2 ≥ 0.8) with these 13 SNPs (Table S4, http://links.lww.com/MD/A526). Ten of these 13 SNPs overlapped with those from Model 1, including the 2 annotated eQTLs at chromosome 17: rs199443 (Pmeta = 3.02 × 10−6) and rs2732614 (Pmeta = 6.65 × 10−6). In contrast, the signal for rs2075650 at TOMM40 was drastically diminished (Pmeta = 0.09) after adjustment for ApoE ε4. For these 13 SNPs, we also repeated the meta-analyses using random-effect model and observed similar results. For example, the Pmeta was 3.73 × 10−6 for rs199443 and 8.25 × 10−6 for rs2732614 under the random-effect model.
TABLE 2.
Top Ranked SNPs (Pmeta < 1 × 10−5) From the Genome-Wide Meta-Analysis on the Sense of Smell in Model 2
We conducted gene-based analysis using VEGAS28 to identify genes that harbor multiple association signals based on Pmeta values from the model that adjusted for ApoE. The SNPs were mapped to 17,693 genes. No genes were statistically significant after Bonferroni correction (P < 2.8 × 10−6 = 0.05/17,693). Genes with P < 1.0 × 10−3 were listed in Table S5, http://links.lww.com/MD/A526. Interestingly, MAPT (Pgene = 9.4 × 10−5) was among the top genes identified in this analysis along with nearby LRRC37A (Pgene = 2.9 × 10−5) and NSF (Pgene = 1.4 × 10−5), all located on chromosome 17. LRRC37A and NSF each harbor 1 of the 2 annotated eQTLs (rs2732614 and rs199443, respectively) whereas the expression of MAPT is regulated by both SNPs in brain tissues.30
We further conducted 2 pathway enrichment analyses (top 10 identified pathways listed in Table S6, http://links.lww.com/MD/A526). The first analysis was based on results from Model 2 and we observed 21 significantly enriched canonical pathways with the permutation P < 0.05. These include the CDK5 and p38 MAPK signaling pathways both of which are implicated in neurodegeneration and MAPT is a member of both pathways. The second analysis were based on the 164 genes with Pgene < 0.01 identified by VEGAS. This analysis identified 113 enriched canonical pathways with the permutation P < 0.05. The CDK5 signaling pathway was again identified as 1 of the top ranked pathways.
We further examined MAPT expression using RNA-seq data of 447 postmortem frontal-cortex tissues from the ROS/MAP study in order to define the biological consequence of the risk variants of rs2732614 and rs199443. We found strong correlations between genotypes of the risk allele (C allele for both rs2732614 and rs199443) and higher expression level of MAPT (P < 10−15, Figure 2); however, we observed no significant correlation between these 2 SNPs and the expression levels of NSF or LRRC37A (rs2732614: P = 0.62 for NSF and 0.54 for LRRC37A; rs199443: P = 0.79 for NSF and 0.11 for LRRC37A).
FIGURE 2.
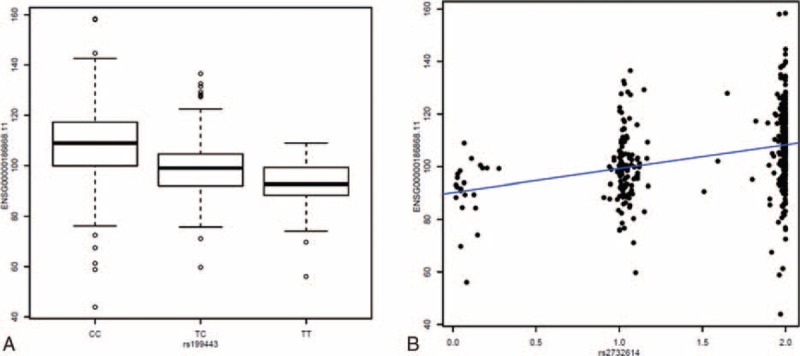
Expression quantitative trait loci analysis for MAPT in 447 human frontal cortex samples. These plots illustrate a dose relationship between allele load at rs199443 (A) and rs2732614 (B) and expression of MAPT; the P values were 2.22 × 10−16 and 4.44 × 10−16, respectively. For rs2732614, genotype data were imputed using MACH which provides an estimated dosage of imputed genotype. The dosage ranges from 0 to 2, with 0 as no copy of the reference allele (C allele) and 2 as 2 copies of the reference allele. MAPT = microtubule-associated protein tau.
As MAPT is 1 of the most important susceptibility genes for PD, we further performed a sensitivity analysis for the 13 SNPs identified in Model 2 by removing potential PD cases based on self-reported physician diagnosis or use of PD medications (90 in ARIC, 14 in Health ABC, and 12 in ROS/MAP) and obtained similar results (Table S7, http://links.lww.com/MD/A526).
DISCUSSION
To the best of our knowledge, this is the first genome-wide meta-analysis on the sense of smell among older adults. The study population includes 3 well-characterized aging cohorts with objective measurements for smell identification. Although no SNP reached genome-wide significance, the preponderance of the evidence suggests MAPT as a potential susceptibility locus for poor sense of smell. This is particular interesting because MAPT is 1 of the most important susceptibility genes for late-onset sporadic PD13,14 and possibly for AD.15
Smell impairment is common among older adults, particularly in patients with neurodegenerative diseases. More importantly, recent data suggest that decreased sense of smell may precede the clinical onset of both PD4,5 and AD.6 For example, in the Honolulu Asia Aging Study, individuals in the lowest quartile of the sense of smell score were 5 times more likely to develop PD in the next 4 years than those in the highest quartile5; further poor sense of smell was associated with higher prevalence of incidental Lewy bodies in postmortem brains, a condition that may represent prodromal PD.32 Similar observations have also been made for both mild cognitive impairment and AD.6,33 Therefore, the declining sense of smell in older adults may indicate underlying neurodegenerative processes. Characterizing the genetic and environmental determinants for the sense of smell in the context of aging may provide targets for intervention with enormous potential translational impact.
The human MAPT locus has long been associated with the risk for PD.13,14 A recent meta-analysis of GWAS data further implicated MAPT as a susceptibility locus for late onset sporadic AD.15 The human MAPT encodes the microtubule-associated protein tau. It is notable that tau-related pathology was found in the bulbar component of the anterior olfactory nucleus only in patients of neurodegenerative disorders with marked loss of the sense of smell, such as AD, PD, and Lewy body dementia.34,35 Experimental studies further suggest that overexpression of tau may contribute to olfactory dysfunction in vivo.36,37 These findings indicate that overexpression or abnormal aggregation of tau may represent a common mechanism between olfactory dysfunction and several neurodegenerative diseases.
The genomic architecture in the region spanning MAPT is highly complex. It covers ∼1.8 Mb block of LD encompassing not only MAPT but also several other genes that have been linked to neurodegenerative diseases, such as STH (Saitohin) and NSF (N-ethylmaleimide sensitive factor). We, however, in the RNA-seq data of 447 postmortem frontal-cortex tissues, found that rs2732614 and rs199443 had statistically significant cis effects on the expression of MAPT (P < 10−15), but not on NSF or LRRC37A. Additional gene-based and pathway-enrichment analyses also showed the potential of MAPT as a candidate susceptibility gene for poor sense of smell. Although our study provides fairly consistent evidence that MAPT may be a candidate susceptibility locus for the sense of smell in older adults of European descent, fine-scale mapping of this LD block with next generation sequencing will be necessary to identify potential causal locus.
ApoE ε4 is the most important genetic risk factor for late-onset sporadic AD38 and has been linked to olfactory dysfunction in several studies.9 The present study confirms that the ApoE ε4 allele is a risk factor for poor smell identification even after accounting for cognitive function. In our initial analysis, we also identified an intronic SNP in TOMM40 (rs2075650) in association with the sense of smell; but this signal was extinguished after adjustment for the ApoE ε4 allele. Although the mechanisms are yet unknown, preliminary studies showed that ApoE was expressed at high levels in various types of cells in the olfactory epithelium and its underlying lamina propria, and that ApoE might play a critical role in olfactory nerve regeneration in mice.39,40 Furthermore, experimental studies suggest that in contrast to the ApoE ε2 and ε3 alleles, the ApoE ε4 fails to promote neurite outgrowth in olfactory epithelium cultures, which may lead to olfactory dysfunction in AD.41
The main limitation of the current study was the lack of independent confirmation of GWAS signals. GWAS analysis often requires very large sample size to obtain adequate power to detect modest effects of underlying variants in complex diseases. However, few large population-based studies of older adults have both the sense of smell and GWAS data. We were able to identify data from 3 well-established cohorts with similar population characteristics and data collection. We chose to combine our samples for SNP discovery rather than set up the conventional 2-stage design to maximize power to detect signals. We did, however, follow up suggestive signals with functional and expression analyses. Finally, the determinants for the sense of smell are likely complex42,43 and our study was conducted among old White adults, therefore the results should be interpreted in this context. Further studies are needed to confirm our findings and to examine whether they could be generalized to other populations.
In conclusion, we report the results of the first comprehensive GWAS analysis to understand the genetic basis for the sense of smell in older adults of European ancestry. We provide novel evidence that the MAPT locus may regulate the sense of smell in older adults. This finding warrants independent confirmation and further mechanistic investigations into how MAPT region contributes to the loss of the sense of smell and the development of related neurodegenerative diseases.
ACKNOWLEDGMENTS
The authors first thank the staff and participants of the ARIC study, Health ABC study, and the ROS/MAP for their important contributions. Second, we would like to thank Professors Hongbing Shen, Zhibin Hu, and Juncheng Dai to provide important comments on an early version of the manuscript.
Footnotes
Abbreviations: AD = Alzheimer disease, ARIC = Atherosclerosis Risk in Communities study, B-SIT = Brief Smell Identification Test, eQTL = expression quantitative trait loci, GWAS = genome-wide association study, Health ABC = the Health, Aging, and Body Composition study, MAP = the Rush Memory and Aging Project, MMSE = Mini-Mental State Examination, NCS = the ARIC Neuro-Cognitive Study, PD = Parkinson disease, ROS = the Religious Orders Study.
The study was supported by the Intramural Research Program of NIH (Z01 ES101986) and the National Institute of Environmental Health Sciences.
The Atherosclerosis Risk in Communities study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, and R01HL086694, with the ARIC carotid MRI examination funded by U01HL075572-01; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Neurocognitive data are collected by U01 HL096812, HL096814, HL096899, HL096902, and HL096917 with previous brain MRI examinations funded by R01-HL70825. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Additional support for this project includes grants R01-HL093029 and R01-NS087541.
The Health ABC Study research was supported by in part by the Intramural Research Program of the NIH, National Institute on Aging (NIA) and by NIA contracts N01AG62101, N01AG62103, and N01AG62106. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C.
The Religious Orders Study and Rush Memory and Aging Project were supported by grants from the National Institutes of Aging (R01 AG30146, P30 AG10161, R01 AG17917, R01 AG15819), the Illinois Department of Public Health, and the Translational Genomics Research Institute.
JMP was supported by the National Institute on Aging (K23 AG036762), the McHugh Otolaryngology Research Fund, the American Geriatrics Society, the Center on the Demography and Economics of Aging, and the Institute of Translational Medicine at the University of Chicago (KL2RR025000 and UL1RR024999).
XH received research grants and consultation fees from NIEHS, NIH, and GE. Other authors declare no competing financial interests.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Deems DA, Doty RL, Settle RG, et al. Smell and taste disorders, a study of 750 patients from the University of Pennsylvania Smell and Taste Center. Arch Otolaryngol Head Neck Surg 1991; 117:519–528. [DOI] [PubMed] [Google Scholar]
- 2.Wilson RS, Yu L, Bennett DA. Odor identification and mortality in old age. Chem Senses 2011; 36:63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy C, Schubert CR, Cruickshanks KJ, et al. Prevalence of olfactory impairment in older adults. JAMA 2002; 288:2307–2312. [DOI] [PubMed] [Google Scholar]
- 4.Ponsen MM, Stoffers D, Booij J, et al. Idiopathic hyposmia as a preclinical sign of Parkinson's disease. Ann Neurol 2004; 56:173–181. [DOI] [PubMed] [Google Scholar]
- 5.Ross GW, Petrovitch H, Abbott RD, et al. Association of olfactory dysfunction with risk for future Parkinson's disease. Ann Neurol 2008; 63:167–173. [DOI] [PubMed] [Google Scholar]
- 6.Wilson RS, Arnold SE, Schneider JA, et al. Olfactory impairment in presymptomatic Alzheimer's disease. Ann N Y Acad Sci 2009; 1170:730–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braak H, Del Tredici K, Rub U, et al. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging 2003; 24:197–211. [DOI] [PubMed] [Google Scholar]
- 8.Hawkes CH, Del Tredici K, Braak H. Parkinson's disease: a dual-hit hypothesis. Neuropathol Appl Neurobiol 2007; 33:599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olofsson JK, Nordin S, Wiens S, et al. Odor identification impairment in carriers of ApoE-varepsilon4 is independent of clinical dementia. Neurobiol Aging 2010; 31:567–577. [DOI] [PubMed] [Google Scholar]
- 10.Menashe I, Man O, Lancet D, et al. Different noses for different people. Nat Genet 2003; 34:143–144. [DOI] [PubMed] [Google Scholar]
- 11.Pinto JM, Thanaviratananich S, Hayes MG, et al. A genome-wide screen for hyposmia susceptibility loci. Chem Senses 2008; 33:319–329. [DOI] [PubMed] [Google Scholar]
- 12.Knaapila A, Keskitalo K, Kallela M, et al. Genetic component of identification, intensity and pleasantness of odours: a Finnish family study. Eur J Hum Genet 2007; 15:596–602. [DOI] [PubMed] [Google Scholar]
- 13.Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet 2014; 46:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shulman JM, Yu L, Buchman AS, et al. Association of Parkinson disease risk loci with mild Parkinsonian signs in older persons. JAMA Neurol 2014; 71:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desikan RS, Schork AJ, Wang Y, et al. Genetic overlap between Alzheimer's disease and Parkinson's disease at the MAPT locus. Mol Psychiatry 2015; doi: 10.1038/mp.2015.6. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) study: design objectives. Am J Epidemiol 1989; 129:687–702. [PubMed] [Google Scholar]
- 17.Ix JH, Wassel CL, Kanaya AM, et al. Fetuin-A and incident diabetes mellitus in older persons. JAMA 2008; 300:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett DA, Schneider JA, Arvanitakis Z, et al. Overview and findings from the religious orders study. Curr Alzheimer Res 2012; 9:628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett DA, Schneider JA, Buchman AS, et al. The Rush Memory and Aging Project: study design and baseline characteristics of the study cohort. Neuroepidemiology 2005; 25:163–175. [DOI] [PubMed] [Google Scholar]
- 20.Hummel T, Konnerth CG, Rosenheim K, et al. Screening of olfactory function with a four-minute odor identification test: reliability, normative data, and investigations in patients with olfactory loss. Ann Otol Rhinol Laryngol 2001; 110:976–981. [DOI] [PubMed] [Google Scholar]
- 21.Doty RL, Marcus A, Lee WW. Development of the 12-item Cross-Cultural Smell Identification Test (CC-SIT). Laryngoscope 1996; 106:353–356. [DOI] [PubMed] [Google Scholar]
- 22.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 2010; 11:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet 2010; 42:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet 2010; 42:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YR, Li J, Zhao SD, et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat Med 2015; 21:1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu JZ, McRae AF, Nyholt DR, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet 2010; 87:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 2010; 6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 2010; 464:773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross GW, Abbott RD, Petrovitch H, et al. Association of olfactory dysfunction with incidental Lewy bodies. Mov Disord 2006; 21:2062–2067. [DOI] [PubMed] [Google Scholar]
- 33.Lehrner J, Pusswald G, Gleiss A, et al. Odor identification and self-reported olfactory functioning in patients with subtypes of mild cognitive impairment. Clin Neuropsychol 2009; 23:818–830. [DOI] [PubMed] [Google Scholar]
- 34.Tsuboi Y, Wszolek ZK, Graff-Radford NR, et al. Tau pathology in the olfactory bulb correlates with Braak stage, Lewy body pathology and apolipoprotein epsilon4. Neuropathol Appl Neurobiol 2003; 29:503–510. [DOI] [PubMed] [Google Scholar]
- 35.Mundinano IC, Caballero MC, Ordonez C, et al. Increased dopaminergic cells and protein aggregates in the olfactory bulb of patients with neurodegenerative disorders. Acta Neuropathol 2011; 122:61–74. [DOI] [PubMed] [Google Scholar]
- 36.Macknin JB, Higuchi M, Lee VM, et al. Olfactory dysfunction occurs in transgenic mice overexpressing human tau protein. Brain Res 2004; 1000:174–178. [DOI] [PubMed] [Google Scholar]
- 37.Beharry C, Alaniz ME, Alonso Adel C. Expression of Alzheimer-like pathological human tau induces a behavioral motor and olfactory learning deficit in Drosophila melanogaster. J Alzheimers Dis 2013; 37:539–550. [DOI] [PubMed] [Google Scholar]
- 38.Sadigh-Eteghad S, Talebi M, Farhoudi M. Association of apolipoprotein E epsilon 4 allele with sporadic late onset Alzheimer's disease: a meta-analysis. Neurosciences (Riyadh) 2012; 17:321–326. [PubMed] [Google Scholar]
- 39.Nathan BP, Nannapaneni S, Gairhe S, et al. The distribution of apolipoprotein E in mouse olfactory epithelium. Brain Res 2007; 1137:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nathan BP, Nisar R, Short J, et al. Delayed olfactory nerve regeneration in ApoE-deficient mice. Brain Res 2005; 1041:87–94. [DOI] [PubMed] [Google Scholar]
- 41.Hussain A, Luong M, Pooley A, et al. Isoform-specific effects of ApoE on neurite outgrowth in olfactory epithelium culture. J Biomed Sci 2013; 20:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doty RL, Kamath V. The influences of age on olfaction: a review. Front Psychol 2014; 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doty RL, Petersen I, Mensah N, et al. Genetic and environmental influences on odor identification ability in the very old. Psychol Aging 2011; 26:864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]


